Significance
The discovery of the archaic Denisovan hominins is one of the most significant findings in human evolutionary biology in the last decade. However, as of today, we have more questions than answers regarding this mysterious hominin group. This study leverages the information from the well-known example of adaptive introgression on the EPAS1 gene in Tibetans, to gain insight on the history of our species’ interaction with Denisovans. We show that the Tibetan-EPAS1 haplotype came from the East Asian-specific Denisovan introgression event, and it remained selectively neutral for a long time in the population before positive selection occurred, which may be concurrent with the permanent inhabitation of the Tibetan Plateau after the Last Glacial Maximum (LGM).
Keywords: adaptation, archaic introgression, high altitude, natural selection, admixture
Abstract
Recent studies suggest that admixture with archaic hominins played an important role in facilitating biological adaptations to new environments. For example, interbreeding with Denisovans facilitated the adaptation to high-altitude environments on the Tibetan Plateau. Specifically, the EPAS1 gene, a transcription factor that regulates the response to hypoxia, exhibits strong signatures of both positive selection and introgression from Denisovans in Tibetan individuals. Interestingly, despite being geographically closer to the Denisova Cave, East Asian populations do not harbor as much Denisovan ancestry as populations from Melanesia. Recently, two studies have suggested two independent waves of Denisovan admixture into East Asians, one of which is shared with South Asians and Oceanians. Here, we leverage data from EPAS1 in 78 Tibetan individuals to interrogate which of these two introgression events introduced the EPAS1 beneficial sequence into the ancestral population of Tibetans, and we use the distribution of introgressed segment lengths at this locus to infer the timing of the introgression and selection event. We find that the introgression event unique to East Asians most likely introduced the beneficial haplotype into the ancestral population of Tibetans around 48,700 (16,000–59,500) y ago, and selection started around 9,000 (2,500–42,000) y ago. Our estimates suggest that one of the most convincing examples of adaptive introgression is in fact selection acting on standing archaic variation.
The identification of the Denisovan genome using DNA recovered from a phalanx bone is one of the most stunning discoveries in human evolution in the past decade (1, 2). However, many questions remain unanswered regarding the Denisovans. For example: What did they look like? What was their geographical range? What is their genetic legacy to modern humans? Much of the ongoing research investigating the Denisovans focuses on studying the morphological features from dental and cranial samples (3), dating the age of remains from the Denisova Cave (4), and learning about the admixture events that involved Denisovans, Neanderthals, and other unknown archaic populations (1, 5–7). We now know that Denisovans diverged from Neanderthals ∼390 thousand years ago (ka) (8, 9), and both groups inhabited Eurasia until up to 40 ka (4, 10) based on radiocarbon dating of materials from Neanderthal or Denisovan archeological sites.
Although the fossil remains of Denisovans found so far are limited in number and highly fragmented in nature (1, 11, 12), certain aspects of this hominin group have been revealed through studying a single high-coverage genome (2). The occurrence of admixture between archaic hominins and modern humans is undisputed, as it left varying amounts of archaic DNA in our genomes at detectable levels (1, 8, 13). Notably, Papuans and Indigenous Australians harbor the largest genome-wide amount of Denisovan introgression [∼1–5% (1, 6, 14–16)], followed by East and South Asians [∼0.06–0.5% (1, 6, 14)], and Indigenous Americans [∼0.05–0.4% (1, 6, 14)]. Thus, one approach to study the Denisovans is through the surviving Denisovan DNA segments in modern humans.
Examination of Denisovan-like DNA in modern humans revealed a number of candidate genes with robust signatures of adaptive introgression (16–21), among which the most well-known example is found in the Endothelial Pas Domain Protein 1 gene (EPAS1) in modern Tibetans (22–24) that facilitated local adaptation to their high altitude and hypoxic environment. The discovery of adaptive introgression in Tibetans is particularly striking, as they do not carry high amounts of Denisovan ancestry genome-wide, compared to other South Asian and Oceanian populations (6). Conversely, the Oceanian populations—including the Papuans—do not carry the Denisovan EPAS1 haplotype, perhaps because Denisovan populations that introgressed into ancestral Papuan populations did not harbor the EPAS1 adaptive haplotype, or the variant got lost through genetic drift due to the absence of selective pressure outside of the high-altitude environment. The Tibetan Plateau, with an average altitude above 3,500 m and oxygen concentration considerably lower than at sea level, creates a strong physiological stress for most humans. One common acclimatization to the hypoxic environment is an increase in hemoglobin concentration (25), which increases blood viscosity and is associated with increased risk of pregnancy complications and cardiovascular disease (26, 27). Remarkably, Tibetans have a severely blunted acclimatization response compared to lowlanders at high altitudes and tend not to suffer from clinically elevated hemoglobin concentration (28). This presumed adaptive response is directly associated with variants in the EPAS1 gene, which encodes a transcription factor in the hypoxia response pathway.
The remarkable Denisovan connection to Tibetans’ high-altitude adaptation has led to more questions regarding this already mysterious hominin group. For example, why are populations with Denisovan ancestry, including the Tibetans and Oceanians, located far away from the Denisova Cave in Siberia? One explanation for these seemingly puzzling findings is a large Denisovan geographical range. Multiple introgression events or a higher initial proportion of introgression may explain why some human populations exhibit higher levels of Denisovan introgression despite being located far away from the Altai Mountains in Siberia. Indeed, Browning et al. (7) proposed two Denisovan introgressions into modern East Asians, one of which is shared with Papuans and South Asians. More recently, Jacobs et al. (29) proposed an additional introgression event into the ancestral population of Papuans, making a total of three Denisovan introgression pulses in Asia. Their estimates of split times between the Denisovan groups that admixed with modern humans are large enough (∼280–360 ka) to suggest that there were multiple Denisovan-like hominin groups inhabiting diverse locations in Asia.
In this study, we investigate the surviving Denisovan introgressed segments in Tibetans to address the following questions: Do Tibetans exhibit signatures of more than one Denisovan introgression? If so, which introgression event introduced the beneficial EPAS1 haplotype, and when? Did selection act immediately after introgression, or plausibly later when modern humans began inhabiting the Tibetan Plateau? To address these questions, we examined the EPAS1 gene sequences from a combined dataset of 78 Tibetan individuals from two previously published studies (23, 30), among which 38 are high-coverage whole-genome sequences (30). We leveraged information from the introgressed tracts in Tibetans to infer the key time points related to the Denisovan introgression, as well as the onset of selection. We also employed the whole genomes in the combined dataset to demonstrate that the ancestors of modern Tibetans, similar to other East Asian populations (7), experienced two Denisovan introgression events. Our results provide resolution to the East Asian-specific Denisovan admixture event that led to one of the most fascinating stories of human adaptation, and shed light on the effects of different evolutionary processes that shape patterns of adaptive introgression in humans.
Results
Evidence of Three Distinct Archaic Introgression Episodes with Ancestral Tibetans: One from Neanderthals and Two from Denisovans.
