Abstract
The cell plasma membrane provides a highly interactive platform for the information transfer between the inside and outside of cells. The surface glycoprotein interaction network is extremely important in many extracellular events, and aberrant protein interactions are closely correlated with various diseases including cancer. Comprehensive analysis of cell surface protein interactions will deepen our understanding of the collaborations among surface proteins to regulate cellular activity. In this work, we developed a method integrating chemical crosslinking, an enzymatic reaction, and MS-based proteomics to systematically characterize proteins interacting with surface glycoproteins, and then constructed the surfaceome interaction network. Glycans covalently bound to proteins were employed as “baits”, and proteins that interact with surface glycoproteins were connected using chemical crosslinking. Glycans on surface glycoproteins were oxidized with galactose oxidase (GAO) and sequentially surface glycoproteins together with their interactors (“prey”) were enriched through hydrazide chemistry. In combination with quantitative proteomics, over 300 proteins interacting with surface glycoproteins were identified. Many important domains related to extracellular events were found on these proteins. Based on the protein–protein interaction database, we constructed the interaction network among the identified proteins, in which the hub proteins play more important roles in the interactome. Through analysis of crosslinked peptides, specific interactors were identified for glycoproteins on the cell surface. The newly developed method can be extensively applied to study glycoprotein interactions on the cell surface, including the dynamics of the surfaceome interactions in cells with external stimuli.
Proteins interacting with glycoproteins on the cell surface were systematically characterized by integrating chemical crosslinking, enzymatic oxidation, and MS-based proteomics. The surface glycoprotein interaction network was then constructed.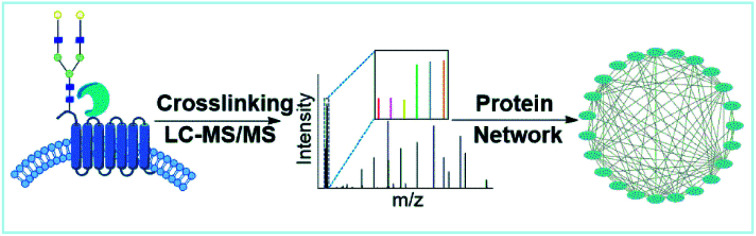
Introduction
Proteins located on the plasma membrane play extremely important roles in cells. They are responsible for many cellular events, such as sensing extracellular signaling molecules and regulating cell–cell adhesion.1,2 These cell surface proteins interact with extracellular matrix, hormones, as well as lateral and intracellular proteins, which creates a highly interactive network.3,4 The interaction network is highly dynamic for cells to adapt to the ever-changing extracellular environment. Moreover, signal transduction is dependent on transient protein–protein interactions.5,6 It has been found that the cell surface is covered by the pericellular matrix called the glycocalyx in which glycans are attached to surface proteins and lipids.7,8 The glycocalyx participates in various biological activities including cell–cell recognition, communication, and cell adhesion.2,9 The majority of cell surface proteins are glycosylated, and the most common ones are protein N- and O-glycosylation.10–14 Aberrant glycosylation can alter protein–protein interactions, which has been correlated with disease initiation and development.15,16 For instance, changes in the glycosylation of CD44, a cell surface receptor involved in cancer proliferation and migration, can significantly affect its binding towards the ligand of hyaluronic acid and thus change cancer cell signaling.17,18 Therefore, studies on the cell surface protein interaction network will advance our understanding of the regulation of cell signaling within specific spatial and temporal organization and promote the discovery of essential proteins in the surface protein network as drug targets.
Despite the importance of the cell surface protein interaction network, systematic investigation of extracellular interactions of surface proteins are still underrepresented.10,19 With the rapid development of modern mass spectrometry (MS), large-scale studies of protein interactions have become feasible. Affinity purification-mass spectrometry (AP-MS) methods have been extensively applied to investigate protein interactions.20,21 However, AP-MS may not be able to detect transient or weak interactions from surface proteins because of the harsh conditions including detergents and high concentration of salts required to dissolve these membrane proteins. Recently, enzyme-catalyzed proximity labeling methods were reported to study membrane protein interactions, in which an enzyme was genetically fused to the protein of interest for catalyzing the transfer of a biotin functionality to nearby proteins.22,23 However, these methods are often employed to tag the intracellular domain of surface proteins, and thus study the interactions between surface proteins and neighboring intracellular proteins or surface proteins with intracellular domains. Although proximity labeling methods were also developed to detect the extracellular interaction network such as EMARS, SPPLAT, and PUP-IT,24–26 they are normally used to construct the interaction network of one specific surface protein.
Similarly, ligand-based receptor capture technology (LRC) including TRICEPS and HATRIC were designed to study the interactions between one specific extracellular ligand and its surface receptors.27,28 Photo-crosslinking approaches were capable of studying glycan-binding protein interactions.29,30 With this method, a photo-reactive sugar analog was used to metabolically label glycans and study glycan–protein interactions upon the UV exposure. Although the photo-reactive sugar analog can be incorporated into different glycans on the cell surface, this method is usually used to study the binding partners of one glycoprotein. Moreover, the functional group in sugar analogs may intervene the interactions between proteins and glycans. Recently, a beautiful method integrating metabolic and chemical proximity labeling was reported to study the binding between proteins and surface sialic acids on a large scale. A clickable probe was synthesized and attached to modified sialic acids on the cell membrane, which functioned as a catalyst for the formation of radicals from hydrogen peroxide. The proteins in the sialic acid environment were labeled through amino acid oxidation, and were categorized into three groups including sialylated proteins, non-sialylated proteins with transmembrane domains, and proteins that are associated with the membrane with neither sialylated nor transmembrane domains.31 Considering the importance and complexity of surface glycoproteins and their interactors, effective methods are critical to decipher the surfaceome interaction network.
