Abstract
Germline editing, the process by which the genome of an individual is edited in such a way that the change is heritable, has been applied to a wide variety of animals [D. A. Sorrell, A. F. Kolb, Biotechnol. Adv. 23, 431–469 (2005); D. Baltimore et al., Science 348, 36–38 (2015)]. Because of its relevancy in agricultural and biomedical research, the pig genome has been extensively modified using a multitude of technologies [K. Lee, K. Farrell, K. Uh, Reprod. Fertil. Dev. 32, 40–49 (2019); C. Proudfoot, S. Lillico, C. Tait-Burkard, Anim. Front. 9, 6–12 (2019)]. In this perspective, we will focus on using pigs as the model system to review the current methodologies, applications, and challenges of mammalian germline genome editing. We will also discuss the broad implications of animal germline editing and its clinical potential.
Keywords: germline genome editing, pig, disease model, agriculture, xenotransplantation
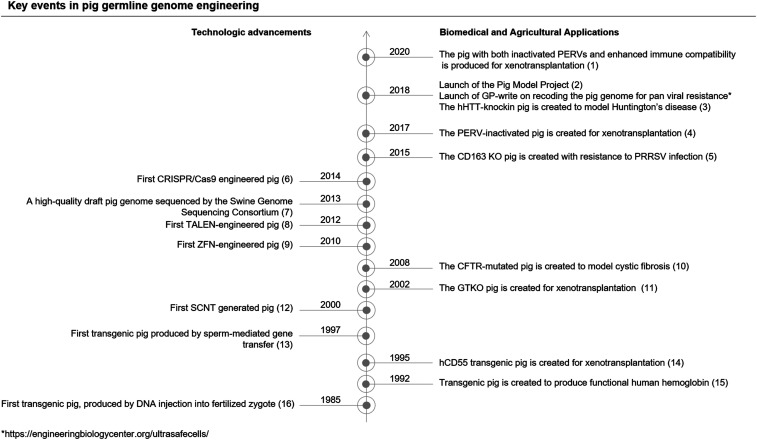
Genomic changes in pigs were traditionally accomplished using selective breeding, which is not only time consuming but also restricted to only existing genomic traits. Germline engineering, on the other hand, by transgene integration or direct genome modification, can quickly introduce new traits. We highlight some key events in pig germline genome engineering in Fig. 1 (1–16). The first transgenic pig was established through microinjection of purified DNA into fertilized zygotes (Fig. 1) (16). Later, gamete genetic modification, such as sperm-mediated gene transfer (13) and retroviral-mediated transduction of oocytes (17), offered alternative routes to produce transgenic pigs. The establishment of the somatic cell nuclear transfer (SCNT) technique was a significant breakthrough in the field of transgenic pig production (12). With SCNT, genetic manipulation is conducted on somatic cells either through the introduction of a transgene (18) or the knockout (KO) of an endogenous gene (11); the modified somatic cell is then selected and transferred into an enucleated oocyte to reconstitute a zygote, which can develop into a live piglet after transfer into a surrogate pig.
Fig. 1.
Key events in pig germline genome engineering.
Concurrently, the development of customized endonucleases, such as zinc-finger nucleases (19, 20), transcription activator-like effector nucleases (8, 21), and CRISPR/CRISPR-Cas9 system (22), have facilitated the production of germline genome-modified pigs. While these tools hold great promise to effectively edit the genome, potential off-target mutations that can lead to genomic instability are still a major concern. Off-target mutations arise from the binding and cutting of the genome editing endonuclease at cognate sites that resemble the intended target sequence. Off-target mutations are not uncommon in gene-edited cells (23–25), and it can be difficult to eliminate these cells with off-target mutations during ex vivo and in vivo genome editing. With germline editing, however, it is feasible to generate single cell clones and then apply whole genome sequencing to select clones with the least number of off-target mutations; these single cell clones are then used for SCNT and embryo transfer, thereby minimizing the potential detrimental effects of off-target mutations.