To characterize the genomic landscape of archaic introgression in Tibetans and to determine the number of introgression pulses, we applied the method developed in Browning et al. (7), SPrime. This is a reference-free method that detects sets of diagnostic single-nucleotide polymorphisms (SNPs) that tag putatively archaic-introgressed segments in different regions of the genome. Applying SPrime to the autosomes of 38 Tibetan genomes (30), we inferred 1,426 regions, each containing a set of diagnostic, putatively archaic-introgressed SNPs using Africans (YRI) (31) as an outgroup. The remnants of archaic introgression in Tibetans are spread widely across the genome, illustrated by the presence of SPrime-inferred segments on all 22 autosomes (SI Appendix, Fig. S1). Following Browning et al. (7), for each segment, we computed the match rate of Tibetans against the Altai Neanderthal and Denisovan genomes at the different sets of diagnostic SNPs. The match rate distribution is visualized as a contour plot in Fig. 1. Most regions detected show high affinity to Neanderthal (∼80% matching) and low affinity to Denisovan representing the highest peak (colored in red) in the plot. This is consistent with the observation of higher rate of Neanderthal introgression compared to Denisovan introgression in all Eurasian populations (13). We also observe two additional peaks representing segments of the genome that have low (∼10%) affinity with Neanderthals and higher (∼50% and ∼80%) affinity with Denisovans. These two peaks represent putative Denisovan introgressed segments, and the bimodal distribution of Denisovan match rates is concordant with the hypothesis of two pulses of admixture with Denisovan-like archaic humans in East Asia (7), as only one peak in the match rate (with the Altai Denisovan) distribution is expected under a single pulse of introgression.
Fig. 1.
Introgressed segments in EPAS1 and the genome-wide match rate with archaic individuals. This figure shows the density distribution of match rate to archaic individuals (Altai Denisovan or Neanderthal) in putatively archaic introgressed segments in 38 Tibetans, inferred by the SPrime program using Africans (YRI) as the outgroup. The match rate is defined as the proportion of alleles at the SPrime diagnostic polymorphic sites in a putatively introgressed segment that are present in the genome of archaic individuals at those positions (7). For a given segment, a match rate of 0 denotes that at a given set of diagnostic sites inferred by SPrime, none of the alleles at those sites match the corresponding alleles in the sequenced archaic human. The color range denotes the density of the contours, with red indicating high density and yellow indicating low density. The red star represents the matching coordinates for the introgressed segment within the EPAS1 gene.
Near EPAS1, two putatively archaic introgressed segments are inferred within 200 kb upstream and downstream of the EPAS1 gene region, with a match rate to the Altai Denisovan of 72% and 46%, respectively (Table 1 and SI Appendix, Fig. S2A). The previously identified segment within the EPAS1 gene (23) that harbors the adaptive allele was not detected by SPrime using YRI as the outgroup population. This is likely due to the fact that Yorubans carry a small number of the archaic alleles in the EPAS1 region (23, 32), which could occur from mechanisms such as shared ancestry with archaic humans, unknown archaic admixture in Africa (33–35), or backward gene flow from non-Africans to Africans (36). The latter two, however, are unlikely because 1) inspection of archaic introgression maps in Africans (33, 37) do not detect this region as being introgressed, 2) visual inspection of the haplotypes shows that the Africans do not harbor the adaptive EPAS1 haplotype, and 3) allele sharing between African and Denisovans is similar to other non-Tibetan populations (SI Appendix, Fig. S3 and Table S4). Therefore, the most parsimonious explanation is shared ancestry with archaic humans, but the presence of a few archaic alleles in this region hinders the detection of introgressed segments using algorithms such as SPrime. In fact, repeating the SPrime analysis using modern Europeans (CEU, who do not harbor the Denisovan variants at EPAS1) as an outgroup population does detect the putatively adaptive archaic segment in the core region of EPAS1 (chr2:46,550,132–46,600,661, hg19) that matches with high affinity to the Altai Denisovan (82.14%) but not the Altai Neanderthal (28.57%; Table 1). In this region, the SPrime-inferred variants in Tibetans are more similar to the Denisovan (SI Appendix, Fig. S3), and as expected, exhibit high genetic differentiation between Tibetans and Han Chinese (as measured by FST; see SI Appendix, Fig. S4). Using Europeans (CEU) as an outgroup for detecting introgression in Tibetans results in a similar genome-wide distribution of match rates as observed earlier, but with fewer inferred segments in general, indicating that these are primarily a subset of the ones we obtained when using the YRI as an outgroup (SI Appendix, Fig. S2B).
Table 1.
Archaic introgression segments within 200 kb of the EPAS1 gene region inferred by SPrime
| Upstream region | Core region | Downstream region | |
| Segment length | 140.9 kb | 50.5 kb | 150.9 kb |
| Positions (hg19) | chr2:46,317,587–46,458,516 | chr2:46,550,132–46,600,661 | chr2:46,657,114–46,808,047 |
| No. of archaic-specific alleles | 63 | 29 | 81 |
| Match rate with Altai Neanderthal | 13.6% | 28.57% | 22.53% |
| Match rate with Altai Denisovan | 72.73% | 82.14% | 46.48% |
| Reference outgroup | YRI, CEU | CEU | YRI, CEU |
| Input data | 38 Tibetan whole genomes (30) | ||
The SPrime program infers three introgressed segments at or within 200-kb range of the EPAS1 gene in Tibetans. One segment is within the gene (core region), another segment is upstream of EPAS1, and the third segment is downstream of EPAS1. Table shows the match rates to the Altai Denisovan and Neanderthal, the length of each segment, the chromosome and position range, number of diagnostic SNPs detected by SPrime, and the outgroup used by SPrime. The match rate is defined as the proportion of alleles at the SPrime diagnostic SNPs that are present in the sequenced archaic individual.
The East Asian-Specific Denisovan Introgression Event Introduced the Beneficial EPAS1 Haplotype to Ancestral Tibetans.
We showed in the previous section that, similar to other East Asians (7, 29), Tibetans also display evidence of two Denisovan introgression events, with one being unique to East Asians. Next, we tried to determine which of these two admixture events introduced the beneficial haplotype in EPAS1. To do so, we compared the introgressed segments in the 38 Tibetans at EPAS1 to the SPrime-inferred regions that exhibit the highest (>60%) Denisovan match rate and a low (<40%) Neanderthal match rate (peak from Fig. 1 and SI Appendix, Fig. S5). These segments were likely introduced via an East Asian-specific introgression event with a Denisovan population more closely related to the Altai Denisovan; other populations (e.g., South Asians, Oceanians) lack introgressed segments with this level of affinity to the Altai Denisovan (7).
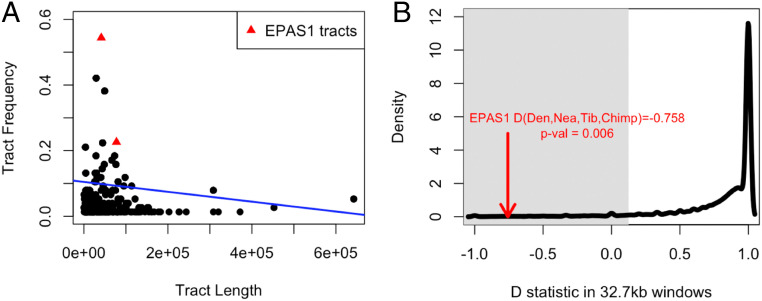
Since SPrime does not infer the introgressed segments for each individual chromosome, we applied a hidden Markov model (HMM) (13, 17, 38) to infer the Denisovan-introgressed tracts in each Tibetan haplotype (Methods). We show that the introgressed segments in EPAS1 exhibit high Denisovan affinity, as shown in Fig. 1, and are of similar length as other segments with the highest Denisovan affinity (SI Appendix, Fig. S6 A and B). Segments in EPAS1 are only outliers in terms of the tract frequency (Fig. 2A), which is in concordance with the expectation of positive selection acting on this region. Based on these observations, we propose that the EPAS1 haplotype in Tibetans was introduced through the pulse of East Asian-specific Denisovan introgression.