In this work, we developed a new method integrating chemical crosslinking, an enzymatic reaction, and MS-based proteomics to study the surfaceome interaction network. A crosslinker was employed to capture proteins that interact with surface glycoproteins. Then, galactose oxidase (GAO) was used to generate a chemical handle on glycans, and the tagged glycoproteins together with crosslinked interactors were enriched through hydrazide chemistry. The captured proteins were enzymatically digested and identified through quantitative proteomics. With the identification of proteins that interact with surface glycoproteins using MS-based proteomics, we were able to construct the surfaceome interaction network. The current method does not require genetic engineering or metabolic labeling, and thus it can be extensively applied to study cell surface glycoprotein interactions.
Results and discussion
Principle of an integrative method to study the surface glycoprotein interaction network
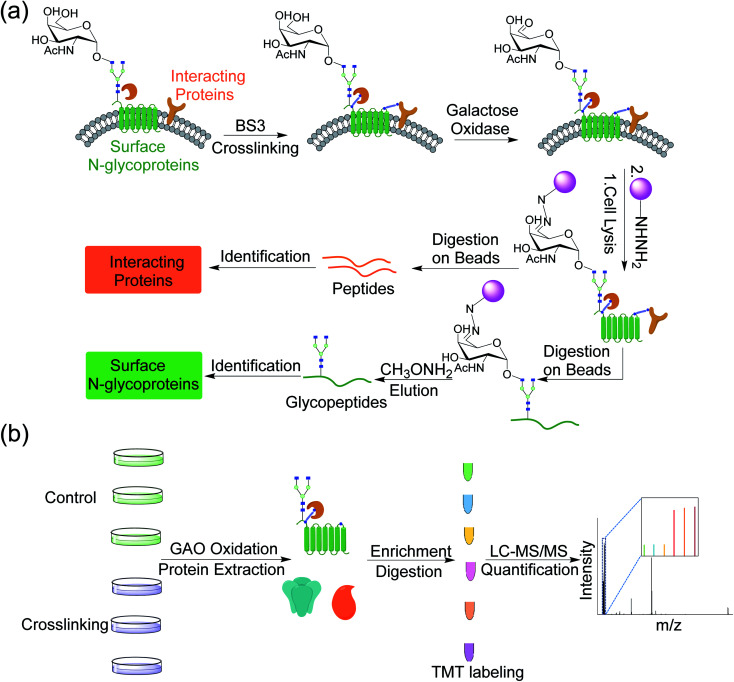
Although AP-MS has been widely applied to study protein–protein interactions, it is still challenging to investigate membrane protein interactions due to detergents and high concentration of salts normally required to dissolve membrane proteins, which disrupt the interactions. Chemical crosslinking can be used to study membrane protein interactions, which preserves transient or labile protein interactions.32,33 In order to capture proteins that interact with surface glycoproteins, we employed bis(sulfosuccinimidyl)suberate (BS3), a membrane impermeable crosslinker, to covalently bind surface glycoproteins with their interactors (Fig. 1a). After chemical crosslinking, the interaction partners can be preserved through the covalent interactions even if proteins were denatured in different detergent- and salt-containing buffers. Surface glycoproteins with their interactors can then be pulled down and separated through enzymatic oxidation and chemical enrichment. A galactose oxidase (GAO)-based method has been demonstrated to effectively identify glycoproteins only located on the cell surface.34 GAO can efficiently convert the hydroxyl group at C6 of galactose (Gal)/N-acetylgalactosamine (GalNAc) to the aldehyde group under mild conditions.35 The aldehyde group as a chemical handle enables us to selectively enrich surface glycoproteins and their interactors connected by the chemical crosslinker.
Fig. 1. (a) Experimental procedure for investigating the cell surface glycoprotein interactions by integrating chemical crosslinking, an enzymatic reaction, and MS-based proteomics. (b) Detailed procedure for identification and quantification of proteins interacting with surface glycoproteins using multiplexed MS-based proteomics.
After protein digestion, peptides were labeled with the tandem mass tag (TMT) reagents for their quantification (Fig. 1b). In this work, biological triplicate experiments were performed. To eliminate the effects of non-specific binding proteins, a control group without chemical crosslinking was also included. Peptides from each sample were labeled with each channel of the six-plex TMT reagents, respectively, and all labeled peptides were then mixed before LC-MS/MS analysis. Based on the intensity ratios of the reporter ions generated from the TMT tags under HCD between the crosslinking and control samples, the enriched proteins from the chemical crosslinking samples were selectively identified. As a commonly used cell line, MCF7 served as a model for investigating the surface interactome by integrating chemical crosslinking and MS-based proteomics.
Identification of proteins that interact with surface glycoproteins
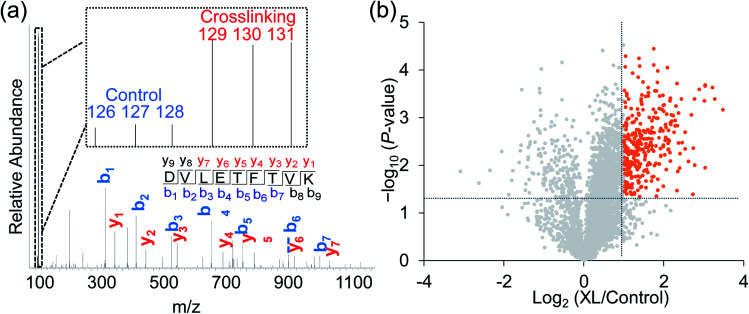
Taking advantage of the six-plex isobaric labeling reagents that can label peptides from six samples, we performed biological triplicate experiments to identify proteins involved in the surfaceome interaction network. The peptides labeled by each isobaric reagent will generate a unique reporter ion in the tandem MS for relative quantitation of peptide abundance from each sample. An example of peptide identification and quantification is shown in Fig. 2a. The peptide DVLETFVK was confidently identified with an Xcorr of 3.9 and a mass accuracy of 0.38 ppm. The intensities of the reporter ions clearly demonstrate that the peptide was enriched in the chemical crosslinking sample compared with the control group. This peptide is from CD9, which is a known surface protein with four transmembrane domains. It was reported that CD9 can interact with many signaling and adhesion surface proteins and establish a compact web over the cell surface.36,37
Fig. 2. (a) Identification and quantification of an example peptide. The inset shows the TMT reporter ion intensities, which are used for the quantification. (b) Volcano plot illustrating proteins with statistically significant abundance differences between the crosslinking and control samples. Significant differences are defined with the abundance change >2 fold and the P value <0.05.
