Significance
This work provides a report of accumulation of unconventional CD56−CD16+ NK cells in nonvirally induced malignancies. Increased frequency of CD56−CD16+ NK cells is associated with adverse clinical outcome in AML, as well as other maturation defects, and might contribute to a defective control of AML progression. Pseudotime analysis highlights a disruption in the maturation process of conventional NK cells in AML patients, leading to a bifurcation point absent in healthy subjects. This analysis, combined with the reduced frequency of conventional NK cells observed in AML patients, suggests that unconventional CD56−CD16+ NK cells derive from an aberrant maturation of conventional NK cells. Overall, accumulation of CD56−CD16+ NK cells could be an important feature of immune escape from innate immunity.
Keywords: AML, natural killer cells, CD56−CD16+ NK cells, mass cytometry
Abstract
Natural killer (NK) cells are major antileukemic immune effectors. Leukemic blasts have a negative impact on NK cell function and promote the emergence of phenotypically and functionally impaired NK cells. In the current work, we highlight an accumulation of CD56−CD16+ unconventional NK cells in acute myeloid leukemia (AML), an aberrant subset initially described as being elevated in patients chronically infected with HIV-1. Deep phenotyping of NK cells was performed using peripheral blood from patients with newly diagnosed AML (n = 48, HEMATOBIO cohort, NCT02320656) and healthy subjects (n = 18) by mass cytometry. We showed evidence of a moderate to drastic accumulation of CD56−CD16+ unconventional NK cells in 27% of patients. These NK cells displayed decreased expression of NKG2A as well as the triggering receptors NKp30 and NKp46, in line with previous observations in HIV-infected patients. High-dimensional characterization of these NK cells highlighted a decreased expression of three additional major triggering receptors required for NK cell activation, NKG2D, DNAM-1, and CD96. A high proportion of CD56−CD16+ NK cells at diagnosis was associated with an adverse clinical outcome and decreased overall survival (HR = 0.13; P = 0.0002) and event-free survival (HR = 0.33; P = 0.018) and retained statistical significance in multivariate analysis. Pseudotime analysis of the NK cell compartment highlighted a disruption of the maturation process, with a bifurcation from conventional NK cells toward CD56−CD16+ NK cells. Overall, our data suggest that the accumulation of CD56−CD16+ NK cells may be the consequence of immune escape from innate immunity during AML progression.
Natural killer (NK) cells are critical cytotoxic effectors involved in leukemic blast recognition, tumor cell clearance, and maintenance of long-term remission (1). NK cells directly kill target cells without prior sensitization, enabling lysis of cells stressed by viral infections or tumor transformation. NK cells are divided into different functional subsets according to CD56 and CD16 expression (2–4). CD56bright NK cells are the most immature NK cells found in peripheral blood. This subset is less cytotoxic than mature NK cells and secretes high amounts of chemokines and cytokines such as IFNγ and TNFα. These cytokines have a major effect on the infected or tumor target cells and play a critical role in orchestration of the adaptive immune response through dendritic cell activation. CD56dimCD16+ NK cells, which account for the majority of circulating human NK cells, are the most cytotoxic NK cells. NK cell activation is finely tuned by integration of signals from inhibitory and triggering receptors, in particular, those of NKp30, NKp46 and NKp44, DNAM-1, and NKG2D (5). Upon target recognition, CD56dimCD16+ NK cells release perforin and granzyme granules and mediate antibody-dependent cellular cytotoxicity through CD16 (FcɣRIII) to clear transformed cells.
NK cells are a major component of the antileukemic immune response, and NK cell alterations have been associated with adverse clinical outcomes in acute myeloid leukemia (AML) (6–9). Therefore, it is crucial to better characterize AML-induced NK cell alterations in order to optimize NK cell–targeted therapies. During AML progression, NK cell functions are deeply altered, with decreased expression of NK cell–triggering receptors and reduced cytotoxic functions as well as impaired NK cell maturation (6, 9–13). Cancer-induced NK cell impairment occurs through various mechanisms of immune escape, including shedding and release of ligands for NK cell–triggering receptors; release of immunosuppressive soluble factors such as TGFβ, adenosine, PGE2, or L-kynurenine; and interference with NK cell development, among others (14).
Interestingly, these mechanisms of immune evasion are also seen to some extent in chronic viral infections, notably HIV (2). In patients with HIV, NK cell functional anergy is mediated by the release of inflammatory cytokines and TGFβ, the presence of MHClow target cells, and the shedding of ligands for NK cell–triggering receptors (2). As a consequence, some phenotypical alterations described in cancer patients are also induced by chronic HIV infections, with decreased expression of major triggering receptors such as NKp30, NKp46, and NKp44 (15, 16); decreased expression of CD16 (17); and increased expression of inhibitory receptors such as T cell immunoreceptor with Ig and ITIM domains (TIGIT) (18) all observed. In addition, patients with HIV display an accumulation of CD56−CD16+ unconventional NK cells, a highly dysfunctional NK cell subset (19, 20). Mechanisms leading to the loss of CD56 are still poorly described, and the origin of this subset of CD56− NK cells is still unknown. To date, two hypotheses have been considered: CD56− NK cells could be terminally differentiated cells arising from a mixed population of mature NK cells with altered characteristics or could expand from a pool of immature precursor NK cells (21). Expansion of CD56−CD16+ NK cells is mainly observed in viral noncontrollers (19, 20). Indeed, CD56 is an important adhesion molecule involved in NK cell development, motility, and pathogen recognition (22–27). CD56 is also required for the formation of the immunological synapse between NK cells and target cells, lytic functions, and cytokine production (26, 28). As a consequence, CD56−CD16+ NK cells display lower degranulation capacities and decreased expression of triggering receptors, perforin, and granzyme B, dramatically reducing their cytotoxic potential, notably against tumor target cells (2, 19, 20, 29, 30). In line with this loss of the cytotoxic functions against tumor cells, patients with concomitant Burkitt lymphoma and Epstein-Barr virus infection display a dramatic increase of CD56−CD16+ NK cells (30), which could represent an important hallmark of escape to NK cell immunosurveillance in virus-driven hematological malignancies.
To our knowledge, this population has not been characterized in the context of nonvirally induced hematological malignancies. In the present work, we investigated the presence of this population of unconventional NK cells in patients with AML, its phenotypical characteristics, and the consequences of its accumulation on disease control. Finally, we explored NK cell developmental trajectories leading to the emergence of this phenotype.
Results
Accumulation of CD56−CD16+ NK Cells in Patients with AML.