Fig. 2.
Introgressed tract length and frequency and D statistics. A shows the lengths (x axis) and frequencies (y axis) of introgressed tracts inferred by an HMM applied to the high Denisovan affinity regions detected with SPrime in 38 Tibetans. These regions have a match rate <40% to Neanderthals and >60% to Denisovans (Fig. 1 and Methods). The red triangles represent the introgressed tracts at EPAS1, including a long and a short segment (80 and 40 kb, respectively). The tract frequency is the number of haplotypes harboring the tract of a specific length divided by the total number of haplotypes. B shows the distribution of divergence between two Neanderthals captured by the ABBA-BABA (D) statistic in the form of (Denisovan, Altai Neanderthal, Vindija Neanderthal, Chimp) in nonoverlapping 32.7-kb windows (black solid curve). The shaded gray area is defined by the lower 5% percentile of the distribution (to the Left of D = 0.125). The red arrow points to the value of D(Denisovan, Neanderthal, Tibetan, Chimp) at the 32.7-kb window within EPAS1 identified in Huerta-Sánchez et al. (23). The value of D(Denisovan, Neanderthal, Tibetan, Chimp) in the adaptive 32.7-kb region in EPAS1 is statistically significant (P = 0.006).
High affinity to a single Denisovan genome alone, however, does not necessarily mean the introgressed segment originated from Denisovans. To examine the possibility of the beneficial haplotype in EPAS1 instead originating from Neanderthals, we obtained the distribution of Neanderthal–Neanderthal divergence captured by computing the D statistic (39) in the form of D(Denisovan, Altai Neanderthal, Vindija Neanderthal, Chimp) in nonoverlapping 32.7-kb windows (SI Appendix, Methods) to match the length of the previously identified adaptive EPAS1 haplotype in Tibetans (23). This distribution, as expected, has the highest density at 1, as the two Neanderthals are more genetically related to each other (Fig. 2B). If the Tibetan EPAS1 haplotype was introduced by Neanderthals instead of Denisovans, we would expect the value of D(Denisovan, Neanderthal, Tibetans, Chimp) to be within the distribution because we would expect more sharing of derived alleles between Neanderthal and the Tibetan EPAS1 haplotype. However, instead, we found that the value of D(Denisovan, Neanderthal, Tibetans, Chimp) at EPAS1 is significantly lower (P = 0.006), indicating that the Tibetan haplotype does not originate from a Neanderthal population, and that a Denisovan origin for the adaptively introgressed EPAS1 haplotype in Tibetans has the greatest support.
The Denisovan Introgression Introducing the Tibetan EPAS1 Haplotype Occurred More than 48,000 y Ago.
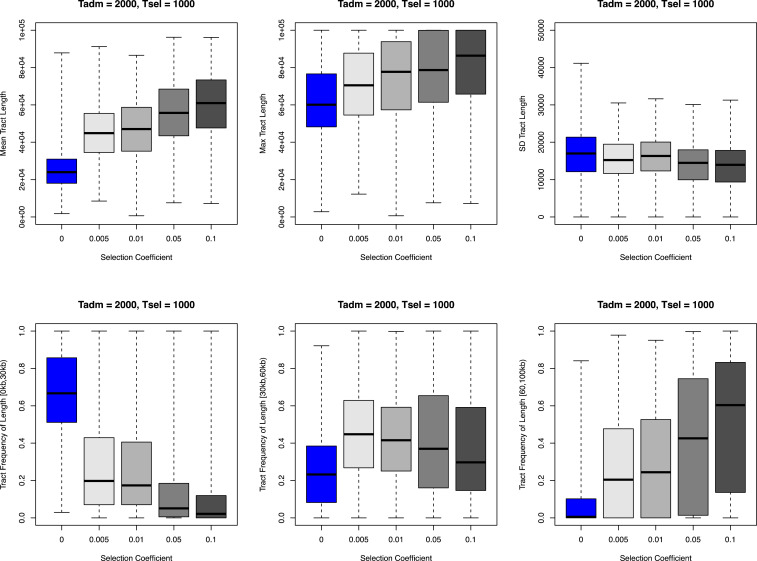
We next sought to infer the timing of Denisovan admixture and positive selection acting on EPAS1. Since the introgressed haplotypes generally become fragmented over time due to recombination (40), the distribution of introgressed tract lengths in an admixed population can be computed across the genome, and is commonly used to infer the time of admixture (41–43). For example, simulations show that, as expected, a more recent admixture time leads to higher mean introgressed tract length (42, 44). However, some studies have suggested that selection also affects the mean introgressed tract length differently depending on whether one conditions on the present-day allele frequency (42). Here, with simulations we confirm that the mean introgressed tract length increases with stronger positive selection when not conditioning on the current allele frequency (Fig. 3 and SI Appendix, Fig. S10). This is because, under positive selection, the tract reaches high frequencies sooner while it is still long and has not been broken up by recombination. As the process of recombination then continues to break up the haplotype into more fragments, the probability that recombination results in the merger of two introgression tracts increases. In other words, the effect of selection is mediated by the allele frequency increase, which elevates the probability of back-recombination between introgression fragments. Since selection acted on EPAS1 variants, we need to account for both positive selection and archaic admixture in the modeling of the system.
Fig. 3.
Relationship between introgressed tract length and selection coefficient, admixture time, and selection start time. We show the relationship between introgressed tract length (summarized by six statistics) and selection coefficient with simulations. In the simulations shown here, the admixture time (Tadm) is fixed at 2,000 generations ago, and the selection time (Tsel) starts at 1,000 generations ago (standing archaic variation). The introgressed tract lengths are tracked directly from the simulation program SLiM. Each data point in the box plot represents the statistic in all individuals from the admixed population per simulation. Each combination of evolutionary parameters (selection time, selection coefficient) was repeated 5,000 times in simulations. The demography for the simulations is model D in SI Appendix, Fig. S7.
