Significance
Fifty percent of diffuse large B cell lymphoma (DLBCL) evade immune-surveillance via somatic genetic lesions abrogating the expression of the class I major histocompatibility complex (MHC-I) complex on the cell surface, thus preventing the presentation of tumor neoantigens to the immune system. The results herein significantly extend these findings by showing that an additional 40% of DLBCL cases, despite expressing MHC-I, carry monoallelic HLA-I genetic alterations that limit the repertoire of neoantigens for presentation to immune cells. Both MHC-INEG and MHC-IPOS/monoallelically disrupted cases have significantly higher mutational load. Notably, homozygosis of HLA-I loci is significantly and preferentially enriched in the germline of DLBCL patients, suggesting a stepwise process by which limited neoantigen presentation is selected during DLBCL development.
Keywords: DLBCL, immune evasion, HLA
Abstract
Fifty percent of diffuse large B cell lymphoma (DLBCL) cases lack cell-surface expression of the class I major histocompatibility complex (MHC-I), thus escaping recognition by cytotoxic T cells. Here we show that, across B cell lymphomas, loss of MHC-I, but not MHC-II, is preferentially restricted to DLBCL. To identify the involved mechanisms, we performed whole exome and targeted HLA deep-sequencing in 74 DLBCL samples, and found somatic inactivation of B2M and the HLA-I loci in 80% (34 of 42) of MHC-INEG tumors. Furthermore, 70% (22 of 32) of MHC-IPOS DLBCLs harbored monoallelic HLA-I genetic alterations (MHC-IPOS/mono), indicating allele-specific inactivation. MHC-INEG and MHC-IPOS/mono cases harbored significantly higher mutational burden and inferred neoantigen load, suggesting potential coselection of HLA-I loss and sustained neoantigen production. Notably, the analysis of >500,000 individuals across different cancer types revealed common germline HLA-I homozygosity, preferentially in DLBCL. In mice, germinal-center B cells lacking HLA-I expression did not progress to lymphoma and were counterselected in the context of oncogene-driven lymphomagenesis, suggesting that additional events are needed to license immune evasion. These results suggest a multistep process of HLA-I loss in DLBCL development including both germline and somatic events, and have direct implications for the pathogenesis and immunotherapeutic targeting of this disease.
Diffuse large B cell lymphoma (DLBCL), the most common B cell lymphoma (1), is a genetically, phenotypically, and clinically heterogeneous disease that can occur de novo or upon histologic transformation of indolent lymphomas (2, 3). The updated World Health Organization classification recognizes two phenotypic subtypes of DLBCL that display common as well as subtype-specific genetic alterations (4–8) and are associated with different response to standard treatment (9–11): the germinal center (GC) B cell–like DLBCL, and the activated B cell–like (ABC) DLBCL, while ∼30% of cases remain unclassified. Moreover, at least five DLBCL genetic subsets have been recently defined based on the presence of concurrent genetic alterations, which were shown to correlate with distinct prognosis (12–14).
Genetic analysis of DLBCL has also provided initial clues as to how this tumor may escape immune surveillance. The B2M gene, encoding for an invariable subunit necessary for the assembly of the class I major histocompatibility complex (MHC-I), undergoes inactivating mutations and focal deletions in up to 29% of DLBCLs, constituting one of the most common altered genes in this malignancy (12, 13, 15, 16). Furthermore, the comparison of sequential biopsies obtained from follicular lymphoma (FLs) and their transformation into aggressive DLBCL (tFL) revealed the acquisition of B2M genomic aberrations with loss of B2M protein expression in 13% of tFL cases (17, 18), suggesting that disruption of the MHC-I complex plays a role in the progression from indolent disease to high-grade lymphoma.
The MHC-I complex is involved in the presentation of antigenic peptides derived from the degradation of self- and nonself-proteins, including viral- and tumor-associated antigens (19–21). The complex is a heterodimer expressed on the membrane of most nucleated cells and formed by the product of the B2M gene and one of the HLA-I heavy-chain (hcHLA) molecules, with HLA-A, -B, and -C being the most common. MHC-I complexes present nonself-antigens on the cell surface, where they are recognized by the αβ receptors of cytotoxic CD8+ T lymphocytes, resulting in the destruction of the target cell (22). Consistent with these notions, DLBCL cases carrying biallelic genetic inactivation of B2M lack cell surface expression of the MHC-I protein (15). However, the fraction of cases that fail to express surface MHC-I (43 to 75% of de novo DLBCL) is significantly higher than what can be explained by the presence of B2M genetic lesions, suggesting the existence of additional (genetic or epigenetic) mechanisms of inactivation (15, 16). Indeed, HLA-I gene mutations have been observed in large DLBCL whole-exome sequencing (WES) studies, and recent work reported that ∼70% of EZH2 mutated cases are negative for MHC-I expression, which could be restored by the use of EZH2 inhibitors in an Ezh2 mutant mouse model (15, 16). Nonetheless, the mechanisms leading to MHC-1 loss remain unknown for the majority of cases.
In this study, we addressed the role and specificity of MHC-I loss among B cell malignancies, the genetic mechanisms underlying MHC-I loss in DLBCL, and the genetics of MHC-I–positive (MHC-IPOS) tumors. Additionally, we investigated the contribution of MHC-I loss to the neoplastic transformation of GC B cells in vivo, alone, or in combination with BCL6 oncogene activation.
Results
Loss of HLA-I Protein Expression Is Preferentially Associated with DLBCL among Mature B Cell Malignancies.
In order to investigate whether the loss of surface MHC-I is a common phenomenon across B cell malignancies, we performed immunohistochemistry analysis with antibodies against B2M and HLA-I in a multiinstitutional panel of 657 lymphoma biopsies obtained at diagnosis. This panel was representative of the most common types of mature B cell lymphoma, comprising 422 DLBCL, 25 tFL, 43 Burkitt lymphomas (BL), 54 FL, 38 mantle cell lymphomas (MCL), 39 marginal zone lymphomas (MZL), and 36 chronic lymphocytic leukemia/small lymphocytic lymphomas (CLL/SLL).