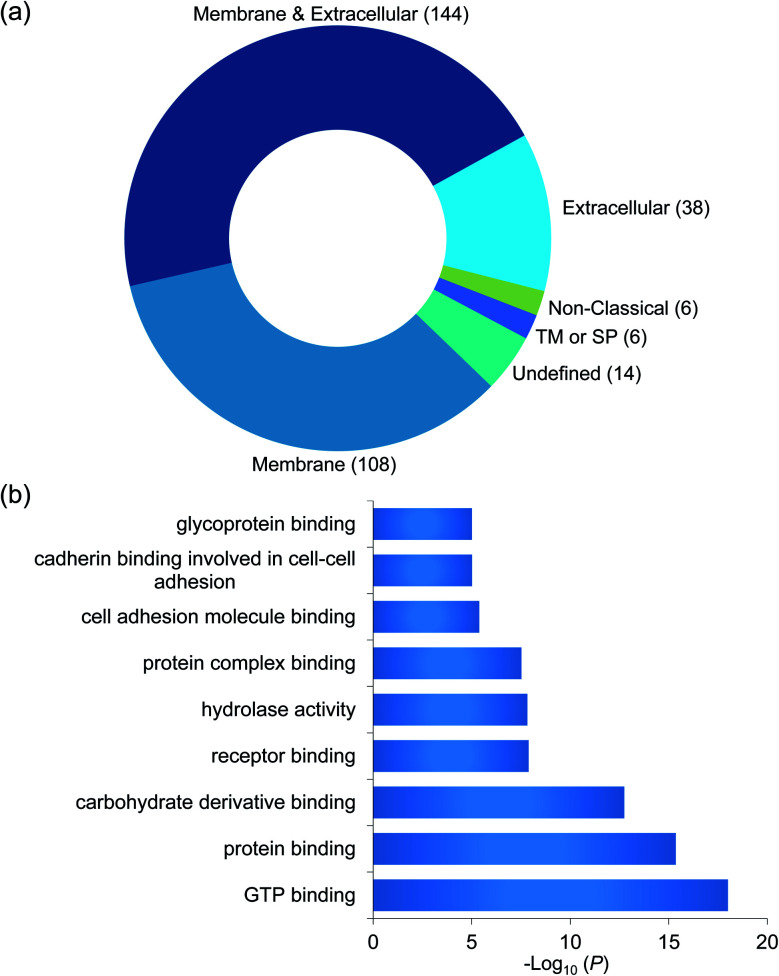
To confidently identify proteins that interact with surface glycoproteins, we applied these criteria: the abundance change of a protein between the chemical crosslinking samples and the control ones must be at least 2-fold, and the P value determined from a t test must be <0.05. In total, 316 proteins meet these criteria (Fig. 2b, Table S1†). Among those, 144 proteins are annotated as both membrane and extracellular proteins, 108 proteins are annotated as only membrane proteins and 38 proteins are only extracellular ones according to the GO cellular component annotation from UniProt (Fig. 3a). Phobius and SecretomeP were used to predict the location of the remaining 26 proteins. Six proteins contain a transmembrane domain or a signal peptide, and six may be non-classically secreted. The results indicate that nearly all the identified proteins are potentially located in the extracellular region and thus may participate in the surfaceome interaction network. Clustering of all these proteins based on biological process showed that proteins related to exocytosis, secretion, membrane organization, signaling, and cell–cell communication were overrepresented (Fig. S1†). Based on molecular function, the highly enriched categories include protein and carbohydrate derivative binding, receptor binding, and glycoprotein binding, which are correlated very well with the functions of proteins located at the cell surface and in the extracellular space (Fig. 3b). All these results demonstrate that the current method can effectively analyze proteins that interact with surface glycoproteins.
Fig. 3. (a) Classification of the identified proteins interacting with surface glycoproteins based on cellular component. (b) Clustering of the identified proteins based on molecular function.
The proteomic workflow also allowed us to site-specifically identify surface N-glycoproteins (Fig. 1a). From the control and chemical crosslinking samples, over 600 N-glycosylation sites were identified on 328 surface glycoproteins (Table S2†). For all identified N-glycoproteins, over half are type I membrane proteins (170) where their N-termini are located in the extracellular space (Fig. S2a†). Only 16 proteins belong to type II membrane proteins where the C-termini are located outside of cells. About one third are multi-pass membrane proteins, and 5 proteins are peripheral membrane proteins. For undefined proteins (41), nearly all of them (40) contain a transmembrane domain or a signal peptide based on the prediction by Phobius. The N-glycosylation site localization on type I and type II membrane proteins is illustrated in Fig. S2b.† All glycosylation sites are found in the extracellular space, which is consistent with the common belief that glycans on surface proteins are located outside of cells. The results demonstrated that the GAO-based method can specifically target surface glycoproteins and thus enable us to selectively enrich proteins that interact with surface glycoproteins. Overall, integrating chemical crosslinking, enzymatic oxidation, and hydrazide chemistry, the current method can identify not only proteins interacting with surface glycoproteins, but also surface glycoproteins themselves.