We used an approximate Bayesian computation (ABC)-based inference framework (45–47) and a set of summary statistics to infer three parameters: selection coefficient (s); the timing of selection (Tsel) and admixture (Tadm). We used the program SLiM 3.2.0 (48) to simulate forward in time the evolution of a 100-kb genomic segment representing the EPAS1 gene under a human demographic model that considers three populations (Denisovans, Tibetans and an outgroup population; see model A in SI Appendix, Fig. S7). At a given admixture time (Tadm), a single pulse of admixture is introduced from Denisovans to the ancestral population of Tibetans at a fixed proportion of 0.1%. We chose 0.1% as previous genome-wide estimates of Denisovan ancestry in modern-day East Asians range from 0.06% (6) to 0.5% (14). Subsequently, the adaptive mutation that arose in the Denisovan population remains neutral in the Tibetan population until the selection onset time (Tsel). Additionally, the Tibetan population experienced two bottlenecks: one representing the out-of-Africa bottleneck (Ne = 1,860), and a second bottleneck (Ne = 1,000) around the time of the European–Asian split. After the second bottleneck, the population size recovers to a size of Ne = 7,000 (see SI Appendix, Methods and model A in SI Appendix, Fig. S7). We chose these sizes based on estimates of the ancestral population of East Asians and Europeans (49, 50) and based on pairwise sequentially Markovian coalescent (PSMC) results for the Tibetans studied here (30). In the simulations, the admixture time and selection coefficient were drawn from a uniform prior. The onset of selection is bounded above by the drawn admixture time, resulting in a prior that is uniform when conditioned on the admixture time.
We computed six summary statistics in both the simulations and the observed data that summarize the distribution of introgressed tract lengths. These statistics include the mean, the SD, the max, and the number of tracts with length within the following three intervals: [0, 30 kb), [30 kb, 60 kb), [>60 kb]. Tract lengths were inferred by an HMM for each simulated chromosome and for each chromosome in the observed data at the EPAS1 sequences from the combined dataset of 78 Tibetans (SI Appendix, Figs. S8 and S9; Methods). We first confirmed that the summary statistics chosen were informative about the parameters, especially under the demographic model we simulated. By directly tracking the introgressed segments in SLiM, we see a correlation between the statistics describing the distribution of tract length and the selection coefficient (s), the time of admixture (Tadm), as well as the standing variation period from admixture to selection (Tadm – Tsel) (Fig. 3 and SI Appendix, Fig. S10).
We obtained a total of 400,000 simulation replicates using parameters drawn from the prior distributions and their summary statistics for the ABC inference. We used the program ABCToolBox (51) with a rejection algorithm to retain the best-fitting 1,000 simulations for the posterior distributions. We estimated the admixture time (Tadm) at 1,950 generations ago (48,760 y ago, assuming 25 y/generation; mode of posterior density; 95% credible interval [15,987–59,500 y ago]). The selection time estimate (Tsel) is 357 generations ago (8,930 y ago; mode of posterior density; 95% credible interval [2,500–42,563 y ago]; SI Appendix, Fig. S11, model A; Table 2). Furthermore, by comparing a model of selection on standing archaic variation with selection on newly introduced archaic variants, we find more support for selection acting on standing archaic variants (Bayes factor = 5.04; SI Appendix, Table S1 and Methods), indicating that selection did not act immediately after introgression. The selection coefficient of the EPAS1 haplotype (s) was estimated to be 0.018.
Table 2.
ABC estimates of EPAS1 introgression time, selection start time, and selection strength in modern Tibetans
| Parameter | Estimate | Credible interval, 95% |
| Tadm, generations, ka | 1,950.303 (48.76 ka) | 639.500–2,380.000 |
| Tsel, generations, ka | 357.033 (8.93 ka) | 100.000–1,702.500 |
| Selection coefficient | 0.018 | 0.005–0.099 |
We used an approximate Bayesian computation (ABC) approach to estimate three parameters related to the evolutionary history of EPAS1 in Tibetans, including the admixture (Tadm) and positive selection start time (Tsel), and the selection coefficient. We show the point estimates of parameters using the mode of the posterior distributions, and 95% credible intervals. We convert times to year units by assuming that one generation is 25 y. These estimates assume demographic model A (SI Appendix, Fig. S7).
The bias of our ABC-based estimation method was assessed by computing the distribution of relative errors (Methods) using 1,000 randomly sampled simulation replicates. We found that our method had highest accuracy estimating the gap period between admixture and selection start time (Tadm – Tsel; SI Appendix, Fig. S12). The summary statistics also show high agreement between the observed data and the retained simulations (SI Appendix, Fig. S13). To evaluate the goodness-of-fit of our inference, we performed posterior predictive simulations (SI Appendix, Fig. S14), which showed that the observed statistics are within the range of newly simulated summary statistics using parameters drawn from the posterior distribution.
While our primary demographic model described above has two bottlenecks, prior demographic inference on Tibetans alone has not indicated a bottleneck in Tibetan populations around the Eurasian–East Asian split time (22, 52), unlike for example Han Chinese. As there is no clear consensus on the demographic history of Tibetan samples, and the number of plausible models is large, we considered three other models to investigate how distinct demographic scenarios change our conclusions. Specifically, we consider a model with a second bottleneck happening more recently in the past (model B in SI Appendix, Fig. S7), a model with a single bottleneck (model C), and a model with a single bottleneck and higher introgression proportion (model D). When the introgression proportion is 0.1% (models A–C) our point estimates range from [43.5–48.7 ka] for the time of introgression and [7.5–12.3 ka] for the time of selection. In model D where the introgression proportion is 1.0%, our point estimates are 52.7 ka for the time of introgression and 9.2 ka for the time of selection. All these estimates are consistent with selection of EPAS1 occurring on standing archaic variation.
To contextualize our estimates from our primary model (model A), we created a figure similar to figure 4 in Jacobs et al. (29) describing evolutionary events and relationships between Denisovan populations. Our point estimates suggest that the East Asian-specific Denisovan introgression occurred at an earlier time (∼48 ka) than the Papuan-specific Denisovan introgression (∼30 ka) that they report.
Archaic Introgression Affects Multiple Genes in Other Biological Pathways.
Last, we investigated whether other genomic regions that were influenced by archaic introgression show signals of positive selection in Tibetans. We first asked whether the SPrime-inferred segments overlap with other high-altitude adaptation candidate genes (22, 28, 53, 54) (SI Appendix, Table S3). We found a total of 11 unique regions harboring archaic segments that overlap with either a candidate gene core region, or within 200 kb of the gene’s flanking region (SI Appendix, Table S5 and Fig. S2 A and B). However, most of these segments do not show signals of positive selection—for example, most SPrime alleles except those associated with EPAS1 and FANCA were not significantly differentiated between Tibetans and Han Chinese compared to their genome-wide mean FST of 0.02 (22) (SI Appendix, Fig. S16). This is also true for another well-known gene associated with high-altitude adaptation, the EGLN1 (22) gene on chromosome 1, which shows elevated FST across the gene region, and harbors archaic alleles from Neanderthals, but shows no evidence that these archaic variants are under positive selection (low FST on the archaic alleles). Given the evidence so far, in terms of high-altitude adaptation, only the EPAS1 gene region shows a clear adaptive introgression signal.
Next, we examined whether other biological pathways received contributions from archaic introgression that facilitated positive selection. We considered all diagnostic SNPs identified from SPrime using YRI as outgroup, among which most presumably originated from Neanderthal, Denisovan, or other unknown archaic populations. The introgressed segment in the EPAS1 region is not included in this analysis due to the concern that its exceptionally strong selection signal may dampen the weaker signals in other pathways.