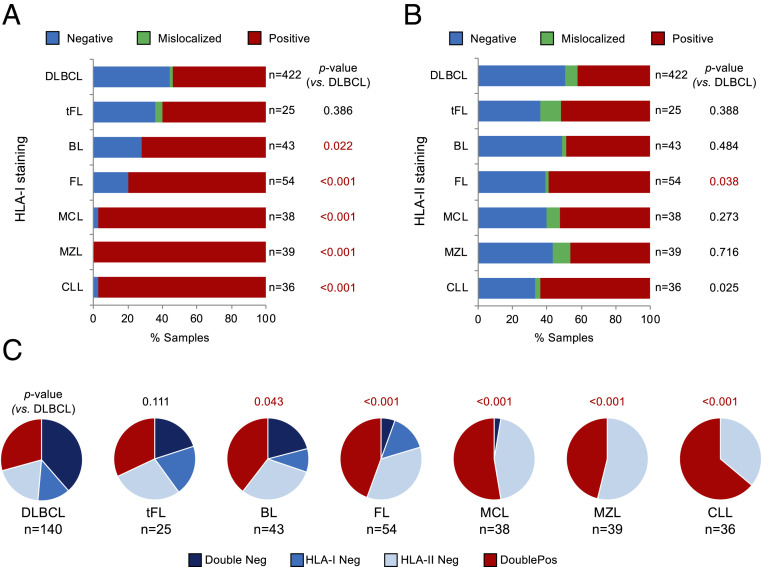
In line with previous reports, 46.2% (195 of 422) of de novo DLBCL cases and 40.0% (10 of 25) of DLBCLs derived from FL transformation were MHC-I cell surface-negative due to the complete lack of HLA-I protein expression or to aberrant cytoplasmic localization, confirming the high incidence of MHC-I loss in this disease (Fig. 1A). MHC-I–negative (MHC-INEG) DLBCLs included both GC B cell–like (21 of 33; 66.6%) and ABC-like (12 of 33; 36.4%) cases, and analogous distribution was observed in an independent series (GC B cell–like: 86 of 127; 67.7%; ABC-like: 41 of 127; 32.3%) (16). In contrast, loss of MHC-I expression was significantly less common in BL (n = 12 of 43 cases, 27.9%, P = 0.024) and FL (11 of 54, 20.4%, P < 0.001), and virtually absent in MCL (1 of 38 cases, 2.6%, P < 0.001), CLL/SLL (1 of 36, 2.8%, P < 0.001), and MZL (0 of 39, 0.0%, P < 0.001) (Fig. 1A). Within each sample, the pattern of MHC-I expression was highly uniform across the tumor cell population, with >70% of tumor cells displaying B2M and HLA-I membrane staining in cases scored as MHC-IPOS, and >90% of tumor cells lacking expression of these two proteins in cases scored as MHC-INEG.
Fig. 1.
Loss of MHC-I expression is significantly more frequent in DLBCL than in other mature B cell neoplasms. (A) Distribution and pattern of HLA-I protein expression in 657 B cell non-Hodgkin lymphomas. The total number of cases in each lymphoma subtype is indicated on the right; in red, statistically significant differences compared to DLBCL. (B) Pattern of HLA-II protein expression in the same cases. (C) Relationship between HLA-I and HLA-II protein expression in the indicated lymphomas.
The preferential association of MHC-I loss with DLBCL and tFL was not paralleled by the loss of MHC-II expression, observed in 30 to 50% of cases independent of lymphoma entity (81 of 140 DLBCL [57.8%], 12 of 25 tFL [48.0%], 22 of 43 BL [51.2%, P = 0.48], 22 of 54 FL [40.7%, P = 0.04], 18 of 38 MCL [47.4%, P = 0.27], 21 of 39 MZL [53.8%, P = 0.72], and 13 of 36 CLL/SLL [36.1%, P = 0.03]) (Fig. 1B).
Integrated analysis of MHC-I and MHC-II expression showed concurrent loss of these two proteins in 54 of 140 (38.6%) DLBCL biopsies analyzed, compared to 5 of 25 tFL (20%, not significant) and 9 of 43 BL (20.9%, P = 0.04); only 3 of 54 FL (5.6%, P < 0.001), 1 of 38 MCL (2.6%, P < 0.001), and none of the 39 MZL and 36 CLL/SLL (P < 0.001) were negative for both proteins, largely reflecting the preferential association of MHC-I loss with DLBCL and tFL (Fig. 1C).
Together, these data point to a selective role for loss of MHC-I and combined MHC-I/MHC-II in the pathogenesis of DLBCL, but not of more indolent or non-GC–derived mature B cell neoplasms.
Both B2M and HLA-I Gene Inactivation Contribute to Loss of MHC-I Expression in DLBCL.
To comprehensively investigate the genetic mechanisms underlying the loss of MHC-I membrane expression in DLBCL, we analyzed a panel of 74 previously untreated DLBCL samples with matched normal DNA (n = 32 MHC-IPOS and 42 MHC-INEG) by integrating WES or whole-genome sequencing (WGS) with targeted deep sequencing of the hcHLA-I locus, performed in a subset of cases (Dataset S1). For the identification of lesions in the highly polymorphic HLA loci, we first genotyped the 74 patients by applying the Polysolver algorithm to both normal and tumor-derived DNA sequences, and then compared the tumor alleles to the corresponding germline hcHLA-I alleles (two each for HLA-A, -B, and -C). The HLALOH computational tool, which allows monitoring allele-specific changes in HLA gene copy number, was used to uncover allelic imbalances due to genetic deletion of one allele or copy-neutral loss of heterozgyosity (cnLOH).
Consistent with previous reports (16), 17 of 42 MHC-INEG DLBCLs harbored biallelic (n = 11) or monoallelic (n = 6) mutations and deletions inactivating B2M (Fig. 2A and Datasets S2 and S3). Four additional cases (9.4%) showed biallelic disruption of one or more of the main hcHLA-I genes (Datasets S3–S5). Analogously, two B2M-WT DLBCL cell lines with aberrant MHC-I cytoplasmic localization were found to carry biallelic truncating mutations in both HLA-A and HLA-B (SI Appendix, Fig. S1). Biallelic genetic alterations of hcHLA-I were mutually exclusive with biallelic B2M disruption, together accounting for 35.7% (15 of 42) of MHC-INEG DLBCLs (Fig. 2A).
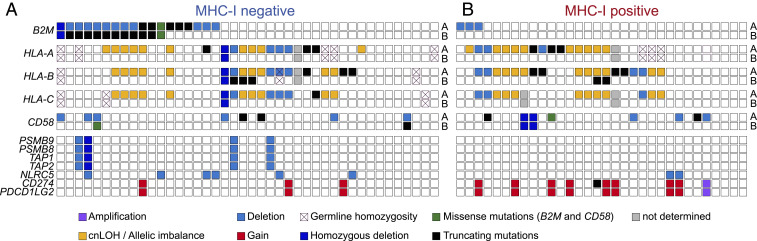
Fig. 2.
Mutations and deletions of the hcHLA-I genes in MHC-INEG and MHC-IPOS DLBCL. Genetic lesions affecting B2M and other genes implicated in antigen presentation in 42 MHC-INEG (A) and 32 MHC-IPOS (B) DLBCL primary cases, color-coded according to mutation type. Cross marks indicate germline homozygous HLA-I alleles, where allele-specific LOH cannot be predicted; on the right side, “A” and “B” denote separate alleles (see Materials and Methods and SI Appendix, Tables S2–S4 for details on mutation type).