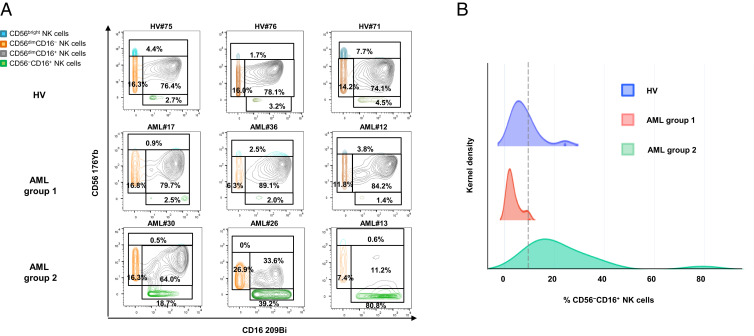
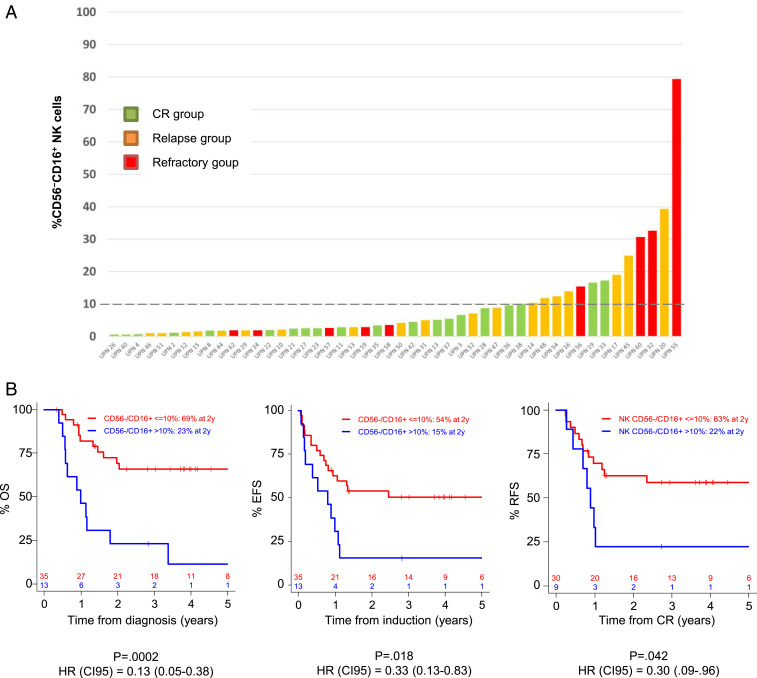
Peripheral blood mononuclear cells (PBMCs) from 48 newly diagnosed AML patients were analyzed by mass cytometry. Based on expression of CD56 and CD16, we defined four NK cell subsets: CD56bright NK cells, CD56dimCD16− NK cells, CD56dimCD16+ NK cells, and CD56−CD16+ NK cells (SI Appendix, Fig. S1). In 13 out of 48 patients (27.1%), we observed an accumulation of CD56−CD16+ NK cells, which represented up to 80.8% of the total NK cells (Fig. 1A). A threshold of 10% was defined on the basis of the maxstat algorithm (SI Appendix, Fig. S2). Patients with ≤10% CD56−CD16+ NK cells are referred to as group 1 (n = 35), and patients with >10% CD56−CD16+ NK cells are referred to as group 2 (n = 13) in the rest of the article (Fig. 1A). This threshold is illustrated on the intensity distribution graph using a kernel density estimation (Fig. 1B).
Fig. 1.
Accumulation of unconventional CD56−CD16+ NK cells in AML. PBMC from 48 newly diagnosed AML patients and 18 HV were phenotyped by mass cytometry. (A) NK cell phenotype by CD56 and CD16 expression; representative examples of HV and AML patients without (group 1) or with (group 2) accumulation of CD56−CD16+ NK cells. (B) Threshold visualization and value distribution illustrated by Kernel density estimation. HV, healthy volunteer. *P < 0.05, **P < 0.01, and ***P < 0.001.
More than 95% of cells expressed the transcription factors T-bet and Eomes across the four NK cell subsets (SI Appendix, Fig. S3A). However, as group 1 innate lymphoid cells (ILC1) share expression of T-bet with NK cells (31), we assessed expression of the ILC markers CD25, CD127 (IL7R), and CD278 (ICOS) to exclude the hypothesis that ILC1s were accumulated in patients from group 2. As expected, CD25 and IL7R expression was mostly restricted to CD56bright NK cells, whereas expression was reduced in CD56dim and CD56− NK cells compared with other clusters of conventional CD56+ NK cells (SI Appendix, Fig. S3B). In addition, more than 95% of NK cells were ICOS−, including those within the CD56−CD16+ subset, further confirming that CD56−CD16+ cells were not ILC1s (SI Appendix, Fig. S3B). We also excluded the possibility of contamination of the gate of unconventional NK cells by CD56+ leukemic blasts, as evidenced by the absence of CD13/CD33/CD34 expression on NK cell subsets (SI Appendix, Fig. S3C). Transcripts of genes coding CD3δ and CD3γ (CD3D and CD3G) and TCRα and TCRβ sequences (TRAC and TRBC1), as well as TCRγδ sequences (TRVD1, TRVD2, and TRVD3), were absent in both healthy volunteers (HV) and AML patients in the different subsets of NK cells (SI Appendix, Fig. S4 A–C). Gene transcripts of markers specific of mucosal-associated invariant T cells (Vα7.2, TRAV1-2) and invariant natural killer T cells (Vβ11, TCRBV11S1; Vα24, TCRAV24S1) were also absent (SI Appendix, Fig. S4D). Finally, we analyzed expression of genes transcripts of markers that differentiate monocytes/DC-like cells from unconventional NK cells (32). As expected, all subsets of NK cells displayed transcripts of genes coding CD11b (ITGAM), CD11c (ITGAM), and CD7 (CD7), whereas transcripts of the gene coding CD14 (CD14) were absent in both HV and AML patients in the different subsets of NK cells (SI Appendix, Fig. S4E). Taken together, these elements provide evidence that CD3−CD56−CD16+ cells are a subset of NK cells rather than another population expressing CD16 that would have contaminated the gate of unconventional NK cells.
High-Dimensional Analysis of CD56−CD16+ NK Cells’ Phenotypical Characteristics.
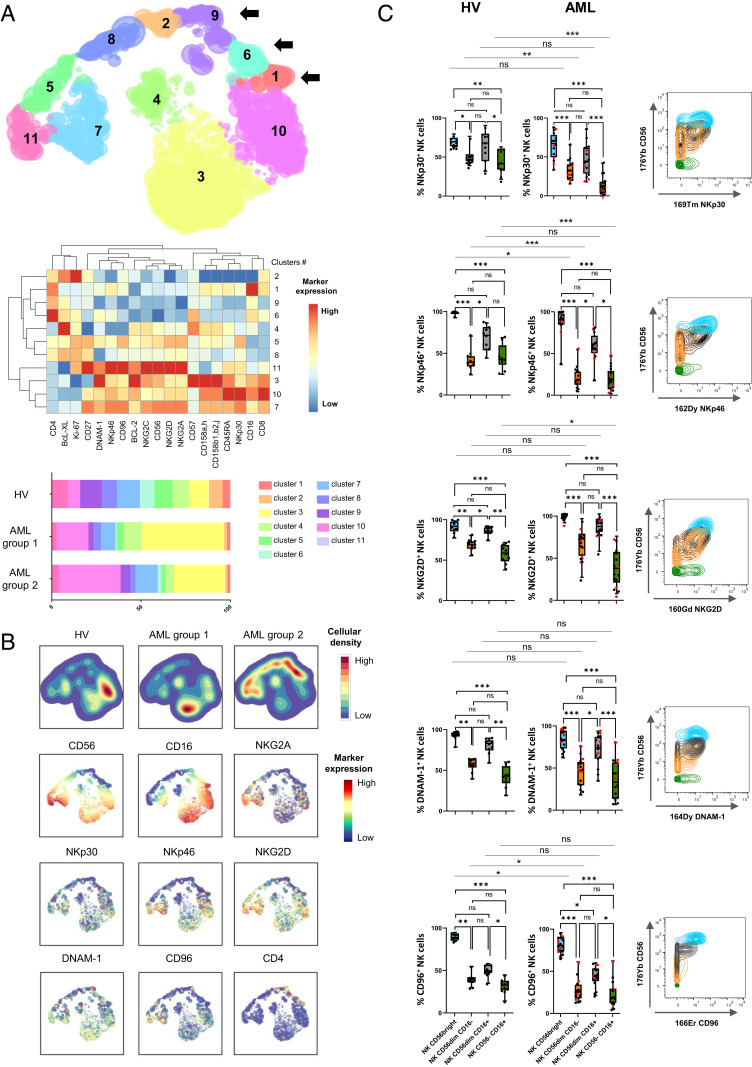
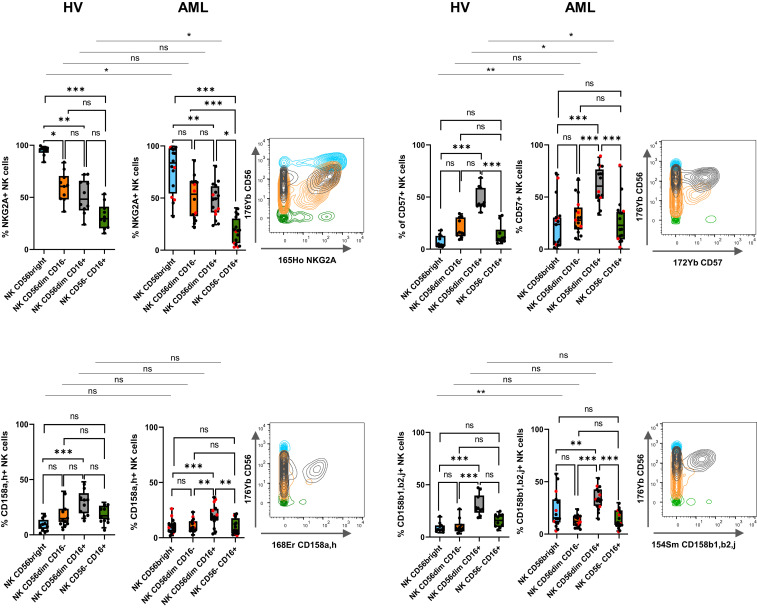
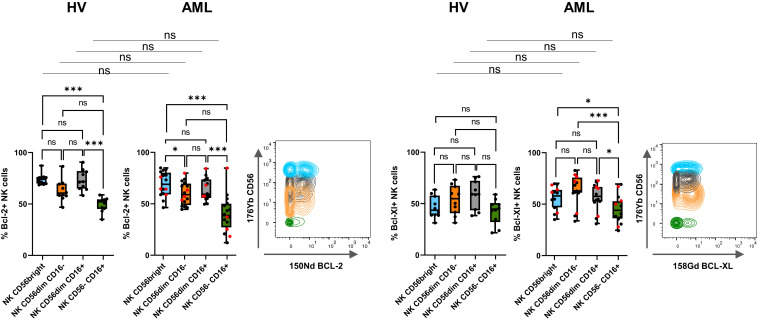
Loss of CD56 expression on NK cells has been associated with a loss of two of the main NK cell–triggering receptors, NKp30 and NKp46, as well as NKG2A in HIV-infected patients (20). We used the hierarchical stochastic neighbor embedding (h-SNE) algorithm to explore NK cell alterations associated with accumulation of CD56−CD16+ NK cells in AML. Interestingly, cluster 11 was enriched in AML group 2 compared to HV and group 1 (9% versus 4% and 3%, respectively) (Fig. 2A). This cluster displays hallmarks of immature stage 4B NK cells, with a CD56brightCD16−NKG2A+ phenotype. Cluster 5, which displayed a CD56dimCD16− phenotype, was also enriched in AML group 2 compared to HV and group 1 (10% versus 3% and 4%, respectively). As described in HIV infection, CD56−CD16+ NK cells from patients with AML also exhibited a loss of NKp30, NKp46, and NKG2A (Fig. 2 A and B). In addition, h-SNE