Domain analysis for proteins interacting with surface glycoproteins
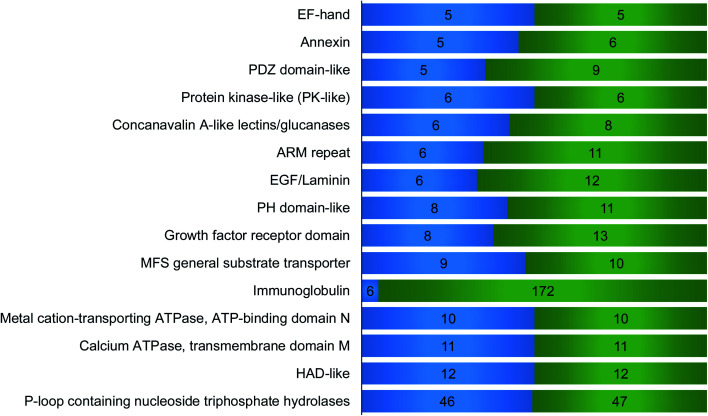
We then performed domain analysis to investigate the functions of the proteins interacting with surface glycoproteins and their correlation with the surfaceome interaction network. About 160 different types of protein domains were found on the 316 identified proteins (Table S3†). These domains are involved in various functions, indicating that proteins with different biological functions participate in the interaction network. All the domains that were found in at least five proteins are displayed in Fig. 4. For example, six identified proteins contain concanavalin A-like lectins/glucanases domain, including galectin-1, galectin-3, and galectin-8, which are known to bind to the carbohydrate part of glycoproteins and glycolipids on the cell surface.38 Galectins have been reported to form the interaction lattice on the plasma membrane through their binding to carbohydrates, which confines specific surface domains to control signaling of surface receptors including the T cell receptor (TCR), B cell receptor (BCR), and specific cytokine receptors.39,40 We also identified proteins with C-type lectin-like domain including DEC-205 and tetranectin. DEC-205 is a glycan-binding receptor located on the plasma membrane and can capture the glycosylated antigens in the extracellular space to initiate endocytosis or immune response.41,42 The identification of different types of lectins indicates that the chemical crosslinking method can keep the glycan–protein interactions. It is well-known that global analysis of glycan-binding proteins on the cell surface is very challenging. The current method offers a great opportunity to identify surface glycan-binding proteins on a large scale.
Fig. 4. Domain analysis for the identified proteins that interacted with surface glycoproteins. The number of proteins containing each domain is on the left side and the total hit number of each domain is on the right. For example, 6 proteins contain immunoglobulin (Ig) domain, and in total, these proteins have 172 Ig domains. The domains appeared in at least five proteins are shown here.
Other important domains related to cell surface activities were also found. For example, the EGF-laminin domain is mainly from extracellular matrix proteins such as FBLN1, COMP, and LTBP1, which can interact with surface proteins and participate in the extracellular interaction network. EF-hand, a calcium dependent domain, may interact with the extracellular matrix. For example, S100A13, an EF-hand domain-containing secreted protein, has been proved to play a key role in the interaction with FGF1 based on the synergistic binding to Ca2+ and Cu2+, which is the initial step in non-classical release of FGF1.43 Six annexin proteins have been identified, which may interact with sialic acid-containing carbohydrates on the plasma membrane with the help of calcium.44 Previous studies have shown that NHERF1 containing PDZ domain binds to a multitude of ligands including ion transporters, tyrosine kinase receptors, and G protein-coupled receptors (GPCRs).45 Tyrosine-protein kinase Lyn, which contains protein kinase-like domain, is a non-receptor tyrosine-protein kinase on the cell membrane and has shown binding capacity to receptors. Moreover, it plays an important role in immune response.46 Among the proteins containing ARM repeat domain, CLTCL1 interacts with plasma membrane proteins and mediates the clathrin-dependent endocytosis.47 PH domain is capable to interact with membrane proteins to modulate protein and cell localization.48 A previous report demonstrated that RDX with PH domain was a cadherin-binding protein and regulated cell junctions and localization.49 Eight proteins contain growth factor receptor domain including IGFBP2 and IGFBP5, which are involved in the growth factor receptor signaling pathway. Identified surface protein transporters with MFS general substrate transport domain include SLC49A4, SLC16A10 and SLC16A1. Metal cation-transporting ATPase and calcium ATPase membrane domain is a superfamily of membrane proteins responsible for transportation of lipid or cations powered by hydrolysis of adenosine triphosphate (ATP). HAD-like domain is the catalytic domain of metal-cation ATPase and promotes the transportation of molecules across the plasma membrane.50 Interestingly, p-loop containing nucleoside triphosphate hydrolase domain was found on 46 proteins, and most of these proteins are G-proteins and small GTPases. Many of these proteins can be attached to the membrane through lipidation. These membrane proteins normally reside in the cytosolic side, but may be transported to the extracellular region through exocytosis or secretion.
Construction of the protein interactome on the cell surface
Proteins usually cooperate with each other to participate in numerous biological processes. This can often be achieved through protein–protein interactions. With the development of high-throughput methods including AP-MS, numerous protein–protein interactions have been experimentally identified, which helps construct protein–protein interaction network. In the network, proteins are represented by nodes and their interactions are connected by edges in a graphical view.51,52 Protein interaction network offers a new approach to study biological pathways, and reveals protein functions.53
The interaction network of membrane protein is of critical importance to cells as they participate in various biological processes including molecular transport, signal transduction, and cell–cell communication. With the identification of proteins that interact with surface glycoproteins in this work, we aim to construct the surface protein interaction network based on the existing protein–protein interaction data and gain insights into their roles in the network. We submitted a query of the 316 identified proteins to IntAct, a public database deposited with protein–protein interactions determined from many previous experiments, and 4347 interactions were extracted after the removal of non-human proteins and their corresponding interactions (Table S4†). In order to have a better understanding of the network's functions, we analyzed the topology of the reconstructed network using Network Analyzer in Cytoscape (Fig. 5). The characteristic parameters of the network included clustering coefficient (0.015), network density (0.001), and characteristic pathway length (4.852). The results are consistent with the power law of typical biological networks.54,55
Fig. 5. Interaction network of the identified proteins that interacted with surface glycoproteins. All the interactions were downloaded from the IntAct database and visualized by Cytoscape. The proteins with ≥20 interactions were enlarged. The proteins that overlap with those interacting with surface glycoproteins identified in this work are shown in blue and the rest are in gray.
