Significance
Since 2015, Zika virus (ZIKV) has been recognized as a serious public health concern due to devastating birth defects caused by ZIKV infection of pregnant women. An improved understanding of ZIKV employment of host factors to facilitate infection would enhance our knowledge of the subcellular events driving severe clinical outcomes of ZIKV infection and provide potential avenues for therapeutic and vaccine development to combat this virus. We reveal that ZIKV exploits the host RNase L protein to assist in generation of new virus, a feature that is not shared during infection with other flaviviruses, thus identifying a unique host interaction possibly facilitating ZIKV pathogenesis.
Keywords: Zika virus, flavivirus, RNase L, replication factories, OAS3
Abstract
Infection with the flavivirus Zika virus (ZIKV) can result in tissue tropism, disease outcome, and route of transmission distinct from those of other flaviviruses; therefore, we aimed to identify host machinery that exclusively promotes the ZIKV replication cycle, which can inform on differences at the organismal level. We previously reported that deletion of the host antiviral ribonuclease L (RNase L) protein decreases ZIKV production. Canonical RNase L catalytic activity typically restricts viral infection, including that of the flavivirus dengue virus (DENV), suggesting an unconventional, proviral RNase L function during ZIKV infection. In this study, we reveal that an inactive form of RNase L supports assembly of ZIKV replication factories (RFs) to enhance infectious virus production. Compared with the densely concentrated ZIKV RFs generated with RNase L present, deletion of RNase L induced broader subcellular distribution of ZIKV replication intermediate double-stranded RNA (dsRNA) and NS3 protease, two constituents of ZIKV RFs. An inactive form of RNase L was sufficient to contain ZIKV genome and dsRNA within a smaller RF area, which subsequently increased infectious ZIKV release from the cell. Inactive RNase L can interact with cytoskeleton, and flaviviruses remodel cytoskeleton to construct RFs. Thus, we used the microtubule-stabilization drug paclitaxel to demonstrate that ZIKV repurposes RNase L to facilitate the cytoskeleton rearrangements required for proper generation of RFs. During infection with flaviviruses DENV or West Nile Kunjin virus, inactive RNase L did not improve virus production, suggesting that a proviral RNase L role is not a general feature of all flavivirus infections.
The flavivirus genus contains arthropod-transmitted viruses with a positive-sense single-stranded RNA (ssRNA) genome, including Zika virus (ZIKV), dengue virus (DENV), and West Nile virus (WNV). These viruses are transmitted by mosquitos and globally distributed, with high associated morbidity and mortality in humans (1–3). More recently, ZIKV sexual and vertical transmission has been recognized, the latter involving transplacental migration of the virus, potentially resulting in fetal microcephaly (4–9). Due to diversity in ZIKV tissue tropism, disease, and route of transmission as compared with other flaviviruses, it is possible that variances in ZIKV infection at the molecular level confer the observed shifts in clinical outcome at the organismal level. For this reason, we are interested in host machinery that supports the ZIKV replication cycle but not that of other flaviviruses, as this may improve our understanding of the molecular determinants of ZIKV pathogenesis.
After entry into the host cell, the flavivirus genome, which also serves as the messenger RNA (mRNA), is directly translated at the endoplasmic reticulum (ER). Proximal to sites of translation, flaviviruses create replication factories (RFs) through extensive cytoskeletal rearrangements that generate invaginations in the folds of the ER membrane, within which new genome synthesis occurs (10–15). These RFs contain ZIKV replication complex proteins, including the NS3 protease, the replication intermediate double-stranded RNA (dsRNA), as well as template genomic ssRNA. New genome is packaged into compartments at opposite ER folds, and new virions traffic through the transgolgi network and eventually bud from the plasma membrane (16). RFs therefore enable efficient throughput of key viral processes as centers of new genome synthesis linked with viral protein translation as well as new virus assembly. In addition, RFs serve as a protective barrier to impede cytosolic innate immune sensing, as flavivirus RNA predominantly resides within RFs during the bulk of the intracellular replication cycle.
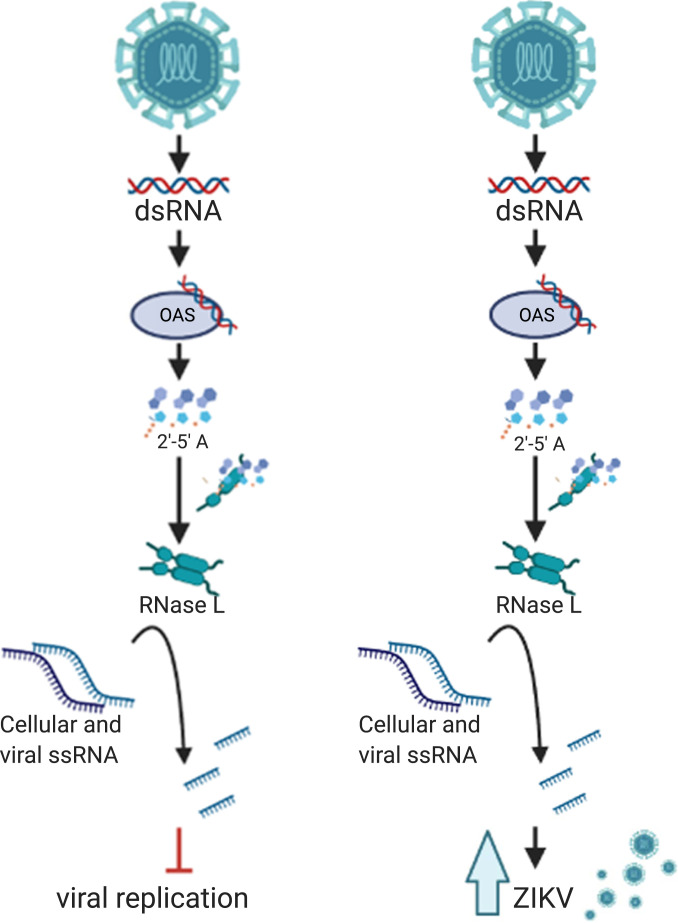
Innate immune sensors within the cytoplasm of the infected cell detect viral RNA to activate antiviral responses, including the type I interferon (IFN) and oligoadenylate synthetase/ribonuclease L (OAS/RNase L) pathways. Extensive research has demonstrated that flaviviruses, including ZIKV, have evolved strategies for counteracting the type I IFN response (17–22). Since OAS genes are IFN-stimulated genes and therefore up-regulated by type I IFN signaling, the OAS/RNase L pathway can also be potentiated by type I IFN production. However, activation of RNase L can occur in the absence of type I IFN responses when basal OAS expression is sufficient (23, 24). In either event, OAS sensors detect viral dsRNA and generate 2'-5'-oligoadenylates (2-5A). RNase L, which is constitutively expressed in an inactive form, homodimerizes upon 2-5A binding to become catalytically active (25). Active RNase L cleaves both host and viral ssRNA within the cell (Fig. 1). While there are three OAS isoforms, we have shown that the OAS3 isoform is the predominant activator of RNase L during infection with a variety of viruses including ZIKV (23, 26). Activated RNase L cleavage of host ribosomal RNA (rRNA) and mRNA as well as viral ssRNA ultimately inhibits virus infection (26–31).
Fig. 1.
Noncanonical RNase L function promotes infectious ZIKV production. (Left) Canonical RNase L antiviral activity. Viral dsRNA is detected by OAS3, which produces the small molecule 2-5A that binds inactive RNase L, inducing its homodimerization and catalytic activation, resulting in cleavage of host and viral ssRNA, leading to inhibition of viral infection. (Right) RNase L activity during ZIKV infection. ZIKV dsRNA is recognized by OAS3, which activates RNase L resulting in ssRNA cleavage; however, ZIKV production is improved with RNase L expression.
