Abstract
Crop improvement relies heavily on genetic variation that arises spontaneously through mutation. Modern breeding methods are very adept at combining this genetic variation in ways that achieve remarkable improvements in plant performance. Novel traits have also been created through mutation breeding and transgenesis. The advent of gene editing, however, marks a turning point: With gene editing, synthetic variation will increasingly supplement and, in some cases, supplant the genetic variation that occurs naturally. We are still in the very early stages of realizing the opportunity provided by plant gene editing. At present, typically only one or a few genes are targeted for mutation at a time, and most mutations result in loss of gene function. New technological developments, however, promise to make it possible to perform gene editing at scale. RNA virus vectors, for example, can deliver gene-editing reagents to the germ line through infection and create hundreds to thousands of diverse mutations in the progeny of infected plants. With developmental regulators, edited somatic cells can be induced to form meristems that yield seed-producing shoots, thereby increasing throughput and shrinking timescales for creating edited plants. As these approaches are refined and others developed, they will allow for accelerated breeding, the domestication of orphan crops and the reengineering of metabolism in a more directed manner than has ever previously been possible.
Keywords: gene editing, plant breeding, crop improvement
For centuries, humans have been developing new plant varieties to ensure that their dinner plates are full and their food is palatable (1, 2). By iteratively selecting for desired traits, such as increased seed or fruit size, species have been phenotypically transformed over time. A modern crop, such as maize, barely resembles its ancestral, grassy relatives. Traits that provide color and flavor have also been selected, giving rise to enormous variation in species such as pepper (3). An extreme example of phenotypic diversity through selection is the spectrum of vegetables that come from a single species: Brassica oleracea. Cabbage, broccoli, cauliflower, Brussel sprouts, kohlrabi, and kale were all derived by selectively choosing plants with different leaf sizes, floret shapes, and stem architectures (4). What is the origin of this enormous phenotypic plasticity? It arises from changes in the plant’s genome brought about by spontaneous mutation, DNA transposition, polyploidization, or crosses with distant relatives. The diverse array of fruits and vegetables in the produce aisle of our grocery stores is the result of human selection imposed upon naturally occurring genetic diversity.
Beginning in the latter part of the last century, knowledge of plant genomes increasingly guided efforts to create new crop varieties. Modern breeding, however, still relies heavily on genetic diversity that occurs spontaneously in nature. Thousands of DNA polymorphisms distinguish one plant variety from the next. Some desired traits are associated with genetic markers. The progeny of crosses between varieties can be quickly surveyed for these markers to determine those that have a desired combination as a result of genome shuffling during meiosis (5). In most cases, the underlying genetic basis for the trait is unknown, but that often doesn’t matter as long as the desired mixture of traits can be assembled by following the markers.
Also, in the last century, methods were developed to deliberately create genetic variation of value, thereby no longer relying upon variation that occurs by happenstance. On one extreme is transgenesis, involving the addition of new genes to plant genomes that confer traits such as insect tolerance or herbicide resistance (6). With few exceptions, however, transgenesis has been predominantly deployed in only a handful of high-value row crops. Novel genetic variation can also be created using mutagenic chemicals such as ethyl methanesulfonate or high energy radiation to alter the genome of the given crop (7). A drawback of mutation breeding is that it provides no control; large mutant populations must be generated and screened in the hope of identifying genetic variation of value (8). Additionally, mutagens typically alter multiple loci, creating unwanted genetic changes and necessitating multiple rounds of crossing to isolate the desired mutation in a preferred genetic background.
The Advent of Gene Editing
With the development of gene-editing technologies over the past 20 y, it is now possible to create genetic variation with a high degree of precision and specificity (9). Gene editing relies on DNA targeting: that is, our ability to deliver molecular reagents to specific sites in complex genomes. DNA targeting was first achieved by engineering DNA binding proteins, such as zinc finger proteins, to recognize specific DNA sequences (10). The discovery of the mechanism by which transcription activator-like effectors (TALEs) bind DNA was a key advance because DNA targeting with engineered TALE proteins is both highly effective and reliable (11). Finally, the discovery of RNA-guided DNA targeting by CRISPR systems made DNA targeting even simpler, less expensive, and easier to achieve (12, 13).