To visualize the interactome of the identified proteins as a function of connectivity, we arranged the interactors into concentric circles based on the number of interactions per protein. Proteins with many interactions (at least 20) in the center can be considered as the hubs in the interaction network, which play a critical role in maintaining the stability of the biological network.56 Most of the hub proteins (61 of 63) are from the proteins identified in this work, and these proteins have various functions including cadherin binding, enzyme binding, and Ras GTPase binding. Manipulation of the hub proteins in the network may have higher possibility to disrupt the cell activity, which offers potential ways for the discovery of drug targets.57 For example, CD81 is one important component of tetraspanin-enriched microdomains (TERMs) on the plasma membrane that act as a platform for receptor clustering and signal transduction.58,59 Here, CD81 was identified as a hub protein, which suggests that this protein may serve as a promising therapeutic target. It participates in many important cellular processes such as membrane organization, protein trafficking, cellular fusion, and cell–cell interactions. Recently, a study showed that it was closely related to cancer proliferation and metastasis.60 CD81-knocked-out mice had reduced primary tumor growth and significantly decreased level of lung cancer metastasis.
Identification of crosslinked peptides to further study the surface interactome
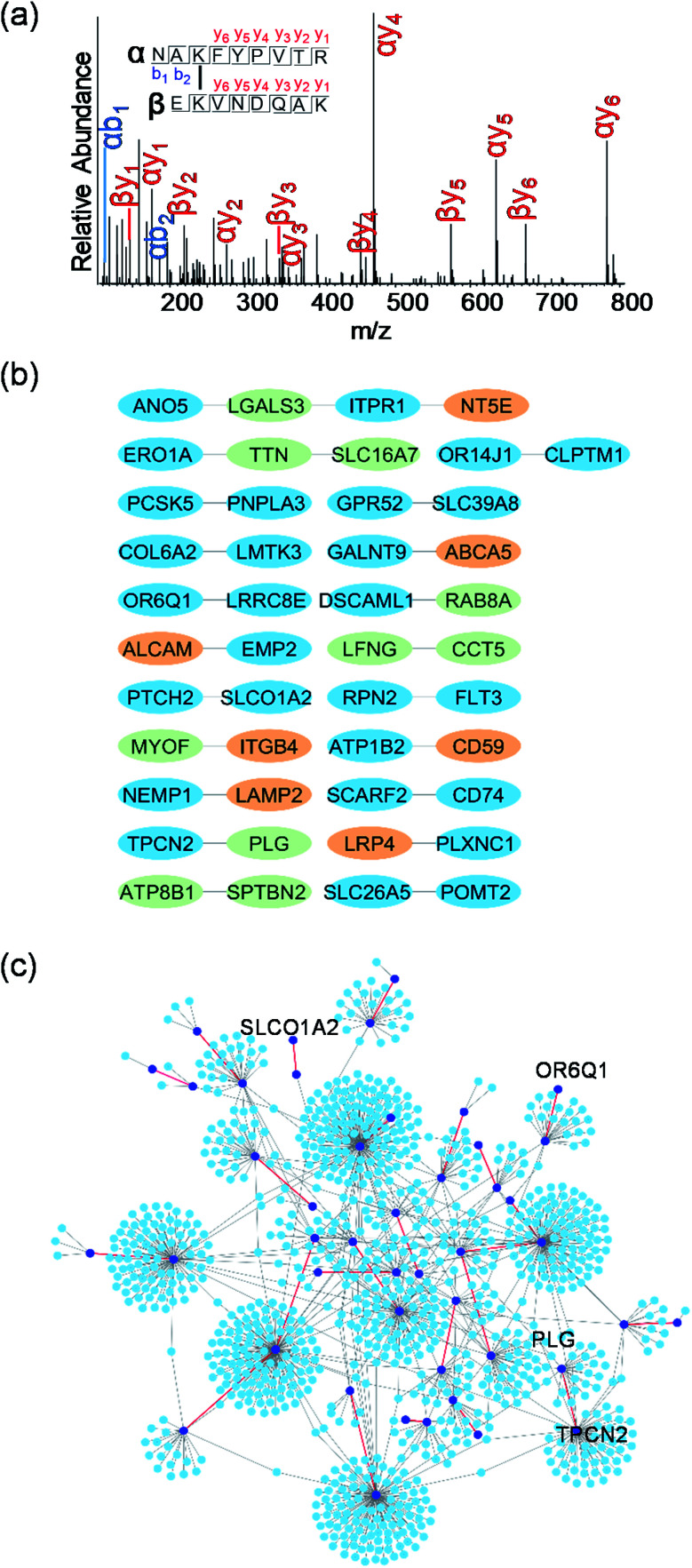
Identification of crosslinked peptides can provide direct experimental evidence for the interactions between two proteins, revealing the interaction partners of surface glycoproteins. After chemical crosslinking and on-bead digestion, crosslinked peptides were analyzed by LC-MS/MS. About 200 total crosslinked peptides were identified (Fig. S3, Table S5†) and one example is displayed in Fig. 6a. This crosslinked peptide was from epithelial membrane protein 2 (EMP2) and CD166 antigen (ALCAM). Both EMP2 and ALCAM are glycoproteins located on the plasma membrane and are involved in regulating cell adhesion and signal transduction. Based on the identification of the crosslinked peptides, some confirmed protein–protein interactions on the cell surface are shown in Fig. 6b. For each pair of interactions, one or both proteins are glycosylated except TTN-SLC16A7 (“–” refers to protein–protein interaction based on the identified crosslinked peptides). For TTN-SLC16A7, we also found that TTN interacted with ERO1A, which is a surface glycoprotein (Fig. 6b). The results indicated that TTN and ERO1A may form a protein complex with SLC16A7.
Fig. 6. (a) An example tandem MS of the crosslinked peptide, NAKFYPVTR(3)-EKVNDQAK(2). (b) Some protein–protein interactions based on the identified crosslinked peptides. The proteins also found in the data set of the surface glycoprotein interactors from quantitative proteomics were colored in light green and proteins from the data set of surface glycoproteins identified in this work were in orange. (c) Protein–protein interaction network constructed from the IntAct database. Proteins identified based on the crosslinked peptides were in dark blue and the remaining proteins from IntAct were in light blue. The identified interactions were colored in red and the known interactions from IntAct were in gray.