To date, a large number of germline genome-modified animals have been generated (26–28), mostly via SCNT technique (Fig. 2A). As mentioned previously, this process can be highly specific by selecting cells with minimal off-target mutations before producing a reconstituted zygote. However, the application of this procedure is still limited to a few species, due to low cloning efficiency. In addition, animals produced with SCNT frequently suffer from developmental defects associated with epigenetic abnormalities (27, 29). Alternatively, direct injection of genome editing tools into the zygote has been found to be effective in producing genome-modified large animals (Fig. 2B) (6, 8). The pregnancy rate is higher, and the piglets produced in this manner exhibit less procedure-related abnormalities. Direct injection is especially useful when SCNT technique is not available, and generally requires a lower number of oocytes. Despite these advantages, direct injection is less efficient in generating genome-modified pigs over SCNT, as it does not allow preimplantation selection steps. In addition, on-target genetic mosaicism is often observed; tighter control of the presence of genome editing tools may help reduce the occurrence of genetic mosaicism (30–37). Lastly, gamete modification followed by in vitro fertilization can be used but is currently less favored in animal genome modification, as it is technically less favorable and can only change one allele of the resulting zygote. However, spermatogonial stem cell editing is actively explored as a therapeutic approach to cure some paternally inherited genetic disease in humans (38). Of these three approaches, SCNT technique is generally preferred when available to produce transgenic pigs, as it is more scalable and provides more predictable outcomes.
Fig. 2.
Comparison of methods for producing germline genome-engineered pigs. All three methods require production of modified embryos, which are transferred into surrogate sows to give birth to germline-engineered pigs. (A) In the method of somatic cell genome engineering, somatic cells are modified, and the modified cell is fused with an enucleated egg to produce a reconstituted zygote. The reconstituted zygote then develops into an embryo in vitro before being transplanted into a surrogate sow. (B) In the method of zygote modification, gene modification is carried out at the stage of the zygote, which produces genome-modified zygotes. (C) In the method of gamete modification, either sperm or oocyte are modified in the desired loci before producing zygotes via in vitro fertilization.
Applications of Germline Genome-Engineered Pigs
Germline-modified pigs have broad applications in biomedical research. First, due to their similar size and physiology with humans, pigs represent a better animal model to study human diseases than their corresponding mouse models. These pig models closely resemble human neurodegenerative diseases (2), cancer (39), cystic fibrosis (10), retinitis pigmentosa (40), cardiovascular diseases (41), and metabolic diseases (42).
Second, owing to their similar organ size to humans, pigs have long been thought to be a promising source of organs, tissues, and cells for human transplantation. There are two major hurdles, however, that prevent effective pig-to-human organ transplantation: 1) risk of cross-species transmission of the porcine endogenous retroviruses (PERVs) and 2) molecular incompatibilities between the porcine graft and the human recipient. To date, more than 40 genetic modifications have been attempted on pigs, either individually or in combination, with goals of mitigating PERV transmission (4, 43) and molecular incompatibility (44). We have demonstrated simultaneous KO of the 25 copies of PERV in the porcine genome to produce PERV-inactivated pigs (4); these pigs are not only healthy and fertile, but their genomic changes are heritable. With regard to addressing molecular incompatibility, a recent study demonstrated that transgenic pig hearts carrying the KO of major pig antigen, GGTA1, along with the overexpression of two human proteins, hCD46 and hTBM, achieved life-supporting function in baboons for 6 months (45), suggesting the feasibility of long-term graft survival using genetically engineered pig organs. We have since engineered pigs with both PERV-KO and enhanced molecular compatibility (46), representing a significant step toward clinical application of xenotransplantation.
Another interesting application of germline-modified pigs is to produce commercially relevant bioproducts. For example, pigs have been engineered to produce human hemoglobin, which is potentially useful for life-saving transfusions in trauma patients with severe blood loss (15, 47). Similarly, human albumin (48), coagulation factors (49), and protein C (50) have all been produced in pigs.
In addition, for agricultural applications, a number of pigs have been genetically modified to increase litter size (51), enhance meat production (52–55), and achieve pathogen resistance (5, 56) (Fig. 1). While none of these modified pigs have been approved for human consumption, they show great promise to enhance both health and economic benefits to the breeder and the consumer. For example, PRRSV (porcine reproductive and respiratory syndrome virus) is the most economically impactful epidemic disease in the pork industry, with no effective vaccine to date. It has been demonstrated that CD163 KO pigs display normal physiology and gain complete resistance to PRRSV viral infection in vivo (5). Long-term follow-up studies to examine the impact of this modification on subsequent generations are needed to warrant the effective use of this new porcine breed in agricultural practice. Similarly, efforts have been taken to engineer pigs to gain natural resistance to African swine fever virus (57), coronavirus, and numerous bacteria (58). The Genome Project-write consortium, for example, has launched an effort to recode the entire pig genome, replacing at least one redundant genetic codon (i.e., TAG->TAA and/or TTR->CTN) and removing the corresponding translation machinery; such an effort would prevent the survival of any exogenous virus that are dependent on this codon (https://engineeringbiologycenter.org/ultrasafecells/).
