Abstract
CRISPR-Cas9 nuclease-based gene drives have been developed toward the aim of control of the human malaria vector Anopheles gambiae. Gene drives are based on an active source of Cas9 nuclease in the germline that promotes super-Mendelian inheritance of the transgene by homology-directed repair (“homing”). Understanding whether CRISPR-induced off-target mutations are generated in Anopheles mosquitoes is an important aspect of risk assessment before any potential field release of this technology. We compared the frequencies and the propensity of off-target events to occur in four different gene-drive strains, including a deliberately promiscuous set-up, using a nongermline restricted promoter for SpCas9 and a guide RNA with many closely related sites (two or more mismatches) across the mosquito genome. Under this scenario we observed off-target mutations at frequencies no greater than 1.42%. We witnessed no evidence that CRISPR-induced off-target mutations were able to accumulate (or drive) in a mosquito population, despite multiple generations’ exposure to the CRISPR-Cas9 nuclease construct. Furthermore, judicious design of the guide RNA used for homing of the CRISPR construct, combined with tight temporal constriction of Cas9 expression to the germline, rendered off-target mutations undetectable. The findings of this study represent an important milestone for the understanding and managing of CRISPR-Cas9 specificity in mosquitoes, and demonstrates that CRISPR off-target editing in the context of a mosquito gene drive can be reduced to minimal levels.
Keywords: CRISPR-Cas, gene drive, off-target, Anopheles, vector control
The versatility of CRISPR-Cas9 nucleases has expedited the development of homing-based gene-drive systems that hold huge potential for vector control (1–4). Considerable efforts have gone into the development of such gene-drive systems, including assessments of potential obstacles to successful efficacy of this technology, with mitigation of resistance emergence being a major barrier to negotiate (3, 5, 6). Prior to open-field testing, it is also important to assess and manage potential risks that may arise, including those associated with cleavage at off-target sites by the CRISPR construct (7).
In the context of nuclease-based gene drive for vector control, there could be genome-editing events occurring at off-target sites, which are not necessarily harmful, and in most cases mutations arising from these events will remain rare and would likely be of no consequence. It is worthy of notice that a single guide RNA (gRNA) may induce editing events at multiple sites with different frequencies and that these may differ in distribution in each gamete. A nuclease-induced mutation could in principle become common either if it increases the fitness of the mosquito and therefore is positively selected, or if off-target cleavage occurs at such a high rate at that site that the cleavage itself (with homologous or nonhomologous repair) leads to a high frequency of the mutation. Many or most mutations would be expected to have no phenotypic effect, and so be of no consequence. Other mutations may reduce the fitness of the transgenic mosquitoes, impacting the efficacy of gene drive (8, 9). Although not expected, it is in principle possible that mutations derived from off-target cleavage events could affect epidemiologically important traits, such as resistance to insecticides or pathogen susceptibility, and the risk of these unwanted effects should be evaluated on a case-by-case basis.
A further consideration is the sheer genetic diversity that a gene drive would face upon field release. Sequencing of 1,142 wild-caught mosquito specimens sampled across Africa revealed more than 57 million single-nucleotide polymorphisms (SNPs) (10, 11). Such polymorphisms could have the effect of creating or destroying protospacer adjacent motifs (PAMs), as well as altering genomic sites to resemble the gRNA spacer target sequence, with previous studies showing how human genetic variation could alter CRISPR-Cas9 specificity (12–14). As well as the gRNA, the profile of nuclease activity itself—depending on its dosage, specificity, and spatiotemporal regulation—could affect the rate of generation of off-target indels (8, 15).
Extensive effort has gone into nominating off-target cleavage sites using computational (16, 17), cell-based (e.g., GUIDE-seq, BLESS/BLISS, HTGTS, DISCOVER-Seq) (18–22), or in vitro methods (e.g., circular in vitro reporting of cleavage effects by sequencing [CIRCLE-seq], SITE-seq, Digenome-seq) (23–25). The CIRCLE-seq in vitro method, in particular, has demonstrated that poorly chosen CRISPR-Cas9 gRNAs (with a large number of sites in the genome that closely match the desired on-target site) are capable of causing extensive in vivo off-targeting edits in mice (26). However, by careful choice of the gRNA to limit the number of such closely matched sites in the mouse genome, the sites that showed insertions or deletions (indels) at above the detection limit of next-generation sequencing (0.1%) could be reduced to none or very few (26).
In this study, we investigated the off-target activity of four previously developed gene-drive strains that were designed to suppress populations of Anopheles gambiae (SI Appendix, Table S1) (2–4, 27). The strains include a potential candidate for field testing (3) and represent a diversity in construct design that would be expected to affect their potentials to generate off-target mutations, including key differences in spatiotemporal activity of Cas9 (i.e., somatic versus germline), levels of Cas9 expression, and choice of gRNA/genomic target sequence.