DNA targeting is important because it allows us to create DNA damage at specific sites in genomes. Nucleases, whether they be fused to engineered zinc finger and TALE arrays or are inherently a part of CRISPR systems, create double strand breaks (DSBs) (14). The DSBs are most frequently repaired by end-joining repair mechanisms, resulting in targeted insertion/deletion mutations. Alternatively, DNA templates can be provided that carry sequence information to be copied to the break site through homology dependent repair (HDR). Other forms of DNA damage-inducing agents have also been deployed for gene editing, especially cytosine and adenine deaminases (15–18). Deamination of cytosines and adenines prompts base excision repair (19, 20), thus generating targeted mutations without a DSB. Further, Cas proteins have been repurposed to promote high-fidelity repair off an RNA template. These prime editors contain a “nicking” Cas9 and a fused polymerase that will complex with a primer sequence added to the sgRNA. The prime editor uses the sequence encoded within the repair primer to incorporate base changes at the target site (19).
An editor, of course, has no value unless there is text to edit. The advent of gene editing closely followed the next generation sequencing era—a period of more than a decade in which our capacity to sequence genomes grew exponentially. We now have high quality genomes for most domesticated crop species and numerous wild plants (20). Coupled with DNA sequence information are enormous datasets documenting gene expression and metabolite profiles in various tissues throughout the lifecycle of many plant species (21, 22). Gene editing makes it possible to test hypotheses generated from analysis of all of this data to gain a better understanding of how plant genomes dictate growth and development.
It is now more than 10 y since the first endogenous plant gene has been edited, and how has the technology been used since? First of all, gene editing works in diverse plant species, ranging from model plants like Arabidopsis thaliana, to both monocot (e.g., maize, wheat, and rice) and dicot (e.g., tomato, potato, and soybean) crop species (9, 23). Whereas many plants have been edited, the vast majority of the edits have been loss-of-function mutations created by imprecise end-joining of DSBs, and typically only one or a few genes are targeted at a time. In biosynthetic pathways, loss of gene function can prevent production of undesired end-products or enable accumulation of desired precursors. An example of the latter is the first gene-edited food product to enter the food supply, namely a healthier cooking oil produced from a gene-edited soybean variety (24, 25). Inactivation of two genes encoding fatty acid desaturases in soybean causes monounsaturated fatty acids to accumulate in the seed, resulting in an oil that is healthier and that lasts much longer when used for frying. Whereas knockout mutations are useful in some contexts, a binary output is often inadequate to confer the more complex traits needed for crop improvement.
Of course, more sophisticated gene edits can be created by repairing DSBs through HDR or by using tools such as base and prime editors. To this point, applications of base editors have been limited to transition mutations (15, 16), but newly described base editors broaden the potential scope of editing (26, 27). Prime editors offer much promise but have only recently been implemented in plants (28). HDR makes possible the broadest range of DNA sequence alterations, from single-base substitutions to the insertion of large transgene arrays (9). HDR, however, is inefficient in somatic plant cells where gene editing typically occurs. Strategies have been tested to increase HDR, including knocking out genes involved in end-joining to promote HDR and using reagents to promote entry into S phase, where HDR predominates (29). Although some improvements in HDR efficiencies have been realized, HDR is still infrequently pursued to edit plant genomes because of the large investment in time and resources needed to recover rare events (30).
To fully realize the potential of gene editing, methods are needed that enable the production of edited plants faster, with less cost, and at scale. The majority of gene-edited plants are produced through tissue culture (9, 31), where gene editing reagents are delivered to sterile explants and then edited cells are regenerated into whole plants. These two steps—delivery of reagents and regeneration of edited cells into plants—are the primary bottlenecks for gene editing and for realizing its full potential (31). The following two sections outline two rapidly developing areas of research that promise to break these bottlenecks, including contributions made by our laboratory.