Once activated, RNase L can restrict infection of a diverse range of DNA and RNA viruses, including flaviviruses DENV and WNV (30, 32–34). Many viruses have subsequently developed mechanisms for evading RNase L antiviral effects, most of which target this pathway upstream of RNase L activation through sequestration of dsRNA, which prevents OAS activation, or by degradation of 2-5A (32, 35–40). We recently showed that ZIKV avoids antiviral effects of activated RNase L and that this evasion strategy requires assembly of RFs to protect genome from RNase L cleavage (23). Despite substantial RNase L–mediated cleavage of intracellular ZIKV genome, a portion of uncleaved genome was shielded from activated RNase L within RFs. This genome was sufficient to produce high levels of infectious virus particles, as infectious ZIKV released from wild-type (WT) cells was significantly higher than from RNase L knockout (KO) cells. Unlike ZIKV, infectious DENV production was decreased by canonical RNase L antiviral activity (23, 33). These results indicated that RNase L expression was ultimately proviral during ZIKV infection (Fig. 1). As this was the initial report of viral resistance to catalytically active RNase L during infection, we sought to isolate the differences between ZIKV RFs and those constructed by other flaviviruses, to identify factors that enable this ZIKV evasion mechanism.
In this study, we focused on elucidating how RNase L increases ZIKV production. An earlier study reported that an inactive form of RNase L interacts with the actin cytoskeleton to reorganize cellular framework during viral infection (41). Since flaviviruses reorganize the cellular cytoskeletal and organellar network during infection (11), we investigated the possibility that RNase L was exploited by ZIKV to assemble protective RFs that dually serve as a barrier against host sensors in addition to providing sites of replication.
Results
RNase L Improves RF Function to Increase ZIKV RNA and Protein Expression at RFs.
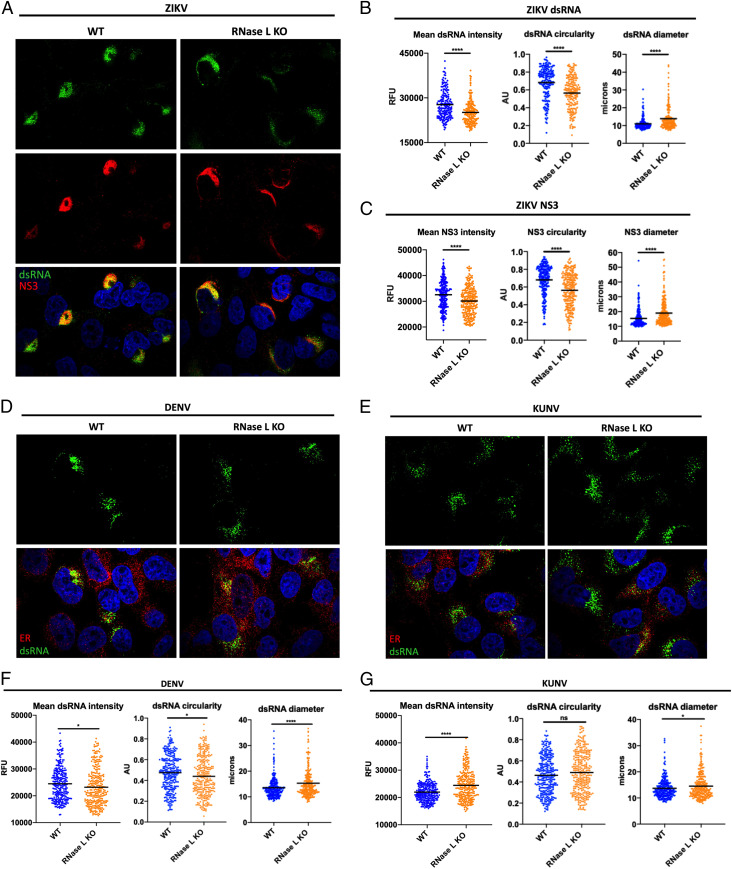
Since infectious ZIKV production was enhanced by RNase L expression, we examined whether RNase L was required for optimal ZIKV replication. We used immunofluorescence assays (IFAs) for detection of the replication intermediate dsRNA and the ZIKV NS3 protein as markers for ZIKV RFs, comparing expression levels in WT and RNase L KO cells. At 20 h postinfection (hpi), expression of both ZIKV dsRNA and NS3 was increased in WT cells compared with RNase L KO cells, as measured by mean dsRNA and NS3 intensity at perinuclear ER sites characteristic of flaviviruses (Fig. 2 A–C). We also observed a change in localization of ZIKV dsRNA and NS3 in RNase L KO cells, both of which were more disseminated around the nucleus and into the cytoplasm. We used circularity and diameter parameters to quantify the spread of viral products throughout the cell at 20 hpi and determined that RNase L deletion decreased circularity and increased diameter of RFs, distinct from the densely concentrated RF phenotype observed in WT cells (Fig. 2 A–C). We observed this trend of viral RF diffusion in RNase L KO cells as early as 16 through 30 hpi in two different RNase L KO clones (SI Appendix, Fig. S1). We performed all future analyses on cells fixed at 20 hpi, which provides adequate infected cells for quantification purposes but is early enough for examination of a uniform stage of the viral replication cycle across each sample. Additionally, since alterations in ZIKV dsRNA and NS3 were nearly identical, we proceeded to measure effects on RFs using either dsRNA or NS3 expression.
Fig. 2.
RNase L improves RF function to increase ZIKV RNA and protein expression at RFs. A549 WT and RNase L cells were infected at a multiplicity of infection of 1, cells were fixed at 20 hpi for IFA. (A) ZIKV-infected cells were stained for dsRNA (green) and ZIKV NS3 (red) with 4′,6-diamidino-2-phenylindole (DAPI, blue) staining of nuclei. Quantification of ZIKV (B) dsRNA and (C) NS3 mean intensity, circularity, and diameter of staining shown in A. (D) DENV- or (E) KUNV-infected cells were stained for dsRNA (green) and ER (PDI, red) with DAPI (blue) staining of nuclei. Quantification of (F) DENV or (G) KUNV dsRNA mean intensity, circularity, and diameter of staining shown in D and E, respectively. The data are representative of at least two independent experiments. Statistical significance was determined by Student’s t test. The black bars represent the mean. ns = not significant, *P < 0.05, and ****P < 0.0001. (Imaged at 100× magnification.) RFU = relative fluorescence units, AU = arbitrary units. See also SI Appendix, Figs. S1 and S2.
Since canonical antiviral RNase L activity restricts flaviviruses DENV and WNV Kunjin strain (KUNV) (SI Appendix, Fig. S2), we examined effects of RNase L on DENV and KUNV RFs. In comparing DENV dsRNA expression and localization in RNase L KO cells to that of WT cells, we found that dsRNA intensity and circularity were decreased while diameter was increased without RNase L, a similar trend as with ZIKV but to a lesser degree (Fig. 2 D and F). However, RNase L restriction of DENV (SI Appendix, Fig. S2) suggests slight changes in RF intensity and shape at the ER do not translate to changes in DENV titers. In contrast, KUNV dsRNA intensity increased in RNase L KO cells compared to WT, with little or no change in diameter or circularity of dsRNA, respectively (Fig. 2 E and G). Therefore, RNase L not only fails to inhibit ZIKV infection but also specifically promotes ZIKV production and normal RF morphology.
Effects of OAS3 KO on ZIKV RF Assembly and Virus Output.