Insights and Challenges of Germline Genome Editing
The wealth of information obtained from genome-modified animals has allowed the scientific community to better understand the biological complexity of germline genome editing. A genome-modified somatic cell must undergo reprogramming (via SCNT), embryogenesis, development, and gametogenesis in order to pass its information onto the resulting animal and its offspring. As such, the engineered genomic information is subject to dynamic tissue-specific regulations in vivo. Here, we lay out a few insights from our practice to illustrate the challenges and considerations to achieve functional germline genome engineering (Fig. 3).
Fig. 3.
Genomic knockout (KO) may not result in complete protein KO in vivo, and transgene knock-in may not result in proper in vivo expression. (A) A normal gene produces mature mRNA transcripts with all introns correctly spliced out. In the scenario with proper gene KO as illustrated here, genomic engineering leads to a frameshift mutation in exon 2. As a result, the resulting mRNA is degraded by nonsense-mediated decay (NMD) with complete loss of protein expression. However, alternative splicing can take place that skips the targeted exon, producing an isoform protein that is potentially functional. In another scenario, translation may be reinitiated at exons after the modified site, and the target gene locus produces a truncated protein which can also be potentially functional. (B) In an ideal knock-in scenario, the transgenes, which are usually used in the form of complementary DNA (cDNA), are correctly transcribed and translated into functional protein. In one scenario, the transgene may be inactivated by epigenetic silencing via promoter CpG methylation or/and enhancer histone deacetylation. In another scenario, the transgene may fail to produce the correct mRNA, due to unpredicted cryptic splicing in the transgene. Subsequently, the incorrect mRNA is degraded by NMD or is translated into a truncated protein. In the last scenario, the transgene mRNA is the target of endogenous microRNA (miRNA) or small interfering RNA, which leads to mRNA degradation and loss of protein expression.
First, the frameshift KO might not result in complete loss of functional protein production (Fig. 3A). It has been reported that CRISPR KO cells and animals can demonstrate rescued targeted protein expression through exon skipping and translation reinitiation mechanisms (59, 60). Such biological plasticity has striking heterogeneity, likely due to varied nonsense-mediated decay (NMD) efficiency or tissue-specific isoform choices. In light of these observations, large deletions (rather than point mutations) are recommended to achieve complete KO in live animals (60).
In addition, consistent transgene expression in an animal requires the comprehensive consideration of epigenetic, posttranscriptional, and posttranslational regulation (Fig. 3B). In vivo gene silencing has been a major problem for transgenic animal production since the 1990s. De novo DNA methylation during early embryogenesis is highly dynamic and stochastic (61) and can lead to permanent silencing of exogenously introduced DNA. The extent of silencing depends heavily on the choices of promoters, expression strategy used, and the genomic location of transgene insertion. For example, targeting genes to the pig Rosa26 site led to consistently high expression levels of the transgene (62). We recently demonstrated that, even though we could detect transgene expression at the bulk tissue level of our engineered pigs, close examination revealed mosaic gene expression, likely derived from early lineage-specific silencing. We initially hypothesized that gametogenesis could reset the epigenetic system, and therefore allow the offspring to exhibit universal transgene expression. However, we found that offspring from transgenic pigs still could not achieve universal transgene expression.
Beyond transcriptional regulation, posttranscriptional modifications, including tissue-specific pre-messenger RNA (pre-mRNA) splicing, mRNA transportation, and RNA interference (RNAi), allow multicellular organisms to create a huge diversity of proteomes from a finite number of genes. Such diversity, however, poses multiple challenges to obtaining consistent transgene expression in an animal. Unpredictable cryptic RNA splicing within the coding region of transgenes has been previously described (63), which abrogates the proper expression of the transgene protein. This cryptic splicing pattern is also tissue specific, and, therefore, difficult to predict with current available algorithms. In addition, RNAi is a well-described protective mechanism in plants and certain organisms (such as Caenorhabditis elegans) to silence exogenous transcripts in a tissue-specific fashion. It remains to be determined the degree to which the RNAi pathway plays a role in regulating transgene expression in the mammalian system.