Breaking the Tissue Culture Bottleneck with Developmental Regulators
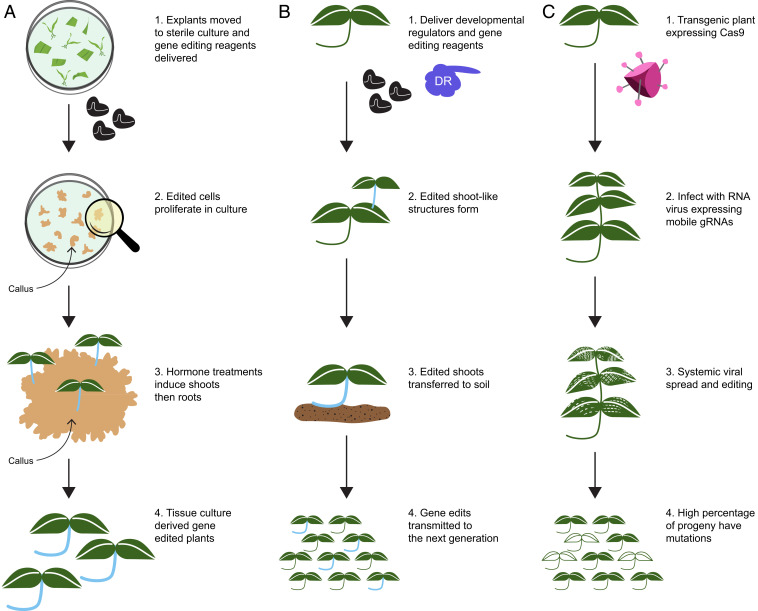
Plant cells are totipotent and can be induced to differentiate into other cell types (32, 33). Tissue culture is used to take advantage of this totipotency to propagate, transform, or edit somatic cells. Prior to inducing differentiation into shoots and roots, somatic cells are typically allowed to grow and divide in an undifferentiated state as a callus (Fig. 1A). Promoting totipotency is not easy. Different plants have different requirements for growth media and hormones, particularly auxin and cytokinin that induce the shoots and roots. For many species, we have not yet defined the appropriate media and hormone regimes needed to regenerate somatic cells, and even within a species, enormous variation is observed between cultivars or land races in terms of the capacity to regenerate plants from somatic cells. Further, it is well-documented that passage of cells through culture is mutagenic and often alters the epigenetic landscape (34, 35). Finally, considerable time is required—from several months to year—to coax an edited cell into a plant (31). Attempts have been made to avoid transformation by delivering transgenes to meristems or egg cells; however, efficiencies are low and only robust in A. thaliana and its close relatives (36).
Fig. 1.
Approaches for creating genetic variation in plants. (A) Gene editing through tissue culture. Most gene-edited plants are produced through tissue culture. Explants (leaf tissue, cotyledons, immature embryos) are sterilized and placed on medium. Gene-editing reagents are then delivered to the somatic cells; edited cells are allowed to proliferate. Addition of plant hormones induces first shoots and then roots, resulting in the edited plant. (B) Gene editing through de novo meristem induction. A seedling is treated with an Agrobacterium strain that delivers gene-editing reagents and developmental regulators. The latter promote the induction of a meristem from edited somatic cells. The meristem ultimately forms a shoot, which is excised, and roots are induced. Progeny of the plant are characterized for mutations of interest. (C) Gene editing with RNA viruses. Because RNA viruses have limited cargo capacity (typically <1 kb), a transgenic plant is created that expresses Cas9 (or a base or prime editor). The plant is infected with viruses that express sgRNAs as they spread throughout the plant. The sgRNAs are augmented to carry sequences that promote movement into the germ line. Infected plants are grown to maturity and seed harvested. Mutant progeny are characterized to identify those with mutations of interest.
Certainly hormones, like auxin and cytokinin, are critical to induce organogenesis from somatic cells, but they work in concert with developmental regulators (37), mostly transcription factors, that help realize totipotency. It was recently shown that ectopic expression of developmental regulators BABYBOOM and WUSCHEL could dramatically promote organogenesis, in this case the formation of somatic embryos, in maize somatic cells (38, 39). When transgenes were codelivered with the developmental regulators, transgenic embryos were formed in 2 wk and plantlets in 2 to 4 wk (39) in contrast to traditional maize transformation, which requires 12–20 wk (40). The developmental regulators also promote somatic embryogenesis in sorghum (41), and if broadly applicable across monocots, the technology will be transformative, allowing rapid production of gene-edited plants with reduced costs and at scale.