In addition to tumors harboring biallelic hcHLA-I lesions, 18 cases showed somatic monoallelic loss of one or more of the main hcHLA-I genes in the absence (n = 11) or presence (n = 7) of B2M lesions, due to a variety of genetic mechanisms that included truncating mutations (n = 5 cases), heterozygous deletions (n = 3), and cnLOH/allelic imbalance (n = 10) (Fig. 2A and Datasets S3 and S5); missense mutations were not considered, as their functional impact is currently unclear (see reconstitution experiments in SI Appendix, Fig. S2).
Analysis of other genes implicated in antigen presentation through MHC-I, among which those encoding for the known MHC-I transactivator NLRC5 (23) and for the transporter associated with antigen-processing complex (TAP1/TAP2), uncovered copy number losses in 9 of 42 cases (Fig. 2A). With one exception, these lesions were heterozygous and were observed both in the presence or absence of B2M and/or hcHLA-I genetic alterations, with no specific distribution.
Together, these results suggest that, in a subset of B2M-WT cases, loss of surface MHC-I expression could be explained by direct biallelic genetic disruption of the hcHLA-I genes. Overall, 76.2% (n = 32 of 42) of MHC-INEG DLBCLs harbor genetic alterations in B2M and/or hcHLA-I genes.
Common Somatic Monoallelic HLA-I Loss in MHC-IPOS Cases.
To explore possible mechanisms of immune escape in DLBCL retaining cell-surface MHC-I expression, we investigated the genetics of B2M and hcHLA-I in the subset of 32 MHC-IPOS cases. As expected, biallelic inactivation of B2M or HLA-I genes was never found in these tumors, and only three cases harbored monoallelic B2M deletions (9.4%; P = 0.002) (Fig. 2B). Conversely, HLA-I allelic imbalance, defined as the monoallelic loss of at least one hcHLA-I gene, was detected in 22 of 32 cases (68.8%), of which 5 harbored genetic deletions encompassing one or more hcHLA-I loci, 8 showed truncating mutations that are predicted to eliminate the protein antigen binding domains, and 10 were cnLOH, with or without a concurrent point mutation (Fig. 2B and Datasets S3–S5).
Thus, monoallelic hcHLA-I loss is a common event in a substantial fraction of MHC-IPOS DLBCL (hereafter referred to as MHC-IPOS/mono), raising the hypothesis that disruption of a single HLA-I allele could interfere with the presentation of specific antigens by the MHC-I complex (Discussion).
MHC-INEG and MHC-IPOS/mono DLBCLs Show Higher Mutation Load and Increased Activation-Induced Cytidine Deaminase-Mediated Aberrant Somatic Hypermutation.
In several cancer types, the loss of MHC-I expression has been associated with increased mutational load, suggesting that the accumulation of DNA damage and the consequent generation of neoantigens are coselected with mechanisms allowing escape from immune recognition (24). To investigate whether MHC-I–defective DLBCLs accumulate a higher number of somatic mutations, we first assessed the nonsilent mutation load of the 74 DLBCL samples by interrogating WES and, in a subset of cases, WGS data.
When focusing on genes expressed in normal or transformed GC B cells, MHC-INEG DLBCLs showed a significantly higher nonsilent mutation load compared with MHC-IPOS cases carrying WT hcHLA-I alleles (MHC-IPOS/WT; average: 73.7 ± 48.0 vs. 33.4 ± 22.3; Mann–Whitney U = 94.5, P = 0.008) (Fig. 3A). Notably, a significantly higher mutational burden was also detected in MHC-IPOS/mono tumors (average 60.1 ± 29.5; Mann–Whitney U = 46.5, P = 0.010), which were in turn indistinguishable from the MHC-INEG group (Mann–Whitney U = 392.0, P = 0.326) (Fig. 3A).
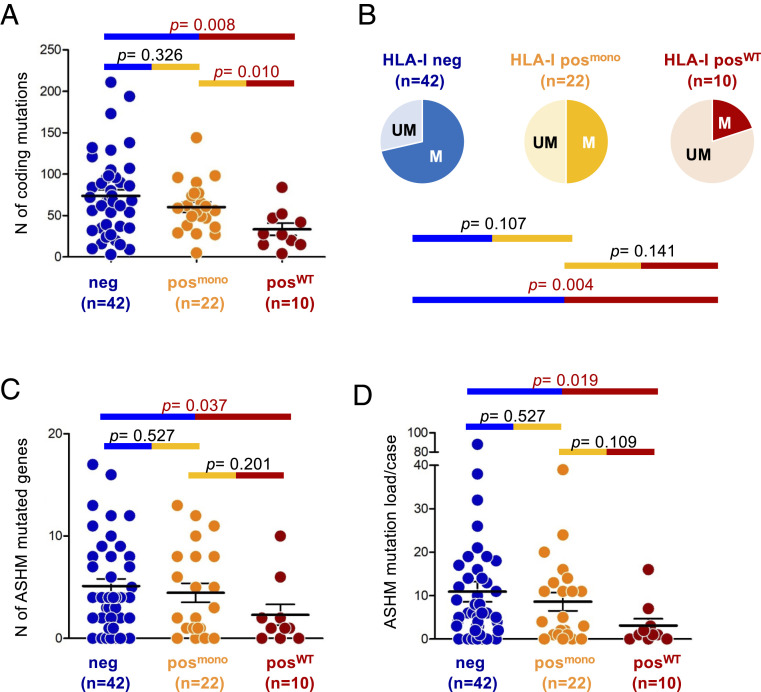
Fig. 3.
MHC-INEG and MHC-IPOS/mono DLBCL show significantly higher coding mutation load. (A) Number of coding somatic mutations in each of the 74 DLBCL samples, subdivided into 3 categories based on MHC-I protein expression and hcHLA-I gene status. Only mutations affecting genes that are expressed in B cells were considered. (B) Proportion of cases displaying ASHM in MHC-INEG, MHC-IPOS/mono and MHC-IPOS/WT DLBCL. Cases were defined as hypermutated (M) if harboring at least three somatic mutations in one or more ASHM genes. UM, unmutated. (C) Number of mutated ASHM-target genes/case in the three MHC-I/HLA-I defined categories. The total n of samples analyzed is indicated below the graph. (D) Number of somatic mutations affecting ASHM-associated genes in each of the 74 DLBCL samples.
We then examined whether the different mutational load across DLBCL subgroups could be driven in part by aberrant somatic hypermutation (ASHM), a mechanism described in over 50% of de novo DLBCL as well as in tFL (13, 18, 25). To this end, we analyzed 126 previously identified target genes (13) for the presence of variants targeting ∼2 kb from the transcription initiation site (the hypermutable domain) (Materials and Methods). We found mutations displaying typical features of activation-induced cytidine deaminase (AID)-mediated ASHM in 30 of 42 (71.4%) MHC-INEG cases, but only 2 of 10 (20.0%) MHC-IPOS/WT samples (P = 0.004) (Fig. 3B). A twofold higher prevalence of ASHM-targeted cases was also detected among MHC-IPOS/mono DLBCL (n = 11 of 22, 50.0%), although the small size of this panel prevents the assessment of statistical significance. Moreover, both MHC-INEG and MHC-IPOS/mono cases carried a larger number of ASHM-mutated genes per case (average, 5.1 and 4.5 vs. 2.3 in MHC-IPOS/WT; P = 0.037 and 0.201, respectively; Mann–Whitney U test), which was paralleled by a higher number of mutations per case (average, 10.9 and 8.6 vs. 4.1; P = 0.019 and 0.109; Mann–Whitney U test) (Fig. 3 C and D).