Last but not least, the majority of natural proteins bear some form of posttranslational modification (PTM), such as glycosylation, carboxylation, hydroxylation, sulfation, and amidation. These modifications influence the stability and biological activity of proteins. For example, tissue-specific down-regulation of the transgene protein has been reported in mice, despite ubiquitous transgene RNA expression (64). Furthermore, whereas the majority of PTM pathways are highly conserved among mammalian species, there are some species-specific pathways that can alter the makeup of the transgene protein. As such, some human proteins produced in nonhuman species have been found to be inactive and unstable (65).
In light of these biological considerations, different strategies have been explored and developed to ensure consistent transgene expression (Fig. 4).
Fig. 4.
Synthetic approaches to achieve programmable transgene expressions in vivo. (A) Self-contained regions with insulators to shield the transgene from epigenetic silencing. (B) Tools such as dCas9-P300 and dCas9-Tet fusions can override the epigenetic status of the transgene genomic context and ensure consistent gene expression. (C) Large-scale chromosomal engineering to graft the entire genomic region of interest to preserve endogenous level of gene regulation for transgene expression.
Self-Contained Regions and Safe “Harbors.”
One strategy to stabilize transgene expression is to incorporate cis-acting DNA elements in the construct to shield the transgene from endogenous regulations. Insulators, such as chicken β-globin 5′ HS4, are the most commonly utilized elements (Fig. 4A) (66). Other elements, such as UCOE, MARs, and STAR (67), have also been shown to be beneficial for long-term transgene expression. One caveat is that naturally occurring insulators are lineage specific. It is unknown if these cis-elements can protect transgene expression on a universal level (66).
Besides adding cis-acting DNA, a different approach is to insert transgenes into “open safe harbors,” which are endogenous sites in the genome where ubiquitous and high gene expression occurs. Multiple safe harbors have been identified in the mouse genome, including ROSA26, ColA1, and TIGRE (68). Studies are underway to determine whether the homology regions in the pig genome can be similarly utilized (62). We envision that a detailed characterization of the epigenetic status and transcriptome of the pig genome will provide further guidance on choosing “open safe harbors.” In fact, such data will soon become available. Pig Model Project (PMP; Fig. 1) is on a mission to generate and interpret consolidated pig genomic, epigenomic, and transcriptomic information (2), ultimately providing tremendous value to both agricultural and biomedical research.
Coengineering the Epigenetic Status.
Dynamic histone modification and DNA methylation are important means of transcriptional regulation (Fig. 4B). Previous studies have suggested that epigenetic modifications are responsible for transgene silencing in vivo. Therefore, it has been postulated that targeted epigenetic modifications can restore transgene expression. Targeted histone acetyltransferase activities have been demonstrated to increase transgene expression in vitro (69). Similarly, recent studies have shown potent local epigenetic modifications with dCas9-p300 (70) and dCas9-Tet1 fusions (71). Future studies are needed to investigate whether such an epigenetic engineering system can override the endogenous silencing mechanism against transgenes in vivo and, if so, what the safety implications in the engineered animals would be given the potential off-target effects.
Large-scale Chromosomal Engineering to Preserve Endogenous Regulation.
An alternative strategy to preserve endogenous regulation of the foreign genetic circuit is to graft the entire genomic region of interest, including enhancers, promoters, introns, exons, and untranslated regions (UTRs), into the new organism (Fig. 4C). Such strategy can help maintain transgene expression by providing a buffer to the neighboring genes, thereby preserving the anticipated activity profile of the target locus. The feasibility of this approach has been established via bacterial artificial chromosome-assisted or yeast artificial chromosome-assisted recombineering (72, 73). Despite these promises, robust application of this approach still requires technology advancement in handling large DNA fragments on the order of 0.1 Mn to 10 Mn base pairs throughout the manipulation process. Recent advancements in artificial chromosome engineering may offer a novel route to engineering heritable large engineered genetic circuits (74, 75).
Broad Implications and the Clinical Prospects
Taken together, predictable germline genome engineering in animals requires careful consideration of various epigenetic, transcriptional, posttranscriptional, and translational regulations. The biological pathways governing these processes also play significant roles in other biological manipulations, such as in vivo gene editing and gene therapy. Therefore, animal germline engineering offers unique scientific and technological insights for other applications.