The overlap between the two data sets of the proteins identified based on the crosslinked peptides and quantitative proteomics are shown in Fig. 6b (colored in light green). Galectin-3 (LGALS3) participating in various interactions as a glycan-binding protein was found to interact with ANO5 and ITPR1, both of which are glycoproteins located on the cell surface. RAB8A, PLG, and MYOF interacted with the surface glycoproteins, DSCAML1, TPCN2, and ITGB4, respectively. Interestingly, four proteins (LFNG, CCT5, ATP8B1, and SPTBN2) were identified as the surface glycoprotein interactors above, and they also had direct interactions (LFNG-CCT5 and ATP8B1-SPTBN2) based on the identified crosslinked peptides. The surface glycoproteins identified through the proteomic workflow shown in Fig. 1a were highlighted in orange (Fig. 6b). Similarly, as described above, the interactors of identified surface glycoproteins, such as ITPR1 for NT5E, ATP1B2 for CD59, and PLXNC1 for LRP4, can also be pinpointed. These results proved that the identification of crosslinked peptides can directly unravel the interactors of glycoproteins on the cell surface.
The identified interactions were further mapped to the known interactions extracted from IntAct (Fig. 6c). Some proteins such as transporter SLCO1A2 and olfactory receptor OR6Q1 have no verified interactions in IntAct, but they were found to participate in the surface interaction network in the current study. The network reveals that one hub protein can interact with another hub protein based on the identified crosslinked peptides such as PLG and TPCN2 (Fig. 6c), which implies more underlying roles of these hub proteins. The combination of the interaction network based on the identified crosslinked peptides (Fig. 6c) and the network constructed from IntAct (Fig. 5) provides more valuable information about protein interactions on the cell surface.
Identification of crosslinked peptides is vital to understand the surface interaction network, but large-scale analysis of crosslinked peptides is still very challenging due to many obstacles such as the low abundance of crosslinked peptides and the high complexity of the fragments in tandem MS.61,62 The identification of crosslinked peptides in the surface protein interaction experiments is even more challenging because the abundances of many surface proteins are normally much lower compared to intracellular proteins, and membrane proteins are difficult to deal with. It has also been reported that the identified crosslinked peptides are biased for highly abundant proteins using the current proteome-wide cross-linking MS.62 With the development of tri-functional63–65 and MS-cleavable crosslinkers66,67 for enhancing MS-based detection of crosslinked peptides, more crosslinked peptides from the cell surface can be identified, which will further deepen our understanding of the surface interactome.
Conclusions
Cell receptors, transporters and adhesion molecules on the plasma membrane interact with many surface proteins, and these interactions modulate various cellular events including signal transduction and cell–cell communications. Growing evidence has indicated that proteins located on the plasma membrane collaborate with each other by forming a microdomain to discriminate the real “input” and then generate the corresponding accurate “output”. Therefore, global analysis of the interactors of glycoproteins on the plasma membrane can advance our understanding of surface protein functions and the overall cell surface protein organization. In this work, integrating chemical crosslinking, enzymatic oxidation, and MS-based proteomics, we systematically investigated the surfaceome interaction network. From the biological triplicate experiments, we identified 316 proteins that interact with surface glycoproteins. The identified proteins contain different types of domains including concanavalin A-like lectins/glucanases domain, C-type lectin-like domain, EGF-laminin domain, and many of them are related to extracellular activities. The surface protein interaction network was constructed based on the proteins identified here and the reported protein–protein interactions. Some hub proteins with at least 20 interactions were identified and they are very important for extracellular activities. These hub proteins can potentially be served as drug targets. Because the current method allows us to globally capture the cell surface protein interactions, it provides an excellent opportunity to study the dynamics of the surface interactome in the future.
Experimental section
Cell culture
MCF7 cells (ATCC) were cultured in Dulbecco's Modified Eagle's Medium (DMEM) (Sigma) containing high glucose and 10% fetal bovine serum (FBS) (Corning). The cells were grown inside a humidified incubator with 5.0% CO2 at 37 °C until the cell confluency reached ∼90%. The cells were washed with ice-cold PBS three times and harvested by scraping. After centrifugation (300g, 5 min), the cell pellets were washed twice with ice-cold PBS (pH = 8).
Chemical crosslinking and oxidation of surface glycoproteins
The cells were suspended in 1 mL ice-cold PBS (pH = 8) containing 2 mM bis(sulfosuccinimidyl)suberate (BS3, Thermo Scientific). For the control group, every step was the same except without the addition of the chemical crosslinking reagent, i.e. BS3. The cells were incubated at 4 °C for 60 min through end-over-end rotation. The reaction was quenched at 4 °C for 30 min by adding Tris to a final concentration of 15 mM. The cells were then washed three times with ice-cold PBS containing 15 mM Tris. Glycoproteins on the cell surface were then oxidized with galactose oxidase (50 U mL−1, Innovative Research) in a solution containing HRP (40 U mL−1, Sigma) and 5% FBS for 60 min at 37 °C.
Cell lysis and enrichment of glycoproteins together with their crosslinked interactors
After oxidation, the cells were washed with ice-cold PBS three times and resuspended in a buffer containing 50 mM HEPES (pH = 7.4), 150 mM NaCl, 25 μg mL−1 digitonin, and 1 tablet per 10 mL protease inhibitor (Roche), and incubated on ice for 10 min. The supernatant was discarded after the solution was centrifuged at 2000g for 10 min. The pellets were lysed in a buffer containing 50 mM HEPES (pH = 7.4), 150 mM NaCl, 0.5% SDC, 0.5% SDS, 10 units per mL benzonase, and 1 tablet per 10 mL protease inhibitor, and incubated at 4 °C for 60 min through end-over-end rotation. The lysates were centrifuged, and the supernatant was collected. The tagged proteins were enriched with hydrazide beads (Thermo Scientific) with aniline (10 mM) as the catalyst at 4 °C for 24 h. Then the beads were washed three times with a buffer containing 8.0 M urea, 0.4 M ammonium carbonate (NH4HCO3), and 0.1% SDS.