More recently, our laboratory used developmental regulators and a cytokinin biosynthesis gene to induce de novo meristems on dicot plants (Fig. 1B) (42). When delivered by Agrobacterium to seedlings or soil-grown plants, shoots formed that were induced by the developmental regulators. Codelivering reporter genes or gene editing reagents made it possible to recover shoots that were either transgenic or gene edited. When the shoots produced flowers and seed, the gene edits and transgenes were transmitted to the next generation. Original proof of concept was carried out in Nicotiana benthamiana, but shoot induction works in other species, including tomato, potato, and grape. The ability to create gene-edited shoots on plants without having to go through tissue culture sidesteps the tissue culture bottleneck and, thereby, accelerates the production of gene-edited plants. Future work will determine if this approach can be more broadly used across dicots.
Breaking the Delivery Bottleneck with RNA Viruses
Methods for delivering DNA to plants have really not changed since the early 1980s. Delivery is most often performed in one of two ways: gene transfer via bacteria, such as Agrobacterium tumefaciens, or biolistic particle bombardment (43, 44). Drawbacks to the use of Agrobacterium include its limited host range and the fact that the DNA that is delivered often integrates into the genome (43, 45). The integrated DNA then needs to be segregated away to produce a nontransgenic, edited plant. Bombardment-based methods often result in multiple and sometimes fragmented transgene insertions (44). More recently, nanoparticles show promise as reagent delivery vehicles; however, work is still in the early stages (46). All of these methods rely on tissue culture in order to generate edited somatic cells into whole plants.
Over the past several years, there has been a growing interest in using plant viruses as vectors to deliver gene-editing reagents. Viruses are rarely used for plant genetic modification, the exception being positive strand RNA viruses, which have been engineered to express proteins or small fragments of endogenous genes that trigger virus induced gene silencing (47). A major drawback of RNA viruses as vectors is their limited cargo capacity, which is typically less than 1 kb, precluding their use for delivering gene-editing reagents such as Cas9 from Streptococcus pyogenes (4.1 kb). To overcome this limitation, transgenic plants have been made that express Cas9, and RNA viruses are used to deliver sgRNAs to these plants through infection (Fig. 1C) (48–52). This approach results in somatic cells being edited; however, recovery of mutant progeny is rare (48). Our laboratory and collaborators recently showed that if RNA sequences that promote cell-to-cell movement are fused to the sgRNAs, heritable gene edits are recovered at high frequency (53). These experiments used Tobacco Rattle Virus as a vector and Cas9-expressing N. benthamiana plants. Most of the seed harvested from infected plants carried one or more gene edits.
RNA virus vectors have been developed for diverse plant species, and the next step will be to test whether mutagenesis through infection works efficiently in other plants, including monocots. Further, if transgenic plants are created that express base or prime editors, then it should be possible to make more precise edits through infection. Although transgenic plants are needed for this approach, tissue culture is only performed once. All subsequent editing occurs with soil grown plants enabling mutagenesis at scale.
Implications for the Future
The technological advances described above and others likely soon to follow will take us from our current state—being able to create a handful of knockout mutations in germplasm amenable to tissue culture—to the ability to efficiently edit multiple genes simultaneously. We will increasingly see natural genetic diversity being supplemented with synthetic diversity. Below are a few glimpses of what the future might look like based on advances already being realized.
As mentioned in the Introduction, since the dawn of agriculture, we have transformed species by selecting desired traits, making continual improvements from season to season. For many species, we now understand the genetic basis for some of the phenotypic changes that have come with crop domestication. Often, only a handful of genes are drivers of domestication and are responsible for increases in yield or alterations in plant architecture that make plants more conducive to cropping systems (1, 2). Modifications to these genes, wrought naturally over time, can now be created in the laboratory through gene editing. As an illustration, in tomato, several loss-of-function mutations impact plant architecture as well as fruit size and quality. We and others inactivated these genes in a wild species of tomato (Solanum pimpinellifolium), such that in one round of editing, the berry-like fruits on the edited plant increased threefold in size and 10-fold in number (54, 55). Further, the vine-like, wild tomato was transformed into the shrub-like plants we are familiar with in our gardens. Some of the same genes were altered in a relative of tomato, ground cherry, to realize improvements in fruit quality (56).