Strategies to detect off-target mutations rely on identifying the products of end-joining mutagenesis, which can generate novel sequences that can be distinguished from naturally occurring genetic variation (26). We have previously demonstrated that fine-tuning the spatiotemporal activity of Cas9, by the use of alternative germline promoters, results in differences in both the rate and type of mutagenesis. These data suggest that cleavage in the early embryo by maternally deposited Cas9 when using the vasa promoter to drive its expression results in DNA repair that is strongly biased toward the mutagenic end-joining pathway (5, 28). The first-generation gene-drive strain, vas-7280CRISPRh, was built using the vasa promoter, which generates high levels of end-joining at the nuclease target site; thus, it is perhaps the most-sensitive platform with which to detect off-target activity. This strain resulted in strong fertility costs and despite a high inheritance rate, was blocked by development of resistance (5). A second gene drive, zpg-7280CRISPRh, targets the same site in the AGAP007280 female fertility gene with expression of Cas9 nuclease under the control of the zero population growth (zpg) promoter, which restricts activity to the germline, where DNA repair is strongly biased toward homology-directed repair (HDR) (3, 27). This promoter is now used in all gene-drive strains currently being developed by our group. Use of the zpg promoter has important implications for the generation and detection of off-target mutations as it may reduce end-joining mutagenesis at off-targets, even when using the same gRNA. A third gene-drive strain, zpg-7280SDGD, was developed to generate a driving male-biased sex-distorter that targets the same AGAP007280 gene and it was designed by linking on the same construct an X-chromosome shredder I-PpoI nuclease to a CRISPR-based gene drive [similarly to Simoni et al. (4)]. The fourth gene-drive strain, zpg-dsxCRISPRh, was developed to target a highly conserved sequence within the doublesex (dsx) gene (AGAP004050) and could quickly spread through caged mosquito populations, reaching 100% frequency and population collapse (3) and it is now being considered for field testing.
Here we present an exploration of off-target mutations in vas-7280CRISPRh, zpg-7280CRISPRh, zpg-7280SDGD, and zpg-dsxCRISPRh mosquito strains to evaluate whether such mutations could hamper the utility of this technology for vector control.
Results
Assessment of Off-Target Mutations in Gene Drives Targeting AGAP007280.
One determinant of CRISPR-Cas9 off-target cleavage activity is the sequence relatedness of a given sequence to the intended on-target site. Computational algorithms designed to predict putative off-targets in the genome consider the number and type of mismatches to a gRNA, as well as their position in the sequence itself; however, they have been shown to have limitations in their ability to identify bona fide sites that actually mutated in cells or organisms. The CIRCLE-seq method identifies and provides semiquantitative assessment of off-target cleavage activity in vitro (23) and these nominated sites can then be assessed to identify off-targets that are mutated in a specific cell- or organism-based context. To identify putative off-targets for a gRNA targeting AGAP007280 (gRNA used in three gene-drive populations: vas-7280CRISPRh, zpg-7280CRISPRh, and zpg-7280SDGD), we performed CIRCLE-seq using wild-type mosquito DNA (G3 laboratory strain) (SI Appendix, Fig. S1 and Table S2 and Dataset S1) and identified 98 off-target sites cleaved in vitro above an arbitrary threshold, 45 of which were in annotated genes.
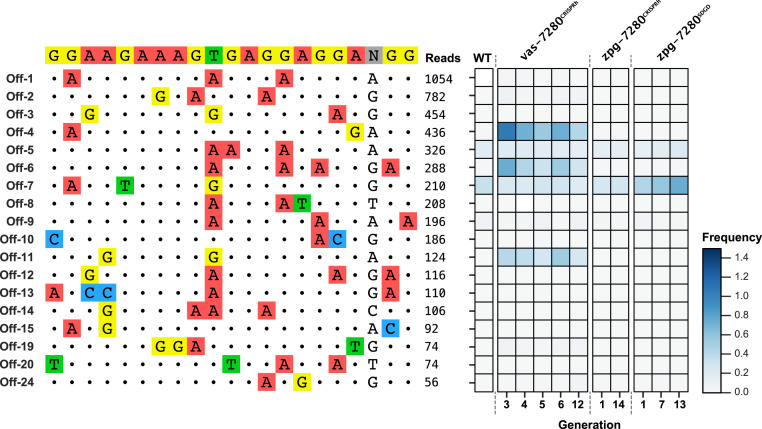
To assess whether sites identified from the CIRCLE-seq assay showed evidence of indels in vivo, we chose the 15 sites with the highest CIRCLE-seq read counts (Materials and Methods), as well as 5 additional sites that were located within annotated genes. We then performed targeted amplicon sequencing from pools of mosquitoes for the intended on-target site and these 20 sites to look for and quantify potential indel mutations. Among the 20 sites we assessed, 19 returned sequencing reads. The remaining site could be amplified but could not be sequenced (no reads were aligned to this site), and this site was therefore excluded from the analysis. An additional site was also removed from analysis due to the highly repetitive nature of the sequence and existing polymorphisms that prevented further analysis. At the on-target site, we observed significant levels of mutations at all generations sequenced, for all three of the strains targeting AGAP007280 (SI Appendix, Fig. S2A and Dataset S2). Using the strain predicted to be most sensitive to off-target mutagenesis, vas-7280CRISPRh (1, 2), we identified indels above threshold frequency at five sites (off-2, -4, -6, -11, and -19) in all five sampled generations (G3 to G6 and G12) of the caged release experiment (minimum of 274 mosquitoes sequenced per cage) (Fig. 1, SI Appendix, Fig. S3, and Dataset S2). Indel frequencies ranged from 0.03 to 1.42% of the total sequencing reads per off-target site and four of five sites were located within exons or introns (AGAP000774, AGAP011092, AGAP000061, and AGAP000042), and all had four or fewer mismatches relative to the on-target site (Fig. 1).
Fig. 1.
Assessment of off-targets indels within gene-drive populations targeting AGAP007280. Targeted amplicon sequencing was conducted for 18 off-target sites predicted from the CIRCLE-seq for gRNA-7280. The frequency (in percent) of reads containing indels are displayed as a heatmap. WT served as the negative control. Three different gene-drive populations were assessed for off-target mutations over several generations (as indicated below the heatmap): vas-7280CRISPRh, zpg-7280CRISPRh, and zpg-7280SDGD. For vas-7280CRISPRh and zpg-7280CRISPRh, each generation was comprised of two biological replicates (cages 1 and 2). For WT and zpg-7280SDGD, each generation comprised one biological replicate only. For zpg-7280SDGD this was due to population collapse of one of the biological replicates (cage 2) after eight generations (which was intended as part of the study).