Proteins on the beads were reduced with 5 mM dithiothreitol (DTT) (37 °C, 45 min), and alkylated with 14 mM iodoacetamide at room temperature for 25 min in the dark. Non-specific binding proteins were removed by washing the beads 4 times with the above buffer. The hydrazide beads were resuspended in 50 mM HEPES (pH = 8.5) with 1.5 M urea, and proteins on the beads were digested with trypsin at 37 °C overnight. The eluent was collected. Then glycopeptides covalently bound on the beads were eluted by incubating with the elution buffer (0.2 M methoxylamine, 1.5 M NaCl, and 0.1 M aniline in 0.1 M sodium acetate solution, pH = 4.5). Peptides and glycopeptides were purified using tC18 Sep-Pak cartridge (Waters) and dried in a SpeedVac system.
Peptide TMT labeling and fractionation
Peptides and glycopeptides from six samples (three crosslinked samples and three control ones) were labeled by the six-plex tandem mass tag (TMT) reagents (Thermo Scientific) according to the manufacturer's protocol. The labeled peptides from all six samples were combined, desalted using a tC18 Sep-Pak cartridge, and further separated into 20 fractions by high pH reversed-phase high-performance liquid chromatography (HPLC) using a 40 min gradient of 5–55% ACN in 10 mM ammonium acetate (pH = 10). Each fraction was purified again by the StageTip method. The labeled glycopeptides were deglycosylated in heavy-oxygen water (H218O) with peptide-N-glycosidase F (PNGase F, Sigma) for 3 h at 37 °C. The deglycosylated peptides were purified and separated into three fractions using the StageTip method. For the identification of crosslinked peptides, the peptides were fractionated and each fraction was also purified using StageTip before MS analysis.
LC-MS/MS analysis
Purified and dried peptide samples were dissolved in a loading buffer containing 5% ACN and 4% formic acid, and 2 μl was injected into a microcapillary column packed with C18 beads using a WPS-3000TPLRS autosampler (UltiMate 3000). After being separated by reversed-phase HPLC using an UltiMate 3000 binary pump, the peptides were detected in a hybrid dual-cell quadrupole linear ion trap-Orbitrap mass spectrometer (LTQ Orbitrap Elite, Thermo Fisher) using a data-dependent Top15 method. A full MS scan (resolution: 60 000) was recorded in the Orbitrap cell with the automatic gain control (AGC) of 106. The peptides were fragmented using higher-energy collision dissociation (HCD) with 40% normalized energy, and fragments were detected in the Orbitrap cell with high resolution and high mass accuracy. Selected precursor ions were excluded from further sequencing for 90 s. Ions with a single or unassigned charge were not sequenced.
Database searching and data filtering
The raw files were converted to the mzXML format and searched against the database containing sequences of all human proteins downloaded from UniProt (Homo sapiens) with the SEQUEST algorithm (version 28).68 The following parameters were used during the search: 20 ppm precursor mass tolerance; 0.025 Da product ion mass tolerance; fully digested with trypsin; up to three missed cleavages; fixed modifications: carbamidomethylation of cysteine (+57.0214); TMT tag of lysine and the peptide N-terminus (+229.1629), variable modifications: oxidation of methionine (+15.9949); 18O tag of Asn (N-glycosylation site) (+2.9883, for glycopeptide identification). The target-decoy method was used to evaluate the false discovery rates (FDRs) for peptide and protein identifications, and linear discriminant analysis (LDA), which integrates several parameters including XCorr, precursor mass error, and charge state, was employed to control the accuracy of peptide identifications.69,70 Peptides with fewer than seven amino acid residues were removed. Peptide were filtered to <1% FDR, and then proteins were further filtered to have FDR <1%. The TMT reporter ion intensities in the tandem mass spectra were employed to quantify the identified peptides. pLink2 was used for the crosslinked peptide search against a database containing sequences of both surface glycoproteins downloaded from Uniprot and proteins that were defined as the surface glycoprotein interactors in the work.71 The settings of pLink2 included: peptide mass: 500–6000; peptide length: 5–60; 20 ppm precursor mass tolerance; 20 ppm product ion mass tolerance; fixed modification: carbamidomethylation of cysteine; variable modification: oxidation of methionine. FDR was set to <5% at the spectral level with 10 ppm MS1 filter tolerance according to the recommendation of the developer.
Bioinformatic analysis
Protein functional annotation was performed with the Database for Annotation, Visualization and Integrated Discovery (DAVID).72 For the volcano plots, P-values were calculated using Perseus, in which a one-sample t-test (S0 = 0) was performed.73 Proteins were considered being enriched when the abundance changed by >2-fold compared to the control group and the P-value was <0.05. Protein localization was downloaded from UniProt.74 Protein network was constructed through the IntAct database and visualized by Cytoscape.75,76
Conflicts of interest
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (R01GM127711).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc06327d
References
- Uings I. J. Farrow S. N. Mol. Pathol. 2000;53:295–299. doi: 10.1136/mp.53.6.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan V. Packer N. H. Thaysen-Andersen M. Expert Rev. Respir. Med. 2013;7:553–576. doi: 10.1586/17476348.2013.837752. [DOI] [PubMed] [Google Scholar]