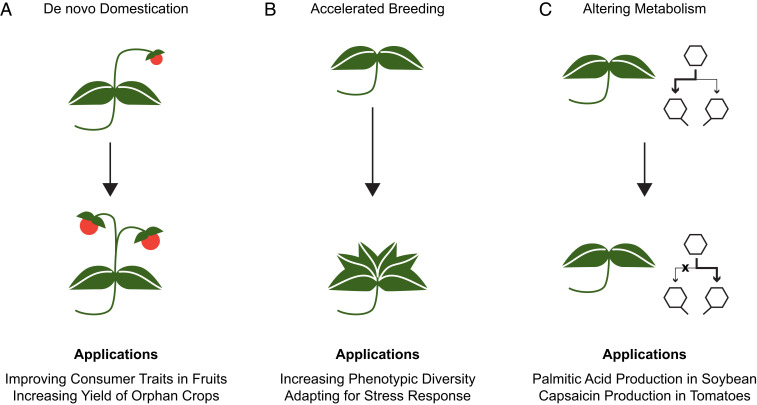
The above studies in tomato and ground cherry suggest that gene editing can be used to rapidly improve so-called orphan crops—crops not been subjected to intensive selection and breeding regimes (Fig. 2A). The list of orphan crops is long and includes dietary staples like cassava, millet, and savory bananas (57). Many of these orphan crops are locally adapted to specific environments and are important to the peoples who grow them, but they nonetheless have undesirable traits that could be addressed through gene editing. One candidate for domestication is the staple crop of Ethiopia and Eritrea, tef. Tef is a cereal used to make the bread, injera, but productivity is hampered by substantial yield losses due to lodging and seed shattering (58, 59). Using gene editing, it should be possible to rapidly introduce semidwarf varieties resistant to lodging or with reduced seed shatter. De novo domestication promises to complement and augment traditional breeding efforts. The end result will be an abundant and more diverse food supply.
Fig. 2.
Future applications for innovative plant design. The ability to edit genes in a programmable fashion, in conjunction with advanced reagent delivery and transformation technologies, makes new methods of crop design possible. (A) Orphan crops can be domesticated through editing to confer traits such as increased fruit size, reduced tillering, or less seed shatter, all of which promise to increase yield and marketability of orphan crops. (B) New breeding methods can accelerate creation of novel genetic variation. (C) Primary metabolic pathways (like fatty acid biosynthesis) or secondary metabolic pathways (like the production of capsaicin) can be harnessed to produce a diverse array of metabolites for food, fuel, and industrial purposes.
Of course, gene editing can also be used to improve highly domesticated species (Fig. 2B). In the experiments with tomato, it was loss-of-function mutations that helped drive domestication, but more commonly, traits important for agriculture are polygenic and quantitative. Further, in species such as maize, many of the key domestication genes are transcription factors that control traits such as plant architecture or flowering time (60). In some cases, changes to cis-regulatory sequences have dialed up or down the expression of these master regulators or altered tissue specificity or responses to environmental cues. Littering the promoters of master regulators with mutations can recapitulate changes that occurred during domestication or create new variation of value. Indeed, this approach has already been demonstrated in tomato, resulting in changes to plant architecture and fruit locule number (61).
As mentioned earlier, a goal of marker-assisted breeding is to combine genetic information for an optimal outcome. As a simple illustration, two parents, each with desired traits and molecular markers linked to those traits, are crossed to make an F1. Meiotic recombination that occurs in the production of eggs and pollen mixes the genomes of the parents (62). By following the markers in the progeny, it is possible to identify individuals that carry the desired complement of markers and, thereby, the desired phenotype. Meiotic recombination is really a process of DSB repair, and it should be possible to target where the breaks occur in meiosis, making the outcome of meiotic recombination predictable. Such an approach has already been demonstrated in yeast, and plants are likely soon to follow (63). The ability to control meiotic recombination could eliminate so-called linkage drag by separating desirable traits from closely linked undesirable ones. In the coming years, systems biology will continually reveal the genes and genetic variation responsible for many polygenic and quantitative traits. Controlling meiotic recombination will make it possible to assemble this variation in new ways to enable complex traits, from increases in photosynthetic efficiency to responses to environmental stresses caused by climate change.