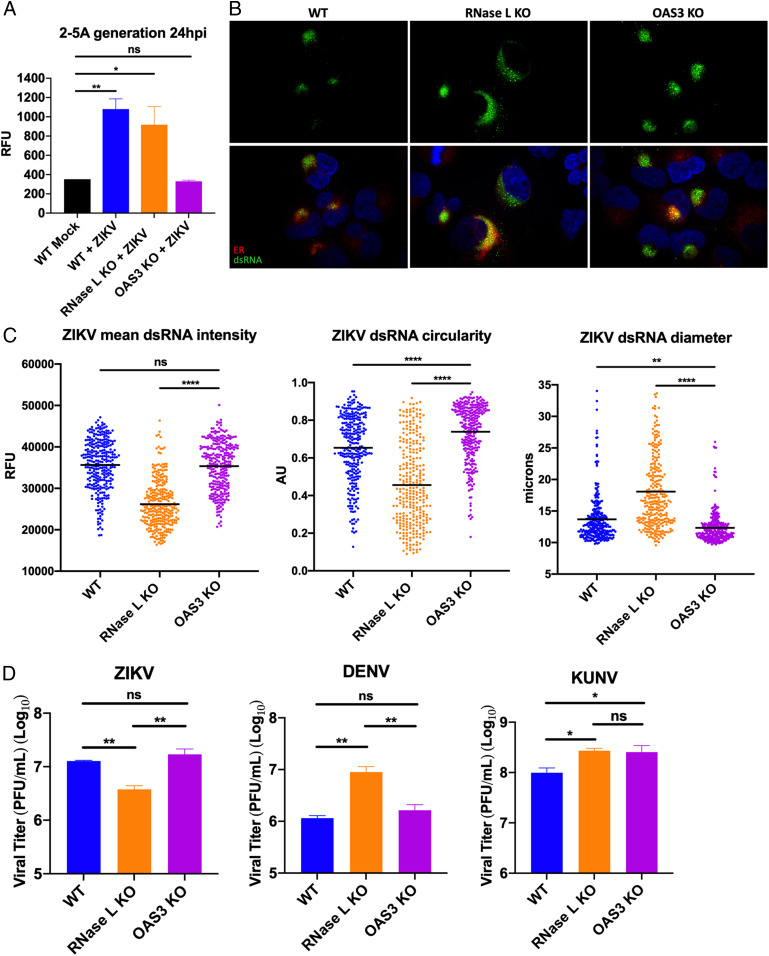
To determine if the proviral effects of RNase L on ZIKV RF structure involve its nuclease function, we examined ZIKV RFs in A549 OAS3 KO cells where RNase L catalytic activity is undetectable. To demonstrate RNase L latency, we measured 2-5A, the activator of RNase L synthesized by OAS enzymes, with a fluorescence resonance energy transfer assay during ZIKV infection of WT, RNase L KO, and OAS3 KO cells. We detected 2-5A generation during ZIKV infection of both WT and RNase L KO cells; however, 2-5A in ZIKV-infected OAS3 KO cells was not increased over levels in mock-infected WT cells (Fig. 3A). Additionally, we show that RNase L activation during ZIKV infection is mediated by the OAS3 isoform specifically using host 28S and 18S rRNA degradation as a measurement for RNase L activation (SI Appendix, Fig. S3). These findings corroborated previous results describing OAS3 as the primary human OAS isoform required for 2-5A synthesis during viral infection (23, 26) and verified that other OAS isoforms did not compensate for loss of OAS3 expression during ZIKV infections. OAS3 KO cells thus provide a useful system for investigating RNase L function independent of its catalytic activity during viral infection.
Fig. 3.
Effects of OAS3 KO on ZIKV RF assembly and virus output. (A) A549 WT, RNase L KO, or OAS3 KO cells were mock infected or infected with ZIKV at a multiplicity of infection (MOI) of 5, 24 hpi lysates were harvested for 2-5A fluorescence resonance energy transfer assay. (B) A549 WT, RNase L KO, or OAS3 KO cells were infected with ZIKV at an MOI of 1, fixed at 20 hpi, and stained for dsRNA (green) and ER (PDI, red) with 4′,6-diamidino-2-phenylindole (blue) staining of nuclei. (C) Quantification of ZIKV mean intensity, circularity, and diameter of staining shown in B. (D) A549 WT, RNase L KO, or OAS3 KO cells were infected with ZIKV, DENV, or KUNV at an MOI of 0.1, supernatants were harvested at 48 hpi for measurement of viral titers by plaque assay, shown as plaque forming units (PFU)/mL virus. The data are representative of at least two independent experiments. Statistical significance was determined by one-way ANOVA. Displayed is the mean of three replicates ± SD for replication assays. ns = not significant, *P < 0.05, **P < 0.01, and ****P < 0.0001. (Imaged at 100× magnification.) RFU = relative fluorescence units, AU = arbitrary units. See also SI Appendix, Figs. S3–S5.
We compared dsRNA expression levels and localization in ZIKV-infected OAS3 KO cells to that of infected WT and RNase L KO cells. We used protein disulfide isomerase (PDI) as a marker for the ER, where RFs reside. At 20 hpi, ZIKV dsRNA intensity in the OAS3 KO cells was rescued to WT levels from that of RNase L KO cells (Fig. 3 B and C). RFs in OAS3 KO cells were more circular and smaller in diameter than in both RNase L KO and WT cells, suggesting enhanced RF formation supported by inactive RNase L. We also performed western blotting and RT-qPCR to measure changes in total intracellular ZIKV protein and genomic RNA, respectively. We found inactive RNase L in OAS3 KO cells increased expression of ZIKV NS3 and genome compared to WT cells (SI Appendix, Fig. S4). While some changes in DENV and KUNV dsRNA expression in OAS3 KO cells displayed a similar trend as that of ZIKV dsRNA, changes were minimal in comparison with alterations in ZIKV dsRNA observed in OAS3 KO cells (SI Appendix, Fig. S5). Additionally, we did not detect any changes in total intracellular NS3 or genomic RNA levels in DENV- or KUNV-infected OAS3 KO cells compared to WT (SI Appendix, Fig. S4). Importantly, OAS3-dependent rRNA degradation was detected during KUNV infection, but we could not detect any rRNA degradation during DENV infection (SI Appendix, Fig. S3). However, rRNA degradation during infection is not the sole marker of RNase L activation, as both viruses were restricted by RNase L activity in WT cells (SI Appendix, Fig. S2).
We found that by 48 hpi, inactive RNase L in the absence of OAS3 restored infectious ZIKV production from reduced levels in RNase L KO cells to that observed during WT cell infection (Fig. 3D). DENV production was not improved by OAS3 KO, suggesting possible OAS3-independent RNase L–mediated inhibition of DENV infection in WT cells (Fig. 3D). In contrast, KUNV production improved in OAS3 KO cells (Fig. 3D). As we observed that RNase L activation during KUNV infection was OAS3-dependent (SI Appendix, Fig. S3), canonical RNase L antiviral activity that restricts KUNV in WT cells is likely absent under these conditions. These results suggest that ZIKV infection may utilize an inactive form of RNase L in a unique manner that results in higher virus production than with RNase L absent.
RNase L Maintains ZIKV RF Structure in the Absence of OAS3 to Enhance Virus Replication.