analyses revealed a loss of three additional major triggering receptors required for NK cell activation: NKG2D, DNAM-1, and CD96 (Fig. 2 A and B). Manual gating confirmed that a significantly lower percentage of CD56−CD16+ NK cells than CD56dimCD16+ NK cells expressed the triggering receptors NKp30, NKp46, NKG2D, DNAM-1, and CD96 compared to CD56dimCD16+ NK cells in patients with AML (NKp30: 14.0 versus 47.4%, P < 0.001; NKp46: 20.5 versus 58.9%, P < 0.05; NKG2D: 36.1 versus 84.7%, P < 0.001; DNAM-1: 35.7 versus 71.8%, P < 0.001; CD96: 21.2 versus 42.7%, P < 0.05) (Fig. 2C). Maturation profiles revealed that CD56−CD16+ NK cells displayed significantly lower expression of maturation markers, such as CD57 and killer cell immunoglobulin-like receptors (KIRs), than conventional CD56dimCD16+ NK cells (CD57: 27.7 versus 61.8%, P < 0.001; CD158a,h: 9.8 versus 19.8%, P < 0.01; CD158b1,b2,j: 13.8 versus 33.7%, P < 0.001), suggesting that these cells do not represent a senescent subset issued from terminally differentiated NK cells (Fig. 3). However, these cells did not belong to an immature subset of NK cells, either, since this population poorly expressed NKG2A compared to immature CD56bright NK cells (17.9 versus 77.1%, P < 0.001) or with CD56dimCD16+ NK cells (17.9 versus 47.5%, P < 0.05) (Fig. 3). Most of the variations between CD56dimCD16+ NK cells and CD56−CD16+ NK cells were also present in healthy volunteers (Figs. 2C and 3). Furthermore, CD56−CD16+ NK cells displayed reduced expression of the antiapoptotic proteins BCL-2 and BCL-XL compared to CD56dimCD16+ NK cells in both healthy volunteers and patients with AML (Fig. 4).
Fig. 2.
Loss of NK cell–triggering receptors in CD56−CD16+ NK cells. (A) Total NK cells from peripheral blood were manually pregated and exported in Cytosplore for h-SNE analysis. Consensus files were generated for each group of patients with fixed number of NK cells. (Upper) h-SNE enables identification of NK cell clusters based on CD56 and CD16 expression; arrows indicate clusters of CD56−CD16+ NK cells. (Middle) The heatmap summarizes phenotypical characteristics of NK cell populations identified by h-SNE. (Lower) The stacked bar plot represents frequencies of NK cells by cluster in each group. (B) Expression of NK cell–triggering receptors projected on h-SNE maps. (C, Left) NK cell–triggering receptor expression profiles in clusters of CD56bright, CD56dimCD16−, CD56dimCD16+, and CD56−CD16+ NK cells were confirmed by manual gating to enable quantification of differences between clusters of NK cells. Results of 16 representative AML patients are presented as interquartile ranges, median, and whiskers from minimum to maximum. Data were analyzed using a Friedman test followed by a Dunn’s test. Comparison between two groups were performed using a Mann–Whitney U test. Red dots indicate AML group 2. (Right) Marker expression by NK cell subset; blue: CD56bright NK cells; orange: CD56dimCD16− NK cells; gray: CD56dimCD16+ NK cells; green: CD56−CD16+ NK cells. *P < 0.05, **P < 0.01, and ***P < 0.001; ns, nonsignificant.
Fig. 3.
CD56−CD16+ NK cell clusters display intermediate maturation profiles. (Left) Expression of maturation markers in clusters of CD56bright, CD56dimCD16−, CD56dimCD16+, and CD56−CD16+ NK cells was analyzed by manual gating. Results of 16 representative AML patients are presented as interquartile ranges, median, and whiskers from minimum to maximum. Differences between clusters were analyzed using a Kruskal–Wallis test followed by a Dunn’s test. Comparison between two groups were performed using a Mann–Whitney U test. Red dots indicate AML group 2. (Right) Marker expression by NK cell subset; blue: CD56bright NK cells; orange: CD56dimCD16− NK cells; gray: CD56dimCD16+ NK cells; green: CD56−CD16+ NK cells. *P < 0.05, **P < 0.01, and ***P < 0.001; ns, nonsignificant.
Fig. 4.
CD56−CD16+ NK cells display decreased expression of antiapoptotic proteins. (Left) Expression of antiapoptotic proteins in clusters of CD56bright, CD56dimCD16−, CD56dimCD16+, and CD56−CD16+ NK cells was analyzed by manual gating. Differences between clusters were assessed using a Friedman test followed by a Dunn’s test. Comparison between two groups were performed using a Mann–Whitney U test. Red dots indicate AML group 2. Results of 16 representative AML patients are presented as interquartile ranges, median, and whiskers from minimum to maximum. (Right) Marker expression by NK cell subset; blue: CD56bright NK cells; orange: CD56dimCD16− NK cells; gray: CD56dimCD16+ NK cells; green: CD56−CD16+ NK cells. *P < 0.05 and ***P < 0.001; ns, nonsignificant.