These data indicate that loss of MHC-I is associated with higher mutational load, reflecting in part the aberrant activity of the somatic hypermutation mechanism. Notably, the similar mutational load of MHC-INEG and MHC-IPOS/mono DLBCL supports the hypothesis that loss of a single and possibly specific hcHLA-I allele might be sufficient to blunt the immunogenic potential of the tumor cells (Discussion).
MHC-INEG and MHC-IPOS/mono DLBCL Exhibit Higher Predicted Neoantigen Load.
Studies conducted on epithelial cancers have shown that the overall mutational load directly correlates with the neoantigen load of a tumor (26). Tumors with higher neoantigen load may thus be under selective pressure to lose MHC-I in order to escape immune surveillance, which could explain their overrepresentation in MHC-I–defective cases. In order to test this hypothesis in DLBCL, we interrogated the WES/WGS data with three well-established algorithms for the prediction of tumor-associated neoantigens (NetMHC, NetMHCpan, and PickPocket) (Materials and Methods) (27–29), and compared the overall neoantigen load in MHC-INEG, MHC-IPOS/WT, and MHC-IPOS/mono cases.
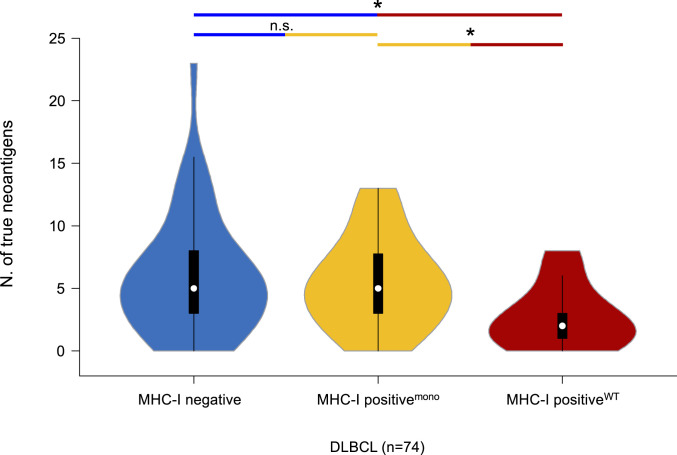
Of 5,128 somatic coding mutations identified across the 74 DLBCL samples, 394 (7.7%) were categorized as predicted tumor neoantigens (pTNA) upon filtering for affinity (≤500 nM), mutant affinity specificity (mutant affinity > WT affinity), neoantigen size (9- to 10mers), expression in DLBCL and normal B cells, and nonhomology to human peptides (Materials and Methods and SI Appendix, Fig. S3). Peptide binding-affinity assays of 12 representative pTNAs validated the in silico prediction, confirming the robustness of the approach (SI Appendix, Fig. S4). We found that the average number of pTNAs was significantly higher in the MHC-INEG cases (n = ∼6 per case; range: 0 to 23) compared to MHC-IPOS/WT DLBCL (n = 3 per case; range: 0 to 8; Mann–Whitney U = 104.0; P = 0.014). Of note, the pTNA load of MHC-IPOS/mono DLBCLs was also significantly higher compared to that of MHC-IPOS/WT cases (∼6 per case; range: 0 to 13; Mann–Whitney U = 52.0; P = 0.019) (Fig. 4 and Dataset S6), further supporting a mechanistic analogy in terms of immune-escape between MHC-INEG and MHC-IPOS/mono DLBCL.
Fig. 4.
MHC-INEG and MHC-IPOS/mono DLBCL show higher neoantigen load. Number of predicted true neoantigens identified in different HLA-defined subsets of DLBCL using NETMHC, NETMHCpan, and Pickpocket. *P < 0.05.
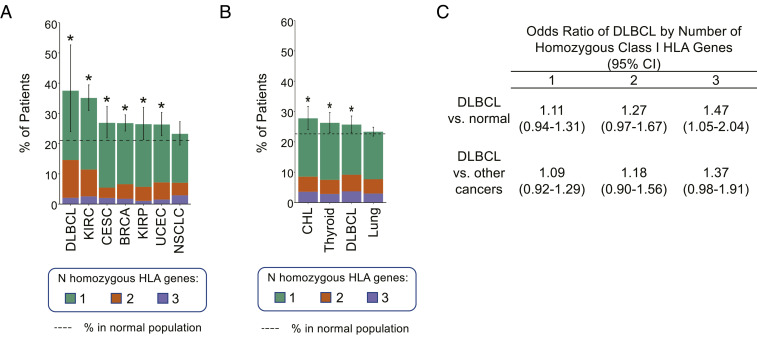
DLBCL Patients Show Significantly Higher Rate of HLA-I Germline Homozygosity.
The similarity between MHC-INEG and MHC-IPOS/mono DLBCL suggested that decreased HLA-I gene diversity may impair the ability of cells to present endogenous and exogenous antigens by lowering the repertoire of functional MHC-I molecules (Discussion). To test whether reduced HLA-I genes diversity may initiate in the germline, we evaluated the percentage of cases harboring one, two, and/or all three classic hcHLA-I (A, B, C) genes in homozygosis in a cohort of 9,623 patients with 30 different types of cancer from The Cancer Genome Atlas (TCGA). We found that 18 of 48 (38%) TCGA DLBCL patients had at least one homozygous HLA-I germline locus, representing the tumor type with the highest frequency of homozygosity across all cancers (Fig. 5A and SI Appendix, Fig. S5A). This frequency was also significantly higher than that observed in a comparable healthy population obtained from the Genotype-Tissue Expression (GTEx) database (21%; P = 0.0117, binomial test). The association of germline HLA-I homozygosity with increased risk of DLBCL was confirmed in a larger panel of cancer patients from the UK BioBank cohort (in total, 502,506 individuals), where DLBCL showed as one of the tumors most strongly associated with germline HLA-I homozygosity (26%, 258 of 1,004), significantly higher than the normal rate in individuals without a cancer diagnosis in the same population (23%, P = 0.0236, binomial test) (Fig. 5B and SI Appendix, Fig. S5B). Finally, we calculated the odds ratio (OR) of DLBCL diagnosis as related to the number of homozygous HLA-I genes in the UK BioBank cohort. We found that the OR of DLBCL increases with the number of homozygous HLA-I genes (1.11, 1.27, and 1.47 for one, two, and three homozygous genes) (Fig. 5C). Collectively, these observations suggest a multistep process of HLA-I loss including both germline and somatic events in DLBCL pathogenesis.