The long-term impact of genome editing tools in vivo remains to be investigated for safe therapeutic applications. Germline-modified large animals provide a good surrogate system to understand the safety aspects of genome editing tools. In our studies for xenotransplantation (4), we closely monitor multiple blood panels, vital organ function, fertility, and genetic heritability of the genome-modified pigs. We are mindful that cloned animals may have epigenetic abnormalities despite having a normal phenotype at a young age. Therefore, we plan to follow our genome-modified animals for multiple generations to identify the potential impact of genome editing tools.
We also envision germline editing in humans as a possible therapeutic approach, complementing somatic gene editing approaches in humans (Fig. 5). Admittedly, safety and ethical issues need to be closely examined and answered before any clinical application. A recent report from an international commission of the US National Academy of Medicine, US National Academy of Sciences, and the UK’s Royal Society can be used as general guidelines to consider potential benefits, harms, and uncertainties regarding application of heritable human genome editing. The report also specifies stringent preclinical and clinical requirements for safety, efficacy, and long-term monitoring of such genome editing applications (76). With that said, human germline editing has merits that warrant further discussion and investigation. First, unlike somatic gene editing, where millions and sometimes billions of cells are exposed to the genome editing tool, germline gene editing has millions-fold reduction of off-target mutations, given that only a few cells are exposed to the tool. Furthermore, the clonability of germline cells provides an additional level of safety control. We can perform whole genome sequencing before the modified cells are released into the next stage; this quality control process is not feasible for ex vivo or in vivo gene editing. Similarly, it is technically feasible to deliver genome editing tools to single germline cells to achieve 100% efficiency, while it is nearly impossible to obtain such efficiency for somatic genome editing. In addition, as most genome editing tools have foreign protein elements, immunogenicity commonly develops for in vivo gene therapy; conversely, germline editing does not expose the editing tool to the human immune system. Last but not least, from a health economics perspective, germline editing allows for a cure that is heritable, without any manipulation and cost needed for subsequent generations.
Fig. 5.
Germline (vs. somatic) genome engineering.
Acknowledgments
We would like to thank John Murphy from Pfizer for feedbacks on the manuscript, Meng Yang at Qihan Bio Inc. for the illustration, and other team members at Qihan Bio Inc. for discussions.
Footnotes
Competing interest statement: L.Y. and Y.G. are employees of Qihan Bio. G.C. is the cofounder and advisor to Qihan Bio.
This paper results from the NAS Colloquium of the National Academy of Sciences, “Life 2.0: The Promise and Challenge of a CRISPR Path to a Sustainable Planet,” held December 10–11, 2019, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. NAS colloquia began in 1991 and have been published in PNAS since 1995. The complete program and video recordings of presentations are available on the NAS website at http://www.nasonline.org/CRISPR. The collection of colloquium papers in PNAS can be found at https://www.pnas.org/page/collection/crispr-sustainable-planet.
This article is a PNAS Direct Submission.
Data Availability
There are no data underlying this work.