Previous reports described unconventional NK cells as dysfunctional rather than exhausted cells. These cells do not overexpress Tim-3 and PD-1, two markers of NK cell exhaustion, and display altered cytotoxic function and cytokine production associated with a decrease of Siglec-7, an early marker of dysfunctional NK cell subset (20, 33–36). In line with these results, CD56−CD16+ NK cells from AML patients did not overexpress Tim-3 and PD-1 and tended to display lower Siglec-7 expression compared with their CD56+ counterpart, although the difference was not significant (SI Appendix, Fig. S5). In addition, CD56−CD16+ NK cells from AML patients displayed a decreased CD107α response in presence of K562 target cells compared to their CD56+ counterpart; this decrease could be overcome by overnight incubation with IL-15 (SI Appendix, Fig. S6).
However, CD56−CD16+ NK cells retained ability to respond to IL-15 stimulation with an increase of perforin and granzyme B expression (SI Appendix, Fig. S7A). CD56−CD16+ NK cells also responded to IL-2/IL-15/IL-18 stimulation, with increased granzyme B expression in HV; CD56−CD16+ NK cells from AML displayed lower granzyme B expression compared with HV upon IL-2/IL-15/IL-18 stimulation (SI Appendix, Fig. S7B). In addition, CD56−CD16+ NK cells poorly expressed IFNγ, which was not up-regulated upon IL-12/IL-18 stimulation in AML, yet levels of expression were similar to their CD56dimCD16+ counterpart (SI Appendix, Fig. S7C).
Evolution at Complete Remission.
When material was available, we then assessed variations of the frequency of CD56−CD16+ NK cells at complete remission compared with baseline frequencies (SI Appendix, Fig. S8). A normal frequency of CD56−CD16+ NK cells was not systematically restored after induction chemotherapy. After induction chemotherapy, 7 out of 16 patients (43.8%) displayed less than 10% of CD56−CD16+ NK cells. Among them, five patients maintained long-term complete remission and two patients relapsed. A total of 9 out of 16 patients (56.2%) displayed more than 10% of CD56−CD16+ NK cells after induction chemotherapy. Among them, three patients maintained long-term complete remission, and six patients relapsed.
High Frequency of CD56−CD16+ NK Cells at Diagnosis Is Associated with Adverse Clinical Outcome.
Patients were classified into two groups according to the frequency of CD56−CD16+ NK cells, using the thresholds of 10% defined above. The 24-mo clinical outcome of the patients with AML was analyzed after stratification for the CD56−CD16+ NK cells frequency at diagnosis. After induction therapy, complete remission rates were 85.7% and 69.2% in groups 1 and 2, respectively. Among patients without accumulation of CD56−CD16+ NK cells (group 1), 18 out of 35 (54.3%) patients were in continuous complete remission after 24 mo of follow up, whereas among patients with accumulation of CD56−CD16+ NK cells (group 2), 2 out of 13 patients were in continuous complete remission (15.4%) (Fig. 5A). Once in first complete remission, 2-y relapse-free survival (RFS) was 58.8% and 22.2% in group 1 and 2, respectively (P = 0.042, Fig. 5B). Overall survival (OS) (hazard ratio [HR] = 0.13, P < 0.001) and event-free survival (EFS) (HR = 0.33, P < 0.05) were significantly reduced in patients with high frequency of CD56−CD16+ NK cells as compared to patients that displayed a conventional NK cell profile, with 3-y OS and EFS rates of 23.7% versus 65.9%, and 15.4% versus 50.4%, respectively (Fig. 5B). In multivariate Cox regression models, the frequency of CD56−CD16+ NK cells retained statistical significance in both OS and EFS (HR = 1.11, P = 0.0001; HR = 1.04, P = 0.004, respectively) (Table 1).
Fig. 5.
High frequency of CD56−CD16+ NK cells at diagnosis is associated with adverse clinical outcome. (A) Frequency of CD56−CD16+ NK cells in AML patients at diagnosis according to clinical outcome after 24 mo of follow up. (B) Patients were stratified according to the frequency of CD56−CD16+ NK cells (group 1: CD56−CD16+ NK cells ≤10%; group 2: CD56−CD16+ NK cells >10%). The impact of the accumulation of CD56−CD16+ NK cells on OS, EFS and relapse-free was assessed using a log-rank test. CI95, 95% confidence interval; CR, complete remission; HR, hazard ratio; and UPN, unique patient number.
Table 1.
Baseline patient characteristics
| Characteristic | All | CD56−CD16+ NK cells ≤10% | CD56−CD16+ NK cells >10% | P |
| Patients, No. (%) | 48 (100) | 35 (72.9) | 13 (27.1) | |
| Age at diagnosis, years, mean (SD) | 54.0 (14.0) | 54.4 (14.2) | 53.2 (14.0) | 0.871 |
| Patient sex | ||||
| Female | 28 (58.3) | 18 (51.4) | 10 (76.9) | 0.188 |
| Male | 20 (41.7) | 17 (48.6) | 3 (23.1) | |
| Median white blood cells, 109 cells/L (SD) | 11.0 (40.5) | 12.4 (43.5) | 6.0 (31.1) | 0.476 |
| Cytogenetic prognosis, No. (%) | ||||
| Favorable | 10 (20.8) | 7 (20.0) | 3 (23.1) | 0.342 |
| Intermediate | 26 (54.2) | 21 (60.0) | 5 (38.5) | |
| Adverse | 12 (25.0) | 7 (20.0) | 5 (38.5) | |
| Mutations in intermediate cytogenetic group, No. (%) | ||||
| Analyzed | 24/26 | 19/21 | 5/5 | NA |
| FLT3 ITDmut | 6 (25.0) | 3 (15.8) | 3 (60.0) | |
| NPM1mut | 13 (54.2) | 9 (47.4) | 4 (80.0) | |
| CEBPαmut/FLT3wt NPM1wt | 3 (12.5) | 3 (15.8) | 0 (0.0) | |
| ELN, No. (%) | ||||
| Favorable | 19 (39.6) | 15 (42.9) | 4 (30.8) | 0.417 |
| Intermediate | 17 (35.4) | 13 (37.1) | 4 (30.8) | |
| Adverse | 12 (25.0) | 7 (20.0) | 5 (38.5) | |
| Postinduction CR, No. (%) | 39 (81.3) | 30 (85.7) | 9 (69.2) | 0.228 |
| No postinduction CR | 9 (18.8) | 5 (14.3) | 4 (30.8) | |
| Induction death | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| No CR achieved | 9 (18.8) | 5 (14.3) | 4 (30.8) | |
| Relapse, No. (%) | 19 (39.6) | 12 (34.3) | 7 (53.8) | 0.065 |
| No relapse | 20 (41.7) | 18 (51.4) | 2 (15.4) | |
| Consolidation, No. (%) | ||||
| Chemotherapy + Allo-SCT | 16 (33.3) | 13 (37.1) | 3 (23.1) | NA |
| in CR1 | 8 (16.7) | 7 (20.0) | 1 (7.7) | |
| in CR > 1 | 8 (16.7) | 6 (17.1) | 2 (15.4) | |
| Blasts (blood) at diagnosis, mean (SD) | 50.7 (32.9) | 52.6 (34.3) | 45.1 (29.0) | 0.540 |
| Blasts (BM) at diagnosis, mean (SD) | 60.6 (25.8) | 64.1 (27.3) | 51.2 (19.3) | 0.104 |
BM, bone marrow; CR, complete remission; ELN, European Leukemia Net genetic classification 2010 (doi: 10.1182/blood-2009-07-235358); ITD, internal tandem duplication; and NA, not available.