Sequencing of the 18 sites identified by CIRCLE-seq was performed on the zpg-based gene-drive strains previously shown to generate substantially reduced end-joining mutations, zpg-7280CRISPRh (27) and zpg-7280SDGD. These strains harbor the same gRNA but show no evidence of Cas9 deposition, the major cause of end-joining mutations (28). Neither of these strains showed evidence of off-target mutagenesis above the threshold frequency at any of the 18 selected sites (minimum of 300 mosquitoes sequenced per cage) (Fig. 1). However, we detected indels at 0.02% frequency at off-target site 4 at generation 7 (G7) of the zpg-7280SDGD population that appeared consistent with Cas9-induced end-joining mutagenesis (neither G1 nor G13 of this population showed such indels) (Dataset S2). This was 15 times lower than the indel frequencies seen at the same off-target site for the vas-7280CRISPRh. Analysis for off-target 7 did identify indels; however, these same mutations were present across all populations, including the wild-type (negative control), and therefore may represent sequencing error (or a sequence variant not present in the reference wild-type sequence) rather than a Cas9-induced event (SI Appendix, Fig. S4).
Screening for Off-Target Mutations within a Gene Drive Considered for Field Studies.
A gene drive designed for field release would contain a gRNA that targets a sequence highly constrained and conserved in the mosquito genome, so as to mitigate against target site resistance. To that end, we analyzed the level of off-target mutations in an additional gene drive developed targeting the exon 5 of the doublesex (dsx) gene (3). The gRNA that targets the dsx gene (gRNA-dsx) was designed to contain fewer closely related sites across the mosquito genome than gRNA-7280 for its respective target site. In silico analysis showed that the gRNA-dsx had no closely related genomic sites (three or fewer mismatches) in the reference mosquito genome (SI Appendix, Table S2). CIRCLE-seq performed using gRNA-dsx identified a smaller number of off-target cleavage sites than was observed for the gRNA-7280 (30 and 98, respectively) (SI Appendix, Fig. S5). In addition, the only site with a high number of reads in the CIRCLE-seq experiment was the on-target site (accounting for 88.8% of all reads), with the next highest site accounting for only 1.5% of the on-target reads. Of the 30 sites identified by CIRCLE-seq for the gRNA-dsx, 28 harbored at least 6 mismatches to the dsx target, a finding consistent with the high orthogonality of the gRNA-dsx target site to the mosquito genome (Dataset S3).
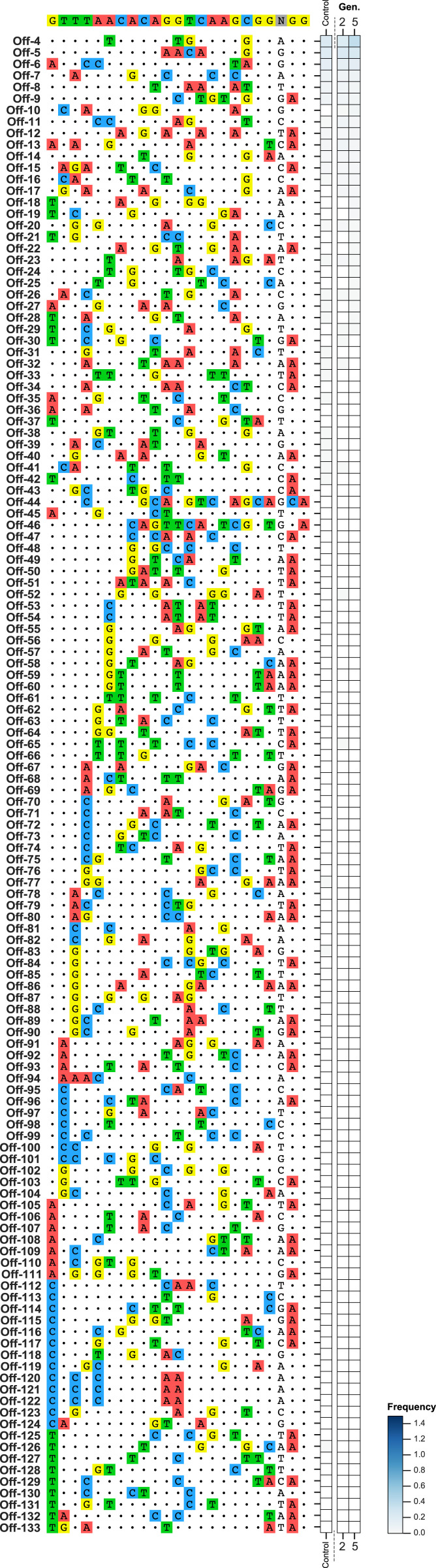
Because CIRCLE-seq did not identify any genomic sites that looked plausible to be cleaved in vivo at levels above the limit of detection [given that bona fide off-targets for gRNA-7280 contained no more than four mismatches to the target site, and as previously reported (26)], we used an in silico off-target analysis approach (16) to identify sites in the mosquito genome that had four or five mismatches to the gRNA-dsx on-target site. Two of these 133 sites had also been identified by CIRCLE-seq. While the gene-drive strain may generate and accumulate off-target mutations over time, population-release experiments (where gene drive invades a target wild-type caged population) can saturate these mutations, and thus facilitate the process of identifying off-targets. We performed deep sequencing of all in silico-predicted sites from two distinct generations (G2 and G5) of the zpg-dsxCRISPRh gene-drive population experiment (3). For this the on-target exhibited significant levels of indels generated compared to the wild-type control (SI Appendix, Fig. S2B), while among the 133 predicted off-target sites, none showed evidence of significant Cas9-induced indels relative to the negative control (Fig. 2 and Dataset S4).