The reason we grow many plants, particularly commodity crops, is because they provide an ample supply of primary metabolites like fatty acids, carbohydrates, and protein, which we use for food and fuel. In general, we understand the genetics and biochemistry of primary metabolism, which should make it easier to manipulate these pathways through gene editing, in contrast to some of the more poorly understood polygenic and quantitative traits described above that affect crop performance and yield. It is really no surprise, then, that the first gene-edited food ingredient to be commercialized is a trait that makes soybean oil more similar in fatty acid composition to sunflower oil (25). Globally, we invest enormously in agricultural enterprises and supply chains to produce and distribute food oils. The production of some oils, like palm, has been criticized because of the deforestation of the tropical rain forests that take place to increase productivity (64). With a plant like soybean, why not dial up and down the composition of certain fatty acids to make a palm oil equivalent? When grown locally, such a soybean variety could help reduce deforestation of tropical lands and obviate the need to transport oil around the world. Although the socioeconomic impacts of oil choice and production are complex (65), it is intriguing to think that soybean, or perhaps another oil seed crop such as canola, could become the genetic chassis used to produce diverse oils for a more planet-friendly means of food and fuel production (Fig. 2C).
If soybean is a chassis for producing food oils, other plants can be chassis for the production of other metabolites. Plants make a diverse array of secondary metabolites of value as industrial chemicals, pharmaceuticals, flavors, and fragrances. Solanaceous plants, like tobacco and tomato, are particularly known for the diverse array of secondary metabolites they produce. These biosynthetic pathways could be repurposed to produce key secondary metabolites of value. Secondary metabolites could also be altered to enhance food flavor and color. The enzymatic pathway that produces capsaicin in peppers (66, 67) is also present in tomato species, but is expressed at a very low level (68). Imagine a spicier tomato created with gene edits that increase promoter activity of capsaicin biosynthetic genes. Plants possess a variety of additional flavoring compounds that could be manipulated through editing (69–71) to enhance taste and create exciting new food cultivars.
Conclusion
The gene-editing revolution has been slow to be realized in plants, due to inefficient methods of reagent delivery and the reliance on tissue culture. Here, we presented two technological approaches being worked on in our laboratory, namely the de novo induction of gene-edited meristems and the use of RNA viruses, to create gene edits through infection. Will these approaches overcome current bottlenecks in the production of gene edited plants? It is still too early to tell, but they are a starting point as we work toward the next era of plant gene editing. Improvements to the technology described here and technological advances under development elsewhere will gradually open the bottlenecks. The consequent ability to efficiently edit plant genomes at scale will irrevocably alter our approach to trait development and enable us to truly harness the biosynthetic capacity of plants to meet our growing need for plant-based products.
Acknowledgments
Work in the D.F.V. laboratory is supported by National Science Foundation Grant 1833402 and the Department of Energy Grant DE-SC0018277. We thank M. Leffler for help with the figures.
Footnotes
The authors declare no competing interest.
This paper results from the NAS Colloquium of the National Academy of Sciences, “Life 2.0: The Promise and Challenge of a CRISPR Path to a Sustainable Planet,” held December 10–11, 2019, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. NAS colloquia began in 1991 and have been published in PNAS since 1995. The complete program and video recordings of presentations are available on the NAS website at http://www.nasonline.org/CRISPR. The collection of colloquium papers in PNAS can be found at https://www.pnas.org/page/collection/crispr-sustainable-planet.
This article is a PNAS Direct Submission.
Data Availability
There are no data underlying this work.