References
- 1.Yue Y., et al., Extensive germline genome engineering in pigs. Nat. Biomed. Eng., 10.1038/s41551-020-00613-9 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Wu S., 160 The pig model project of China. J. Anim. Sci. 96, 291–292 (2018). [Google Scholar]
- 3.Yan S., et al., A Huntingtin knockin pig model recapitulates features of selective neurodegeneration in Huntington’s disease. Cell 173, 989–1002.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niu D., et al., Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 357, 1303–1307 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitworth K. M., et al., Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 34, 20–22 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Hai T., Teng F., Guo R., Li W., Zhou Q., One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 24, 372–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groenen M. A. M., et al., Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491, 393–398 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson D. F., et al., Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. U.S.A. 109, 17382–17387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe M., et al., Knockout of exogenous EGFP gene in porcine somatic cells using zinc-finger nucleases. Biochem. Biophys. Res. Commun. 402, 14–18 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Rogers C. S., et al., The porcine lung as a potential model for cystic fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 295, L240–L263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai L., et al., Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 295, 1089–1092 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Polejaeva I. A., et al., Cloned pigs produced by nuclear transfer from adult somatic cells. Nature 407, 86–90 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Lavitrano M., et al., Sperm-mediated gene transfer: Production of pigs transgenic for a human regulator of complement activation. Transplant. Proc. 29, 3508–3509 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Cozzi E., White D. J. G., The generation of transgenic pigs as potential organ donors for humans. Nat. Med. 1, 964–966 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Swanson M. E., et al., Production of functional human hemoglobin in transgenic swine. Bio Technol. 10, 557–559 (1992). [DOI] [PubMed] [Google Scholar]
- 16.Hammer R. E., et al., Production of transgenic rabbits, sheep and pigs by microinjection. Nature 315, 680–683 (1985). [DOI] [PubMed] [Google Scholar]
- 17.Cabot R. A., et al., Transgenic pigs produced using in vitro matured oocytes infected with a retroviral vector. Anim. Biotechnol. 12, 205–214 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Ourednik V., et al., Segregation of human neural stem cells in the developing primate forebrain. Science 293, 1820–1824 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Shi Y., Berg J. M., A direct comparison of the properties of natural and designed zinc-finger proteins. Chem. Biol. 2, 83–89 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Hauschild J., et al., Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc. Natl. Acad. Sci. U.S.A. 108, 12013–12017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boch J., et al., Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Ran F. A., et al., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattanayak V., Ramirez C. L., Joung J. K., Liu D. R., Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 8, 765–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guilinger J. P., et al., Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat. Methods 11, 429–435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai S. Q., et al., GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang L., González R., Dobrinski I., Germline modification of domestic animals. Anim. Reprod. 12, 93–104 (2015). [PMC free article] [PubMed] [Google Scholar]
- 27.Galli C., et al., Somatic cell nuclear transfer and transgenesis in large animals: Current and future insights. Reprod. Domest. Anim. 47 (suppl. 3), 2–11 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Lotti S. N., Polkoff K. M., Rubessa M., Wheeler M. B., Modification of the genome of domestic animals. Anim. Biotechnol. 28, 198–210 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Proudfoot C., Lillico S., Tait-Burkard C., Genome editing for disease resistance in pigs and chickens. Anim. Front. 9, 6–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burkard C., et al., Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 13, e1006206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chuang C. K., et al., Generation of GGTA1 mutant pigs by direct pronuclear microinjection of CRISPR/Cas9 plasmid vectors. Anim. Biotechnol. 28, 174–181 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Petersen B., et al., Efficient production of biallelic GGTA1 knockout pigs by cytoplasmic microinjection of CRISPR/Cas9 into zygotes. Xenotransplantation 23, 338–346 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Sato M., et al., Direct injection of CRISPR/Cas9-related mRNA into cytoplasm of parthenogenetically activated porcine oocytes causes frequent mosaicism for indel mutations. Int. J. Mol. Sci. 16, 17838–17856 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., et al., Efficient generation of gene-modified pigs via injection of zygote with Cas9/sgRNA. Sci. Rep. 5, 8256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitworth K. M., et al., Zygote injection of CRISPR/Cas9 RNA successfully modifies the target gene without delaying blastocyst development or altering the sex ratio in pigs. Transgenic Res. 26, 97–107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu H. H., et al., Porcine zygote injection with Cas9/sgRNA results in DMD-modified pig with muscle dystrophy. Int. J. Mol. Sci. 17, E1668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou X., et al., Efficient generation of gene-modified pigs harboring precise orthologous human mutation via CRISPR/Cas9-Induced homology-directed repair in zygotes. Hum. Mutat. 37, 110–118 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Park K., et al., Successful genetic modification of porcine spermatogonial stem cells via an electrically responsive Au nanowire injector. Biomaterials 193, 22–29 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Kalla D., Kind A., Schnieke A., Genetically engineered pigs to study cancer. Int. J. Mol. Sci. 21, 488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petters R. M., et al., Genetically engineered large animal model for studying cone photoreceptor survival and degeneration in retinitis pigmentosa. Nat. Biotechnol. 