Pseudotime Analysis.
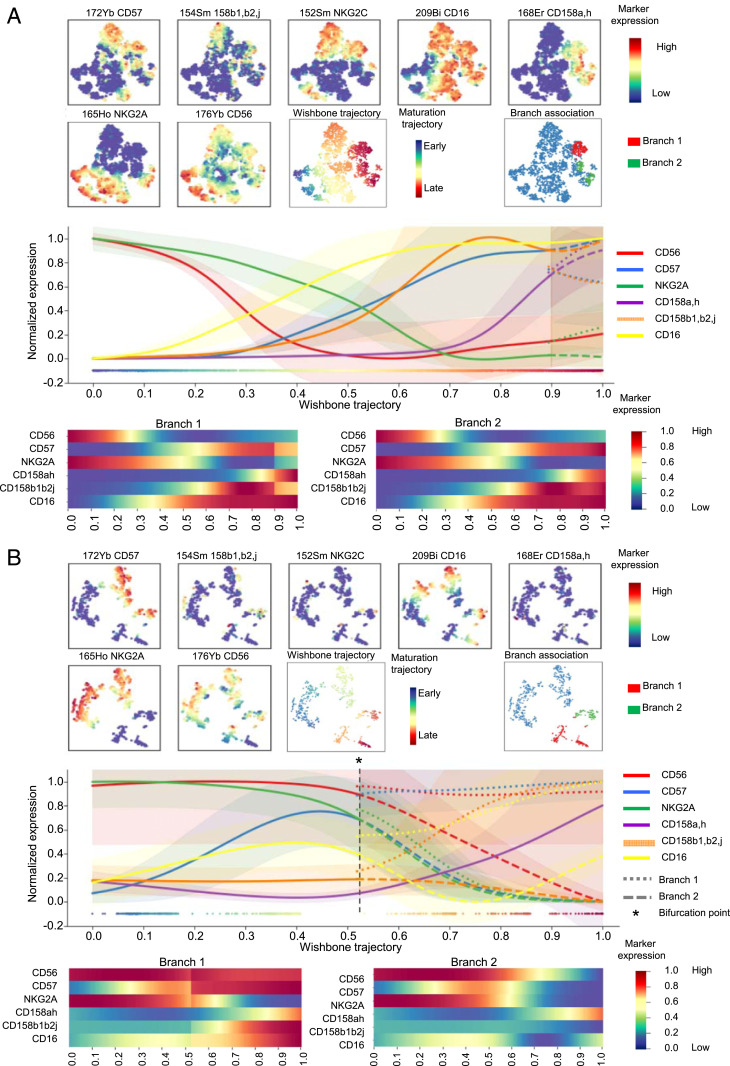
The important question of the origin of these CD56− NK cells remained. Interestingly, in patients with AML, accumulation of CD56−CD16+ NK cells was associated with a decreased frequency as well as a decreased absolute number of conventional (CD3−CD56+) NK cells, thus suggesting that these NK cells are derived from the pool of conventional NK cells rather than expanded from the pool of immature precursor NK cells (SI Appendix, Fig. S9). To probe this hypothesis, we performed a trajectory inference of NK cell maturation using the Wishbone algorithm. Wishbone is a recent algorithm used for analysis of developmental pathways in high-dimensional single-cell datasets. This algorithm positions single cells along bifurcating developmental trajectories on a k-nearest neighbor graph and pinpoints bifurcation points (37). The rise and fall of markers acquired or lost during the course of NK cell development (y-axis) is represented as a function of pseudotime (x-axis) (Fig. 6 A and B). Each branch after the bifurcation point represents a distinct differentiation trajectory. Wishbone recovers hallmarks of NK cell maturation in healthy volunteers. NK cells initially highly express CD56 and NKG2A. CD56bright NK cells expressing low levels of CD16 correspond to a transition between early immature CD56brightCD16− NK cells and CD56dimCD16+ NK cells. Subsequently, NK cells lose expression of NKG2A and sequentially acquire KIRs (CD158a,h and CD158b1,b2,j) and, finally, CD57, which marks the acquisition of high cytotoxic potential (4) (Fig. 6A). In AML, the maturation process is disrupted and leads to a bifurcation point; the first branch displays a normal-like maturation profile, whereas the second branch displays altered maturation features, with decreased CD57 and CD158b1,b2,j expression as well as loss and reacquisition of CD16 (Fig. 6B). These observations further support the hypothesis of a bifurcation from conventional NK cells toward CD56−CD16+ NK cells during the maturation process.
Fig. 6.
Differentiation trajectories are distinct for CD56dimCD16+ NK cells and CD56−CD16+ NK cells. NK cells were manually pregated and exported for differentiation trajectory inference using the Wishbone algorithm in HV (A) and AML patients at diagnosis (B). Wishbone enables identification of the branch point that gives rise to the population of CD56−CD16+ NK cells. Ordering and branching of cells along bifurcating developmental trajectories was performed using t-distributed stochastic neighbor embedding (t-SNE) and diffusion maps. The resulting trajectory and branches are used to visualize the dynamics of maturation markers (CD56, CD57, NKG2A, and KIRs) and of CD16 during NK cell differentiation.
Discussion
Recent clinical trials highlight the central role of NK cells for the control of AML (38). Since NK cell alterations are expected to impact response to NK cell-based immunotherapies, a better characterization of evasion from NK cell surveillance is therefore an absolute prerequisite to design the next generation of therapeutic strategies based on NK cell manipulation. In the present article, we highlight the presence of unconventional CD56− NK cells in 27% of patients with AML, which is associated with adverse clinical outcomes. The impact on survival was probably linked with early relapse rather than a lack of achievement of morphologic remission, as evidenced by the marked difference in RFS after achievement of a first complete remission. This variable appears to be an independent prognostic factor since it retains statistical significance in multivariate analysis. However, allogeneic stem cell transplantation was not equally distributed within groups 1 and 2, with 7 out of 35 patients (20%) versus 1 out of 13 (7.7%) receiving allogeneic stem cell transplantation (allo-SCT), respectively. This difference was not significant and could be partially explained by the lower complete remission rates in group 2. Further exploration of NK cell alterations that have been previously linked with adverse clinical outcome in AML, such as decreased NKp30 expression on conventional NK cells (8, 9) or decreased frequency of mature NK cells (6), revealed that these classical NK cell alterations were not significantly increased in AML group 2 compared to AML group 1 (SI Appendix, Fig. S10); this observation suggests that the mechanisms involved in the increase of CD56− NK cells might differ from those implicated in NCR down-regulation and NK cell maturation blockade.
CD56− NK cells represent a minor subset of NK cells under physiological conditions. Their relative proportion increases in chronically infected HIV viremic patients, in which the CD56−CD16+ subset can represent up to 50% of total NK cells (19, 20, 39, 40). More recently, this population has been described in subjects coinfected with cytomegalovirus (CMV) and Epstein-Barr virus (EBV), with a moderate increase in frequency (33). Elevated frequencies of CD56− NK cells have also been described to a lesser extent in other viral chronic infections, such as hepatitis C or hantavirus infections (41, 42), and autoimmune ocular myasthenia (43). In these pathological situations, functional characterization of these CD56− NK cells revealed profound dysfunctions. In HIV-infected subjects, CD56−CD16+ NK cells are reported to be highly dysfunctional, with reduced natural cytotoxicity, defective antibody-dependent cellular cytotoxicity, and low IFNγ production capacity, although they retain chemokine secretion capacities (20, 21). Phenotypical characterization of CD56−CD16+ NK cells highlighted a decreased expression of the NK cell–triggering receptors NKp30 and NKp46, as well as a decreased expression of NKG2A (20). Whether this population of NK cells is induced by chronic exposure to antigens is unclear, and mechanisms involved in CD56−CD16+ NK cell accumulation remain to be elucidated. Interestingly, an important down-regulation of CD56 could be obtained in vitro using sera from patients with chronic lymphoid leukemia added to NK cells from healthy subjects, suggesting that this phenotype emerges in the presence of an immunosuppressive milieu (44). Of note, BCL-2 and BCL-XL, two critical proteins for the maintenance of homeostasis and survival in noncycling NK cells, were poorly expressed by CD56− NK cells. In a murine model of BCL-2 deficiency, mice displayed a decreased absolute number of NK cells as well as a disruption of NK cell maturation. Impaired NK cell survival in this model was linked with altered antitumor function (45). However, the parallel between decreased NK cell absolute number, maturation defects, and decreased BCL-2 expression in AML patients remains hypothetic, notably since BCL-2 expression is still detectable in human samples.