References
- 1.Doebley J. F., Gaut B. S., Smith B. D., The molecular genetics of crop domestication. Cell 127, 1309–1321 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Meyer R. S., Purugganan M. D., Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Carrizo García C., et al., Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann. Bot. 118, 35–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson K. F., “Cabbages, kales etc. Brassica oleracea (Cruciferae)” in Evolution of Crop Plants, Simmonds N. W., Ed. (Longman, 1976), pp. 49–52. [Google Scholar]
- 5.Collard B. C., Mackill D. J., Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 557–572 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamthan A., Chaudhuri A., Kamthan M., Datta A., Genetically modified (GM) crops: Milestones and new advances in crop improvement. Theor. Appl. Genet. 129, 1639–1655 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Sikora P., Chawade A., Larsson M., Olsson J., Olsson O., Mutagenesis as a tool in plant genetics, functional genomics, and breeding. Int. J. Plant Genomics 2011, 314829 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCallum C. M., Comai L., Greene E. A., Henikoff S., Targeted screening for induced mutations. Nat. Biotechnol. 18, 455–457 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Chen K., Wang Y., Zhang R., Zhang H., Gao C., CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Carroll D., Genome engineering with zinc-finger nucleases. Genetics 188, 773–782 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogdanove A. J., Voytas D. F., TAL effectors: Customizable proteins for DNA targeting. Science 333, 1843–1846 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Doudna J. A., Charpentier E., Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Hsu P. D., Lander E. S., Zhang F., Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jasin M., Haber J. E., The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair (Amst.) 44, 6–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komor A. C., Kim Y. B., Packer M. S., Zuris J. A., Liu D. R., Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudelli N. M., et al., Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roldán-Arjona T., Ariza R. R., Córdoba-Cañero D., DNA base excision repair in plants: An unfolding story with familiar and novel characters. Front Plant Sci 10, 1055 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beard W. A., Horton J. K., Prasad R., Wilson S. H., Eukaryotic base excision repair: New approaches shine light on mechanism. Annu. Rev. Biochem. 88, 137–162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anzalone A. V., et al., Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kersey P. J., Plant genome sequences: Past, present, future. Curr. Opin. Plant Biol. 48, 1–8 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Yonekura-Sakakibara K., Fukushima A., Saito K., Transcriptome data modeling for targeted plant metabolic engineering. Curr. Opin. Biotechnol. 24, 285–290 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Kumar R., Bohra A., Pandey A. K., Pandey M. K., Kumar A., Metabolomics for plant improvement: Status and prospects. Front Plant Sci 8, 1302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malzahn A., Lowder L., Qi Y., Plant genome editing with TALEN and CRISPR. Cell Biosci. 7, 21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haun W., et al., Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Splitter J., The Latest Gene-Edited Food Is A Soybean Oil That Comes With Zero Trans Fats. Forbes (September 21, 2020).
- 26.Zhao D., et al., Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol., 10.1038/s41587-020-0592-2 (2020). Erratum in Nat. Biotechnol., 10.1038/s41587-020-0648-3 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Kurt I. C., et al., CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol., 10.1038/s41587-020-0609-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Q., et al., Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Yeh C. D., Richardson C. D., Corn J. E., Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 21, 1468–1478 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Li S., Xia L., Precise gene replacement in plants through CRISPR/Cas genome editing technology: Current status and future perspectives. aBIOTECH 1, 58–73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altpeter F., et al., Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasil I. K., Vasil V., Totipotency and embryogenesis in plant cell and tissue cultures. In Vitro 8, 117–127 (1972). [DOI] [PubMed] [Google Scholar]
- 33.Verdeil J.-L., Alemanno L., Niemenak N., Tranbarger T. J., Pluripotent versus totipotent plant stem cells: Dependence versus autonomy? Trends Plant Sci. 12, 245–252 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Phillips R. L., Kaeppler S. M., Olhoft P., Genetic instability of plant tissue cultures: Breakdown of normal controls. Proc. Natl. Acad. Sci. U.S.A. 91, 5222–5226 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang D., et al., Tissue culture-induced heritable genomic variation in rice, and their phenotypic implications. PLoS One 9, e96879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clough S. J., Bent A. F., Floral dip: A simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Pierre-Jerome E., Drapek C., Benfey P. N., Regulation of division and differentiation of plant stem cells. Annu. Rev. Cell Dev. Biol. 34, 289–310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowe K., et al., Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowe K., et al., Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell. Dev. Biol. Plant 54, 240–252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishida Y., Hiei Y., Komari T., Agrobacterium-mediated transformation of maize. Nat. Protoc. 