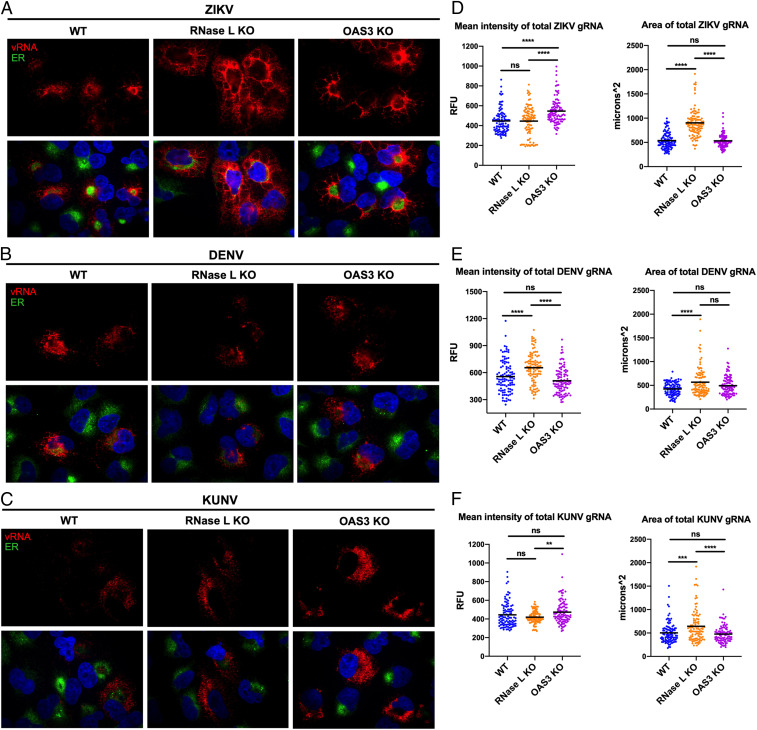
Viral dsRNA expression and localization informs on replication levels and RF location in the cell, respectively, but does not directly reflect changes in viral genome generated by RFs. While dsRNA activates the OAS/RNase L pathway, ssRNA including viral genome is the target of RNase L nucleolytic activity. Using fluorescence in situ hybridization (FISH) to visualize ZIKV genome, we have described an RNase L–dependent absence of ZIKV genome outside of RFs as early as 20 hpi, which we attributed to RNase L cleavage of genome (23). To understand RNase L noncatalytic effects on viral genome, we examined ZIKV genome in OAS3 KO cells using PDI staining of ER as a marker for RF sites. At 20 hpi, ZIKV genomic RNA in WT cells was only detectable at or near the ER. Genome in RNase L KO cells was at the ER but also dispersed throughout the cell. In OAS3 KO cells, genome remained at or proximal to the ER as observed in WT cells, despite the absence of activated RNase L (Fig. 4A). While some genome was detected outside of the ER in OAS3 KO cells, it was minimal compared to that of RNase L KO cells and possibly genome otherwise cleaved by activated RNase L in WT cells (Fig. 4A). Quantification of total genome staining within infected cells revealed that deletion of RNase L did not alter intensity of genome expression at RFs when compared to WT cells. Since genome staining was not circular in shape, we instead used total area to measure increased dissemination of genome throughout the infected cell without RNase L (Fig. 4D). Inactive RNase L in OAS3 KO cells restored the area of genome expression from that of RNase L KO cells back to the area measured in WT cells (Fig. 4D). Furthermore, genome confined to RFs in OAS3 KO cells correlated with higher mean intensity of genome compared to WT and RNase L KO cells (Fig. 4D), suggesting that genome location within the cell dictates new virus release (Fig. 3D). This is corroborated by similar levels of total intracellular ZIKV genome in OAS3 KO and RNase L KO cells, both increased over that in WT cells, suggesting disseminated localization of ZIKV genome outside of RFs in RNase L KO cells results in defective virus assembly and/or release (SI Appendix, Fig. S4B). Enhanced genome intensity in OAS3 KO cells over that of WT cells is thus possibly due to inactive RNase L availability for RF support as opposed to in WT cells wherein RNase L is activated and performs other functions, namely degrading RNA.
Fig. 4.
RNase L maintains ZIKV RF structure in the absence of OAS3 to enhance virus replication. A549 WT, RNase L KO, or OAS3 KO cells were infected at a multiplicity of infection of 1 with (A) ZIKV, (B) DENV, or (C) KUNV. Cells were fixed at 20 hpi and stained with a virus genome–specific probe (red) using FISH before staining ER (PDI, green) and nuclei with 4′,6-diamidino-2-phenylindole (blue) by IFA. Quantification of the mean intensity and area of (D) ZIKV, (E) DENV, or (F) KUNV genome staining. The data are representative of at least two independent experiments. Statistical significance was determined by one-way ANOVA. ns = not significant, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (Imaged at 100× magnification.) RFU = relative fluorescence units. See SI Appendix, Fig. S4B.
In contrast to ZIKV, DENV, and KUNV, genome staining and cellular localization was similar in WT, RNase L KO, and OAS3 KO cells, with genome residing at the ER regardless of RNase L presence or activation status (Fig. 4 B and C). Quantification of DENV total genome staining determined that RNase L deletion increased the area and intensity of genome expression over that in cells lacking OAS3-dependent RNase L activity (Fig. 4E), suggesting OAS3-independent antiviral effects of RNase L on DENV new genome synthesis, which correlates with increased DENV titers in RNase L KO cells but not OAS3 KO cells (Fig. 3D). Quantification of KUNV genome staining exhibited only minor alterations in intensity, with genome in OAS3 KO cells only increased over that in RNase L KO cells but not increased over that in WT cells as was observed for ZIKV (Fig. 4F). Similarly, both DENV and KUNV total intracellular genome was unchanged in OAS3 KO cells compared to in WT cells (SI Appendix, Fig. S4B). Since KUNV does not activate RNase L until 48 hpi by rRNA degradation assay (SI Appendix, Fig. S3), we had not expected a change in genome expression between cell lines at 20 hpi. Additionally, the area of KUNV genome expression was increased in RNase L KO cells compared to both WT and OAS3 KO cells but to a lesser degree than during ZIKV infection (Fig. 4F). Overall, absence of OAS3-dependent RNase L activity had a greater impact on ZIKV genome localization and expression compared to that of DENV and KUNV, which agrees with its minimal effects on DENV and KUNV RFs and viral titers.
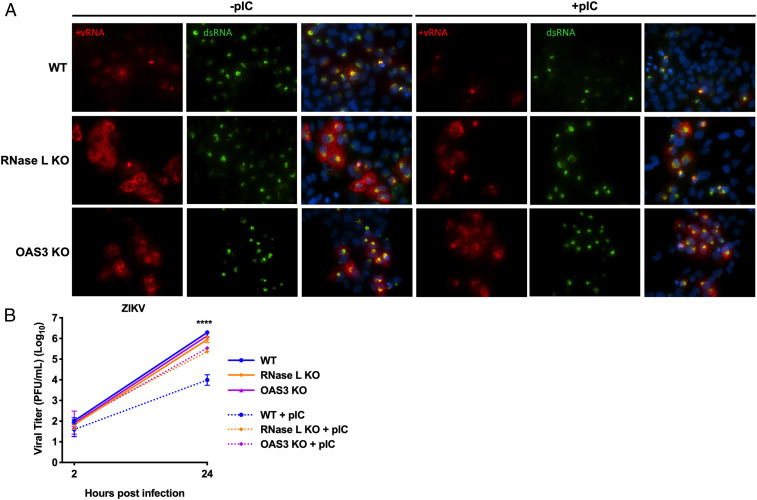
ZIKV RFs Are Unaffected by pIC Treatment in the Absence of OAS3.
