Significance
Aedes aegypti are mosquitoes that transmit dengue and other viruses that infect millions of people annually. One approach to control Ae. aegypti is through the release of sterile males, which suppresses fertilization by fertile males following mating with sterile males. To generate sterile males, the current approach is to mutagenize mosquitoes nonspecifically, which reduces the health of the males. Here, we used genome editing to selectively disrupt a gene (B2t) that specifically affects male fertility. The mutant males were healthy and were effective in suppressing females from producing progeny with fertile males. These studies raise the possibility that the B2t mutation can be employed to improve the sterile insect technique and reduce diseases spread by Ae. aegypti.
Keywords: Aedes aegypti, mosquitoes, SIT, sterility, dengue
Abstract
Aedes aegypti spread devastating viruses such as dengue, which causes disease among 100 to 400 million people annually. A potential approach to control mosquito disease vectors is the sterile insect technique (SIT). The strategy involves repeated release of large numbers of sterile males, which reduces insect populations because the sterile males mate and thereby suppress the fertility of females that would otherwise mate with fertile males. While SIT has been successful in suppressing certain agricultural pests, it has been less effective in depressing populations of Ae. aegypti. This limitation is in part because of the fitness effects resulting from mutagenizing the mosquitoes nonspecifically. Here, we introduced and characterized the impact on female fertility of an Ae. aegypti mutation that disrupts a gene that is specifically expressed in testes. We used CRISPR/Cas9 to generate a null mutation in the Ae. aegypti β2-tubulin (B2t) gene, which eliminates male fertility. When we allowed wild-type females to first mate with B2t mutant males, most of the females did not produce progeny even after being subsequently exposed to wild-type males. We also introduced B2t mutant and wild-type males simultaneously with wild-type females and found that a larger number of B2t mutant males relative to the wild-type males was effective in significantly suppressing female fertility. These results raise the possibility of employing B2t sterile males to improve the efficacy of SIT in suppressing populations of Ae. aegypti through repeated releases and thereby reduce the transmission of viruses by these invasive mosquitoes.
Aedes aegypti transmit dengue and Zika and other viral pathogens that cause disease among many tens of millions of people each year (1). Moreover, the incidence of diseases spread by this invasive mosquito continues to be on the rise (2). Only females spread disease, and this occurs because they take a blood meal to obtain nutrients needed for egg production. One potential approach for controlling Ae. aegypti is the sterile insect technique (SIT) (3, 4). SIT involves inundating a local population with large excesses of sterile males. Sterile males are thought to cause sterility in females because the initial mating prevents successful insemination by a wild-type male (1, 3). SIT involves repeated release of large numbers of males in a defined geographical region until the number of females falls below a critical threshold necessary for sustaining the population at a high level.
To create sterile males for implementation of SIT, male mosquitoes are exposed to high doses of chemicals or radiation (5). These approaches suffer from the limitation that they do not specifically target male fertility genes (5). Rather, they induce mutations in many genes. This leads to a fitness deficit, reducing the overall robustness of the mosquitoes and the ability of the sterilized males to compete with males in the wild.
In addition to classical SIT, many other approaches are being pursued including the release of fertile male insects with a dominant lethal mutation that kills females (6, 7). A very exciting new technology is the use of CRISPR/Cas9 to create gene drives that suppress mosquito populations (8–12). Despite the great promise of these strategies, the release of fertile transgenic mosquitoes creates public concern due to the potential for unintended consequences resulting from these insects lingering in the environment (13).
In addition to the classical SIT approach, sterility can also be induced by release of male mosquitoes infected with endosymbiotic strains of Wolbachia (14). The bacteria are present in the testis and ovaries and can disrupt reproductive capacity through multiple mechanisms including cytoplasmic incompatibility (incompatible insect technique; IIT), which occurs following mating between an infected male and a female that is not infected with the same strain of Wolbachia (15). While IIT is a very exciting approach with examples of success (16–18), Wolbachia could spread through an indigenous population, thereby interfering with its efficacy, or get introduced into areas that are not intended to be targeted.
The classical SIT approach that involves release of sterile males has multiple advantages that motivate continuing interest in improving this strategy. It has been highly effective in controlling insect pests over the course of 65 y (19, 20), the strategy is environmentally sound (3), and SIT does not suffer from a high level of concern about introduction of transgenes into native insect populations since the released males are sterile. Nevertheless, due to the adverse effects of nonspecific chemical- or radiation-induced sterility, the efficacy of SIT in suppressing mosquito vectors is limited (5). Potentially, SIT could be improved by an alternative, genetic approach using sterile males with a targeted mutation in a single gene that is specifically required for male fertility. However, no male fertility gene has been defined experimentally in mosquitoes.
To identify an Ae. aegypti gene likely to cause male sterility, we considered Drosophila male fertility genes that are conserved in these mosquitoes. The Ae. aegypti β2 tubulin (B2t) gene was a prime candidate for eliminating male fertility, since Drosophila B2t is required for male fertility, is specifically expressed in Drosophila sperm (21–23), and the Drosophila and Ae. aegypti proteins are 96% identical. Moreover, the Ae. aegypti B2t gene appears to be expressed specifically in testes (24). In this study, we used CRISPR/Cas9 to knock out the B2t gene. The Ae. aegypti B2t mutant males were sterile and were as robust as wild type in terms of all parameters measured, except for the sterility. Of primary importance, the B2t mutant males were effective in suppressing female fertility. These data indicate that mutation of B2t provides a targeted genetic strategy for improving SIT.
Results
Creation of a Mutation in Ae. aegypti B2t Gene Using CRISPR/Cas9.
To identify a candidate gene in Ae. aegypti that might cause male sterility, we considered genes in Drosophila melanogaster that are specifically expressed in testis, are required for male sterility, and conserved in Ae. aegypti. A Drosophila gene that fits these criteria is the β2-tubulin at 85D (B2t) gene (21–23). Loss of Drosophila B2t causes sterility due to a failure to produce sperm (21–23). The Ae. aegypti AAEL019894 gene was a prime candidate for causing male sterility since it encodes a protein that is ∼96% identity to Drosophila B2t. We refer to AAEL019894 as B2t.
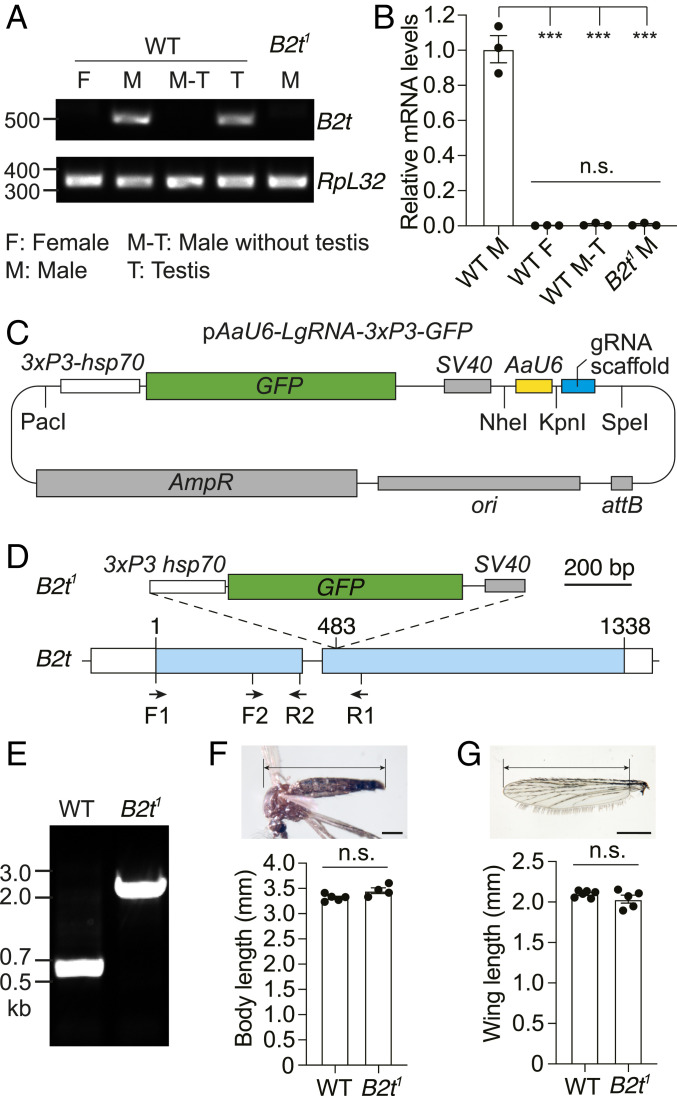
To confirm that the Ae. B2t gene was expressed specifically in testes (24), we performed RT-PCR. We prepared RNA from males and females and observed a PCR product exclusively in males (Fig. 1A). Moreover, the signal was eliminated when we removed testes from the males but was evident in RNA prepared from testis alone (Fig. 1A). In contrast, expression of a control gene (RpL32) (25, 26) was similar in all samples (Fig. 1A). To perform the assay quantitatively, we performed real-time RT-PCR and detected a signal in intact males but not males in which we removed the testis (Fig. 1B). Therefore, within the resolution of this sensitive analysis, we confirm that B2t is expressed specifically in testes.