Fig. 2.
Assessment of off-target indels within a gene-drive population targeting dsx. Amplicon sequencing was conducted for 133 in silico-predicted off-target sites for gRNA-dsx. The percentage of reads containing indels are displayed. A wild-type mosquito population served as control. Individuals from two generations of a zpg-dsxCRISPRh population experiment were assessed. For each generation two biological replicates were sequenced. Three further off-target sites were sequenced, but not displayed here due to high levels of genetic polymorphism at the cleavage site across both the control and experimental populations.
While the zpg promoter does not prevent end-joining entirely, we showed previously that it can reduce off-target mutagenesis to undetectable frequencies with the gRNA-7280 (Fig. 1). In an attempt to reveal putative off-targets of the gRNA-dsx, we aimed to substantially enhance end-joining by crossing gRNA-dsx–expressing males to vasa-Cas9–expressing females known to deposit Cas9 into the embryo, where end-joining rates are higher than in the germline (SI Appendix, Fig. S6). Among the progeny, we selected the fraction of females that had been heavily mutated at dsx, as evidenced by a mosaic intersex phenotype, and subjected these to targeted amplicon sequencing of the 30 sites identified by CIRCLE-seq. Although mutagenic events occurring in the early embryo could differ from adult germline mutations (i.e., timing of nuclease expression, chromatin structure differences), this system provides a good proxy to investigate off-target effects. In two separate crosses tested (from gRNA-dsx–expressing lines integrated in different genomic locations), the on-target site showed greater than 93% frequency of reads containing indels, indicating high activity of the Cas9/gRNA combination. However, there was no evidence of off-target indels caused by Cas9 cleavage at any of the 30 potential off-target sites sequenced (SI Appendix, Fig. S7 and Dataset S5). We observed a putative off-target site (off-11) containing an indel that was at higher frequency in the sample populations (mutated progeny-A and -C), >10%) compared with the wild-type control (∼1%), suggesting that this allele was preferentially selected in the sample population. A further site (off-19) also contained an indel in the sample populations (that was not in the wild-type), but again was not indicative of mutations introduced by Cas9 cleavage.
Genetic Variation Impacts upon Off-Target Editing.
Recent studies indicate that wild Anopheles mosquito populations exhibit high degrees of polymorphisms across their genomes (10, 11). Several sites that showed off-target mutations within the vas-7280CRISPRh gene-drive population contained allele variants (in the laboratory wild-type population). This could impact upon off-target cleavage by creating sites that more closely resemble the on-target sequence, either within spacer and/or PAM sequences. To test the hypothesis that different natural polymorphisms can impact mutation frequencies at off-target sites, we investigated the relationship between indel frequencies and the presence of SNPs in sites identified by CIRCLE-seq.
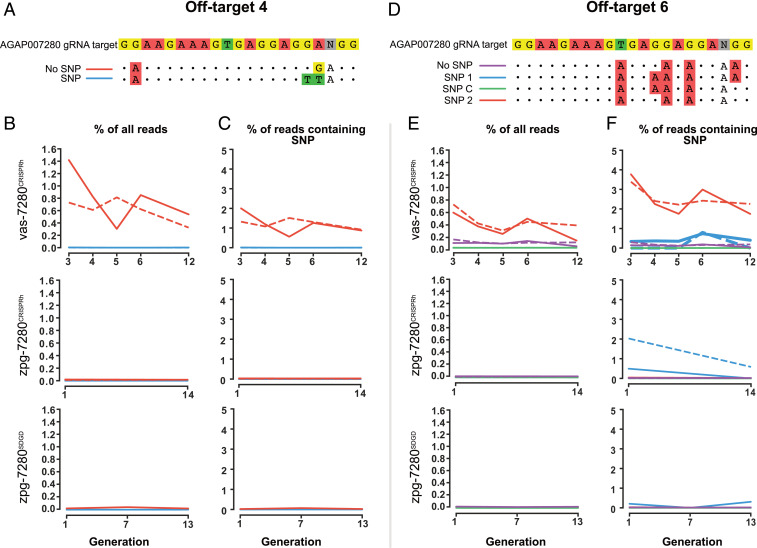
First, amplicon sequencing at the off-4 site revealed two alleles, with the reference allele having two mismatches to gRNA-7280, while an alternative allele had three mismatches to gRNA-7280 (Fig. 3A). When analyzing the frequency of mutations for each allele individually, we observed that only the reference allele showed evidence of cleavage (Fig. 3B and Dataset S6), while no indels were found in the variant allele with three mismatches to the on-target. Frequency of indels detected were normalized according to proportion of each allele within the population (Fig. 3C). Similarly, we identified three variants at off-11, with off-target cleavage occurring exclusively in the alleles that contained fewer mismatches to the on-target site (SI Appendix, Fig. S8).
Fig. 3.
The impact of genetic variation upon CRISPR-Cas9 off-target cleavage. (A) Off-target 4 contained two alleles. (B) Indels were detected in one allele (No SNP) within vas-7280CRISPRh population, with no significant indels detected for zpg-7280CRISPRh or zpg-7280SDGD. This represents editing as a percentage of all reads that align to the off-target site (C) Levels of indels detected are displayed as a percentage of reads that align to each allele, respectively. (D) Off-target 6 contained four alleles. (E) Indels were detected in one allele (SNP 2) which contained an NGG PAM. (F) SNP 2 showed indels for vas2-CRISPRh. Indels found for SNP 1 in zpg-7280CRISPRh are not evidence of genuine off-targeting editing, but artifacts of very low number of reads aligning to this allele. Cage 1 (solid line) and cage 2 (dash line), with zpg-7280SDGD containing one biological replicate only (due to the other cage’s population crashing at G8, as intended as part of the study).