Since archaic introgressed alleles are preserved in mosaic patterns on the genome, we looked for subtle signals of positive selection in subsets of a pathway by detecting enrichment of high-frequency archaic alleles in genes contained in each pathway. Using the R package signet (55), we identified five pathways from the National Cancer Institute/Nature Pathway Interaction Database (NCI) (56) where the archaic alleles are enriched and potentially under positive selection (P < 0.05; SI Appendix, Table S6), among which two are insulin-related pathways that both contain the gene RHOQ. Interestingly, this gene is downstream (155 kb) of EPAS1.
Discussion
Previous studies have shown that archaic introgression contributed to a range of phenotypic variation in modern humans (5, 57, 58), and that a number of introgressed genes were plausibly subject to positive selection (17, 18). Here, we used sequencing data of the most convincing example of adaptive introgression, EPAS1 in Tibetans, to address a series of questions regarding the origin and the timing of Denisovan introgression in East Asia. Our work supports the two-pulse Denisovan admixture model proposed by Browning et al., and our analysis suggests that the beneficial haplotype of EPAS1 in Tibetans originated from the East Asian-specific Denisovan introgression, involving a Denisovan group that is more closely related to the Altai Denisovan individual from the Denisova Cave. Besides EPAS1, archaic introgression has left segments in various genes across the genome, and affected multiple biological pathways including hypoxia.
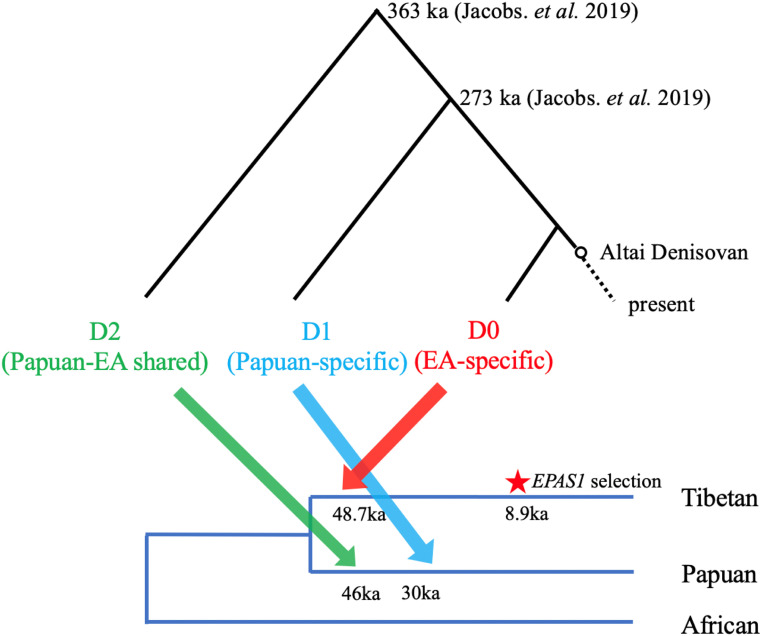
This work provides a timing estimate of the East Asian-specific Denisovan introgression, which we inferred at around 48 ka, and is consistent with archaeological evidence showing Denisovan ancestry in modern human individuals from 34 to 40 ka (59, 60). Our point estimate suggests that the East Asian-specific admixture event is more ancient (48 ka) than the Papuan-specific pulse (30 ka), and closer to the first Denisovan introgression that is shared by Asian and Oceanian lineages (45 ka, Fig. 4) (29). Interestingly, the low mismatch observed in Jacobs et al. between Altai Denisovan and East Asian-specific Denisovan introgressed segments suggests that the Denisovan population that introgressed uniquely into East Asians (D0 in Fig. 4) was more closely related to the Altai population (that the sequenced Altai Denisovan belonged to) compared to the other two Denisovan populations (D1 and D2 in Fig. 4) that introgressed into humans.
Fig. 4.
Timeline of three Denisovan introgression events in Asia, and the selection of the adaptive EPAS1 allele. This figure is inspired by figure 4 in Jacobs et al. (29) that portrays three distinct Denisovan lineages (D0, D1, and D2), their inferred introgression times, and the split times (measured from the present) between them. Time estimates for D1 and D2 are from Jacobs et al. (29) using Papuan data, and the estimate for the East Asian-specific (D0) introgression time and the selection time in Tibetans come from this study.
The timing of human settlement in the Tibetan Plateau, including archaic hominins, remains under investigation. The discovery of a partial mandible from the Middle Pleistocene (Xiahe Denisovan) in Baishiya Karst Cave (BKC), located at 3,280-m altitude in the Tibetan Plateau suggests that Denisovan-like archaic hominins may have been present at high altitude at least 160 ka (61). More recently, analysis of mtDNA recovered from sediment excavated in this cave (inferred to be from ∼100 to ∼60 ka and maybe as recently as 45 ka) revealed that it grouped most closely with Denisovan mtDNA (62), suggesting that Denisovan-like populations may have inhabited this region for a long period of time. In contrast, evidence for modern human activity has been found on the interior of the Tibetan plateau as early as 40 ka from the Nwya Devu site (63), although long-term human settlements on the high-altitude plateau are believed to be rare at that time. Currently, existing archaeological evidence generally supports two settlement scenarios. The archaeological sites from middle to late Holocene (62–65) indicate that year-round large-scale settlements of people on the plateau started after 3.6 ka facilitated by the advent of agriculture, while analyses on the mobility of hunter-gatherers (66–68) suggest that permanent inhabitation (most likely on a smaller scale) may have occurred more distantly in the past. Our estimate of the Denisovan East Asian-specific admixture time (48 ka) from tract lengths surrounding the EPAS1 gene is larger than most estimates of when modern humans permanently settled in the Tibetan Plateau, suggesting that the admixture most likely occurred outside of this region.
Furthermore, our estimate of the onset time of positive selection on EPAS1 (∼8.9 ka) suggests that selection did not target the Denisovan introgressed alleles immediately after introgression, and possibly coincides with the time of permanent hunter-gatherer Tibetan settlements of populations from lowland East Asia during the Late Pleistocene or early Holocene (64). While evidence of even earlier arrivers exists (e.g., Denisovan from BKC and modern humans at the Nwya Devu site, 40 ka or earlier), it is unclear how long they survived in the Tibetan Plateau, whether they were genetically adapted to the hypoxic environment, or if modern Tibetans are their direct descendants. Only one study has reported ancient DNA from the Himalayas (the Nepalese side) where the oldest samples date to 3.15 ka (65). Interestingly, only the more recent samples (dated to 1.75–1.25 ka) exhibit the EPAS1 alleles present in modern Tibetans. Future studies of ancient humans in this region will help provide additional context and finer resolution to the population history on the Tibetan Plateau.
Previous studies have estimated the time of Denisovan introgression in Asia (SI Appendix, Table S7), but there are multiple differences between our analyses and theirs. The first relates to the underlying assumption of a single introgression event into East Asia (1, 6). Those studies used all of the surviving Denisovan segments (in Papuans or Tibetans), and it is unclear whether that estimate is an average of the two Denisovan introgression events (7, 29) into East Asians, or if the estimate is closer to one of the introgression events. By contrast, we are using the data of a single gene that clearly has been the target of selection, having the advantage that, because it is a small local region of the genome, it is highly likely that the fragments are the remnants of archaic DNA introduced by a single admixture event. The second difference is that we account for positive selection in our inference since we show that selection affects the tract length. The other estimates assume neutrality, and it is unclear whether adaptively introgressed loci could change or bias estimates from genome-wide summary statistics of introgression (e.g., the distribution of introgressed tract lengths, linkage disequilibrium decay pattern). Finally, we use an ABC framework for parameter estimation, while the estimation methods used by others (30, 32, 52) could also lead to some differences in the inferences.