Fig. 1.
Generation of the B2t1 knock-in mutant and examination of testis-specific expression of B2t. (A) RT-PCR showing B2t mRNA expression in the indicated samples and sexes from wild type (WT) and B2t1. The locations of the primers used (F1 and R1) are illustrated in D. RpL32 is used as an internal control. The expected PCR products are 527 bp for B2t and 345 bp for RpL32. F, female; M, male; M-T, male without testis; T, testis. (B) Real-time RT-PCR showing relative B2t transcript levels in the indicated samples. The primer pair used (F2 and R2) is presented in D. Three biological replicates, each with three technical replicates, were used for each sample. Means ± SEMs. Statistics were performed using one-way ANOVA with the Tukey’s multiple comparisons test. n.s., not significant; ***P < 0.001. (C) Schematic of the pAaU6-LgRNA-3xP3-GFP plasmid used for engineering the knock-in constructs for CRISPR/Cas9-mediated genome editing of B2t. The PacI and NheI restriction sites were used to introduce the 5′ and 3′ homologous arms in B2t, respectively. The KpnI and SpeI restriction sites were used to clone the gRNA. The vector contains the following components: 3xP3-hsp70, three P3 binding elements and a minimal promoter from the Drosophila hsp70 gene; GFP, coding sequence for green fluorescent protein; SV40, SV40 transcriptional terminator; AaU6, Ae. aegypti U6 promoter; gRNA scaffold; attB sequence; ori, origin of replication; and ampR, ampicillin-resistant gene. (D) Schematic showing the 3xP3-GFP-SV40 knocked into the B2t gene to create the B2t1 allele. The location of the primers used in A, B, and E (F1, F2, R1, and R2) are indicated. (Scale bar, 200 bp.) (E) PCR genotyping of the B2t1 allele. The locations of the primers used are F1 and R1 and are presented in D. The expected PCR products are 584 bp in WT and 1.9 kb in B2t1. (F) Body sizes of WT and B2t1 mutant males. The image illustrates the area used for the length measurements. n = 4 to 5 groups. Each group included ≥5 mosquitoes. The total number of mosquitoes measured was 60 for WT and 32 for B2t1. Means ± SEMs are indicated. Statistics were performed using Mann–Whitney U test. n.s., not significant. (Scale bar, 500 μm.) (G) Wing lengths of WT and B2t1 mutant males. The image illustrates the area used for the length measurements. n = 5 groups. Each group included ≥10 wings. The total number of wings measured was 71 for WT and 101 for B2t1. Means ± SEMs are indicated. Statistics were performed using Mann–Whitney U test. n.s., not significant. (Scale bar, 500 μm.)
To investigate whether the Ae. aegypti B2t homolog is required for male fertility, we used CRISPR/Cas9 genome editing to generate a B2t1 allele. To do so, we first created a vector (pAaU6-LgRNA-3XP3-GFP), which encodes an eye-specific GFP marker (3xP3-hsp70-GFP-SV40) to identify mosquitoes with the insertion and a guide RNA (gRNA) scaffold expressed under control of an Ae. aegypti U6 (AAEL017774) promoter (27, 28) (Fig. 1C). The gRNA scaffold was modified from a standard sequence (28) to reduce premature termination and improve the gRNA stability controlled by an Ae. aegypti U6 (AAEL017774) promoter (27, 28). We identified an effective gRNA with no predicted off-target sequences using CRISPR Optimal Target Finder (targetfinder.flycrispr.neuro.brown.edu). We then introduced the gRNA in the vector flanked by two genomic DNA fragments from the B2t gene. The B2t1 knockout included the gene coding for GFP so that it interrupts the protein-coding sequence in the second exon after amino acid 160 of B2t (Fig. 1D). We injected the vector into a line that expresses Cas9 in the germline (29) and identified putative knockouts on the basis of GFP expression in the eyes. We then confirmed the B2t knockout by PCR (Fig. 1E) and DNA sequencing. On the basis of both PCR and RT-PCR using the F1 and R1 primers (Fig. 1D), the B2t RNA was eliminated in the B2t1 mutant (Fig. 1 A and B). Due to the disruption of the coding region by the insertion, and because the levels of the messenger RNA (mRNA) were undetectable, we conclude that the B2t1 mutant is a null allele. We outcrossed this line for six generations to the wild-type control (Liverpool).
The overall sizes and wing lengths of Ae. aegypti and related species have been shown to be excellent indicators of the overall health of the mosquitoes (30–33). Therefore, we measured these parameters and found that the lengths of their bodies and wings of B2t1 males were indistinguishable from wild-type mosquitoes (Fig. 1 F and G). In addition, in crosses between B2t1/+ males and females, the expected percentage of the total progeny larvae, pupae, and adults (∼25%) were B2t1 homozygous mosquitoes (Table 1). We found that the B2t1 mutants displayed similar development times to wild-type mosquitoes. The number of days required for the eggs to develop into pupae and to adults were equivalent to wild type (Table 1). Additionally, the median time for 50% of the group population to die for sex-separated B2t1 males was 51.0 ± 4.0 d, which was not significantly different from wild type (43.3 ± 1.5 d). Thus, based on size, developmental time, and survival, the B2t1 males were healthy.
Table 1.
Life table for B2t1, B2t1/+, and +/+ mosquitoes
| Developmental stage | Genotype | No. | Developmental time (days) |
| Eggs (from B2t1/+ ♀ × ♂) | 400 | − | |
| B2t1 | 85.7 ± 1.3 | − | |
| Fourth instar larvae | B2t1/+ | 164.0 ± 6.1 | − |
| +/+ | 93.7 ± 0.9 | − | |
| B2t1 | 80.3 ± 2.3 | 6.2 ± 0.2 | |
| Pupae | B2t1/+ | 155.7 ± 8.7 | 6.4 ± 0.2 |
| +/+ | 90.0 ± 1.5 | 6.5 ± 0.2 | |
| B2t1 | 78.7 ± 3.5 (♀ to ♂ ratio: 0.873 ± 0.024) | 8.4 ± 0.4 | |
| Adults | B2t1/+ | 155.3 ± 8.4 (♀ to ♂ ratio: 0.897 ± 0.045) | 8.5 ± 0.4 |
| +/+ | 91.7 ± 1.3 (♀ to ♂ ratio: 0.887 ± 0.067) | 8.5 ± 0.2 |
A total of 400 eggs from the cross of B2t1/+ heterozygous males and females were hatched for each group. Three groups were assayed. B2t1/+, B2t1, and +/+ progeny were distinguished and sorted using the eye fluorescent marker during the fourth instar larval stage. The number of pupae and adults, the genotypes, and the time of development are indicated. Means ± SEMs. There were no significant differences between the various genotypes in development times, adult female to male ratios, and in the number of Bt21 and Bt21/+ genotypes at any developmental stage. Statistics were performed using one-way ANOVA with the Tukey’s multiple comparisons test.