Notably, we identified four alleles at off-6, with a combination of SNPs either modifying the PAM (NAG to NGG), or increasing the number of mismatches to the on-target site (Fig. 3D). The majority of indels were in the allele that restored a canonical NGG PAM, and had fewer mismatches to the on-target site (Fig. 3E). Normalized frequency of indels at this off-target site ranged from 1.75 to 3.75% for the vas-7280CRISPRh populations (Fig. 3F), while no indication of CRISPR-induced indels was observed in zpg-7280CRISPRh or zpg-7280SDGD.
No Evidence Supporting Hypothesized “Drag-Along” Drive of Mutations.
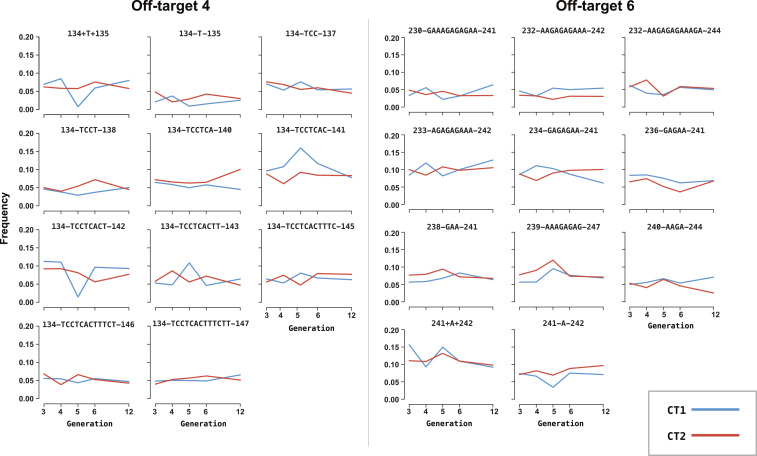
We hypothesized that off-target mutations could potentially increase in frequency over generations and spread within a population alongside the gene drive if the nuclease promotes the off-target mutation to be “dragged-along” by HDR (homing). The detection of bona fide off-target mutations within the vas-7280CRISPRh gene-drive populations allowed for more in depth study as to whether off-target drag-along occurs in vivo. By sequencing multiple consecutive generations (G3, G4, G5, and G6) and a later generation (G12) of a population exposed to the gene drive, we could monitor if any of the off-target indels increased in frequency over time, suggesting a drag-along drive of the mutation via homing at the off-target loci, due to the action of the Cas9 (SI Appendix, Fig. S9). Analyzing the individual frequency of the 10 most-abundant indels found at the earliest generation sequenced (G3), for 3 off-target sites (off-4, off-6, and off-11), and following their frequency over consecutive generations we did not observe any consistent increase in frequency of individual indels (Fig. 4 and SI Appendix, Fig. S10). This suggested that these off-target mutations are not driven through a mosquito population exposed to a gene drive, or if they are, they do not increase in frequency to a level high enough to be distinguished above indels generated at each generation.
Fig. 4.
Indel frequency at off-target 4 and off-target 6 for vas-7280CRISPRh. The top 10 indels witnessed in the G3 were individually followed through subsequent generations to assess if frequency changed over time. The frequencies of the indels were normalized to the total number of reads containing indels for that sample. There are 11 indels displayed, as the top 10 indels in CT1 (cage trial 1) were not identical to those witnessed for CT2 (cage trial 2).
Discussion
Our results show that a homing-based gene drive in An. gambiae can be appropriately designed to show no detectable genomic off-target mutations (i.e., no alterations that rise above the ∼0.1% detection limit of next-generation sequencing-based assays). Following the previously robustly tested methodology for detecting off-target mutations known as VIVO (combining CIRCLE-seq nomination followed by in vivo-targeted deep sequencing) (26), we demonstrated that it was possible to identify and predict off-target edits within the An. gambiae. To do this, a gRNA was used with Cas9 that targeted the AGAP007280 gene and for which there were many sites present in the mosquito genome with three or fewer mismatches. Notably, gRNA-7280 was designed with a truncated spacer of 18 bp, which has been suggested to reduce off-target activity (29), although previous studies have shown that a truncated guide can still induce off-target mutations (18, 29, 30). This was combined with a Cas9 whose expression was regulated by a promoter (vasa), which was not tightly restricted to germline expression and is maternally deposited into the early zygote (5, 27, 31). This maternal deposition of Cas9 has been shown to cause considerable indels at the on-target site for the AGAP007280 gene, due to the accumulation of ribonucleoprotein complexes at a stage when nonhomologous end-joining is favored over HDR (5).