Fig. 5.
Increased risk of DLBCL in individuals with homozygous germline HLA-I genes. (A and B) Percentage of patients harboring homozygous germline HLA-I genes in the indicated cancers (A, TCGA dataset; B, UK BioBank dataset). Dotted line indicates the percentage in a matched normal population (23% for UK BioBank samples and 21% for TCGA samples, based on GTEx; *P < 0.05, binomial test, 95% CI). NSCLC and lung cancer are shown as a reference category for lack of enrichment. See also SI Appendix, Fig. S5. (C) ORs of DLBCL in patients with homozygous germline HLA-I genes as compared to normal individuals or to patients with other cancer diagnoses. BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHL, classical Hodgkin lymphoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; NSCLC, nonsmall cells lung cancer; UCEC, uterine corpus endometrial carcinoma.
Consequences of MHC-I Loss on Lymphomagenesis.
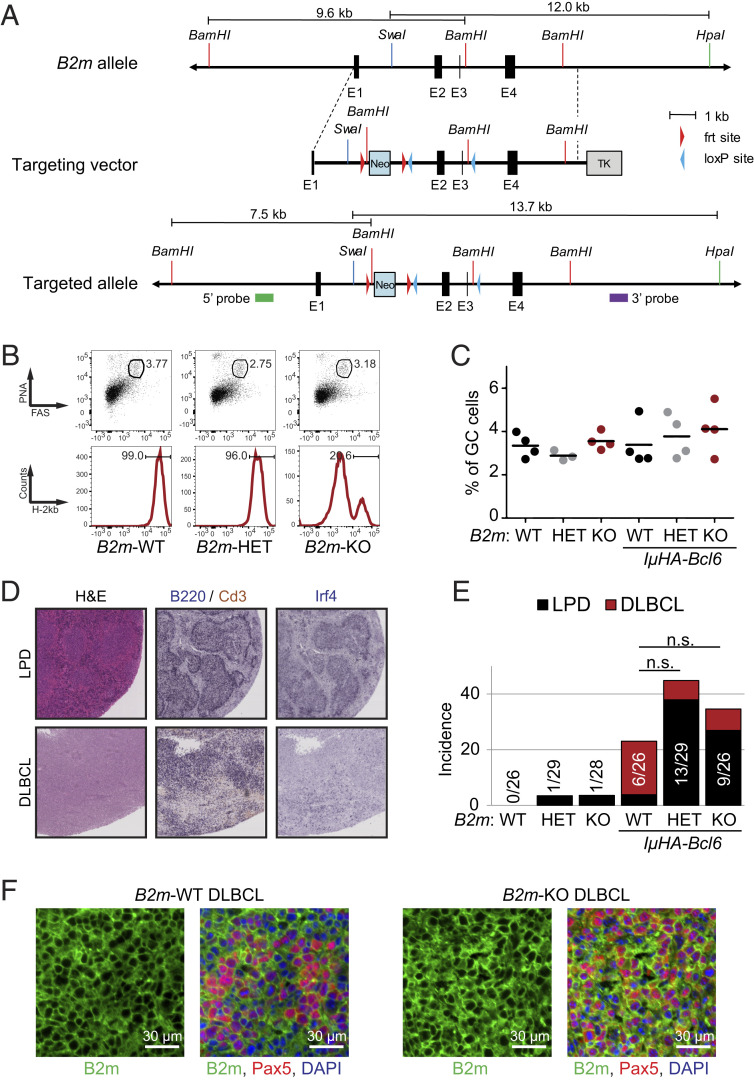
In order to investigate the effect of MHC-I loss in the malignant transformation process in vivo, we analyzed the GC response and lymphomagenesis in mice genetically engineered to lack B2m and thus MHC-I expression specifically in GC B cells, either as a single lesion or together with oncogenic Bcl6 expression via the IμHA-Bcl6 allele (30), which recapitulates the cooccurrence of BCL6 translocations reported in 30 to 50% of B2M mutated human DLBCL (12, 13).
To this end, we engineered the murine chr2qE5 locus with loxP sites flanking the B2m gene exons 2 and 3 (Fig. 6A), and crossed the resulting mice with the GC-specific Cγ1-Cre deleter strain (31). Southern blot analysis confirmed the correct targeting of the locus (SI Appendix, Fig. S6A), and FACS analysis using antibodies against the H-2Kb haplotype documented the loss of MHC-I surface expression in >80% of B220+PNAhighCd95+ GC B cells isolated from B2mfl/fl/Cγ1cre/+ (B2m-KO [knockout]) mice 10 d after sheep red blood cell immunization (Fig. 6B). Despite the absence of surface MHC-I, the percentage of GC B cells was comparable in both single and compound B2m-KO, B2mfl/+/Cγ1cre/+ (B2m-HET), and B2m+/+/Cγ1cre/+ (B2m-WT) littermates (Fig. 6 B and C and SI Appendix, Fig. S6 B and C). The GC structures appeared normal also in terms of dark-zone/light-zone ratio, indicating that acute loss of B2m in GC B cells is compatible with cell proliferation and survival.
Fig. 6.
Tumors developing in compound IμHA-Bcl6/B2mfl/fl mice retain MHC-I expression. (A) Schematic representation of the B2m targeting strategy. The restriction sites used for Southern blot analysis are indicated, with the expected restriction fragment size. Symbols depict the 5′ and 3′ probes. See also SI Appendix, Fig. S6 for verification of correct targeting. (B) Representative flow cytometric analysis of splenic B220+ cells from B2m+/+/Cγ1cre/+ (B2m-WT), B2mfl/+/Cγ1cre/+ (B2m-HET), and B2mfl/fl/Cγ1cre/+ (B2m-KO) mice killed 10 d after SRBC immunization. (Upper) Percentage of Cd95+PNA+ GC B cells in the B220+ gate. (Lower) Percentage of surface MHC-IPOS cells in the GC B cell gate, determined using the anti-H-2Kb antibody. (C) Cumulative percentage of splenic GC B cells in B2m-WT, B2m-HET, B2m-KO, and compound IμHA-Bcl6 littermates, measured by flow cytometry as indicated in B. (D) H&E and immunohistochemical staining of B220, Cd3, and Irf4 in representative spleens from mice diagnosed with LPD or DLBCL. Magnification, 20×. (E) Disease incidence in the indicated cohorts. LPD, defined as in Materials and Methods); n.s., not significant. (F) Immunofluorescence staining for B2m and the B cell marker Pax5 in two representative DLBCL from IμHA-Bcl6/B2m-WT and IμHA-Bcl6/B2m-KO mice.