Besides the question of the mechanisms involved in the emergence of CD56−CD16+ NK cells, the important question of the origin of this subset remained to be explored. We used a pseudotime algorithm to explore differentiation trajectories that give rise to CD56−CD16+ NK cells. These algorithms were developed to investigate the fundamental questions of development processes from individual cells into different cell types. Interestingly, we could reconstitute the theoretical scheme of NK cell maturation in healthy subjects. Identification of a bifurcation occurring during the course of NK cell maturation in patients with AML provides answers regarding the possible origin of CD56−CD16+ NK cells, suggesting that accumulation of CD56− NK cells would be the consequence of disruption of the maturation process. However, while the Wishbone algorithm accounts for the nonlinear relationships between markers and cell maturity, which is a major advantage compared with other pseudotime methods, one major limitation is the fact that it assumes the existence of exactly one branching point leading to two developmental pathways. The exact developmental pathways leading to unconventional NK cells might be more complex, and it will require further exploration using animal models or single-cell RNA sequencing analysis in order to dissect maturation trajectories more accurately. Yet, this observation provides bases for further exploration of the mechanisms involved in the emergence of this population, which will enable us to better define therapeutic strategies likely to restore physiological frequencies of CD56− NK cells. These elements must be considered in regards to other maturation anomalies our group described in AML, with a maturation blockade that affects ∼10% of patients, for whom the clinical outcome is dramatic (6). As with maturation blockade, bifurcation toward CD56− NK cells has major consequences on clinical outcome. Hence, a normal NK cell maturation appears to be critical to control the disease, and these anomalies urgently need to be further explored in AML. Interestingly, it has been reported that this phenotype is reversed when the viral load of patients with HIV was lowered below detectable levels after 12 mo of antiretroviral therapy, a situation associated with considerable improvement in NK cell function, increased NK cell receptor expression, and restoration of normal CD56 expression (21). In the context of AML, the normalization of the frequency of CD56− NK cells was observed only in some patients at the time of complete remission. This persistence of CD56− NK cells 30 d after induction chemotherapy should be explored at later time points to confirm these results.
To our knowledge, accumulation of CD56− NK cells has not been reported in hematological or solid malignancies, except in chronic NK cell large granular lymphocytosis (LGL), in which there is a massive clonal expansion of CD56dim/− NK cells in a subset of patients (46). According to the authors, CD56− NK cell expansion in LGL leukemia might be the consequence of an activating stimulus such as a retroviral infection, notably because sera from patients with LGL leukemia frequently react with HTLV-I/II p21 envelope proteins. Whether CMV infection could induce this aberrant phenotype has not been determined in our cohort, as we do not have CMV status data for all our patients. However, the increase in CD56− NK cells described in CMV and EBV coinfected subjects is moderate (33) and therefore does not explain the massive accumulation we observe in AML, although a synergistic mechanism cannot be excluded. Finally, we observed a higher prevalence of CD56− NK cell accumulation in AML with inversion 3 (inv 3); in three out of four patients, CD56−CD16+ NK cells represented more than 20% of the total NK cell population. This association might represent an interesting feature of immune alteration in the group of patients with inv 3 for whom clinical outcome is extremely unfavorable and requires confirmation in a larger cohort of patients (47).
In conclusion, loss of CD56 expression by the NK cells of patients with AML might represent a feature of immune evasion from NK cell control. Lastly, exploration of the mechanisms involved in down-regulation of CD56 will provide opportunities to restore NK cell functions in this specific subgroup of patients.
Patients and Methods
Patients and Study Design.
This monocentric study (Paoli-Calmettes Institute) included 48 patients with newly diagnosed nonacute promyelocytic leukemia AML from the HEMATOBIO cohort (NCT02320656). Patients were aged 19 to 80 y (mean ± SD = 54.0 ± 14.0). All patients received cytarabine- and anthracycline-based induction chemotherapy as previously described (48). A total of eight patients (16.7%) were treated with allogeneic stem cell transplantation as a consolidation therapy. Patients’ characteristics are summarized in Table 2.
Table 2.
Cox regression
| Multivariate HR for OS | Multivariate HR for EFS | |||||
| Variable | HR | 95% CI | P | HR | 95% CI | P |
| Age at diagnosis, years | 1.05 to 1.17 | 0.0003 | 1.06 to 1.03 | 0.0009 | ||
| ≥50 | Reference | Reference | ||||
| <50 | 1.11 | 1.07 | ||||
| ELN | ||||||
| Adverse | Reference | 0.21 to 1.84 | 0.393 | Reference | 0.32 to 2.23 | 0.746 |
| Intermediate | 0.62 | 0.18 to 2.06 | 0.425 | 0.85 | 0.17 to 1.46 | 0.206 |
| Favorable | 0.61 | 0.51 | ||||
| %CD56−CD16+ NK cells | 0.0001 | 1.01 to 1.07 | 0.004 | |||
| Reference | 1.05 to 1.17 | Reference | ||||
| 1.11 | 1.04 | |||||
Multivariate Cox regression models were used to assess the prognostic value of the frequency of CD56−CD16+ NK cells while adjusting for the prognostic factors in the population (age at diagnosis and ELN). ELN, European Leukemia Net genetic classification.
Ethics Statement.
All participants gave written informed consent in accordance with the Declaration of Helsinki. The entire research procedure was approved by the institutional review board of the Paoli-Calmettes Institute.
Clinical Samples.
PBMCs cryopreserved in 90% albumin/10% dimethyl sulfoxyde were obtained before (n = 48) and after induction chemotherapy (n = 16, paired samples) and from age-matched healthy volunteers (n = 18). Handling, conditioning, and storage of samples were performed by the Paoli-Calmettes Tumor bank, which operates under authorization #AC-2007–33 granted by the French Ministry of Research.
Mass Cytometry Analysis.
PBMCs were thawed and processed as previously described (6). PBMCs were washed with Roswell Park Memorial Institute (RPMI) medium 1640 with 10% fetal calf serum (FCS) and incubated in RPMI 1640 with 2% FCS and 1/10,000 Pierce Universal Nuclease 5 kU (Thermo Fisher Scientific) at 37 °C with 5% CO2 for 30 min. Cells were centrifuged and incubated with cisplatin 0.1 M to stain dead cells. Aspecific epitopes were blocked with 0.5 mg/mL Human Fc Block (BD Biosciences). A total of 2 million PBMCs were stained for 45 min at 4 °C with the extracellular antibodies (SI Appendix, Table S1). Cells were centrifuged and barcoded with the Cell-ID 20-Plex Pd Barcoding Kit (Fluidigm) according to the manufacturer's recommendations. Cells were washed and samples were combined and stained with metal-labeled anti-phycoerythrin secondary antibodies for 30 min at 4 °C. After centrifugation, cells were washed and permeabilized with the Foxp3 Staining Buffer Set (eBioscience) for 40 min at 4 °C. Intracellular aspecific epitopes were blocked with 0.5 mg/mL Human Fc Block for 40 min at 4 °C before incubation with the mix of intracellular antibodies for 40 min at 4 °C in Foxp3 Staining Buffer (SI Appendix, Table S1). Cells were then washed and labeled overnight with 125 nM iridium intercalator (Fluidigm) in Cytofix (BD Biosciences). Finally, cells were diluted in EQ™ Four Element Calibration Beads (Fluidigm) before acquisition on a Helios instrument (Fluidigm).