The lack of rRNA degradation (SI Appendix, Fig. S3) and 2-5A production (Fig. 3A) during ZIKV infection of OAS3 KO cells indicated that OAS1 and OAS2 did not activate RNase L in the absence of OAS3. We wanted to further investigate whether ZIKV genome localization during infection of OAS3 KO cells (Fig. 4 A and D) was due to noncatalytic, proviral effects of RNase L on RF structure and not a result of activated RNase L cleavage of ZIKV genome residing outside of RFs. As an alternative assay for RNase L activation during ZIKV infection, we used the synthetic dsRNA poly(rI):poly(rC) (pIC) to artificially activate RNase L prior to ZIKV infection, which induces RNase L–mediated cleavage of viral genome prior to RF assembly during infection. Thus, we can use dsRNA expression as a readout for RF assembly, which would be reduced after pIC treatment in cells where RNase L is active. We examined RFs at 24 hpi when RNase L is activated by ZIKV infection alone (23). Since we transfect cells with pIC 2 h before infection, we can no longer detect pIC 26 h post-transfection when examining viral dsRNA expression with an anti-dsRNA antibody. In WT cells, pIC treatment before ZIKV infection resulted in less detectable dsRNA compared with WT cells transfected with lipofectamine only (Fig. 5A). In RNase L KO cells, dsRNA expression was unaffected by pIC. Since pIC robustly activates multiple innate immune pathways in addition to RNase L, these results indicate that limited dsRNA expression in pIC-treated WT cells was due to RNase L specifically and not other antiviral responses induced by pIC. In OAS3 KO cells, pIC transfection before ZIKV infection had no effect on dsRNA expression, further demonstrating that RNase L was not active in the absence of OAS3. Interestingly, pIC treatment diminished viral genome expression in both WT and RNase L KO cells, in contrast to limited dsRNA expression in pIC-treated WT cells only, suggesting that RNase L–independent degradation of ZIKV genome outside of RFs does not affect replication within RFs. Furthermore, ZIKV genome was not altered by pIC pretreatment in OAS3 KO cells (Fig. 5A), suggesting that inactive RNase L enhances ZIKV RF protection of RNA.
Fig. 5.
ZIKV RFs are unaffected by pIC treatment in the absence of OAS3. A549 WT, RNase L KO, or OAS3 KO cells were transfected with lipofectamine only or with pIC for 2 h before infection with ZIKV at a multiplicity of infection of 1. (A) Cells were fixed at 24 hpi and stained with a ZIKV positive–strand genome probe (red) using FISH before staining dsRNA (green) and nuclei with 4′,6-diamidino-2-phenylindole (blue) by IFA. (B) Supernatants from infected cells were harvested at 2 or 24 hpi for measurement of viral titers over time by plaque assay, shown as plaque forming units (PFU)/mL virus. The data are representative of at least two independent experiments. Statistical significance was determined by one-way ANOVA. Displayed is comparison of WT ± pIC (****) and the mean of three replicates ± SD; ****P < 0.0001. (Imaged at 60× magnification.).
To corroborate these results quantitatively, we demonstrated that pIC treatment before ZIKV infection of OAS3 KO cells had no effect on infectious ZIKV production. After transfecting cells for 2 h with pIC, cells were infected with ZIKV for 24 hpi, and viral titers were measured by plaque assay. Without pIC, ZIKV titers in all three cell lines were similar at 24 hpi, which agrees with previous data showing decreased ZIKV from RNase L KO cells only after 24 hpi (Fig. 5B). In WT cells with pIC-induced activation of RNase L, infectious ZIKV production was restricted. In RNase L KO and OAS3 KO cells, pIC only slightly inhibited ZIKV production, indicating that restriction of viral titers in WT cells was predominantly due to RNase L antiviral activity. These results correlate with reduced dsRNA detection in only WT cells after pIC transfection (Fig. 5A) and establish that RNase L is not catalytically active in OAS3 KO cells. This also maintains that decreased ZIKV from RNase L KO cells compared to WT and OAS3 KO cells (Fig. 3D) is not due to RNase L–independent antiviral responses that subsequently limit virus production, as responses unrelated to RNase L induced by pIC in RNase L KO or OAS3 KO cells had little to no effect on ZIKV production or replication, respectively (Fig. 5). Finally, these results suggest that inactive RNase L retains ZIKV genome in smaller, more efficient RFs.
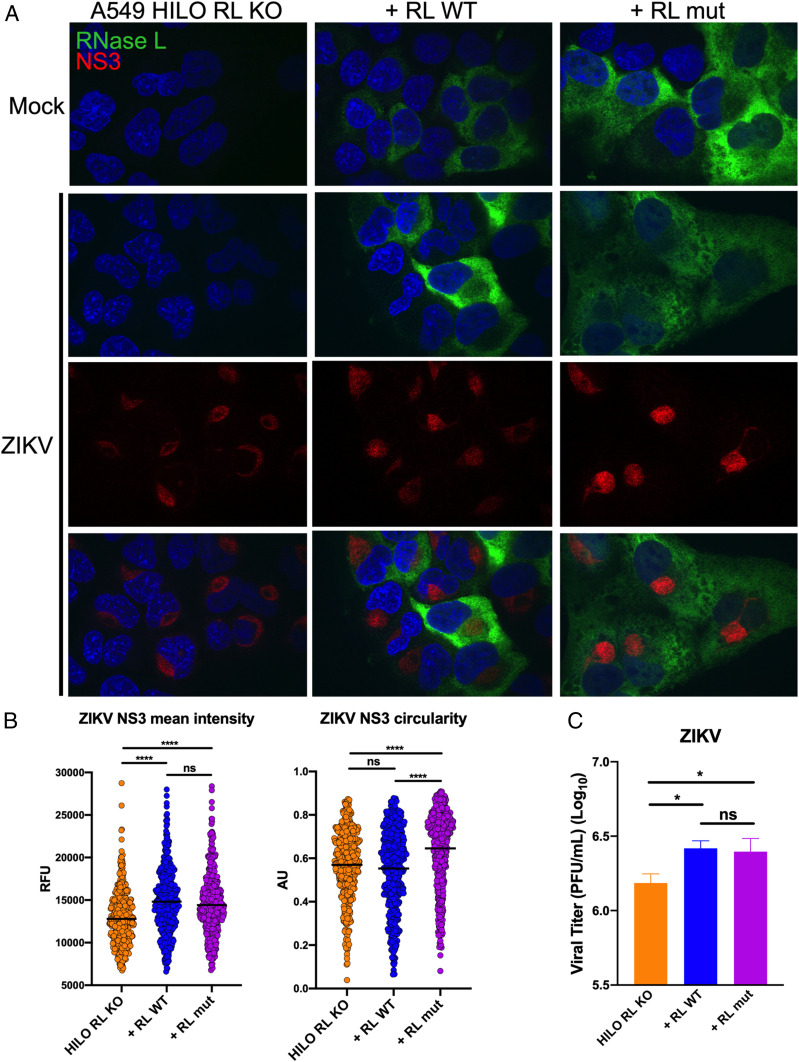
Expression of RNase L R667A Catalytic Mutant in A549 HILO Cells Enhances ZIKV RF Function and Virus Production.
We next directly investigated whether an inactive form of RNase L can enhance ZIKV RF function to augment infectious virus production. We generated A549 HILO cells in which endogenous RNase L is deleted (A549 HILO RL KO), and doxycycline-inducible flag-tagged RNase L WT (+RL WT) or R667A catalytic mutant (+RL mut) are expressed (SI Appendix, Fig. S6) (23, 42). We evaluated effects of RNase L expression on ZIKV RF function and localization compared to that of RNase L KO cells at 20 hpi, using ZIKV NS3 staining to denote RFs. We found that expression of both RNase L WT or mutant increased ZIKV NS3 intensity; however, only the RNase L mutant increased circularity of RFs in comparison to those of RNase L KO cells (Fig. 6 A and B). To determine RNase L effect on infectious ZIKV titer, we measured virus from infected cells at 48 hpi and detected a significant increase in ZIKV from both RL WT– and RL mut–expressing cells compared to RNase L KO cells (Fig. 6C). To verify these results in a different cell line, we infected HeLa M cells, which have undetectable levels of endogenous RNase L, that express either the empty pcDNA3 vector (vector control, VC), +RL WT, or +RL mut and again show that catalytically inactive RNase L can enhance ZIKV RF function and infectious virus production (43) (SI Appendix, Fig. S7).
Fig. 6.