B2t Mutation Eliminates Sperm Production and Causes Male Sterility.
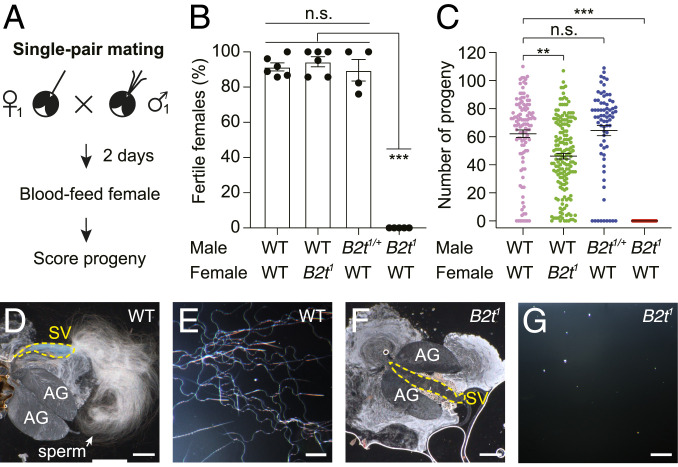
To determine whether the B2t1 mutation disrupted male fertility, we performed single-pair matings. Each assay entailed combining one male and one female in ≥15 individual vials for 2 d. We then allowed the females to blood feed and scored the resulting progeny several days later (Fig. 2A). This assay was effective as the vast majority of vials that contained wild-type males and females contained progeny (91.5 ± 2.3%). Crosses between B2t1/+ heterozygous males and wild-type females produced progeny in a similar percentage of vials (Fig. 2 B and C; 89.6 ± 6.1%). In contrast, when we combined B2t1 mutant males with wild-type females, 0% of the vials contained progeny (Fig. 2 B and C). These results demonstrate that B2t1 male mosquitoes were sterile. Consistent with expression of B2t exclusively in male testes, nearly all B2t1 mutant females were fertile (Fig. 2B; 94.4 ± 2.9%), although in some crosses there were fewer progeny per vial (Fig. 2C). This effect might be due to slight toxicity of the GFP transgene or to a background mutation, although we outcrossed the B2t1 mutant with the control (Liverpool) for six generations.
Fig. 2.
B2t1 mutant males are sterile. (A) Schematic of the single-pair mating assay. One male and one female were allowed to mate for 2 d, and the females were then blood fed. The males were then removed, and the progeny were scored 13 to 15 d later. (B) Percentage of fertile females from the single-pair mating assays. n ≥ 4 groups. Each group consists of ≥15 single-pair matings. Means ± SEMs. Statistics were performed using one-way ANOVA with Tukey’s multiple comparisons tests. (C) Numbers of progeny produced from the single-pair mating assay. The raw data sets are the same as in B. Each data point represents the number of progeny from an individual mating. The number of individual crosses were 113, 168, 72, and 149 with the parents indicated from left to the right, respectively. Means ± SEMs. Statistics were performed using Kruskal–Wallis test with the Dunn’s multiple comparisons test. (D and E) Wild-type (WT) male seminal vesicles contain mature sperm. Seminal vesicles were torn open to release the content inside. AG, accessory gland; SV, seminal vesicle. (F and G) B2t1 seminal vesicles lack mature sperm. (The scale bars in D and F represent 250 μm and the scale bars in E and G represent 50 μm.) n.s., not significant. P > 0.05. **P ≤ 0.01. ***P ≤ 0.001.
In Drosophila, B2t is essential for sperm production (21–23). Therefore, we tested whether the Ae. aegypti mutant males failed to produce sperm. Mature sperm are produced in the testis and transferred to the seminal vesicles, where they are stored prior to ejaculation during mating (34). Juxtaposed to the seminal vesicles are accessory glands that produce the seminal fluid (34). To determine whether there is an impact on sperm production, we opened the seminal vesicles. Seminal vesicles from wild-type males were filled with sperm (Fig. 2 D and E). However, the B2t1 seminal vesicles were devoid of sperm (Fig. 2 F and G), explaining the basis for the sterility of the mutant males. Thus, while the B2t1 mutant males were sterile, within the resolution of the parameters tested as described above, they were as robust as wild-type males.
Prior Mating with B2t Males Suppresses Female Fertility.
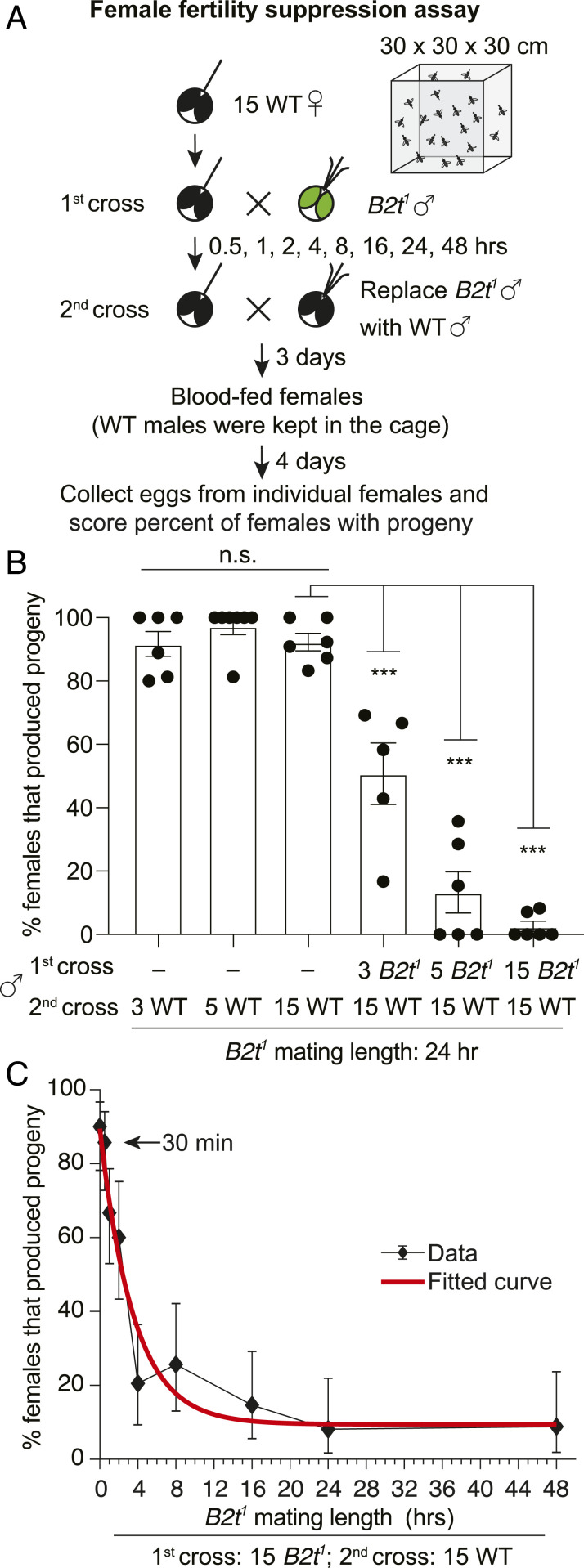
SIT involves inundating a local insect population with sterile males, thereby suppressing female fertility. To address whether prior mating with B2t1 males would inhibit female fertility, we performed assays in 30 × 30 × 30 cm cages (Fig. 3A). We introduced ∼15 wild-type virgin females in each cage for 24 h. As a control, we added either 3, 5, or 15 wild-type males for 24 h, allowed the females to blood feed, and subsequently scored the percentage of females that produced progeny. Under these conditions, addition of 3 males was as effective as 5 or 15 males in yielding a very high percentage of females that produced progeny (Fig. 3B). We did not use single males in these assays since the occasional male that is refractory to mating due to a subtle injury that is difficult to detect would result in 0% of the females yielding progeny.