Transcriptomic Analysis.
CD56bright, CD56dimCD16−, CD56dimCD16+, and CD56−CD16+ subsets of peripheral NK cells from AML were sorted using a FACSAria III cell sorter (BD Biosciences). Control PBMCs were from age-matched HV. For each NK cell subset, RNA was extracted using the RNA extraction kit RNeasy Plus Micro, Qiagen. For a given NK cell subset and a given group, RNA from the different samples were pooled together before analysis. Library preparation and sequencing were performed by Integragen. RNA-sequencing libraries were performed with NEBNext Ultra II Directional RNA Library Prep Kit for Illumina according to supplier recommendations. The capture was then performed on complementary DNA (cDNA) libraries with the Twist Human Core Exome + Custom Integragen V2 Enrichment System according to supplier recommendations (Twist Bioscience). cDNA was then sequenced on an Illumina NovaSeq as paired-end 100-bp reads.
Functional Assays.
PBMCs from AML patients were thawed and incubated in RPMI medium 1640 10% FCS with or without IL-2 100IU/mL, IL-15 10 ng/mL, and/or IL-18 100 ng/mL. GolgiStop was added during incubation. Perforin, Granzyme B, and IFNγ expression was assessed by flow cytometry. For CD107α assays, CD56bright, CD56dimCD16−, CD56dimCD16+, and CD56−CD16+ subsets of peripheral NK cells from AML were sorted using a FACSAria III cell sorter and incubated overnight in RPMI medium 1640 10% FCS, supplemented with IL-2 100IU/mL or IL-2 100IU/mL IL-15 10 ng/mL, and cocultured during 4 h with K562 target cells (1:1 effector-to-target ratio). CD107α expression was assessed by flow cytometry.
Statistical Analysis.
Statistical analyses were carried out using Graph Pad Prism V5.01 and SPSS V9.0. The χ2 or Fisher’s exact test was used to assess association between variables. Comparisons between two groups were performed using a two-tailed Mann–Whitney U test. For multiple comparisons of paired values, a Friedman test was performed followed by a Dunn’s posttest. For multiple comparisons of independent samples, a Kruskal–Wallis test was performed followed by a Dunn’s posttest. Groups of patients were defined according to the frequency of CD56−CD16+ NK cells. The threshold was defined based on optimized cut points using maximally selected log-rank statistics (maxstat package, R software V 3.6.2) (49) and on imposing that no group could represent fewer than 25% of patients. For survival analyses, OS was defined as the time from diagnosis until death from any cause and EFS as the time between induction and relapse, death from any cause, or induction failure—whatever occurred first. RFS was calculated from the time of complete remission achievement to relevant events (i.e., AML relapse or death from any cause). Patients without an event were censored at the time of their last follow up. Survival times were estimated by the Kaplan–Meier method and compared using the log-rank test. A multivariate Cox regression model was used to assess the prognostic value of CD56−CD16+ NK cells while adjusting for other prognostic factors. Candidate variables for the Cox regression were European Leukemia Net genetic classification, age at diagnosis, leukocytosis, and percentage of CD56−CD16+ NK cells. Continuous variables were discretized as follows: age < or ≥50 y old; leukocytosis: < or ≥50 giga/L; the frequency of CD56−CD16+ NK cells was modeled as a continuous variable. All factors with a P value < 0.15 in univariate analysis were considered to be candidates for the backward stepwise Cox regression model. For subgroup analyses, patients were divided into two groups, a relapsed AML group and a long-term complete remission group, according to clinical outcome after 24 mo of follow up. The limit of significance was set at P < 0.05.
Algorithm-Based High-Dimensional Analysis.
NK cells were manually defined as CD13−CD33−CD34−CD45+CD3−CD19−CD56+ or CD13−CD33−CD34−CD45+ CD3−CD19−CD56−CD16+ and exported using FlowJo V10.6.2. The gating strategy is displayed in SI Appendix, Fig. S1. Data were arcsinh-transformed with a cofactor of 5. NK cell populations were automatically defined using h-SNE analysis (Cytosplore V2.2.1) with default settings (30 perplexity and 1,000 iterations) (50). For the h-SNE analyses, consensus files were generated for each group of patients with a fixed number of NK cells in order to obtain a representative and balanced view of all patient groups. Markers used for clustering were CD16, CD56, CD158a,h, CD158b1,b2,j, NKG2A, NKG2C, and CD57. NK cell trajectory inferences were performed using the Wishbone algorithm (37). The library was updated for Python V3.6 and is available at https://github.com/moreymat/wishbone/releases/tag/0.4.2-py.3. Markers used for trajectory calculation were CD56, CD57, NKG2A, KIRs (CD158a,h and CD158b1,b2,j), and CD16. The starting cell was defined as CD56brightNKG2A+CD57−KIR−. Wishbone was run with the following parameter values: 15 nearest neighbors (k = 15), 250 randomly sampled waypoints (num_waypoints = 250), and using diffusion components 2, 3, and 4 (components_list = [1, 2, 3] [in zero-based numbering]).
Supplementary Material
Acknowledgments
We thank the CRCM cytometry core facility as well as the IPC/CRCM/UMR 1068 Tumor Bank, which operates under authorization number AC-2007-33, granted by the French Ministry of Research (Ministère de la Recherche et de l’Enseignement Supérieur). Louise Ball from Angloscribe, an independent scientific language editing service, provided drafts and editorial assistance to the authors during preparation of this manuscript. This work has been financially supported by the Institut National du Cancer (INCa) (Grant 2012-064/2019-038 to D.O. and A.T.), the Fondation de France (Grant 00076207 to A.-S.C.), the Sites de Recherche Intégrée sur le Cancer (SIRIC) Marseille (Grant INCa-DGOS-INSERM 6038), the Cancéropôle Provence-Alpes-Côte d’Azur (Grants K_CyTOF 2014 and AML_CyTOF 2016 to J.A.N.), and the Groupement d’intérêt scientifique -Infrastructures pour la Biologie, la Santé et l’Agronomie (GIS IBiSA) and the Agence Nationale de la Recherche [French National Infrastructure for Mouse Phenogenomics (PHENOMIN) project] to M. Malissen, H.L., and E.G. The team “Immunity and Cancer” was labeled “Equipe Fondation pour la Recherche Médicale (FRM) DEQ 20180339209” (for D.O.). Inserm U1160 and the Paoli Calmettes Institute, Inserm U1068, are members of OPALE Carnot Institute, The Organization for Partnerships in Leukemia, Institut de Recherche Saint-Louis, Hôpital Saint-Louis, 75010 Paris, France.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020459118/-/DCSupplemental.
Data Availability
NK cell trajectory inference data have been deposited in the GitHub repository (https://github.com/moreymat/wishbone/releases/tag/0.4.2-py.3). All other study data are included in the article and/or SI Appendix.