When monitored over 15 mo, chronically immunized B2m-KO/IμHA-Bcl6 mice did not show significant differences in event-free survival correlating with the genotype (SI Appendix, Fig. S6D), except for the reported increased mortality of old IμHA-Bcl6 mice, independent of B2m gene status (30). B2m loss had no significant impact on the overall incidence of lymphoproliferative disorders driven by deregulated BCL6 expression, which were detected in 6 of 26 (23.1%) IμHA-Bcl6/B2m-WT mice, 13 of 29 (44.8%) IμHA-Bcl6/B2m-HET mice, and 9 of 26 (34%) IμHA-Bcl6/B2m-KO mice, although the B2m-KO compound animals showed a higher proportion of oligoclonal, B cell lymphoproliferative diseases (LPD) (Fig. 6 D and E). Notably, however, all seven LPDs and both DLBCLs diagnosed in IμHA-Bcl6/B2m-KO mice showed expression of surface MHC-I by FACS analysis of H-2Kb and B2m immunofluorescence, which revealed only sparse negative cells (Fig. 6F and SI Appendix, Fig. S6E). Thus, the expanded B cell population in these animals must have originated from cells that had escaped B2m deletion. Taken together, these data strongly indicate that B2m-null (MHC-I–null) GC B cells are counter-selected over time. These observations are consistent with the role of natural killer (NK) cells in eliminating MHC-I–null cells (32, 33), and suggest the requirement for additional alterations in order to foster immune escape (Discussion).
Discussion
Loss of surface MHC-I expression has been observed as a common phenomenon across malignancies of different cellular origin, including epithelial and hematologic cancers (34, 35).
The first finding of our study is that, in the context of mature B lymphoid malignancies, and in line with previous studies in Hodgkin lymphoma and primary mediastinal large B cell lymphoma (5, 7, 15, 17, 18, 36–41), down-regulation of MHC-I expression represents a recurrent and specific event in more aggressive diseases, such as DLBCL and tFL, but not in MCL (1%), CLL, and MZL. Within DLBCL, MHC-I loss was observed in both GC B cell (∼60%) and ABC (∼40%) molecular subtypes, with some nonsignificant differences among genetically defined classes (SI Appendix, Fig. S7) (12, 13). These data suggest a specific role for escape from MHC-I–mediated immune-surveillance mechanisms in the pathogenesis of DLBCL, consistent with the notion that immune evasion is linked to the higher mutational burden of these diseases.
We found that, in addition to B2M genetic lesions, biallelic disruption of hcHLA-I genes represents an alternative mechanism to explain the loss of MHC-I expression in B2M unmutated tumors. Of note, only one of our samples showed complete simultaneous loss of all three major HLA-I alleles, suggesting a role for allele dosage, and consistent with the idea that HLA-I genes are not functionally redundant. Conversely, no clear underlying genetic cause was identified for the remaining large fraction of MHC-INEG DLBCLs, which retained either one (43%) or both (26%) intact B2M and hcHLA-I alleles. This negative result is unlikely due to technical issues (e.g., low coverage depth) because analogous results were obtained by using WES or the more robust targeted HLA deep-sequencing approach. A search for mutations in other genes implicated in MHC-I expression revealed recurrent heterozygous deletions of NLRC5 and TAP1/TAP2, but their distribution was independent of surface MHC-I expression and of genetic alterations in B2M and hcHLA-I (Fig. 2) (16). Thus, additional nongenetic mechanisms of (allele-specific) repression, such as epigenetic silencing/DNA hypermethylation or signals from the tumor microenvironment, are likely responsible for the down-regulation of MHC-I expression in this malignancy, as recently reported in solid tumors (42). Among these mechanisms, EZH2 activating mutations were found significantly associated with lack of MHC-I expression, consistent with the increased levels of the repressive H3K27me3 mark observed at the promoter of Nlrc5 upon overexpression of Ezh2 in mouse B cells (16). Indeed, all three EZH2 mutated DLBCL samples in our study were MHC-INEG, although two of them concurrently harbored monoallelic mutations in B2M and hcHLA-I. Thus, future investigations based on comprehensive genetic and epigenetic analysis will be necessary to clarify these issues.
A notable finding of this study was the identification of monoallelic hcHLA-I inactivation (due to genetic lesions or cnLOH) in as many as 69% of MHC-IPOS DLBCL, where these lesions are predicted to cause allele-specific loss of expression. These findings suggest that HLA-I LOH may represent a pervasive mechanism of immune evasion also in MHC-IPOS DLBCL, analogous to what was observed at lower frequency in lung cancer (15, 43). One explanation for this result could be that the loss of specific HLA haplotypes bearing the highest affinity for relevant tumor (neo)antigens is sufficient to escape T cell recognition. The first notable feature of such a model is that these cells would remain MHC-IPOS and therefore evade NK cell-mediated attack otherwise triggered by the loss of MHC-I inhibitory signals (34). Additionally, this model would suggest that attempts to epigenetically reactivate silent HLA-I alleles (44) may not lead to the restoration of tumor immunogenicity, since the most relevant neoantigen presentation may have already been lost via genetic inactivation of the corresponding presenting allele. A confirmation of these notions requires the combined analysis of the genetic HLA-I make-up and the neoantigen allele-specific load of the tumor vis à vis the T cell receptor specificities of autologous T cells.
Our analysis revealed an increased risk of DLBCL in individuals with germline homozygosity for the HLA-I genes (45). The potential relevance of this finding is further emphasized by the significantly higher frequency of DLBCL versus other tumor types, suggesting a specific role in this disease. Consistent with the model suggested above for somatic alterations, these observations lend support to a model of multistep restriction of HLA alleles that would start at the germline level and then progressively reduce the repertoire of antigen-presenting molecules during DLBCL pathogenesis via somatic gene alterations involving HLA-I genes. The specific predisposition for this cancer type could be explained by its intrinsic mutagenic feature due to AID-mediated ASHM (25), which would require progressively increasing protection from immune recognition.
Similar to results in lung cancer (24), MHC-INEG and MHC-IPOS/mono DLBCLs were associated with significantly higher load of nonsilent mutations compared to MHC-IPOS/WT lymphomas, paralleled by an increased number of predicted neoantigens (Fig. 5). These data suggest that the aberrant activity of AID could be a major contributor to the selection of cells capable of evading immune recognition stimulated by increased neoantigen load. Supporting this concept, both ASHM and HLA-I loss are generally absent in the dominant FL clone, while they are commonly acquired upon histologic transformation (18).