Here we demonstrate that the combination of a potentially more promiscuous gRNA with a nongermline-restricted promoter for Cas9 was able to cause off-target editing at frequencies lower than 1.42%; however, there was no evidence that these mutations were able to propagate through the population by a so-called drag-along drive. A concern expressed for gene-drive technology is that off-target mutations could occur and would be able to increase in frequency over generations and alter the fitness of the transgenic mosquitoes, or have undesired effects on disease transmission or insecticide resistance. A reason hypothesized for the lack of such a phenomenon being present here is that the mutations witnessed were somatic, rather than induced in the germline, and so would not be spread to the subsequent generations. Off-target sites that were present in the context of a vasa promoter-driven gene drive did not show evidence of nuclease-induced editing in gene-drive strains using a tighter promoter (zpg) for the Cas9, even when the gRNA was held constant. The use of the zpg promoter was originally designed to improve the invasion dynamics of the CRISPR homing construct in An. gambiae (27). Reducing the potential for nonhomologous end-joining in the germline and early embryo delayed the onset of resistance and also improved the fecundity of females that were heterozygous for the CRISPR-homing construct (27). By extension, this reduction in the generation of indels at the on-target site could also lead to a reduction in indels at off-target sites. In laboratory populations combining the zpg promoter with a gRNA designed to contain no closely related sites across the mosquito genome (gRNA-dsx), no detectable evidence of nuclease-induced editing was found at in silico-predicted off-target sites. Furthermore, we also did not find any detectable indels indicative of Cas9 cleavage for the gRNA-dsx even when combining it with a vasa-driven source of Cas9 (SI Appendix, Table S3).
Notably, we observed enhanced off-target mutagenesis correlating with natural allelic variants that either reduce the number of mismatches to the target site or convert it to a more cleavable site (i.e., mutations that create a PAM site). This suggests that genetic variation for off-target analysis should be considered when assessing the potential impact of gene editing in large natural populations. An important challenge for future work will be to leverage the increasing availability of genomic data from large samples of wild mosquitoes (10, 11) into off-target analysis.
An important consideration of off-target editing, when applied to gene drives, is the likely consequence of the mutations that occur. In the majority of cases, it is reasonable to predict that off-target edits will not cause any phenotypic consequences, and therefore do not automatically equal harm. The essential risk assessment for the development of gene drive has been proposed to be evaluated on a case-by-case basis for each mosquito strain bearing a drive construct (32). By assessing the transgenic strain for insecticide resistance, reproductive fitness, and vector competence (compared to the wild-type control), one can identify whether the transgenic strain differs in these key areas. In this context, off-target mutations are not necessarily harmful, and the likelihood is that most off-target mutations would be lost naturally from the gene drive if they did not confer any advantage.
This study is, to our knowledge, unique in being an in vivo analysis of off-target mutations for CRISPR-Cas9–based gene drives at the whole-genome level. In summary, we demonstrate that a CRISPR-Cas9 homing gene drive is capable of inducing off-target editing within the malaria mosquito vector An. gambiae. Although off-target effects are dependent on individual gRNA design, our results provide some general considerations that we expect will advance gene-drive research and will help the translation from the laboratory to the field. With prudent selection of the gene-drive target site and restricting nuclease activity to the germline, the propagation of off-target edits within caged mosquito populations can be minimized to undetectable levels.
While it cannot be concluded that these steps alone will completely prevent the generation of off-target mutations in a field-release setting due to the large genetic diversity of wild populations, the potential of effects arising that are harmful to the environment or health, or reduce the efficacy or spread of a drive, can be purposefully minimized using the strategies outlined in this report.
Materials and Methods
In Silico Cas-OFFinder Predictions.
Cas-OFFinder is an in silico tool that was used to identify potential off-targets (Webtool: http://www.rgenome.net/cas-offinder/) (16).This tool functions by assembling all 21- to 23-bp DNA sequences (depending on your query spacer length), that consists of the gRNA sequence (18 to 20 bp) and an NRG PAM. The NGG PAM is canonical; however, research has shown that CRISPR-Cas9 is also able to cleave NAG PAMs, although at around one-fifth of the efficiency of an NGG PAM (33). The algorithm then compares those sequences with your queried sequence, and counts the mismatched bases in the gRNA sequence (16). The parameters used were an NGG or NAG PAM, with up to seven mismatches to the spacer sequence. This was conducted for both the gRNA-7280 (GGAAGAAAGTGAGGAGGA) and the gRNA-dsx (GTTTAACACAGGTCAAGCGG).
Reference Genome for CIRCLE-Seq, CRISPResso, and Cas-OFFinder.
Cas-OFFinder, CRISPResso, and CIRCLE-seq utilized the An. gambiae PEST genome (AgamP4.9) (34) as the reference genome, which was sequenced by shotgun sequencing (35).
In Vitro CIRCLE-Seq.
A full description of the CIRCLE-seq methodology is described in the literature (23), and a brief description is provided here. Genomic DNA was extracted from 800 wild-type mosquitoes using the Wizard Genomic DNA Purification Kit (Promega) and was sheared to an average length of 300 bp (using a Covaris S200 instrument) before being end-repaired, A-tailed, and adaptors ligated. USER treatment of the adapter-ligated precircularization library converts adapter hairpin structures to 4-bp single-stranded overhangs that promote library circularization in the following step of the CIRCLE-seq protocol [for more details, see Tsai et al. (23)]. These DNA fragments are circularized with T4 ligase and the remaining linear DNA is degraded. The in vitro cleavage reactions contained SpCas9, the gRNA of interest (either gRNA-7280 or gRNA-dsx), and the circularized DNA. These reactions were performed at 28 °C [being the temperature the mosquitoes (7, 36) are maintained at in the laboratory]. These digested products were A-tailed and a further adaptor was ligated, before PCR-amplification and library preparation. Libraries were sequenced with 150-bp paired-end reads using an Illumina MiSeq. These data were then processed using v1.1 of the CIRCLE-Seq analysis pipeline (23) (https://github.com/tsailabSJ/circleseq). CIRCLE-seq sequence plots were produced using software published by Tsai et al. (23).
Generation of Mosquitoes Exposed to vasa Cas9 and gRNA-dsx.