Expression of RNase L R667A catalytic mutant in A549 HILO cells enhances ZIKV RF function and virus production. (A) A549 HILO Rnase L KO (RL KO) cells, or RL KO cells expressing doxycycline-inducible RNase L WT (+RL WT), or RNase L R667A nuclease dead mutant (+RL mut) were mock infected or infected with ZIKV at a multiplicity of infection (MOI) of 10. At 20 hpi, cells were fixed and stained for ZIKV NS3 (red) and flag-RNase L (green) with 4′,6-diamidino-2-phenylindole (blue) staining of nuclei. (B) Quantification of ZIKV NS3 mean intensity and circularity shown in A. (C) A549 HILO RL KO cells, +RL WT, and +RL mut were infected with ZIKV at an MOI of 0.1, supernatants were harvested at 48 hpi for measurement of viral titers by plaque assay, shown as plaque forming units (PFU)/mL virus. The data are representative of at least two independent experiments. Statistical significance was determined by one-way ANOVA. Displayed is the mean of three replicates ± SD for replication assays. ns = not significant, *P < 0.05, and ****P < 0.0001. (Imaged at 100× magnification.) RFU = relative fluorescence units, AU = arbitrary units. See also SI Appendix, Figs. S6 and S7.
Effects of RNase L Deletion on ZIKV RFs Resemble Antiviral Effects of Microtubule Stabilization on ZIKV RFs.
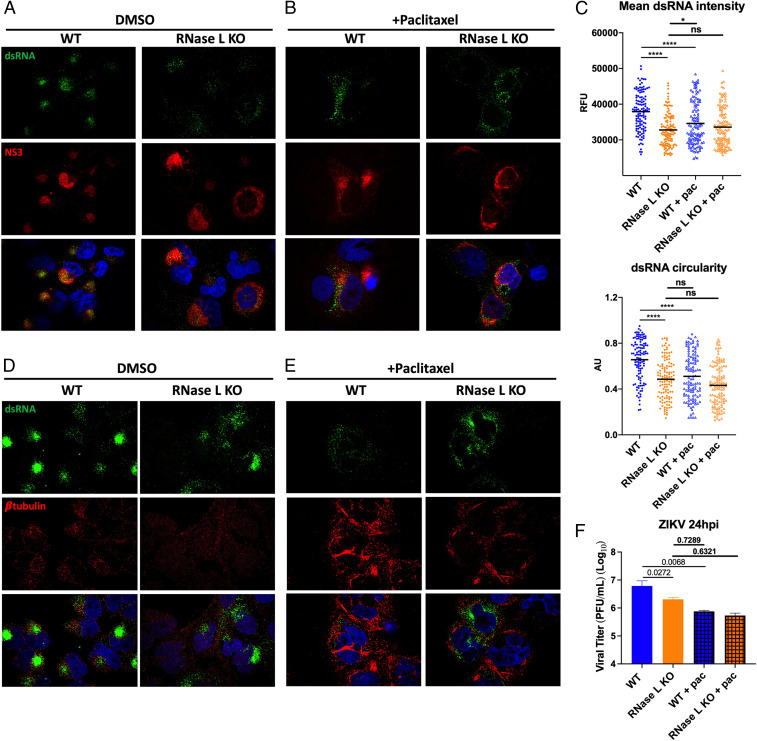
There have been multiple reports of RNase L interaction with the host cytoskeleton, some of which are contingent upon RNase L catalytic latency (41, 44). Since ZIKV rearrangement of the host cytoskeleton to form RFs within ER invaginations is crucial to ZIKV infection, we hypothesized that inactive RNase L association with the cytoskeleton is exploited by ZIKV to form RFs. To demonstrate that proviral RNase L function in ZIKV RF formation depends on cytoskeletal associations, we treated ZIKV-infected cells with paclitaxel, a microtubule-stabilizing drug, and compared its effects on ZIKV RFs with effects of RNase L deletion. Paclitaxel has been shown to inhibit ZIKV titers (12), likely by blocking ZIKV-induced microtubule rearrangements for establishing RFs, although the mechanism by which paclitaxel inhibits ZIKV infection has not been shown. Microtubule stabilization mediated by paclitaxel would prohibit rearrangements necessary for forming ZIKV RFs. We predicted that RNase L deletion similarly impaired ZIKV-stimulated cytoskeletal rearrangements for RF assembly, thereby reducing virus production.
A549 cells were treated with paclitaxel or vehicle dimethyl sulfoxide (DMSO) 3 h after infection with ZIKV. Cells were fixed at 20 hpi and stained for ZIKV dsRNA and NS3 by IFA. Expression of ZIKV dsRNA and NS3 in DMSO-treated RNase L KO cells was again dimmer and more dispersed throughout the cell compared to in WT cells (Fig. 7A). Quantification confirmed that the absence of RNase L lowered ZIKV dsRNA expression and concentration, as measured by intensity and circularity of dsRNA staining, respectively (Fig. 7C). In paclitaxel-treated WT cells, ZIKV dsRNA and NS3 staining resembled that of RNase L KO with or without paclitaxel (Fig. 7B). Quantification maintained a decrease in dsRNA intensity and circularity in WT cells treated with paclitaxel, closer to levels of RNase L KO cells, compared to DMSO-treated WT cells. Additionally, paclitaxel had no effect on dsRNA intensity and minimal impact on circularity in RNase L KO cells (Fig. 7C). We validated effects of paclitaxel treatment on microtubule stability by staining β-tubulin, which together with α-tubulin makes up microtubules. In ZIKV-infected WT cells, β-tubulin localized to dsRNA staining, which agrees with previous findings of ZIKV-induced microtubule rearrangements surrounding RFs (Fig. 7D and SI Appendix, Fig. S8) (12). In RNase L KO cells, β-tubulin was no longer concentrated around RFs, similar to dissemination of ZIKV dsRNA or NS3 without RNase L expressed (Fig. 7D and SI Appendix, Fig. S8). Paclitaxel treatment of both ZIKV-infected WT and RNase L KO cells resulted in β-tubulin aggregation due to inhibition of microtubule tractability (Fig. 7E). Finally, we treated cells with paclitaxel 3 h after infecting with ZIKV to measure changes in infectious virus production at 24 hpi as a result of microtubule stabilization in WT and RNase L KO cells. Paclitaxel in WT cells reduced infectious ZIKV titers to the level of RNase L KO cells (Fig. 7F). Moreover, the reduction in ZIKV titers caused by RNase L deletion is lost in cells treated with paclitaxel (Fig. 7F). This suggests that the means by which RNase L aids in ZIKV production is absent in paclitaxel-treated cells where microtubule networks are stabilized. Together with the effects of paclitaxel on dsRNA expression, these results offer a mechanism for RNase L–mediated increased ZIKV production through ZIKV employment of RNase L–cytoskeleton interactions during formation of RFs.
Fig. 7.
Effects of RNase L deletion on ZIKV RFs resemble antiviral effects of microtubule stabilization on ZIKV RFs. A549 WT or RNase L KO cells were infected with ZIKV at a multiplicity of infection of 1, treated with 12.5 μM paclitaxel or DMSO 3 h after infection. For IFA staining, cells were fixed at 20 hpi. Cells treated with (A) DMSO or (B) paclitaxel were stained for dsRNA (green) and ZIKV NS3 (red) with 4′,6-diamidino-2-phenylindole (DAPI, blue) nuclei staining, mean dsRNA intensity and circularity was quantified in C. Cells treated with (D) DMSO or (E) paclitaxel were stained for dsRNA (green) and β-tubulin (red) with DAPI (blue) nuclei staining. (F) For infectious virus production, supernatants were harvested at 24 hpi for measurement of viral titers by plaque assay, shown as plaque forming units (PFU)/mL virus, displayed are P values. The data are representative of at least two independent experiments. Statistical significance was determined by one-way ANOVA. Displayed is the mean of three replicates ± SD for replication assays. ns = not significant, *P < 0.05, and ****P < 0.0001. (Imaged at 100× magnification.) RFU = relative fluorescence units, AU = arbitrary units. See also SI Appendix, Fig. S8.