Fig. 3.
Mating with B2t1 mutant males suppresses female fertility. (A) Schematic of the female fertility suppression assay performed in a 30 × 30 × 30 cm cage. B2t1 males were allowed to mate with 15 wild-type (WT) females for different lengths of time (first cross). The males were then removed. In some experiments, 15 WT males were then added (second cross) and retained for the duration of the experiment. After 3 d, the females were allowed to blood feed. They were then kept in the cage for an additional 4 d before transferring individual females to vials so that progeny could be scored. (B) The percentage of females that produced progeny following the first and second crosses with the indicated numbers of WT or B2t1 males. n ≥ 5. Means ± SEMs. Statistics were performed using one-way ANOVA with Tukey’s multiple comparisons tests. ***P < 0.001. (C) The percentage of females that produced progeny following the first cross with 15 B2t1 males for different lengths of time and a second cross (of the same females) with 15 WT males. n ≥ 3. Means ± 95% CI. The red curve indicates the fitted curve.
To determine if prior mating by B2t1 males suppresses female fertility, we performed cage assays. We allowed 15 wild-type virgin females to acclimate to the cage for 24 h and then added either 3, 5, or 15 B2t1 males for 24 h (Fig. 3A). We then replaced the mutants with 15 wild-type males, which we retained in the cage for 7 d until we moved individual females to vials so that we could score the percentage of females that produced progeny. We found that addition of three B2t1 males suppressed the number of females bearing progeny from >90% to 50.7 ± 9.7%. Increasing the number of B2t1 males to 5 resulted in greater suppression (13.3 ± 6.5%), while preexposure of the females to 15 B2t1 males either eliminated female fertility or resulted in very low percentages of females that produced progeny (2.6 ± 1.6%; Fig. 3B). The profound effect on fertility occurred even though we found that most of the wild-type males copulated with females within 30 min of their addition to the cages containing the females that were preexposed to the B2t1 males. These data demonstrate that the B2t1 males are able to suppress female fertility, especially if the females are first exposed to large numbers of the mutant males.
We then tested the relationship between the time that the B2t1 males were allowed to first mate with wild-type females and the percentage of females with progeny. We added 15 B2t1 males to cages with 15 wild-type females for different lengths of time (t = 0, 0.5, 1, 2, 4, 8, 16, 24, and 48 h) (Fig. 3 A and C) before replacing them with 15 wild-type males. In these experiments, >90% of the females produced progeny if they were not exposed to B2t1 males (Fig. 3C; t = 0; 90.8 ± 4.1%). Upon addition of the B2t1 males to a cage with females, the males quickly began copulating with the females. We counted the number within the first 10 min and found that they copulated 26.5 ± 1.9 times (SEM, n = 4 cages), which averaged ∼1.8 copulations per female. This value was not statistically different from wild-type males (33.7 ± 2.4 times, SEM, n = 4 cages, nonparametric Mann–Whitney U test). While B2t1 males repeatedly mated with the females, we surprisingly found that 30 min of preexposure of wild-type females to B2t1 males had virtually no effect on female fertility (Fig. 3C). Two h of exposure to B2t1 males was required to significantly reduce female fertility to 59.5 ± 10.2%, while 4 h with B2t1 was needed to further reduce the female fertility to 20.5 ± 9.2% (Fig. 3C). The suppression reached saturation by 24 h, at which point the female fertility had decreased to 7.7 ± 4.4%, which was virtually the same as the suppression of 7.5 ± 4.1% after 48 h (Fig. 3C). We found that the relationship of female fertility and the time of preexposure to B2t1 males to the females fit an exponential decrease (red curve; Fig. 3C).
Suppression of Female Fertility by Cointroduction of B2t1 and Wild-Type Males.
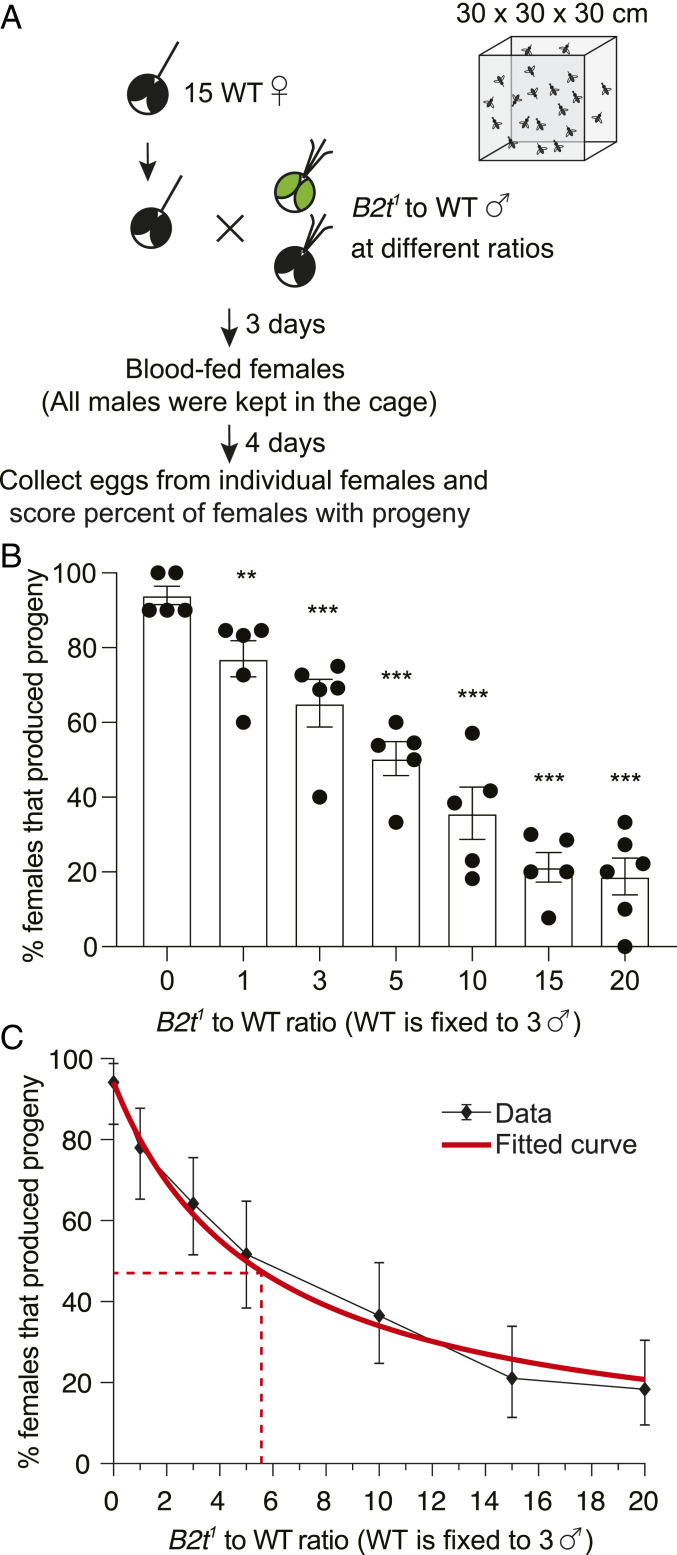
Next, we tested the effectiveness in suppressing female fertility by exposing females to different ratios of B2t1 and wild-type males introduced at the same time (Fig. 4A). We inserted ∼15 virgin wild-type females in cages along with a mixture of B2t1 and wild-type males (Fig. 4A). The males were retained in the cage for 7 d until we moved individual females to vials for egg collections. We included three wild-type males in each experiment and added increasing numbers of B2t1 males to obtain different ratios of B2t1 to wild-type males. Addition of a 1:1 ratio resulted in a modest but significant decrease in in the number of fertile females (Fig. 4B; no B2t1, 0:1 ratio, 94.0 ± 2.4%; 1:1 ratio, 77.1 ± 4.8%), while a 5:1 ratio reduced the percent of fertile females to 50.3 ± 4.5%. A 15:1 ratio suppressed the percent of fertile females to 21.2 ± 4.0%, and a further ratio increase to 20:1 caused only a small additional decline of fertile females to 18.8 ± 4.9%, which was not significantly different from with the 15:1 ratio (Fig. 4B). While the number of fertile females was suppressed, the numbers of eggs and larvae produced per fertile female were similar in each of these experiments (Table 2). We performed curve fitting and found that an average of 5.6 B2t1 males was needed to compete with one wild-type male to achieve a 50% reduction in female fertility from 94 to 47% (Fig. 4C).