References
- 1.Morvan M. G., Lanier L. L., NK cells and cancer: You can teach innate cells new tricks. Nat. Rev. Cancer 16, 7–19 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Lucar O., Reeves R. K., Jost S., A natural impact: NK cells at the intersection of cancer and HIV disease. Front. Immunol. 10, 1850 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanier L. L., Le A. M., Civin C. I., Loken M. R., Phillips J. H., The relationship of CD16 (Leu-11) and Leu-19 (NKH-1) antigen expression on human peripheral blood NK cells and cytotoxic T lymphocytes. J. Immunol. 136, 4480–4486 (1986). [PubMed] [Google Scholar]
- 4.Cooper M. A., Fehniger T. A., Caligiuri M. A., The biology of human natural killer-cell subsets. Trends Immunol. 22, 633–640 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Vivier E., et al., Innate or adaptive immunity? The example of natural killer cells. Science 331, 44–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chretien A.-S., et al., Natural killer defective maturation is associated with adverse clinical outcome in patients with acute myeloid leukemia. Front. Immunol. 8, 573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chretien A.-S., et al., NKp46 expression on NK cells as a prognostic and predictive biomarker for response to allo-SCT in patients with AML. OncoImmunology 6, e1307491 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chretien A.-S., et al., NKp30 expression is a prognostic immune biomarker for stratification of patients with intermediate-risk acute myeloid leukemia. Oncotarget 8, 49548–49563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fauriat C., et al., Deficient expression of NCR in NK cells from acute myeloid leukemia: Evolution during leukemia treatment and impact of leukemia cells in NCRdull phenotype induction. Blood 109, 323–330 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Fauriat C., et al., Natural killer cell-triggering receptors in patients with acute leukaemia. Leuk. Lymphoma 44, 1683–1689 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Chretien A.-S., et al., Increased NK cell maturation in patients with acute myeloid leukemia. Front. Immunol. 6, 564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khaznadar Z., et al., Defective NK cells in acute myeloid leukemia patients at diagnosis are associated with blast transcriptional signatures of immune evasion. J. Immunol. 195, 2580–2590 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Dulphy N., et al., Underground adaptation to a hostile environment: Acute myeloid leukemia vs. natural killer cells. Front. Immunol. 7, 94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chretien A.-S., et al., Cancer-induced alterations of NK-mediated target recognition: Current and investigational pharmacological strategies aiming at restoring NK-mediated anti-tumor activity. Front. Immunol. 5, 122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucar O.et al.; ANRS CO5 IMMUNOVIR-2 Study group , B7-H6-mediated downregulation of NKp30 in natural killer cells contributes to HIV-2 immune escape. AIDS 33, 23–32 (2019). [DOI] [PubMed] [Google Scholar]
- 16.De Maria A., et al., The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44). Eur. J. Immunol. 33, 2410–2418 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Zhou J., et al., An NK cell population lacking FcRγ is expanded in chronically infected HIV patients. J. Immunol. 194, 4688–4697 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Vendrame E., et al., TIGIT is upregulated by HIV-1 infection and marks a highly functional adaptive and mature subset of natural killer cells. AIDS 34, 801–813 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mavilio D., et al., Natural killer cells in HIV-1 infection: Dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc. Natl. Acad. Sci. U.S.A. 100, 15011–15016 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mavilio D., et al., Characterization of CD56-/CD16+ natural killer (NK) cells: A highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc. Natl. Acad. Sci. U.S.A. 102, 2886–2891 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Björkström N. K., Ljunggren H.-G., Sandberg J. K., CD56 negative NK cells: Origin, function, and role in chronic viral disease. Trends Immunol. 31, 401–406 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Taouk G., et al., CD56 expression in breast cancer induces sensitivity to natural killer-mediated cytotoxicity by enhancing the formation of cytotoxic immunological synapse. Sci. Rep. 9, 8756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanier L. L., et al., Molecular and functional analysis of human natural killer cell-associated neural cell adhesion molecule (N-CAM/CD56). J. Immunol. 146, 4421–4426 (1991). [PubMed] [Google Scholar]
- 24.Chen L., et al., CD56 expression marks human group 2 innate lymphoid cell divergence from a shared NK cell and group 3 innate lymphoid cell developmental pathway. Immunity 49, 464–476.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mace E. M., Gunesch J. T., Dixon A., Orange J. S., Human NK cell development requires CD56-mediated motility and formation of the developmental synapse. Nat. Commun. 7, 12171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunesch J. T., et al., CD56 regulates human NK cell cytotoxicity through Pyk2. eLife 9, e57346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziegler S., et al., CD56 is a pathogen recognition receptor on human natural killer cells. Sci. Rep. 7, 6138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nitta T., Yagita H., Sato K., Okumura K., Involvement of CD56 (NKH-1/Leu-19 antigen) as an adhesion molecule in natural killer-target cell interaction. J. Exp. Med. 170, 1757–1761 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong H. S., et al., Phenotypically and functionally distinct subsets contribute to the expansion of CD56-/CD16+ natural killer cells in HIV infection. AIDS 24, 1823–1834 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Forconi C. S., et al., Poorly cytotoxic terminally differentiated CD56negCD16pos NK cells accumulate in Kenyan children with Burkitt lymphomas. Blood Adv. 2, 1101–1114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simoni Y., et al., Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity 46, 148–161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milush J. M., et al., Functionally distinct subsets of human NK cells and monocyte/DC-like cells identified by coexpression of CD56, CD7, and CD4. Blood 114, 4823–4831 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller-Durovic B., Grählert J., Devine O. P., Akbar A. N., Hess C., CD56-negative NK cells with impaired effector function expand in CMV and EBV co-infected healthy donors with age. Aging (Albany NY) 11, 724–740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunetta E., et al., The decreased expression of Siglec-7 represents an early marker of dysfunctional natural killer-cell subsets associated with high levels of HIV-1 viremia. Blood 114, 3822–3830 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forconi C. S., et al., A new hope for CD56negCD16pos NK cells as unconventional cytotoxic mediators: An adaptation to chronic diseases. Front. Cell. Infect. Microbiol. 10, 162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez V. D., et al., Expansion of functionally skewed CD56-negative NK cells in chronic hepatitis C virus infection: Correlation with outcome of pegylated IFN-α and ribavirin treatment. J. Immunol. 183, 6612–6618 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Setty M., et al., Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 34, 637–645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimasaki N., Jain A., Campana D., NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 19, 200–218 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Tarazona R., et al., Selective depletion of CD56(dim) NK cell subsets and maintenance of CD56(bright) NK cells in treatment-naive HIV-1-seropositive individuals. J. Clin. Immunol. 22, 176–183 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Alter G., et al., Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood 106, 3366–3369 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez V. D., et al., Expansion of CD56- NK cells in chronic HCV/HIV-1 co-infection: Reversion by antiviral treatment with pegylated IFNalpha and ribavirin. Clin. Immunol. 128, 46–56 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Björkström N. K., et al., Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J. Exp. Med. 208, 13–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen S., et al., Persistence of CD16+/CD56-/2B4+ natural killer cells: A highly dysfunctional NK subset expanded in ocular myasthenia gravis. J. Neuroimmunol. 179, 117–125 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Reiners K. S., et al., Soluble ligands for NK cell receptors promote evasion of chronic lymphocytic leukemia cells from NK cell anti-tumor activity. Blood 121, 3658–3665 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viant C., et al., Cell cycle progression dictates the requirement for BCL2 in natural killer cell survival. J. Exp. Med. 214, 491–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lima M., et al., Clinicobiological, immunophenotypic, and molecular characteristics of monoclonal CD56-/+dim chronic natural killer cell large granular lymphocytosis. Am. J. Pathol. 165, 1117–1127 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wanquet A., et al., Azacitidine treatment for patients with myelodysplastic syndrome and acute myeloid leukemia with chromosome 3q abnormalities. Am. J. Hematol. 90, 859–863 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Devillier R., et al., Prognostic significance of myelodysplasia-related changes according to the WHO classification among ELN-intermediate-risk AML patients. Am. J. Hematol. 90, E22–E24 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Lausen B., Hothorn T., Bretz F., Schumacher M., Assessment of optimal selected prognostic factors. Biom. J. 46, 364–374 (2004). [Google Scholar]
- 50.van Unen V., et al., Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat. Commun. 8, 1740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NK cell trajectory inference data have been deposited in the GitHub repository (https://github.com/moreymat/wishbone/releases/tag/0.4.2-py.3). All other study data are included in the article and/or SI Appendix.