2, 1614–1621 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Mookkan M., Nelson-Vasilchik K., Hague J., Zhang Z. J., Kausch A. P., Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. Plant Cell Rep. 36, 1477–1491 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maher M. F., et al., Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gelvin S. B., Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorence A., Verpoorte R., “Gene transfer and expression in plants” in Recombinant Gene Expression: Reviews and Protocols, Balbás P., Lorence A., Eds. (Methods in Molecular Biology, Humana Press, 2004), pp. 329–350. [DOI] [PubMed] [Google Scholar]
- 45.Latham J. R., Wilson A. K., Steinbrecher R. A., The mutational consequences of plant transformation. J. Biomed. Biotechnol. 2006, 25376 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cunningham F. J., Goh N. S., Demirer G. S., Matos J. L., Landry M. P., Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 36, 882–897 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burch-Smith T. M., Anderson J. C., Martin G. B., Dinesh-Kumar S. P., Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39, 734–746 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Ali Z., et al., Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant 8, 1288–1291 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Cody W. B., Scholthof H. B., Mirkov T. E., Multiplexed gene editing and protein overexpression using a Tobacco mosaic virus viral vector. Plant Physiol. 175, 23–35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ali Z., Eid A., Ali S., Mahfouz M. M., Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res. 244, 333–337 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Gao Q., et al., Rescue of a plant cytorhabdovirus as versatile expression platforms for planthopper and cereal genomic studies. New Phytol. 223, 2120–2133 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Hu J., et al., A barley stripe mosaic virus-based guide RNA delivery system for targeted mutagenesis in wheat and maize. Mol. Plant Pathol. 20, 1463–1474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ellison E., et al., Highly efficient, heritable, multiplexed gene editing using RNA viruses and mobile sgRNAs. Nat. Plants, 10.1038/s41477-020-0670-y (2020). [DOI] [PubMed] [Google Scholar]
- 54.Zsögön A., et al., De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36, 1211–1216 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Li T., et al., Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol., 10.1038/nbt.4273 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Lemmon Z. H., et al., Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4, 766–770 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Tadele Z., Orphan crops: Their importance and the urgency of improvement. Planta 250, 677–694 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Cheng A., Mayes S., Dalle G., Demissew S., Massawe F., Diversifying crops for food and nutrition security–A case of teff. Biol. Rev. Camb. Philos. Soc. 92, 188–198 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Eshed Y., Lippman Z. B., Revolutions in agriculture chart a coursresee for targeted breeding of old and new crops. Science 366, eaax0025 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Dong Z., et al., The regulatory landscape of a core maize domestication module controlling bud dormancy and growth repression. Nat. Commun. 10, 3810 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodríguez-Leal D., Lemmon Z. H., Man J., Bartlett M. E., Lippman Z. B., Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Wang Y., Copenhaver G. P., Meiotic recombination: Mixing it up in plants. Annu. Rev. Plant Biol. 69, 577–609 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Sarno R., et al., Programming sites of meiotic crossovers using Spo11 fusion proteins. Nucleic Acids Res. 45, e164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vijay V., Pimm S. L., Jenkins C. N., Smith S. J., The impacts of oil palm on recent deforestation and biodiversity loss. PLoS One 11, e0159668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meijaard E., Sheil D., The moral minefield of ethical oil palm and sustainable development. Front. For. Glob. Change 2, 22 (2019). [Google Scholar]
- 66.Kim S., et al., Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Zhang Z.-X., et al., Discovery of putative capsaicin biosynthetic genes by RNA-Seq and digital gene expression analysis of pepper. Sci. Rep. 6, 34121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naves E. R., et al., Capsaicinoids: Pungency beyond capsicum. Trends Plant Sci. 24, 109–120 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Ayseli M. T., İpek Ayseli Y., Flavors of the future: Health benefits of flavor precursors and volatile compounds in plant foods. Trends Food Sci. Technol. 48, 69–77 (2016). [Google Scholar]
- 70.Wang D., Seymour G. B., Tomato flavor: Lost and found? Mol. Plant 10, 782–784 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Tieman D., et al., A chemical genetic roadmap to improved tomato flavor. Science 355, 391–394 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no data underlying this work.