Fig. 4.
B2t1 mutant males compete with wild-type (WT) males to suppress female fertility. (A) Schematic of the mating competition assay performed in a 30 × 30 × 30 cm cage. B2t1 and WT males were introduced simultaneously to mate with 15 WT females. The males were kept in the cages with females until egg collection. (B) The percentage of females that produced progeny following mating with males of indicated B2t1 to WT ratios. n ≥ 5. Means ± SEMs. Statistics were performed using one-way ANOVA with Tukey’s multiple comparisons tests. ***P < 0.001. (C) Curve fitting for the same dataset from B. n ≥ 5. Means ± 95% CI. The dashed lines indicate the point at which the B2t1 to WT ratio (∼5.6) leads to a 50% decrease in the number of fertile females (47%) from the 94% female fertility obtained when the females are exposed to WT males only.
Table 2.
Egg, larval production, and hatch rate of individual females from matings that included addition of wild-type and Bt21 mutant males at the same time
| B2t1 to wild-type male ratio | No. of eggs per female | No. of larvae per fertile female | Hatch rate of eggs from fertile females (%) |
| 0:1 | 75.1 ± 2.5 | 40.0 ± 3.5 | 30.3 ± 2.3 |
| 1:1 | 63.0 ± 4.8 | 36.2 ± 4.5 | 33.8 ± 3.2 |
| 3:1 | 62.5 ± 4.1 | 37.2 ± 5.0 | 32.6 ± 2.5 |
| 5:1 | 74.2 ± 2.0 | 57.6 ± 5.9 | 41.3 ± 1.6 |
| 10:1 | 61.7 ± 5.1 | 35.6 ± 7.0 | 31.9 ± 4.1 |
| 15:1 | 66.2 ± 4.3 | 45.5 ± 6.0 | 35.4 ± 3.1 |
| 20:1 | 64.8 ± 3.8 | 29.6 ± 4.7 | 29.6 ± 4.8 |
Shown are the raw data from the mating assays presented in Fig. 4 in which the B2t1 and wild-type males were introduced to the females simultaneously. The number of eggs produced by individual females, the number of larvae from each fertile female, and the egg hatch rate of the fertile female were calculated. There were no significant differences between the control (0:1) and other B2t1 to wild-type ratio conditions in terms of egg production, the number of larvae per fertile female, and the hatch rate of eggs from fertile females. Statistics were performed using one-way ANOVA with the Tukey’s multiple comparisons test.
Discussion
SIT has been successfully used for decades to control agricultural pests such as the New World screwworm and the Mediterranean fruit fly (35, 36). Attempts to use SIT to control mosquitoes have been ongoing since the 1950s (4). However, the use of nonspecific chemical and radiation approaches to create sterile male mosquitoes mutagenizes many genes and consequently has fitness effects that have limited this approach from achieving a high level of success (1, 5, 37).
In this work, we used CRISPR/Cas9 to target the B2t gene in Ae. aegypti. The mutant Ae. aegypti males are sterile, and this occurs due to lack of sperm production, similar to the effect of the mutation in Drosophila (21–23). When we allowed wild-type females to mate first with the B2t mutant males, the females were rendered sterile since they did not produce progeny. Due to the absence of sperm, it was initially unclear how effective B2t mutant males would be in causing female sterility. In mosquitoes, there are indications that sperm from sterile male mosquitoes might have to be transferred to suppress fertility since the sperm may be needed to compete with wild-type sperm and prevent fertilization (4, 38). Nevertheless, multiple studies indicate that transfer of seminal fluid from Ae. aegypti males is sufficient to cause female sterility (39–41). This is distinct from Drosophila, since sperm have to be transferred during the first mating to maintain a female’s resistance to remating for more than 1 d (42, 43). Thus, even if the Ae. aegypti females were initially resistant to fertilization of eggs by wild-type sperm following an initial mating with sperm-free males, it was unclear if this female sterility would persist.
The cage assays described in this study were conducted by combining the females with B2t mutant males for 1 d and then subsequently exposing the females to wild-type males until the end of the assay (7 d) when the females were transferred to individual vials to determine whether they produced progeny. Thus, our findings not only strongly support the conclusion that sperm transfer is not required for inducing sterility in female Ae. aegypti (39–41) but demonstrate that the female sterility persists for at least 7 d. Thus, as previously indicated, transfer of accessory gland proteins to the females from the B2t1 males appears to be sufficient to prevent the females from producing progeny even if they copulate with wild-type males.
SIT involves inundating a local environment with an indigenous population of mosquitoes, such as Ae. aegypti, with a large excess of sterile males. Therefore, both wild-type and sterile males are present and simultaneously competing for female mates. We found that a 15:1 excess of the B2t1 mutant males suppressed the percentage of fertile wild-type females to ∼20%. By curve fitting the experimental data, an average of 5.6 B2t1 males reduced female fertility by 50%. These data demonstrate that an excess of B2t1 males suppresses female fertility in the presence of wild-type males.
We suggest that more than five sterile males are required for a 50% reduction in female fertility and a 15-fold excess for suppression to 20% fertility because the females have to mate many times with sterile males before they become refractory to remating. In support of this proposal, we found that when we combine B2t1 males only with wild-type females for just 30 min, the females mate multiple times with the sterile males. However, upon replacing the sterile males with wild-type males, the 30-min exposure to the B2t1 males was not sufficient for any notable suppression of female fertility, even though the females already mated repeatedly with the sterile males. Rather, 4 h of preincubation with the B2t1 males lead to a large reduction in female fertility. We propose that the 4-h time window and an average of 5.6 sterile males per female is required for a 50% suppression since females need to mate multiple times with sterile males over the course of several hours before they resist mating with a wild-type male. Since SIT involves the repeated release of excess males, and a 15:1 excess of Bt21 males results in 80% suppression of female fertility, these findings suggest that successive release cycles of B2t mutant males would be effective in reducing the levels of Ae. aegypti to below a critical threshold. Since mutation of Bt2 only affects male fertility, while irradiated sterile Ae. aegypti males have fitness costs (5), we suggest that fewer B2t sterile males would need to be released than irradiated males to achieve effective suppression. However, the precise ratios of released irradiated Ae. aegypti males relative to indigenous males have been difficult to establish due to challenges in establishing accurate numbers of Ae. aegypti males in the wild.