Males of two strains, A and C (100 males per strain), containing a randomly integrated U6::gRNA (RFP+) cassette in heterozygosity, were each crossed to 100 female heterozygotes for vas2::Cas9 (YFP+) (2). The RFP+YFP− quarter of the progeny that had inherited the U6::gRNA cassette, but not vas2::Cas9, were selected on the basis of their fluorescence using a COPAS (complex object parametric analyzer and sorter) larval sorter, as in Marois et al. (37). The ubiquitously expressed gRNA present in both strains is complementary to the gene-drive target site on the doublesex gene (3), and if combined with vas2-expressed Cas9, present in the embryo through maternal deposition, they can form an active riboendonuclease complex that can introduce double-stranded breaks that are predominantly repaired through end-joining in the early embryo (28). As a result, female progeny (minimum 48 mosquitoes) showed a partially intersex phenotype due to mosaicism for a doublesex knockout and were selected for pooled amplicon sequencing for on-target and off-target detection of end-joining mutations.
Containment of Gene-Drive Mosquitoes.
All work using GM mosquitoes was performed according to previously described guidelines and protocols (3). The ecological and physical containment of mosquitoes housed in the insectary facility at Imperial College London, meets established safeguards to follow when working with synthetic gene-drive mosquitoes (38).
In Vivo Amplicon Sequencing of vas-7280CRISPRh, zpg-7280CRISPRh, zpg-7280SDGD Populations and vas-dsx (Mutated Progeny-A and -C).
Genomic DNA was extracted using Wizard Genomic DNA Purification Kit from massed pools of mosquitoes (49 to 425 adult). Amplicon sequencing was performed as previously described (3), with the following changes to account for the necessary multiplexing of amplicons for sequencing. Individual PCR reactions were performed per amplicon under nonsaturating conditions (23 cycles), before purification using AMPure XP beads and validated using the fragment Analyzer (High Sensitivity Small Fragment Analysis Kit). The amplicons were then normalized individually at 0.54 nM concentration and pooled into equal volumes. A second PCR step attached the dual indices and Illumina sequencing adapters using the Nextera XT Index kit, with a final AMPure XP beads purification step, with validation and quantification of the final libraries. Each library contained between 10 and 16 amplicons and was sequenced using an Illumina MiSEq. 2 with 2 × 250-bp v2 paired-end run. The wild-type mosquito population was used as the negative control for the amplicon sequencing. We aimed for a minimum of 10,000 sequencing reads per site. Occasionally sites proved tricky to sequence, probably due to the highly repetitive nature of certain loci (X:1286129–1286150 being an example) and fell below the desired minimum of 10,000 reads.
In Vivo zpg-dsxCRISPRh Gene-Drive Amplicon Sequencing.
Genomic DNA from zpg-dsxCRISPRh and wild-type mosquitoes was harvested and used as a template for targeted amplicon sequencing of in silico-predicted off targets. To detect mosaicism down to 1%, we used an average of at least 100 genomes per mosquito as our genomic DNA template and allotted at least 10,000 sequencing reads per site to detect low-frequency events (≤0.01%). Amplicon sequencing of wild-type mosquito gDNA was used as our negative control.
PCRs were performed using Phusion Hot Start Flex DNA polymerase (New England Biolabs). PCR products were then purified using homebrew magnetic beads, quantified using a QuantiFlor dsDNA System kit (Promega), normalized to 10 ng/μL per amplicon and pooled. Pooled samples were end-repaired and A-tailed using an End Prep Enzyme Mix and reaction buffer from NEBNext Ultra II DNA Library Prep Kit for Illumina, and ligated to Illumina TruSeq adapters using a ligation master mix and ligation enhancer from the same kit. Library samples were purified with homebrew magnetic beads, size-selected using PEG/NaCl SPRI solution (KAPA Biosystems), quantified using droplet digital PCR (Bio-Rad), and loaded onto an Illumina MiSeq for deep sequencing.
Amplicon-Sequencing Analysis.
The amplicon-sequencing analysis was performed using the command line version of CRISPRessoPooled 2 software (39) (https://github.com/pinellolab/CRISPResso2) using default parameters. Haplotypes carrying common individual insertion or deletion events at the cut site were grouped together to calculate overall frequency at which a given indel occurred. Furthermore, edited alleles were grouped based on the presence of existing variation around the cut site and their frequency was calculated. The calculations, analysis, and plots were done by using custom Python scripts on data tables with results produced by CRISPResso2.
Statistical Analysis of Amplicon Sequencing.
A two-tailed Fisher exact test was used for comparison between wild-type and nuclease-exposed samples at each off-target site. Indel and nonindel read counts were derived from pooled samples of the same condition and used in the test. Benjamini–Hochberg procedure was used for P value adjustment to account for multiple comparison.
Supplementary Material
Acknowledgments
We thank Tony Nolan for helpful feedback on the manuscript. This work was supported by Grant OPP1159968 from the Bill & Melinda Gates Foundation. J.K.J. and colleagues were supported by additional funding by National Institutes of Health Maximizing Investigators’ Research Award (R35 GM118158). The authors received funding from Defense Advanced Research Projects Agency Safe Genes program (HR0011-17-2-0042) for this research. The views, opinions and/or findings expressed should not be interpreted as representing the official views or policies of the Department of Defense or the US Government. J.K.J. is additionally supported by the Desmond and Ann Heathwood Massachusetts General Hospital Research Scholar Award.