15, 965–970 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Xiangdong L., et al., Animal models for the atherosclerosis research: A review. Protein Cell 2, 189–201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X., Lerman L. O., Investigating the metabolic syndrome: Contributions of swine models. Toxicol. Pathol. 44, 358–366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L., et al., Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 350, 1101–1104 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Puga Yung G. L., Rieben R., Bühler L., Schuurman H. J., Seebach J., Xenotransplantation: Where do we stand in 2016? Swiss Med. Wkly. 147, w14403 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Längin M., et al., Consistent success in life-supporting porcine cardiac xenotransplantation. Nature 564, 430–433 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Yue Y., et al., Extensive mammalian germline genome engineering. bioRxiv:876862 (19 December 2019). [Google Scholar]
- 47.Sharma A., et al., An isologous porcine promoter permits high level expression of human hemoglobin in transgenic swine. Bio Technol. 12, 55–59 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Peng J., et al., Production of human albumin in pigs through CRISPR/Cas9-mediated knockin of human cDNA into swine albumin locus in the zygotes. Sci. Rep. 5, 16705 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paleyanda R. K., et al., Transgenic pigs produce functional human factor VIII in milk. Nat. Biotechnol. 15, 971–975 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Van Cott K. E., et al., Affinity purification of biologically active and inactive forms of recombinant human protein C produced in porcine mammary gland. J. Mol. Recognit. 9, 407–414 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Xu P., et al., BAC mediated transgenic Large White boars with FSHα/β genes from Chinese Erhualian pigs. Transgenic Res. 25, 693–709 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Bi Y., et al., Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP. Sci. Rep. 6, 31729 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qian L., et al., Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci. Rep. 5, 14435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanihara F., et al., Somatic cell reprogramming-free generation of genetically modified pigs. Sci. Adv. 2, e1600803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang K., et al., Efficient generation of myostatin mutations in pigs using the CRISPR/Cas9 system. Sci. Rep. 5, 16623 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu S., et al., Transgenic shRNA pigs reduce susceptibility to foot and mouth disease virus infection. eLife 4, e06951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lillico S. G., et al., Mammalian interspecies substitution of immune modulatory alleles by genome editing. Sci. Rep. 6, 21645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan X., Yang Y., Zhang J.-R., Molecular basis of host specificity in human pathogenic bacteria. Emerg. Microbes Infect. 3, e23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smits A. H., et al., Biological plasticity rescues target activity in CRISPR knock outs. Nat. Methods 16, 1087–1093 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Sui T., et al., CRISPR-induced exon skipping is dependent on premature termination codon mutations. Genome Biol. 19, 164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song Y., et al., Dynamic enhancer DNA methylation as basis for transcriptional and cellular heterogeneity of ESCs. Mol. Cell 75, 905–920.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X., et al., Rosa26-targeted swine models for stable gene over-expression and Cre-mediated lineage tracing. Cell Res. 24, 501–504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jaganathan K., et al., Predicting splicing from primary sequence with deep learning. Cell 176, 535–548.e24 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Ambartsumian N., et al., Tissue-specific posttranscriptional downregulation of expression of the S100A4(mts1) gene in transgenic animals. Invasion Metastasis 18, 96–104 (1998-1999). [DOI] [PubMed] [Google Scholar]
- 65.Walsh G., Jefferis R., Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 24, 1241–1252 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Chung J. S., Dircksen H., Webster S. G., A remarkable, precisely timed release of hyperglycemic hormone from endocrine cells in the gut is associated with ecdysis in the crab Carcinus maenas. Proc. Natl. Acad. Sci. U.S.A. 96, 13103–13107 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwaks T. H., Otte A. P., Employing epigenetics to augment the expression of therapeutic proteins in mammalian cells. Trends Biotechnol. 24, 137–142 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Madisen L., et al., Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwaks T. H., et al., Targeting of a histone acetyltransferase domain to a promoter enhances protein expression levels in mammalian cells. J. Biotechnol. 115, 35–46 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Hilton I. B., et al., Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X. S., et al., Editing DNA methylation in the mammalian genome. Cell 167, 233–247.e217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abbasi M. N., et al., Recombineering for genetic engineering of natural product biosynthetic pathways. Trends Biotechnol. 38, 715–728 (2020). [DOI] [PubMed] [Google Scholar]
- 73.Copeland N. G., Jenkins N. A., Court D. L., Recombineering: A powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2, 769–779 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Annaluru N., et al., Total synthesis of a functional designer eukaryotic chromosome. Science 344, 55–58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Logsdon G. A., et al., Human artificial chromosomes that bypass centromeric DNA. Cell 178, 624–639.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.National Academy of Medicine, National Academy of Sciences, Royal Society , Heritable Human Genome Editing (The National Academies Press, Washington, DC, 2020). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no data underlying this work.