A key next step to employ B2t mutant males for SIT is to overcome the challenge that the mutant males cannot be maintained as a homozygous line since they are sterile. We propose that this issue can be circumvented by inclusion of a drug-inducible wild-type B2t transgene so that large numbers of B2t homozygous mutant males can be propagated in the laboratory. The B2t sterile males could also be combined with transgenic approaches used in other insects to produce only males (44, 45). An additional question is whether mutation of B2t can be employed to improve SIT in other mosquitoes. Along these lines, the B2t proteins are highly conserved in Anopheles species that spread malaria, including ∼94 to 95% identities in Anopheles gambiae and Anopheles stephensi B2t. Thus, we propose that mutation of B2t offers great potential for improving the efficacy of SIT in a wide range of mosquito disease vectors.
Materials and Methods
Mosquito Rearing.
Ae. aegypti strains were reared at 28 °C, 80% humidity, and under 14-h light/10-h dark cycles. The adult mosquitoes were maintained with 10% sucrose in 15 × 15 × 15 cm cages (BugDorm-4M1515 insect rearing cage, MegaView Science Co., Ltd.). For egg production, females were fed defibrinated sheep blood (HemoStat Laboratories, DSB250) at 37 °C using the Hemotek membrane blood-feeding system (Hemotek, SP6W1-3). Eggs were hatched, and larvae were grown in deionized water with fish food (TetraMin Tropical Granules, Tetra, 98531). The wild-type line used in the study was the Ae. aegypti Liverpool strain.
Molecular Cloning.
To create the construct used for CRISPR/Cas9-mediated genome editing, we first engineered a plasmid (pAaU6-LgRNA-3xP3-GFP; Fig. 1C) that encodes an eye-specific GFP marker (3xP3-hsp70-GFP-SV40) to identify the edited mosquitoes and a gRNA scaffold expressed under control of an Ae. aegypti U6 (AAEL017774) promoter (27, 28). The gRNA scaffold was slightly modified from a standard sequence (46) to reduce premature termination and improve the gRNA stability (N indicates target sequences, underline indicates modifications): NNNNNNNNNNNNNNNNNNNNGTCTTAGAGCTATGCTGGAAACAGCATAGCAAGTTAAGATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGC.
To generate the plasmid construct for creating the B2t1 allele, we first introduced the B2t gRNA target sequence (5′-AATGTGTTCATAATACGATC-3′) into pAaU6-LgRNA-3XP3-GFP. To achieve this, we PCR-amplified the gRNA containing the target sequence and scaffold using pAaU6-LgRNA-3xP3-GFP as a template with the following primers (gRNA target sequence is underlined): forward: 5′-AAGAGTAGTGAAATGAATGTGTTCATAATACGATCGTCTTAGAGCTATGCTGGAAACAG-3′; and reverse: 5′-GCCGCTCTAGAACTAGTGGATCCCCC-3′. We then subcloned this gRNA into the pAaU6-LgRNA-3xP3-GFP digested with KpnI and SpeI (the digestion removes the gRNA scaffold from the vector) using the In-Fusion Cloning system (Takara, In-Fusion HD Cloning Plus kit). Next, we PCR-amplified two homologous arms from genomic DNA prepared from adult Ae. aegypti (left arm, 1.06 kb upstream of the target site; right arm, 0.86 kb downstream of the target site) and subcloned them in the PacI and NheI sites in the vector, respectively. The primer sequences are as follows: Left_Arm-F, 5′-CGCGCACATTTCCTTAATTAAAGGACTTGCATGTTTCCCCTAATGTGG-3′ and Left_Arm-R, 5′- TCTTAACGCGAGTTAATCTGGATACTCTTCTCGAATCTTCGAA-3′; Right_Arm-F, 5′-GGTATCGATAAGCGCTAGGATCGTATTATGAACACATTTTCTGTCG-3′ and Right_Arm-R, 5′-GGCGATTTCATTCGCTAGCTATTCCTCGCCACCCTCCTCTTCTTC-3′.
Generation of B2t1 Mutant Ae. aegypti.
To generate the Ae. aegypti B2t1 strain, we injected the pB2t1 plasmid at a concentration of 500 ng/μL into ∼700 ubiL40-Cas9 embryos that express Cas9 (29). A total of 7 female and 11 male G0 mosquitoes survived. We then crossed individual G0s to the wild type of the opposite sex and harvested the F1 eggs. We hatched the eggs and screened the larvae for transformants on the basis of GFP expression in the eyes using a stereo fluorescence microscope (Zeiss SteREO Discovery. V8). We obtained 69 positive F1 progeny (from three female G0s). We established three transgenic lines (line #1, #2, and #3), each originated from one G0 female. We confirmed that all three lines had the desired mutation by PCR and DNA sequencing. The primers used for the PCR were F1, 5′-ATGCGTGAAATCGTTCACATTCAAGCCG-3′ and R1, 5′-GACACCTTCGGCGAAGGAACGACAGAA-3′. Since all three lines were identical, we arbitrarily selected line #1 to outcross to the wild-type Ae. aegypti Liverpool strain for six generations. We used the resulting outcrossed line for all experiments in this study. The B2t1 strain is kept as a mixture of heterozygotes and homozygotes, since homozygous males are sterile. The homozygous B2t1 mosquitoes were distinguished from heterozygotes on the basis of GFP intensity.
Imaging of Seminal Vesicles.
For visualization of mature sperm, seminal vesicles were dissected and torn open to release the contents. The samples were imaged using a Zeiss Axio Zoom.V16 stereomicroscope, and the images were processed with Zeiss ZEN software.
RT-PCR and qRT-PCR.
RNA was isolated from mosquitoes using TRIzol Reagent (Thermo Fisher Scientific). cDNAs were generated using the SuperScript First-Strand Synthesis System (Thermo Fisher Scientific) and then treated with DNase I (Thermo Fisher Scientific). For RT-PCR, we used the F1 and R1 primers (Fig. 1D), performed 37 PCR cycles (95 °C for 25 s, 62 °C for 30 s, 72 °C for 30 s), and the DNA products were detected on an agarose gel. To perform real-time qRT-PCR, we used the F2 and R2 primers (Fig. 1D) and performed 41 cycles using the LightCycle 480 SYBR Green I Master Mix (Roche). Each of the three biological replicates for the qRT-PCR included three technical replicates. The internal control for both the RT-PCR and qRT-PCR was RpL32 (AAEL003396) (25, 26). Primer sequences and corresponding produce sizes are as follows: B2t-F1, 5′-ATGCGTGAAATCGTTCACATTCAAGCCG-3′ and B2t-R1, 5′-GACACCTTCGGCGAAGGAACGACAGAA-3′, 527 base pairs (bp); B2t-F2, 5′-ATGCGTGAAATCGTTCACATTCAAGCCG-3′ and B2t-R2, 5′-GACACCTTCGGCGAAGGAACGACAGAA-3′, 148 bp; and RpL32-F, 5′-CCAAGATCGTCAAGAAGCGG-3′ and RpL32-R, 5′-GGTTGGTCACAGCGATGG-3′, 345 bp.
Body Size and Wing Size Measurements.
To measure the body and wing sizes of mosquitoes, we captured images using a Zeiss Axio Zoom.V16 stereomicroscope imaging system and performed the measurements using Zeiss ZEN software. To obtain the body size measurements, each group was the average length of ≥5 mosquitoes cultured in the same batch. Four to five groups were measured. For wing length measurements, each group was the average length of ≥12 wings from mosquitoes cultured in the same batch. Five groups were measured.
Assessment of the Life Cycle of the B2t1 Mutant.