Footnotes
Competing interest statement: J.K.J. holds equity in Chroma Medicine and SeQure Dx, Inc. J.K.J. has financial interests in Beam Therapeutics, Editas Medicine, Excelsior Genomics, Pairwise Plants, Poseida Therapeutics, Transposagen Biopharmaceuticals, and Verve Therapeutics (formerly known as Endcadia). J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict-of-interest policies. K.P., V.P., and J.K.J. are coinventors on various patents and patent applications that describe gene editing and epigenetic editing technologies, including the CIRCLE-seq (circular in vitro reporting of cleavage effects by sequencing) assay used in this study.
This paper results from the NAS Colloquium of the National Academy of Sciences, “Life 2.0: The Promise and Challenge of a CRISPR Path to a Sustainable Planet,” held December 10–11, 2019, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. NAS colloquia began in 1991 and have been published in PNAS since 1995. The complete program and video recordings of presentations are available on the NAS website at http://www.nasonline.org/CRISPR. The collection of colloquium papers in PNAS can be found at https://www.pnas.org/page/collection/crispr-sustainable-planet.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004838117/-/DCSupplemental.
Data Availability
All raw amplicon sequencing files have been deposited in the National Center for Biotechnology Information (NCBI) BioProject (accession code PRJNA665154).
Change History
June 10, 2021: The Acknowledgments have been updated.
References
- 1.Gantz V. M., et al., Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. U.S.A. 112, E6736–E6743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond A., et al., A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyrou K., et al., A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simoni A., et al., A male-biased sex-distorter gene drive for the human malaria vector Anopheles gambiae. Nat. Biotechnol. 38, 1054–1060 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond A. M., et al., The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 13, e1007039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champer J., et al., Reducing resistance allele formation in CRISPR gene drive. Proc. Natl. Acad. Sci. U.S.A. 115, 5522–5527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Academies of Sciences, Engineering, and Medicine , Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values (National Academies Press, 2016). [PubMed] [Google Scholar]
- 8.Hammond A. M., Galizi R., Gene drives to fight malaria: Current state and future directions. Pathog. Glob. Health 111, 412–423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esvelt K. M., Smidler A. L., Catteruccia F., Church G. M., Concerning RNA-guided gene drives for the alteration of wild populations. eLife 3, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anopheles gambiae 1000 Genomes Consortium , Genetic diversity of the African malaria vector Anopheles gambiae. Nature 552, 96–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Anopheles gambiae 1000 Genomes Consortium et al., Genome variation and population structure among 1, 142 mosquitoes of the African malaria vector species Anopheles gambiae and Anopheles coluzzii. bioRxiv:10.1101/864314 (9 December 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lessard S., et al., Human genetic variation alters CRISPR-Cas9 on- and off-targeting specificity at therapeutically implicated loci. Proc. Natl. Acad. Sci. U.S.A. 114, E11257–E11266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L., et al., Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat. Commun. 5, 5507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott D. A., Zhang F., Implications of human genetic variation in CRISPR-based therapeutic genome editing. Nat. Med. 23, 1095–1101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X.-H., Tee L. Y., Wang X.-G., Huang Q.-S., Yang S.-H., Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4, e264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae S., Park J., Kim J. S., Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Concordet J. P., Haeussler M., CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai S. Q., et al., GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ran F. A., et al., In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan W. X., et al., BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 8, 15058, 10.1038/ncomms15058 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frock R. L., et al., Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33, 179–186, 10.1038/nbt.3101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wienert B., et al., Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364, 286–289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai S. Q., et al., CIRCLE-seq: A highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cameron P., et al., Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat. Methods 14, 600–606 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Kim D., et al., Digenome-seq: Genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Akcakaya P., et al., In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561, 416–419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond A., et al., Improved CRISPR-based suppression gene drives mitigate resistance and impose a large reproductive load on laboratory-contained mosquito populations. bioRxiv:10.1101/360339 (1 July 2018).
- 28.Hammond A., et al., Regulation of gene drive expression increases invasive potential and mitigates resistance. bioRxiv, 1–32 (2020). [Google Scholar]
- 29.Fu Y., Sander J. D., Reyon D., Cascio V. M., Joung J. K., Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matson A. W., Hosny N., Swanson Z. A., Hering B. J., Burlak C., Optimizing sgRNA length to improve target specificity and efficiency for the GGTA1 gene using the CRISPR/Cas9 gene editing system. PLoS One 14, e0226107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papathanos P. A., Windbichler N., Menichelli M., Burt A., Crisanti A., The vasa regulatory region mediates germline expression and maternal transmission of proteins in the malaria mosquito Anopheles gambiae: A versatile tool for genetic control strategies. BMC Mol. Biol. 10, 65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James S., et al., Pathway to deployment of gene drive mosquitoes as a potential biocontrol tool for elimination of malaria in sub-Saharan Africa: Recommendations of a scientific working group. Am. J. Trop. Med. Hyg. 98 (6_Suppl), 1–49 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu P. D., et al., DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharakhova M. V., et al., Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 8, R5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holt R. A., et al., The genome sequence of the malaria mosquito Anopheles gambiae. Science 298, 129–149 (2002). [DOI] [PubMed] [Google Scholar]
- 36.NEPAD and the African Union , Gene Drives for Malaria Control and Elimination in Africa (African Union Development Agency, 2018). [Google Scholar]
- 37.Marois E., et al., High-throughput sorting of mosquito larvae for laboratory studies and for future vector control interventions. Malar. J. 11, 302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akbari O. S., et al., BIOSAFETY. Safeguarding gene drive experiments in the laboratory. Science 349, 927–929 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clement K., et al., CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw amplicon sequencing files have been deposited in the National Center for Biotechnology Information (NCBI) BioProject (accession code PRJNA665154).