Discussion
In this study, we present findings supporting a proviral role of the host RNase L protein, which has otherwise been considered strictly antiviral. RNase L nucleolytic activity is highly effective at restricting replication of a diverse range of DNA and RNA viruses (32). However, catalytic activation of RNase L by ZIKV does not lead to lower virus production, and instead ZIKV titers are paradoxically increased with RNase L expression. In particular, a catalytically inactive form of RNase L improved ZIKV RF assembly and function, thereby boosting virus production. Since several studies report interactions between inactive RNase L and the cytoskeleton, our findings suggest that ZIKV repurposes the interaction between RNase L and the cytoskeleton to facilitate rearrangement of the ER for establishment of ZIKV RFs. While cytoskeletal arrangements for RF assembly within ER folds are characteristic of flaviviruses, we found that RNase L expression during DENV and KUNV infection had only minimal effects on RF formation. Furthermore, RNase L was ultimately restrictive of both viruses, operating in its canonical antiviral manner to decrease DENV and KUNV titers through its nuclease activity. Therefore, we propose that recruitment of RNase L for enhanced RF function and virus release is a feature presently observed only during ZIKV infection.
Interestingly, DENV production in WT cells was limited by RNase L, as titers increased significantly in RNase L KO cells (SI Appendix, Fig. S2), although we were unable to detect any rRNA degradation in DENV-infected WT cells (SI Appendix, Fig. S3). Unlike other viruses we have examined, RNase L antiviral effects on DENV were not dependent on OAS3, as viral titers in WT and OAS3 KO cells were similar (Fig. 3D). Moreover, RNase L deletion enhanced DENV genome expression (Fig. 4E and SI Appendix, Fig. S4B), which correlated with increased titers in RNase L KO cells and suggests that RNase L is enzymatically active during DENV infection. Indeed, we previously reported RNase L–mediated degradation of DENV RNA during infection (23). Another study showed that overexpression of OAS1 or OAS3 in A549 cells infected with DENV2 resulted in rRNA degradation, suggesting that higher basal levels of OAS proteins may be required for detection of rRNA degradation during DENV-induced RNase L activation (33). While rRNA degradation is a hallmark of RNase L nucleolytic activity and therefore a commonly used marker for RNase L activation, another possibility is that host rRNA effectively avoids activated RNase L while viral genome is primarily targeted during DENV infection in particular (45–47). Further investigation is required for elucidation of the mechanism behind RNase L restriction of DENV.
We used pIC to activate RNase L prior to ZIKV generation of protective RFs as another measurement confirming RNase L catalytic inactivity in OAS3 KO cells (Fig. 5) in addition to rRNA degradation (SI Appendix, Fig. S3) and 2-5A generation (Fig. 3A) assays. OAS3 KO cells therefore provided an elegant tool for individually assaying RNase L catalytic and noncatalytic functions during ZIKV infection (Figs. 3 and 4). We corroborated findings in OAS3 KO cells demonstrating proviral RNase L activity was independent of enzymatic function by generating A549 HILO RNase L KO cells expressing +RL WT or +RL mut (Fig. 6). In addition, we used HeLa M cells to confirm findings in a different cell line (SI Appendix, Fig. S7). One caveat we encountered in HeLa M cells was the variance in RNase L WT and R667A protein levels, which occurred when overexpressing these constructs in several cell types. However, HeLa M cells were effective for confirming that noncatalytic RNase L activity enhances ZIKV production in another cell line. In addition to rescuing ZIKV RF function and virus production to A549 WT levels, the flag-tag expressed on RNase L in A549 HILO +RL WT and +RL mut cells allowed for visualization of RNase L in both active and inactive forms during virus infection, which had not been previously shown. We found that RNase L WT was more concentrated near RFs, possibly as a result of catalytic activation, while inactive RNase L R667A mutant was dispersed throughout the cell (Fig. 6A). Interestingly, both forms of RNase L are notably absent in and around ZIKV RFs, indicating that RNase L does not support RF assembly internally but instead appears to support a structural network throughout the cytoplasm that enhances RF assembly and function.
After establishing that inactive RNase L improved ZIKV RF formation, we next sought to link RNase L proviral effects on ZIKV infection with RNase L–cytoskeleton interactions. Interest in RNase L–cytoskeleton interaction stemmed from an earlier study describing inactive RNase L interacted with the cytoskeletal protein Filamin A for inhibition of viral entry. That study determined that this interaction required RNase L latency and was dissociated upon catalytic activation of RNase L (41), which correlates with our proposed function of inactive RNase L during ZIKV infection. Another study described the cytoskeleton-containing cellular fraction as also comprising a form of RNase L that was unresponsive to catalytic activation by 2-5A (44). Proteomics analysis in a human cell line identified IQGAP1 (an IQ [isoleucineglutamine] motif containing GTPase-activating protein 1), a mediator of actin cytoskeleton reorganization, as an RNase L binding partner (48), and another report revealed 62% of RNase L interactions in mouse spleens were with cytoskeleton and motor proteins (49). Additionally, RNase L interacts with a regulator of tight junctions, ligand of nump protein X (50), although none of these latter findings were in the context of virus infection.
While RNase L function during ZIKV infection is the first suggestion of a proviral role for inactive RNase L, the inactive RNase L interaction with Filamin A indicated that RNase L could actively remodel cytoskeleton during viral infection, which is consistent with reports of virus manipulation of cytoskeletal components to facilitate virus replication cycle events (51, 52). It is known that ZIKV induces extensive cytoskeletal remodeling during infection to establish RFs within invaginations of the ER (12). Since we found that RNase L deletion altered the localization of RF constituents (i.e., ZIKV dsRNA and NS3) and ER proteins to less-concentrated areas that generated less virus, we hypothesized that this was due to defective cytoskeletal remodeling without RNase L. Indeed, we found that absence of RNase L resulted in defective remodeling of microtubules for RF support during ZIKV infection (SI Appendix, Fig. S8). We used the microtubule-stabilizing drug paclitaxel to recapitulate the ZIKV RF phenotype of RNase L KO cells. Paclitaxel-mediated loss of microtubule flexibility disturbed ZIKV RF assembly and function and consequently reduced virus production, as was observed in RNase L KO cells (Fig. 7). Furthermore, in paclitaxel-treated cells, deletion of RNase L no longer reduced ZIKV titers as in untreated cells (Fig. 7). These results support the model that ZIKV employs the association of RNase L with cytoskeleton to generate RFs. Alternatively, as peak effects of RNase L deletion on ZIKV RFs and infectious virus production occur later on during infection, RNase L may aid in maintenance of ZIKV RFs rather than in their initial formation. Distinctions in host protein composition of ZIKV RFs compared to that of other flaviviruses have been described (12). Since RNase L improves production of ZIKV but not DENV or KUNV, it is possible that RNase L is another ZIKV RF component not shared among all flaviviruses. Inactive RNase L participation in ZIKV RF formation also implies lower availability of activated RNase L proximal to ZIKV RFs, which offers explanation for ZIKV protection from antiviral effects of RNase L, while DENV and KUNV are sensitive to RNase L activity.
Despite clear alterations in ZIKV RF shape and function without RNase L facilitation of cellular remodeling, ZIKV RFs still form and function to a degree in the absence of RNase L. This suggests redundancy in host protein manipulation for cellular remodeling by ZIKV and is supported by the existence of RFs even after RNase L is activated and cleaving ZIKV genome outside RF sites. Additionally, the dissemination of ZIKV RFs in RNase L KO cells occurred in a majority of infected cells but not in every infected cell, indicating that other host proteins may compensate for the loss of RNase L. Further investigation identifying specific host cytoskeletal components that interact with RNase L or ZIKV proteins will elucidate how RNase L facilitates assembly of ZIKV RFs.