Due to the sterility of the B2t1 males, we performed the analysis of the B2t1 life cycle using eggs obtained from inter se crosses using B2t1/+ heterozygous males and females. In each group, 400 eggs that were 4-d postcollection were hatched in 1 L deionized water supplemented with fish food. Surviving larvae were sorted during the fourth instar stage using the eye-specific GFP marker. The +/+ wild-type mosquitoes did not express GFP while the GFP intensity was stronger in the B2t1 homozygotes relative to the B2t1/+ heterozygotes. The number of mosquitoes of the indicated genotypes during the pupal and adult stages were counted, and the time for pupal and adult development was recorded.
Time for 50% Survival of Adult Mosquitoes.
To assess the viability of the B2t1 mutant relative to wild type, ∼30 newly enclosed males of each genotype were placed in a 15 × 15 × 15 cm cage (BugDorm-4M1515 insect rearing cage, MegaView Science Co., Ltd.) supplemented with 10% sucrose. The numbers of dead mosquitoes were recorded every 2 to 3 d. We then recorded the number of days that elapsed for 50% of the mosquitoes in the cage to die. Three groups of mosquitoes were assayed for each genotype.
Single-Pair Mating Assays.
The assays were performed using 5- to 8-d old sex-separated mosquitoes. One male and one virgin female were transferred to a vial (Narrow Drosophila Vials, Genesee Scientific, catalog no. 32-109) padded with wet cotton at the bottom. The mosquitoes were allowed to mate for 2 d, and the females were released into 15 × 15 × 15 cm cages (BugDorm-4M1515 insect rearing cage, MegaView Science Co., Ltd.) for blood feeding. We then transferred the females into individual vials and collected eggs and scored the number of progeny (larvae). Females that did not blood feed well (did not show an engorged abdomen) were excluded from the study. Each group (≥4), which consisted of ≥15 single-pair mating crosses, was assayed per the indicated condition. The percent fertility of each group was calculated.
Testing for Suppression of Female Fertility after Incubating Wild-Type Females with B2t1 Males before Replacing Them with Wild-Type Males.
The assays were performed using 5- to 8-d old sex-separated mosquitoes. A total of 15 wild-type virgin females were transferred by aspiration into a 30 × 30 × 30 cm cage (BugDorm-1 insect rearing cage, MegaView Science Co., Ltd.). Then, the indicated number of B2t1 males were combined with the females. The mosquitoes were allowed to mate for the indicated times and then the B2t1 males were replaced with the indicated number of wild-type males. After 3 d, the females were blood fed in the same cage, and wild-type males were retained in the cage for 4 more days until the end of the experiment. Females were transferred to individual vials, eggs were collected, and the number of hatched mosquitoes were scored. Females that did not blood feed well (absence of an engorged abdomen) or died during the egg collections were excluded from the study. Per condition, ≥3 independent assays were performed, and the percent fertility of the females was calculated.
Testing for Suppression of Female Fertility after Incubating Wild-Type Females with B2t1 Males and Wild-Type Males at the Same Time.
The assays were performed using 5- to 8-d old sex-separated mosquitoes. A total of 15 wild-type virgin females were transferred by aspiration into a 30 × 30 × 30 cm cage (BugDorm-1 insect rearing cage, MegaView Science Co., Ltd.). Then, the indicated numbers of B2t1 males and wild-type males were introduced simultaneously to the cage with the females. After 3 d, the females were blood fed in the same cage, and all males were retained in the cage for 4 more days until the end of the experiment. Individual females were transferred to separate vials for egg collections. Females that did not blood feed well (absence of an engorged abdomen) or died during the egg collections were excluded from the study. The numbers of eggs and hatched larvae (third or fourth instar) from individual female were counted, the egg hatch rate for each fertile female was measured, and the percent fertility of the females was calculated. Per condition, ≥5 independent assays were performed.
Curve Fitting of Experimental Data for Suppression of Female Fertility by B2t1 Males.
The maximum likelihood estimation of the parameters for the curves in Figs. 3C and 4C were performed as follows. The fertility rate of the experimental condition i is . In the experimental condition i and biological repeat j, we observed Nij females and found that nij out of them were fertile. The number of fertile females followed a binomial distribution . The likelihood of the experimental data are as follows:
In fitting the experimental data obtained from prior mating with B2t1 males (before adding the wild-type males) in suppressing female fertility, we fit the curve with a semiempirical formula:
Here, the parameter contains pc and kc. pc is the maximum probability of successful copulation under all experimental conditions. kc is the number of females copulated by each B2t1 male per hour. NB2t,i and ti are the number of B2t1 males and their mating length in experimental condition i.
When t = 0 h, the percentage of females that produced progeny reached the maximum probability of copulation, pc × 100%. When t is large enough, the percentage of females that produced progeny approached (1 – pc)pc × 100%, which means these females did not copulate with B2t1 males but copulated with LVP males later. We estimated the two parameters pc and kc in the equation using the maximum likelihood estimation and computed the confidence interval from the Fisher information matrix. Their values are pc = 0.895 ± 0.047 (95% CI) and kc = 0.0189 h−1 ± 0.0057 h−1 (95% CI).
For the female fertility data derived from the experiments in which we added the B2t1 and wild-type males at the same time, we fitted the experimental data with the following equation:
Here, the parameter contains pc and Kc. pc is the maximum probability of successful copulation under all experimental conditions. Kc is the competitiveness of B2t1 males normalized to wild-type males. NB2t,i and NWT,i are the number of B2t1 males and wild-type males in experiment condition i.
We assumed that B2t1 mutant males are able to compete with Kc wild-type males for copulation. The addition of B2t1 males dilutes the chance for wild-type females to copulate with wild-type males. Therefore, only NWT/(NWT + KcNB2t) of the wild-type females had the opportunity to copulate with wild-type (Liverpool) males. We estimated the parameters pc and Kc using the maximum likelihood estimation. Their values are pc = 0.941 ± 0.060 (95% CI) and Kc = 0.179 ± 0.051 (95% CI).
Statistics.
All error bars represent SEMs except for the curve fitting (Figs. 3C and 4C). The error bars in Figures 3C and 4C are 95% CI. We use 95% CI in these two figures to indicate that the fitted curves are in the range of the error bars. The number of times each experiment was performed (n) and statistical analysis are indicated in the figure legends. For real-time RT-PCR, n represents the number of biological replicates, and each biological sample was subjected to qPCR with three technical replicates. Each data point in the graph indicates the average from three technical replicates. For the measurements of body size and wing length, each data point represents a group of ≥5 male mosquitoes or ≥12 wings from males, respectively. n represents the number of groups measured. For the life table assessments, n represents the number of groups that were assayed, with each group starting with 400 eggs. For the assay in which we monitored the number of days required for 50% of the mosquitoes to die, n represents the number of groups of mosquitoes that were assayed. For the experiments showing the percent of fertile females in the single-pair mating assays, each data point represents a group consisting of ≥15 single-pair matings. For the fertility suppression assays, each data point represents one cage assay. For single comparisons, we used the nonparametric Mann–Whitney U test. For multiple comparisons, we conducted parametric one-way ANOVA with the Tukey’s multiple comparisons test or nonparametric Kruskal–Wallis test with the Dunn’s multiple comparisons test. Statistical analyses and graph data visualization were performed using GraphPad Prism 8 for MacOS.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID), which funded an NIH Director’s Pioneer Award to C.M. (DP1AI124453), a grant to C.M. (R56-AI153334) from the NIAID, and support to C.M. from the US Army Research Office and accomplished under cooperative agreement W911NF-19-2-0026 for the Institute for Collaborative Biotechnologies. O.S.A. was supported by a Defense Advanced Research Projects Agency (DARPA) Safe Genes Program Grant (HR0011-17-2-0047).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability
All study data are included in the main text.