We acknowledge that we have made several assumptions and choices in our work. First, we rely on the sequencing data of only a single gene, which is reflected in our large credible intervals. One way to reduce uncertainty might be to use all the putative introgressed segments introduced via the East Asian Denisovan introgression event, but doing so would require making a different set of assumptions regarding how selection is acting on each of those regions. Second, we have assumed a demographic model for Tibetans from estimates of population size changes from PSMC curves. Our conclusion that selection of EPAS1 acted on standing archaic variation also stands true under all scenarios. We also do not know what the real distribution of tract lengths looks like in Tibetans, and we have inferred that using an HMM. How accurately the HMM infers the true tract lengths in Tibetans is unknown, but other methods [e.g., ArchaicSeeker 2.0 (30)] yield similar results (SI Appendix, Methods and Fig. S15). Even if the HMM does not capture the true Tibetan tract lengths, by applying the HMM to both the real data and the simulated data, we hope that the same bias occurs in both, reducing the likelihood of distorting the parameter estimates.
During the last decade, we have begun to appreciate that gene flow between archaic and modern humans played a major role in shaping human evolution as well as our genetic diversity. The introduction of archaic variants evidently facilitated adaptations to local environments in multiple populations. Our results for EPAS1 demonstrate the importance of selection on standing archaic variation, which other studies suggest is widespread (69, 70). However, recent work infers that selection immediately after introgression explains most examples of adaptive introgression from Neanderthals in Europeans (71). More analysis of other adaptively introgressed loci in multiple populations will further elucidate whether and under what conditions selection on standing archaic variation is the primary mode for adaptation. As we continue to sequence the remains of other archaic and modern humans, a high-resolution picture of archaic introgression in modern humans is expected to be revealed.
Materials and Methods
Genomic Data from Tibetan Population.
For the whole-genome analyses in this study, we used 38 Tibetan samples from Lu et al. (30). We phased the data with Beagle 5.0 (72, 73), with the 1,000 genomes worldwide populations as imputation reference. SNPs that were very rare (<5% frequencies) or very common (>95% frequencies) were removed from the phased variant call format files (VCFs) that are used for downstream analyses, including the SPrime analysis. For inferring the timing of admixture and selection as well as the selection strength we combined the sequences from 40 Tibetan individuals from Huerta-Sánchez et al. (23) covering a 120-kb region at the EPAS1 gene, and the 38 Tibetan sequences at the EPAS1 locus from Lu et al. (30).
Genomic Data from Worldwide Population.
This study utilized the following data collections as reference: 1) modern human individuals from 1000 Genomes Project (31); 2) archaic human genomes: Altai Neanderthal (13), Vindija Neanderthal (8), and Altai Denisovan (1).
SPrime Inference of Putative Archaic-Introgressed Genomic Segments.
To infer the introgressed regions in Tibetans, we used SPrime (7). We merged the whole-genome sequences of African individuals (YRI) from the 1000 Genomes Panel data to use as the reference outgroup with the VCFs of 38 Tibetans from Lu et al. (30). We ran SPrime on the combined VCF file with all 22 autosomes included to infer the putative introgressed regions in Tibetans, which are each tagged by a set of sites. Alleles at these sites had maximum frequency no higher than 1% in the YRI population, as that was the threshold specified in SPrime. For each inferred segment, we further filtered out sites that were not biallelic, had low coverage depth in archaic genomes (<10), or low mapping quality score (<25). For the set of sites that passed these filters, we extracted the genotypes of the Altai Neanderthal and Altai Denisovan. For each site, we reported a “match” if the archaic genotype includes the putative introgressed allele and a “mismatch” otherwise. The match rate is calculated as the number of matches divided by the total number of sites compared (matches plus mismatches). Sites that did not pass the filters were excluded in the match rate calculation. We also applied SPrime using the CEU (individuals of European ancestry) from the 1000 Genomes Panel data as the outgroup.
To visualize the densities of match rates between the introgressed segments and the archaic genomes (in Fig. 1 and SI Appendix, Fig. S2), we utilized the function “kde2d” from the MASS package in R with the script from Browning et al. (7).
HMM and the Inference of Introgressed Tracts.
Since SPrime does not identify the introgressed segments within each individual chromosome, we used a HMM to call each introgressed segment in each Tibetan chromosome. We used the same HMM (13, 17, 38) described in Racimo et al. (17) and applied it to both the simulated data (under models described in SI Appendix, Fig. S7) and the observed data. For the observed data, the HMM was used for two analyses: 1) to infer the tract lengths of the 38 Tibetans in the inferred SPrime regions whose match rate to the Denisovan was >60% and <40% to the Altai Neanderthal (Fig. 2 and SI Appendix, Fig. S6), and 2) to infer the introgressed tract lengths at the EPAS1 gene in 78 Tibetans [40 from Huerta-Sánchez et al. (23) and 38 from Lu et al. (30); see SI Appendix, Fig. S9]. In both analyses, we used 176 Yorubans (YRI) as the outgroup. We removed the nonbialleleic sites in the observed data, and formatted variants to their ancestral/derived alleles, and kept only SNPs with derived allele frequency >0 in the Tibetans. We also removed the SNPs that were private in one of the two datasets [the 40 Tibetans from Huerta-Sánchez et al. (23) and the 38 Tibetans from Lu et al. (30)] before joining them for the HMM analysis. We plotted the inferred archaic-introgressed segments from 78 Tibetans as a heat map in the R environment. Each row represented a haplotype from Tibetan individuals, and each column represented a genomic position in EPAS1 core region. The sites that were inferred to be archaic-introgressed were highlighted in yellow, in contrast to blue that denoted nonintrogressed sites.
We further compared the results of the HMM inference on the tract length per haplotype with the inference on the 38 Tibetan individuals published by another method, ArchaicSeeker 2.0 (AS) (30). ArchaicSeeker and the HMM showed high level of agreement (SI Appendix, Fig. S15); both methods inferred a mode in intermediate-length tracts (∼40 kb), while HMM inferred a few more larger tracts (∼80 kb) and AS inferred more shorter tracts (∼10 kb).
ABC Inference.
To infer the parameters, we simulated the evolution of the EPAS1 region under the demographic model described in Results (model A in SI Appendix, Fig. S7). We allow the admixture time (Tadm) to vary between [500, 2,400] generations ago, selection time (Tsel) to vary between [100, Tadm) generations ago, and selection coefficient (s) between (0, 0.1). The simulated segment has length L of 100 kb, which was the approximate size of the EPAS1 region. The mutation rate was set to 1.0e-8 as estimated from Huerta-Sánchez et al. (23), and the recombination rate r is uniform across the segment at 2.3e-8 (23).