Materials and Methods
Human A549 cells (and derived knockout cells) were infected with ZIKV (PRVABC2015 strain) and in some cases with DENV2 or KUNV. Infected cells were analyzed for infectious virus production or for RNA and protein expression by immunofluorescence and FISH. All of these techniques are described in SI Appendix, Materials and Methods. All relevant data are presented in the main text figures and SI figures. The associated protocols are described in Materials and Methods and SI Appendix, Materials and Methods. Any materials can be obtained by contacting the corresponding author.
Supplementary Material
Acknowledgments
We thank Dr. Courtney Comar for her help with image quantification experiments and Dr. Andrea Stout and the Cell and Developmental Biology Microscopy Core at the University of Pennsylvania for confocal microscopy training. We thank BioRender.com, used to create Fig. 1. This work was supported by NIH grants R21NS100182 (to S.R.W.), R01AI104887 (to S.R.W. and R.H.S.), and R01AI135922 (to R.H.S.). J.N.W. was supported in part by NIH grant T32NS007180.
Footnotes
Competing interest statement: S.R.W. is on the scientific advisory board of Immunome, Inc. and Ocugen, Inc. R.H.S. is a consultant to Inception Therapeutics, Inc.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101713118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Wikan N., Smith D. R., Zika virus: History of a newly emerging arbovirus. Lancet Infect. Dis. 16, e119–e126 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Guzman M. G., Harris E., Dengue. Lancet 385, 453–465 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Kaiser J. A., Barrett A. D. T., Twenty years of progress toward West Nile virus vaccine development. Viruses 11, 823 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu K. Y., et al., Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Res. 26, 645–654 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cugola F. R., et al., The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534, 267–271 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miner J. J., et al., Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 165, 1081–1091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Ortenzio E., et al., Evidence of sexual transmission of Zika virus. N. Engl. J. Med. 374, 2195–2198 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Mlakar J., et al., Zika virus associated with microcephaly. N. Engl. J. Med. 374, 951–958 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Cauchemez S., et al., Association between Zika virus and microcephaly in French Polynesia, 2013-15: A retrospective study. Lancet 387, 2125–2132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul D., Bartenschlager R., Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2, 32–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neufeldt C. J., Cortese M., Acosta E. G., Bartenschlager R., Rewiring cellular networks by members of the Flaviviridae family. Nat. Rev. Microbiol. 16, 125–142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortese M., et al., Ultrastructural characterization of Zika virus replication factories. Cell Rep. 18, 2113–2123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aktepe T. E., Liebscher S., Prier J. E., Simmons C. P., Mackenzie J. M., The host protein reticulon 3.1A is utilized by flaviviruses to facilitate membrane remodelling. Cell Rep. 21, 1639–1654 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Neufeldt C. J., et al., ER-shaping atlastin proteins act as central hubs to promote flavivirus replication and virion assembly. Nat. Microbiol. 4, 2416–2429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welsch S., et al., Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5, 365–375 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sager G., Gabaglio S., Sztul E., Belov G. A., Role of host cell secretory machinery in Zika virus life cycle. Viruses 10, 559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miorin L., Maestre A. M., Fernandez-Sesma A., García-Sastre A., Antagonism of type I interferon by flaviviruses. Biochem. Biophys. Res. Commun. 492, 587–596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riedl W., et al., Zika virus NS3 mimics a cellular 14-3-3-binding motif to antagonize RIG-I- and MDA5-mediated innate immunity. Cell Host Microbe 26, 493–503.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y., et al., Zika virus evades interferon-mediated antiviral response through the co-operation of multiple nonstructural proteins in vitro. Cell Discov. 3, 17006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowen J. R., et al., Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLoS Pathog. 13, e1006164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant A., et al., Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19, 882–890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Best S. M., The many faces of the flavivirus NS5 protein in antagonism of type I interferon signaling. J. Virol. 91, e01970-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whelan J. N., Li Y., Silverman R. H., Weiss S. R., Zika virus production is resistant to RNase L antiviral activity. J. Virol. 93, e00313-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birdwell L. D., et al., Activation of RNase L by murine coronavirus in myeloid cells is dependent on basal oas gene expression and independent of virus-induced interferon. J. Virol. 90, 3160–3172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong B., Silverman R. H., 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J. Biol. Chem. 270, 4133–4137 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Li Y., et al., Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. U.S.A. 113, 2241–2246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou A., et al., Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16, 6355–6363 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castelli J. C., et al., A study of the interferon antiviral mechanism: Apoptosis activation by the 2-5A system. J. Exp. Med. 186, 967–972 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakrabarti A., et al., RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe 17, 466–477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherbik S. V., Paranjape J. M., Stockman B. M., Silverman R. H., Brinton M. A., RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80, 2987–2999 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malathi K., Dong B., Gale M. Jr, Silverman R. H., Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448, 816–819 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverman R. H., Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 81, 12720–12729 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin R. J., et al., Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J. Immunol. 183, 8035–8043 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Samuel M. A., et al., PKR and RNase L contribute to protection against lethal West Nile virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80, 7009–7019 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao L., et al., Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11, 607–616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drappier M., et al., A novel mechanism of RNase L inhibition: Theiler’s virus L* protein prevents 2-5A from binding to RNase L. PLoS Pathog. 14, e1006989 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silverman R. H., Weiss S. R., Viral phosphodiesterases that antagonize double-stranded RNA signaling to RNase L by degrading 2-5A. J. Interferon Cytokine Res. 34, 455–463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sánchez-Tacuba L., Rojas M., Arias C. F., López S., Rotavirus controls activation of the 2′-5′-oligoadenylate synthetase/RNase L pathway using at least two distinct mechanisms. J. Virol. 89, 12145–12153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldstein S. A., et al., Lineage A betacoronavirus NS2 proteins and the homologous torovirus berne pp1a carboxy-terminal domain are phosphodiesterases that antagonize activation of RNase L. J. Virol. 91, e02201-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang R., et al., Homologous 2′,5′-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 13114–13119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malathi K., et al., RNase L interacts with Filamin A to regulate actin dynamics and barrier function for viral entry. mBio 5, e02012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khandelia P., Yap K., Makeyev E. V., Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 108, 12799–12804 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong B., Niwa M., Walter P., Silverman R. H., Basis for regulated RNA cleavage by functional analysis of RNase L and Ire1p. RNA 7, 361–373 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tnani M., Aliau S., Bayard B., Localization of a molecular form of interferon-regulated RNase L in the cytoskeleton. J. Interferon Cytokine Res. 18, 361–368 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Li X. L., Blackford J. A., Hassel B. A., RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J. Virol. 72, 2752–2759 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rath S., et al., Human RNase L tunes gene expression by selectively destabilizing the microRNA-regulated transcriptome. Proc. Natl. Acad. Sci. U.S.A. 112, 15916–15921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donovan J., Rath S., Kolet-Mandrikov D., Korennykh A., Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23, 1660–1671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato A., et al., Association of RNase L with a Ras GTPase-activating-like protein IQGAP1 in mediating the apoptosis of a human cancer cell-line. FEBS J. 277, 4464–4473 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Gupta A., Rath P. C., Expression of mRNA and protein-protein interaction of the antiviral endoribonuclease RNase L in mouse spleen. Int. J. Biol. Macromol. 69, 307–318 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Ezelle H. J., Malathi K., Hassel B. A., The roles of RNase-L in antimicrobial immunity and the cytoskeleton-associated innate response. Int. J. Mol. Sci. 17, 74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foo K. Y., Chee H. Y., Interaction between flavivirus and cytoskeleton during virus replication. BioMed Res. Int. 2015, 427814 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walsh D., Naghavi M. H., Exploitation of cytoskeletal networks during early viral infection. Trends Microbiol. 27, 39–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.