References
- 1.Qsim M., et al., Genetically modified Aedes aegypti to control dengue: A review. Crit. Rev. Eukaryot. Gene Expr. 27, 331–340 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Benelli G., Mehlhorn H., Declining malaria, rising of dengue and Zika virus: Insights for mosquito vector control. Parasitol. Res. 115, 1747–1754 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Benedict M. Q., Robinson A. S., The first releases of transgenic mosquitoes: An argument for the sterile insect technique. Trends Parasitol. 19, 349–355 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Dame D. A., Curtis C. F., Benedict M. Q., Robinson A. S., Knols B. G., Historical applications of induced sterilisation in field populations of mosquitoes. Malar. J. 8 (suppl. 2), S2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw W. R., Catteruccia F., Vector biology meets disease control: Using basic research to fight vector-borne diseases. Nat. Microbiol. 4, 20–34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seirin Lee S., Baker R. E., Gaffney E. A., White S. M., Modelling Aedes aegypti mosquito control via transgenic and sterile insect techniques: Endemics and emerging outbreaks. J. Theor. Biol. 331, 78–90 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Thomas D. D., Donnelly C. A., Wood R. J., Alphey L. S., Insect population control using a dominant, repressible, lethal genetic system. Science 287, 2474–2476 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Gantz V. M., Bier E., Genome editing. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science 348, 442–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bier E., Harrison M. M., O’Connor-Giles K. M., Wildonger J., Advances in engineering the fly genome with the CRISPR-Cas system. Genetics 208, 1–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gantz V. M., et al., Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. U.S.A. 112, E6736–E6743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raban R. R., Marshall J. M., Akbari O. S., Progress towards engineering gene drives for population control. J. Exp. Biol. 223, jeb208181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M., et al., Development of a confinable gene drive system in the human disease vector Aedes aegypti. eLife 9, e51701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schairer C. E., Taitingfong R., Akbari O. S., Bloss C. S., A typology of community and stakeholder engagement based on documented examples in the field of novel vector control. PLoS Negl. Trop. Dis. 13, e0007863 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alphey L., Genetic control of mosquitoes. Annu. Rev. Entomol. 59, 205–224 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Pan X. L., Thiem S., Xi Z. Y., “Wolbachia-mediated immunity induction in mosquito vectors” in Arthropod Vector: Controller of Disease Transmission, Vol 1: Vector Microbiome and Innate Immunity of Arthropods, Wikel S. K., Aksop S., Dimopoulos G., Eds. (Academic Press, 2017), pp. 35–58. [Google Scholar]
- 16.Laven H., Eradication of Culex pipiens fatigans through cytoplasmic incompatibility. Nature 216, 383–384 (1967). [DOI] [PubMed] [Google Scholar]
- 17.Bian G., et al., Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science 340, 748–751 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Caragata E. P., et al., Prospects and pitfalls: Next-generation tools to control mosquito-transmitted disease. Annu. Rev. Microbiol. 74, 455–475 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Knipling E. F., Possibilities of insect control or eradication through the use of sexually sterile males. J. Econ. Entomol. 48, 459–462 (1955). [Google Scholar]
- 20.Bushland R. C., Lindquist A. W., Knipling E. F., Eradication of screw-worms through release of sterilized males. Science 122, 287–288 (1955). [DOI] [PubMed] [Google Scholar]
- 21.Kemphues K. J., Raff E. C., Kaufman T. C., Genetic analysis of B2t, the structural gene for a testis-specific β-tubulin subunit in Drosophila melanogaster. Genetics 105, 345–356 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemphues K. J., Kaufman T. C., Raff R. A., Raff E. C., The testis-specific β-tubulin subunit in Drosophila melanogaster has multiple functions in spermatogenesis. Cell 31, 655–670 (1982). [DOI] [PubMed] [Google Scholar]
- 23.Kemphues K. J., Raff E. C., Raff R. A., Kaufman T. C., Mutation in a testis-specific β-tubulin in Drosophila: Analysis of its effects on meiosis and map location of the gene. Cell 21, 445–451 (1980). [DOI] [PubMed] [Google Scholar]
- 24.Smith R. C., Walter M. F., Hice R. H., O’Brochta D. A., Atkinson P. W., Testis-specific expression of the β2 tubulin promoter of Aedes aegypti and its application as a genetic sex-separation marker. Insect Mol. Biol. 16, 61–71 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Dzaki N., Ramli K. N., Azlan A., Ishak I. H., Azzam G., Evaluation of reference genes at different developmental stages for quantitative real-time PCR in Aedes aegypti. Sci. Rep. 7, 43618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentile C., Lima J. B., Peixoto A. A., Isolation of a fragment homologous to the rp49 constitutive gene of Drosophila in the neotropical malaria vector Anopheles aquasalis (Diptera: Culicidae). Mem. Inst. Oswaldo Cruz 100, 545–547 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Dong S., et al., Heritable CRISPR/Cas9-mediated genome editing in the yellow fever mosquito, Aedes aegypti. PLoS One 10, e0122353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konet D. S., et al., Short-hairpin RNA expressed from polymerase III promoters mediates RNA interference in mosquito cells. Insect Mol. Biol. 16, 199–206 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Li M., et al., Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 114, E10540–E10549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeap H. L., Endersby N. M., Johnson P. H., Ritchie S. A., Hoffmann A. A., Body size and wing shape measurements as quality indicators of Aedes aegypti mosquitoes destined for field release. Am. J. Trop. Med. Hyg. 89, 78–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renshaw M., Service M. W., Birley M. H., Size variation and reproductive success in the mosquito Aedes cantans. Med. Vet. Entomol. 8, 179–186 (1994). [DOI] [PubMed] [Google Scholar]
- 32.Armbruster P., Hutchinson R. A., Pupal mass and wing length as indicators of fecundity in Aedes albopictus and Aedes geniculatus (Diptera: Culicidae). J. Med. Entomol. 39, 699–704 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Van Handel E., Day J. F., Correlation between wing length and protein content of mosquitoes. J. Am. Mosq. Control Assoc. 5, 180–182 (1989). [PubMed] [Google Scholar]
- 34.Jones J. C., Spermatocysts in Aedes aegypti (Linnaeus). Biol. Bull. 132, 23–33 (1967). [DOI] [PubMed] [Google Scholar]
- 35.Wyss J. H., Screwworm eradication in the Americas. Ann. N. Y. Acad. Sci. 916, 186–193 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Perez-Staples D., Díaz-Fleischer F., Montoya P., The sterile insect technique: Success and perspectives in the neotropics. Neotrop. Entomol. 50, 172–185 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Benelli G., Jeffries C. L., Walker T., Biological mosquito vectors: Past, present, and future. Insects 7, 52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alphey L., et al., Sterile-insect methods for control of mosquito-borne diseases: An analysis. Vector Borne Zoonotic Dis. 10, 295–311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helinski M. E., Deewatthanawong P., Sirot L. K., Wolfner M. F., Harrington L. C., Duration and dose-dependency of female sexual receptivity responses to seminal fluid proteins in Aedes albopictus and Ae. aegypti mosquitoes. J. Insect Physiol. 58, 1307–1313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Craig G. B. Jr, Mosquitoes: Female monogamy induced by male accessory gland substance. Science 156, 1499–1501 (1967). [DOI] [PubMed] [Google Scholar]
- 41.Thailayil J., Magnusson K., Godfray H. C., Crisanti A., Catteruccia F., Spermless males elicit large-scale female responses to mating in the malaria mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 108, 13677–13681 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chapman T., et al., The sex peptide of Drosophila melanogaster: Female post-mating responses analyzed by using RNA interference. Proc. Natl. Acad. Sci. U.S.A. 100, 9923–9928 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H., Kubli E., Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 100, 9929–9933 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Concha C., et al., A transgenic male-only strain of the New World screwworm for an improved control program using the sterile insect technique. BMC Biol. 14, 72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kandul N. P., et al., Transforming insect population control with precision guided sterile males with demonstration in flies. Nat. Commun. 10, 84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mali P., et al., RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the main text.