In vivo, conditional loss of B2m was not associated with increased penetrance of BCL6-driven lymphomas. However, the LPDs that develop in compound IμHA-Bcl6/B2m-KO mice all retained expression of MHC-I, pointing to a strong selective pressure against the B2m-deficient GC B cell population over time. This antitumor response could be mediated by the activation of NK cell cytotoxicity that is innately elicited by the down-regulation of surface MHC-I, with consequent lack of engagement of NK cell inhibitory receptors (32, 33, 46). Thus, additional pathways may need to be disrupted in order to confer survival advantage to the precursor tumor cell. Indeed, as many as 61% of human DLBCLs concurrently lack the expression of MHC-I and CD58 (15), a ligand for the CD2 receptor required for NK cell–mediated recognition (47). Unfortunately, the role of CD58-mediated NK cell escape could not be addressed in vivo, as no mouse homolog has been identified for this gene.
In conclusion, the results herein broaden the role of (complete or haplotype-specific) MHC-I inactivation in the escape from antitumor immune surveillance during the evolution of DLBCL, with direct clinical implications for the development of therapeutic approaches based on immunomodulatory molecules.
Material and Methods
Study Panel.
A multiinstitutional panel of 657 formalin-fixed paraffin-embedded (FFPE) biopsies obtained at diagnosis from various B cell lymphoma types was used for the analysis of B2M and HLA-I protein expression (422 DLBCL, 25 tFL, 43 BL, 54 FL, 38 MCL, 39 MZL, and 36 CLL/SLL). A subset of 74 DLBCL samples was then selected for molecular studies based on availability of 1) paired normal DNA, and 2) FFPE sections for immunohistochemistry analysis of MHC-I expression (n = 42 MHC-INEG and 32 MHC-IPOS) (Dataset S1). The study was approved by the Columbia University Institutional Review Board as exempt research of anonymized/de-identified existing pathological specimens, under regulatory guideline 45 CFR 46.101(b) (4). Thirty-nine of these cases have been used in previously published genomic analyses of DLBCL (12, 48, 49).
Immunohistochemistry and Immunofluorescence Analysis.
Analysis of human and mouse samples was performed on FFPE tissue sections according to standard protocols, with the antibodies reported in SI Appendix, Supplemental Materials and Methods. Samples were independently scored by two investigators, using the standard cutoff of 20% to discriminate negative vs. positive cases; however, >90% of cases that were scored as MHC-IPOS showed membrane staining in >70% of tumor cells, and all cases scored as MHC-INEG lacked membrane signal in >90% of tumors cells. Only samples with concordant calls were included in the study.
Genomic Analyses.
Purified genomic DNA from matched tumor and normal tissues was used for WES, WGS, targeted sequencing of the HLA regions, and ASHM analysis, as reported in detail in SI Appendix, Supplemental Materials and Methods (see also Datasets S7–S9 for details of the sequencing performance). Somatic variants were identified with the Statistical Algorithm for Variant Identification (SAVI) (50); HLA genotyping and mutation calling were performed by applying the Polysolver computational tool, according to published methods (51), followed by Sanger-sequencing validation. The presence of copy number aberrations was determined by Sequenza (52) and confirmed, in a subset of cases, by SNP6 array or GATK analysis; LOHHLA was used to identify haplotype-specific copy number changes of the HLA locus, as described previously (24).
Assessment of Germline Homozygosity at HLA-I Loci in TCGA and UK BioBank.
Germline homozygosity at the HLA-I loci was assessed in 9,623 patients with 30 different types of cancer from the TCGA project (https://www.cancer.gov/tcga), using previously published HLA-I genotypes (53). In addition, HLA-I genotypes were obtained for 488,265 individuals from the UK BioBank (https://www.ukbiobank.ac.uk/); HLA alleles were imputed as the pair of alleles with maximum posterior probability from HLA*IMP:02, as described previously (54). For both cohorts, individuals whose HLA genotypes showed two identical alleles in at least one HLA-I gene were classified as being homozygous. The rate of homozygosity in the general population was estimated using normal samples from the GTEx project (https://gtexportal.org/home/) (see SI Appendix, Supplemental Materials and Methods for additional details).
Targeting Vector Construction and Generation of B2m Conditional KO Mice.
The B2m targeting vector was constructed by sequential subcloning of PCR-generated fragments into the pEMC1-Neo vector. An frt-flanked neomycin-resistance cassette (Neo) was cloned upstream of two loxP-site flanking exon 2 and 3 of the B2m gene (Fig. 6A). The targeting vector was electroporated in the murine embryonic stem (ES) cell line Sv129, and clones were selected with Neomicin (1 μg/μL). Resistant clones were screened for homologous recombination by Southern blot analysis of BamHI-digested DNA, using a 5′ external probe, and of SwaI/HpaI double-digested DNA using a 3′ external probe (Fig. 6A). Homologous recombinant ES cell clones were injected into blastocysts derived from C57BL/6 mice. Chimeric mice able to transmit the B2m conditional allele through the germline were crossed with a mouse expressing the flippase (Flp) recombinase in order to eliminate the Neomycin-resistence cassette, and then backcrossed onto a C57BL/6 background.
Mice.
Deletion of B2m was directed to GC B cells by breeding the mice with the Cγ1-Cre deleter strain (31). The offspring were bred with IμHA-Bcl6 knockin mice, which carry a BCL6 transgene downstream of the endogenous immunoglobulin Iµ promoter (30), to generate compound mice. Analysis of T cell–dependent immune responses was performed on animals intraperitoneally injected with SRBC (Cocalico Biologicals) (n = 500 million per mouse in PBS) and analyzed 10 d postimmunization at 3 and 6 mo of age. Both genders were included in the experiments. Tumor watch studies were conducted for a minimum of 15 mo on 26 to 29 animals per genotype, which were killed upon evidence of illness or at endpoint. Mice were housed in a dedicated pathogen-free environment, and all animal work was performed according to protocols revised and approved by the National Cancer Institute and Columbia University Institutional Animal Care and Use Committee. Genotyping was performed by PCR analysis, and the protocol is available upon request. Mice were killed according to the regulations of the Department of Comparative Medicine, Columbia University. Details on the flow cytometric and immunohistochemistry analysis of mouse cohorts are reported in SI Appendix, Supplemental Materials and Methods.
Detailed procedures and methods for the in silico neoantigen prediction, neoantigen peptides synthesis, HLA-I binding affinity assays, flow cytometric analysis, and statistical analysis can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank David Dominguez-Sola (Icahn School of Medicine) for expert advice on the immunohistochemical analysis of mouse and human tissues, and Hongyan Tang and Tongwei Mo for technical assistance with the mouse husbandry and analysis. Data from the UK Biobank were accessed under Application Number 47884. This study was supported by NIH Grants R35CA-210105 (to R.D.-F.) and U01CA243073 and U54 CA193313 (to R.R.); the Stand Up to Cancer/Lustgarden Foundation (R.R.); and Associazione Italiana per la Ricerca sul Cancro Grant 5 x 1000 No. 21198 “Molecular bases of disease dissemination in lymphoid malignancies to optimize curative therapeutic strategies” (to G.G). E.L. was supported by National Cancer Institute (NCI) Grant K00CA212478. K.G. was supported by the Medical Scientist Training Program (T32GM007367). The study was funded in part through the NIH/NCI Cancer Center Support Grant P30CA013696 and used the resources of the Cancer Center Flow Core Facility (Columbia University), the Genomics and High Throughput Screening Shared Resource (Columbia University), and the Molecular Pathology Shared Resource at Columbia University Irving Medical Center.