We applied the same HMM framework that we used for the observed data to each simulation replicate, to infer the introgressed tracts on simulated haplotypes. Given the location of HMM-inferred introgressed tract(s) in each simulation, we computed the tract length from each simulated chromosome, and recorded six summary statistics that describe the tract length distribution, including 1) the mean (μ), 2) the SD (σ), 3) the maximum length (max), and 4–6) the number of tracts with length within the following three intervals: [0, 30 kb), [30 kb, 60 kb) and [>60 kb]. The six summaries (K) were computed for the EPAS1 100-kb region in 78 Tibetan individuals. For a given parameter θ, the posterior probability is therefore Pr(θ | K).
We applied the program ABCToolBox (51) to infer parameters by retaining the simulated data that best matched the observed data (as in six summary statistics). The program compares the simulated data with the observed data, and implements a rejection algorithm adjustment on retained simulations. We chose the closest 1,000 retained simulations to generate the posterior, and subsequently plotted the marginal posterior distribution of admixture time and selection start time in an R program, and obtained the mode and the 95% credible intervals (Table 2 and SI Appendix, Fig. S11) from the marginal posterior distribution.
We computed the relative errors [Res, (true-estimate)/true] by randomly sampling 1,000 simulations from the 400,000 set. For each of the 1,000 simulations, we used ABCToolBox to infer the parameters using the rest of the simulations (a total of 399,000). We compared the difference between the inferred parameters and the true parameters (that generated the simulation replicate) for each of the 1,000 randomly drawn simulations. We plotted the distribution of differences as histograms in SI Appendix, Fig. S12. To evaluate the goodness-of-fit in our inferred scenario, we performed posterior predictive checking by randomly sampling sets of parameter values that generated 500 of the 1,000 retained simulations, and used each parameter combination to generate a single simulation, and computed the summary statistics. We plotted the relationship of pairwise summary statistics between the observed data, the sampled 500 retained simulation, and the generated 500 simulation replicates under the 500 parameter combinations, and show that the summary statistics from the observed data are within the range of both retained posterior and the newly simulated data (SI Appendix, Fig. S14).
Last, we performed model selection using ABCToolBox (SI Appendix, Table S1). We reran the inference of ABC using the simulations generated previously under the model of selection on standing archaic variation (M1), together with an additional 400,000 simulations generated under a model of immediate selection on archaic variation (M2), where the only difference is that in M2, selection acts immediately after introgression in the Tibetan population. The Bayes factor (74), or the ratio of posterior probabilities of the two competing models with equal prior is 5.04, suggesting more support for M1 than M2.
Biological Pathway Network Analysis.
We looked for subtle signals that a subset of genes within a pathway network may be selected, by searching for enrichment of archaic alleles in gene networks in biological pathway databases and finding the highest scoring subnetwork (55). First, we searched for overlaps between the putatively introgressed segments inferred by SPrime and protein-coding genes in modern humans using the ENSEMBL database (75, 76), which resulted in 3,292 genes by combining the searches using either the YRI or CEU as outgroup populations. At the overlapping SPrime diagnostic SNPs, we calculated their frequencies in the Tibetan population. We used the maximum archaic allele frequency in each gene as input score for the enrichment test. Alternatively, if no archaic allele was found in a gene, the gene received a score of 0. We then computed the subnetwork scoring using the National Cancer Institute/Nature Pathway Interaction Database (NCI) (56) as reference for signaling and metabolic pathways, using the HSS algorithm provided under R package signet (55). The final score of each subnetwork was normalized using the mean and SD of 10,000 simulated random networks of the same size. We reported only the significant subnetworks with P values less than 0.05 as candidate biological pathways that undergo positive selection because of archaic allele enrichment (SI Appendix, Table S6).
Supplementary Material
Acknowledgments
This study was supported by NSF Grant 1557151 and NIH Grant 1R35GM128946-01 (to E.H.-S.). X.Z. was partially supported by NIH Grant R35GM119856 to Kirk Lohmueller at University of California, Los Angeles (UCLA). M.M.B. is a trainee supported under the Brown University Predoctoral Training Program in Biological Data Science (NIH T32 GM128596). S.X. gratefully acknowledges the support of National Natural Science Foundation of China Grants 31525014, 32030020, 31771388, 31961130380, and 32041008, the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), and the UK Royal Society–Newton Advanced Fellowship (NAF\R1\191094). We thank the E.H.-S. laboratory at Brown University and the Lohmueller laboratory at UCLA for helpful discussions during the development of this work. We also thank Dr. Guy Jacobs at University of Cambridge for kindly sharing msprime simulation scripts.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020803118/-/DCSupplemental.
Data Availability
All scripts necessary to reproduce the ABC and simulation results from this work can be found on GitHub, https://github.com/xzhang-popgen/EPAS1Project. The use of 38 Tibetan whole genomes by this work is permitted by The Ministry of Science and Technology of the People’s Republic of China (permission no. 2020BAT0143) at the National Genomics Data Center (https://bigd.big.ac.cn/search/?dbId=gsa&q=PRJCA000246). The EPAS1 sequences of the 40 Tibetans used here are available at the Sequence Read Archive (accession no. SRR1265938).