Footnotes
Competing interest statement: R.R. is founder of Genotwin and a member of the scientific advisory board of AimedBio. R.D.-F. is a member of the scientific advisory board of Akemara and NeoGenomics, and a consultant of Astra Zeneca. The work reported in this paper has no relation to the current activities in these companies.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2104504118/-/DCSupplemental.
Data Availability
The WES and WGS data for the 74 DLBCL patients are available in the European Genome-phenome Archive (EGA) (accession no. EGAS00001005054) and the National Center for Biotechnology Information (accession no. phs000450.v3.p).
References
- 1.Swerdlow S. H., et al., WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (WHO, 2008). [PubMed] [Google Scholar]
- 2.Pasqualucci L., Dalla-Favera R., Genetics of diffuse large B-cell lymphoma. Blood 131, 2307–2319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunleavy K., Wilson W. H., Appropriate management of molecular subtypes of diffuse large B-cell lymphoma. Oncology (Williston Park) 28, 326–334 (2014). [PMC free article] [PubMed] [Google Scholar]
- 4.Lohr J. G., et al., Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 3879–3884 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morin R. D., et al., Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasqualucci L., et al., Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 471, 189–195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasqualucci L., et al., Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43, 830–837 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morin R. D., et al., Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 122, 1256–1265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright G., et al., A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 100, 9991–9996 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenz G., Staudt L. M., Aggressive lymphomas. N. Engl. J. Med. 362, 1417–1429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alizadeh A. A., et al., Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Chapuy B., et al., Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 24, 679–690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz R., et al., Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 378, 1396–1407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright G. W., et al., A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell 37, 551–568.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Challa-Malladi M., et al., Combined genetic inactivation of β2-microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell 20, 728–740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ennishi D., et al., Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov. 9, 546–563 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Okosun J., et al., Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46, 176–181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasqualucci L., et al., Genetics of follicular lymphoma transformation. Cell Rep. 6, 130–140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Townsend A. R., Gotch F. M., Davey J., Cytotoxic T cells recognize fragments of the influenza nucleoprotein. Cell 42, 457–467 (1985). [DOI] [PubMed] [Google Scholar]
- 20.Zinkernagel R. M., Doherty P. C., Immunological surveillance against altered self components by sensitised T lymphocytes in lymphocytic choriomeningitis. Nature 251, 547–548 (1974). [DOI] [PubMed] [Google Scholar]
- 21.Zinkernagel R. M., Doherty P. C., Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature 248, 701–702 (1974). [DOI] [PubMed] [Google Scholar]
- 22.Yewdell J. W., Reits E., Neefjes J., Making sense of mass destruction: Quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 3, 952–961 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi K. S., van den Elsen P. J., NLRC5: A key regulator of MHC class I-dependent immune responses. Nat. Rev. Immunol. 12, 813–820 (2012). [DOI] [PubMed] [Google Scholar]
- 24.McGranahan N.et al.; TRACERx Consortium , Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171, 1259–1271.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasqualucci L., et al., Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 412, 341–346 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Schumacher T. N., Schreiber R. D., Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Andreatta M., Nielsen M., Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics 32, 511–517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jurtz V., et al., NetMHCpan-4.0: Improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 199, 3360–3368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H., Lund O., Nielsen M., The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: Application to MHC-peptide binding. Bioinformatics 25, 1293–1299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cattoretti G., et al., Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell 7, 445–455 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Casola S., et al., Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. U.S.A. 103, 7396–7401 (2006). Corrected in: Proc. Natl. Acad. Sci. U.S.A. 104, 2025 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ljunggren H. G., Kärre K., In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today 11, 237–244 (1990). [DOI] [PubMed] [Google Scholar]
- 33.Moretta L., et al., Allorecognition by NK cells: Nonself or no self? Immunol. Today 13, 300–306 (1992). [DOI] [PubMed] [Google Scholar]
- 34.Garrido F., MHC/HLA class I loss in cancer cells. Adv. Exp. Med. Biol. 1151, 15–78 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Campoli M., Ferrone S., HLA antigen changes in malignant cells: Epigenetic mechanisms and biologic significance. Oncogene 27, 5869–5885 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beà S., et al., Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 110, 18250–18255 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J., et al., The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood 123, 2988–2996 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabbri G., et al., Analysis of the chronic lymphocytic leukemia coding genome: Role of NOTCH1 mutational activation. J. Exp. Med. 208, 1389–1401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puente X. S., et al., Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475, 101–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wienand K., et al., Genomic analyses of flow-sorted Hodgkin Reed-Sternberg cells reveal complementary mechanisms of immune evasion. Blood Adv. 3, 4065–4080 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chapuy B., et al., Genomic analyses of PMBL reveal new drivers and mechanisms of sensitivity to PD-1 blockade. Blood 134, 2369–2382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berglund A., et al., Methylation of immune synapse genes modulates tumor immunogenicity. J. Clin. Invest. 130, 974–980 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gettinger S., et al., Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov. 7, 1420–1435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dersh D., et al., Genome-wide screens identify lineage- and tumor-specific genes modulating MHC-I- and MHC-II-restricted immunosurveillance of human lymphomas. Immunity 54, 116–131.e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S. S., et al., HLA class I and II diversity contributes to the etiologic heterogeneity of non-Hodgkin lymphoma subtypes. Cancer Res. 78, 4086–4096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pende D., et al., The susceptibility to natural killer cell-mediated lysis of HLA class I-positive melanomas reflects the expression of insufficient amounts of different HLA class I alleles. Eur. J. Immunol. 28, 2384–2394 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Binder C., et al., CD2 Immunobiology. Front. Immunol. 11, 1090 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren W., et al., Genetic landscape of hepatitis B virus-associated diffuse large B-cell lymphoma. Blood 131, 2670–2681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Miranda N. F., et al., Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients. Blood 124, 2544–2553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trifonov V., Pasqualucci L., Tiacci E., Falini B., Rabadan R., SAVI: A statistical algorithm for variant frequency identification. BMC Syst. Biol. 7 (suppl. 2), S2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shukla S. A., et al., Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol. 33, 1152–1158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Favero F., et al., Sequenza: Allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 26, 64–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorsson V.et al.; Cancer Genome Atlas Research Network , The immune landscape of cancer. Immunity 48, 812–830.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bycroft C., et al., The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The WES and WGS data for the 74 DLBCL patients are available in the European Genome-phenome Archive (EGA) (accession no. EGAS00001005054) and the National Center for Biotechnology Information (accession no. phs000450.v3.p).