References
- 1.Reich D., et al., Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer M., et al., A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viola B. T., et al., A parietal fragment from Denisova Cave. Am. J. Phys. Anthropol. 168, 258 (2019). [Google Scholar]
- 4.Douka K., et al., Age estimates for hominin fossils and the onset of the Upper Palaeolithic at Denisova Cave. Nature 565, 640–644 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Dannemann M., Andrés A. M., Kelso J., Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human Toll-like receptors. Am. J. Hum. Genet. 98, 22–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sankararaman S., Mallick S., Patterson N., Reich D., The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr. Biol. 26, 1241–1247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browning S. R., Browning B. L., Zhou Y., Tucci S., Akey J. M., Analysis of human sequence data reveals two pulses of archaic Denisovan admixture. Cell 173, 53–61.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prüfer K., et al., A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer M., et al., Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature 531, 504–507 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Higham T., et al., The timing and spatiotemporal patterning of Neanderthal disappearance. Nature 512, 306–309 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Slon V., et al., A fourth Denisovan individual. Sci. Adv. 3, e1700186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slon V., et al., The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature 561, 113–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prüfer K., et al., The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallick S., et al., The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reich D., et al., Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 89, 516–528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Racimo F., Sankararaman S., Nielsen R., Huerta-Sánchez E., Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 16, 359–371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Racimo F., et al., Archaic adaptive introgression in TBX15/WARS2. Mol. Biol. Evol. 34, 509–524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Racimo F., Marnetto D., Huerta-Sánchez E., Signatures of archaic adaptive introgression in present-day human populations. Mol. Biol. Evol. 34, 296–317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gittelman R. M., et al., Archaic hominin admixture facilitated adaptation to out-of-Africa environments. Curr. Biol. 26, 3375–3382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vernot B., Akey J. M., Resurrecting surviving Neandertal lineages from modern human genomes. Science 343, 1017–1021 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Kim B., Lohmueller K. E., Huerta-Sánchez E., The impact of recessive deleterious variation on signals of adaptive introgression in human populations. Genetics 215, 799–812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi X., et al., Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huerta-Sánchez E., et al., Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huerta-Sánchez E., Casey F. P., Archaic inheritance: Supporting high-altitude life in Tibet. J. Appl. Physiol. (1985) 119, 1129–1134 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Wu T., et al., Hemoglobin levels in Qinghai-Tibet: Different effects of gender for Tibetans vs. Han. J. Appl. Physiol. (1985) 98, 598–604 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Moore L. G., et al., Maternal adaptation to high-altitude pregnancy: An experiment of nature–a review. Placenta 25 (suppl. A), S60–S71 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Witt K. E., Huerta-Sánchez E., Convergent evolution in human and domesticate adaptation to high-altitude environments. Philos. Trans. R Soc. B Biol. Sci. 374, 20180235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beall C. M., et al., Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. U.S.A. 107, 11459–11464 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs G. S., et al., Multiple deeply divergent Denisovan ancestries in Papuans. Cell 177, 1010–1021.e32 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Lu D., et al., Ancestral origins and genetic history of Tibetan highlanders. Am. J. Hum. Genet. 99, 580–594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.1000 Genomes Project Consortium, et al., A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hackinger S., et al., Wide distribution and altitude correlation of an archaic high-altitude-adaptive EPAS1 haplotype in the Himalayas. Hum. Genet. 135, 393–402 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durvasula A., Sankararaman S., Recovering signals of ghost archaic introgression in African populations. Sci. Adv. 6, eaax5097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammer M. F., Woerner A. E., Mendez F. L., Watkins J. C., Wall J. D., Genetic evidence for archaic admixture in Africa. Proc. Natl. Acad. Sci. U.S.A. 108, 15123–15128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu D., et al., Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation. Mol. Biol. Evol. 34, 2704–2715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henn B. M., et al., Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 8, e1002397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L., Wolf A. B., Fu W., Li L., Akey J. M., Identifying and interpreting apparent Neanderthal ancestry in African individuals. Cell 180, 677–687.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durand E. Y., Patterson N., Reich D., Slatkin M., Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28, 2239–2252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill W. G., Disequilibrium among several linked neutral genes in finite population. II. Variances and covariances of disequilibria. Theor. Popul. Biol. 6, 184–198 (1974). [DOI] [PubMed] [Google Scholar]
- 41.Loh P.-R., et al., Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193, 1233–1254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corbett-Detig R., Nielsen R., A hidden Markov model approach for simultaneously estimating local ancestry and admixture time using next generation sequence data in samples of arbitrary ploidy. PLoS Genet. 13, e1006529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gravel S., Population genetics models of local ancestry. Genetics 191, 607–619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shchur V., Svedberg J., Medina P., Corbett-Detig R., Nielsen R., On the distribution of tract lengths during adaptive introgression. G3 (Bethesda) 10, 3663–3673 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beaumont M. A., Zhang W., Balding D. J., Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tavaré S., Balding D. J., Griffiths R. C., Donnelly P., Inferring coalescence times from DNA sequence data. Genetics 145, 505–518 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marjoram P., Approximation Bayesian computation. OA Genet. 1, 853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haller B. C., Messer P. W., SLiM 3: Forward genetic simulations beyond the Wright–Fisher model. Mol. Biol. Evol. 36, 632–637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gravel S., et al., Demographic history and rare allele sharing among human populations. Proc. Natl. Acad. Sci.U.S.A. 108, 11983–11988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ragsdale A. P., Gravel S., Models of archaic admixture and recent history from two-locus statistics. PLoS Genet. 15, e1008204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wegmann D., Leuenberger C., Neuenschwander S., Excoffier L., ABCtoolbox: A versatile toolkit for approximate Bayesian computations. BMC Bioinformatics 11, 116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu H., et al., Evolutionary history of Tibetans inferred from whole-genome sequencing. PLoS Genet. 13, e1006675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bigham A., et al., Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 6, e1001116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simonson T. S., et al., Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Gouy A., Daub J. T., Excoffier L., Detecting gene subnetworks under selection in biological pathways. Nucleic Acids Res. 45, e149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaefer C. F., et al., PID: The pathway interaction database. Nucleic Acids Res. 37, D674–D679 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dannemann M., Kelso J., The contribution of Neanderthals to phenotypic variation in modern humans. Am. J. Hum. Genet. 101, 578–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simonti C. N., et al., The phenotypic legacy of admixture between modern humans and Neandertals. Science 351, 737–741 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Massilani D., et al., Denisovan ancestry and population history of early East Asians. Science 370, 579–583 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Fu Q., et al., DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl. Acad. Sci. U.S.A. 110, 2223–2227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen F., et al., A late Middle Pleistocene Denisovan mandible from the Tibetan Plateau. Nature 569, 409–412 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Zhang D., et al., Denisovan DNA in Late Pleistocene sediments from Baishiya Karst Cave on the Tibetan Plateau. Science 370, 584–587 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Zhang X. L., et al., The earliest human occupation of the high-altitude Tibetan Plateau 40 thousand to 30 thousand years ago. Science 362, 1049–1051 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Chen F. H., et al., Agriculture facilitated permanent human occupation of the Tibetan Plateau after 3600 B.P. Science 347, 248–250 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Jeong C., et al., Long-term genetic stability and a high-altitude East Asian origin for the peoples of the high valleys of the Himalayan arc. Proc. Natl. Acad. Sci. U.S.A. 113, 7485–7490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.d’Alpoim G. J., Did foragers adopt farming? A perspective from the margins of the Tibetan Plateau. Quat. Int. 489, 91–100 (2018). [Google Scholar]
- 67.d’Alpoim G. J., Aldenderfer M., The archaeology of the Early Tibetan Plateau: New research on the initial peopling through the Early Bronze Age. J. Archaeol. Res. 28, 339–392 (2020). [Google Scholar]
- 68.Meyer M. C., et al., Permanent human occupation of the central Tibetan Plateau in the early Holocene. Science 355, 64–67 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Jagoda E., et al., Disentangling immediate adaptive introgression from selection on standing introgressed variation in humans. Mol. Biol. Evol. 35, 623–630 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peter B. M., Huerta-Sanchez E., Nielsen R., Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genet. 8, e1003011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yair S., Lee K. M., Coop G., The timing of human adaptation from Neanderthal introgression. Genetics, 10.1093/genetics/iyab052 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Browning S. R., Browning B. L., Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Browning B. L., Zhou Y., Browning S. R., A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 103, 338–348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leuenberger C., Wegmann D., Bayesian computation and model selection without likelihoods. Genetics 184, 243–252 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Consortium I. H. G. S.; International Human Genome Sequencing Consortium , Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Hubbard T., et al., The Ensembl genome database project. Nucleic Acids Res. 30, 38–41 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts necessary to reproduce the ABC and simulation results from this work can be found on GitHub, https://github.com/xzhang-popgen/EPAS1Project. The use of 38 Tibetan whole genomes by this work is permitted by The Ministry of Science and Technology of the People’s Republic of China (permission no. 2020BAT0143) at the National Genomics Data Center (https://bigd.big.ac.cn/search/?dbId=gsa&q=PRJCA000246). The EPAS1 sequences of the 40 Tibetans used here are available at the Sequence Read Archive (accession no. SRR1265938).