- Martinez-Martin N. Ramani S. R. Hackney J. A. Tom I. Wranik B. J. Chan M. Wu J. Paluch M. T. Takeda K. Hass P. E. Clark H. Gonzalez L. C. Nat. Commun. 2016;7:11473. doi: 10.1038/ncomms11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel D. Comps-Agrar L. Brock C. Rives M.-L. Bourrier E. Ayoub M. A. Bazin H. Tinel N. Durroux T. Prézeau L. Trinquet E. Pin J.-P. Nat. Methods. 2008;5:561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin J. A. Hunter T. Palsson B. O. Subramaniam S. Nat. Rev. Mol. Cell Biol. 2005;6:99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- Paek J. Kalocsay M. Staus D. P. Wingler L. Pascolutti R. Paulo J. A. Gygi S. P. Kruse A. C. Cell. 2017;169:338–349. doi: 10.1016/j.cell.2017.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Seara Monne H. Danne R. Róg T. Ilpo V. Gurtovenko A. Biophys. J. 2013;104:251a. doi: 10.1016/j.bpj.2012.11.1412. [DOI] [Google Scholar]
- Möckl L. Pedram K. Roy A. R. Krishnan V. Gustavsson A.-K. Dorigo O. Bertozzi C. R. Moerner W. E. Dev. Cell. 2019;50:57–72. doi: 10.1016/j.devcel.2019.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudette S. Hughes D. Boller M. J. Vet. Emerg. Crit. Car. :18. doi: 10.1111/vec.12925. [DOI] [PubMed] [Google Scholar]
- Bausch-Fluck D. Milani E. S. Wollscheid B. Curr. Opin. Chem. Biol. 2019;48:26–33. doi: 10.1016/j.cbpa.2018.09.020. [DOI] [PubMed] [Google Scholar]
- Xiao H. Sun F. Suttapitugsakul S. Wu R. Mass Spectrom. Rev. 2019;38:356–379. doi: 10.1002/mas.21586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H. Suttapitugsakul S. Sun F. Wu R. Acc. Chem. Res. 2018;51:1796–1806. doi: 10.1021/acs.accounts.8b00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaysen-Andersen M. Packer N. H. Schulz B. L. Mol. Cell. Proteomics. 2016;15:1773–1790. doi: 10.1074/mcp.O115.057638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttapitugsakul S. Ulmer L. D. Jiang C. D. Sun F. X. Wu R. H. Anal. Chem. 2019;91:6934–6942. doi: 10.1021/acs.analchem.9b01447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho S. S. Reis C. A. Nat. Rev. Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- Chen W. X. Smeekens J. M. Wu R. H. Chem. Sci. 2015;6:4681–4689. doi: 10.1039/C5SC01124H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English N. M. Lesley J. F. Hyman R. Cancer Res. 1998;58:3736. [PubMed] [Google Scholar]
- Katoh S. Zheng Z. Oritani K. Shimozato T. Kincade P. W. J. Exp. Med. 1995;182:419–429. doi: 10.1084/jem.182.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swietlik J. J. Sinha A. Meissner F. Curr. Opin. Cell Biol. 2020;63:20–30. doi: 10.1016/j.ceb.2019.12.002. [DOI] [PubMed] [Google Scholar]
- Huttlin E. L. Bruckner R. J. Navarrete-Perea J. Cannon J. R. Baltier K. Gebreab F. Gygi M. P. Thornock A. Zarraga G. Tam S. Szpyt J. Panov A. Parzen H. Fu S. Golbazi A. Maenpaa E. Stricker K. Thakurta S. G. Rad R. Pan J. Nusinow D. P. Paulo J. A. Schweppe D. K. Vaites L. P. Harper J. W. Gygi S. P. bioRxiv. 2020 doi: 10.1101/2020.01.19.905109. [DOI] [Google Scholar]
- Hein M. Y. Hubner N. C. Poser I. Cox J. Nagaraj N. Toyoda Y. Gak I. A. Weisswange I. Mansfeld J. Buchholz F. Hyman A. A. Mann M. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- Rees J. S. Li X.-W. Perrett S. Lilley K. S. Jackson A. P. Mol. Cell. Proteomics. 2015;14:2848–2856. doi: 10.1074/mcp.R115.052902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon T. C. Bosch J. A. Sanchez A. D. Udeshi N. D. Svinkina T. Carr S. A. Feldman J. L. Perrimon N. Ting A. Y. Nat. Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S. Kotani N. Ohnishi T. Miyagawa-Yamguchi A. Tsuda M. Yamashita R. Ishiura Y. Honke K. Proteomics. 2011;12:54–62. doi: 10.1002/pmic.201100551. [DOI] [PubMed] [Google Scholar]
- Rees J. S. Li X.-W. Perrett S. Lilley K. S. Jackson A. P. Curr. Protoc. Protein Sci. 2017;88:19–.27.11. doi: 10.1002/cpps.27. [DOI] [PubMed] [Google Scholar]
- Liu Q. Zheng J. Sun W. Huo Y. Zhang L. Hao P. Wang H. Zhuang M. Nat. Methods. 2018;15:715–722. doi: 10.1038/s41592-018-0100-5. [DOI] [PubMed] [Google Scholar]
- Sobotzki N. Schafroth M. A. Rudnicka A. Koetemann A. Marty F. Goetze S. Yamauchi Y. Carreira E. M. Wollscheid B. Nat. Commun. 2018;9:1519. doi: 10.1038/s41467-018-03936-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei A. P. Jeon O.-Y. Kilcher S. Moest H. Henning L. M. Jost C. Pluckthun A. Mercer J. Aebersold R. Carreira E. M. Wollscheid B. Nat. Biotechnol. 2012;30:997–1001. doi: 10.1038/nbt.2354. [DOI] [PubMed] [Google Scholar]
- Wu H. Kohler J. Curr. Opin. Chem. Biol. 2019;53:173–182. doi: 10.1016/j.cbpa.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. Collins B. E. Bengtson P. Paulson J. C. Nat. Chem. Biol. 2005;1:93–97. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- Li Q. Xie Y. Xu G. Lebrilla C. B. Chem. Sci. 2019;10:6199–6209. doi: 10.1039/C9SC01360A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corgiat B. A. Nordman J. C. Kabbani N. Front. Pharmacol. 2014;4:171. doi: 10.3389/fphar.2013.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. Huang L. Anal. Chem. 2018;90:144–165. doi: 10.1021/acs.analchem.7b04431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F. Suttapitugsakul S. Wu R. Anal. Chem. 2019;91:4195–4203. doi: 10.1021/acs.analchem.9b00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. N. Xiao H. P. Wu R. H. Angew. Chem., Int. Ed. 2017;56:7107–7111. doi: 10.1002/anie.201702191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai N. Lee Y. Youn B.-S. Fukuda S. Kim Y.-J. Mantel C. Akashi M. Broxmeyer H. E. Blood. 2002;99:4413–4421. doi: 10.1182/blood.V99.12.4413. [DOI] [PubMed] [Google Scholar]
- Horváth G. Serru V. Clay D. Billard M. Boucheix C. Rubinstein E. J. Biol. Chem. 1998;273:30537–30543. doi: 10.1074/jbc.273.46.30537. [DOI] [PubMed] [Google Scholar]
- Rabinovich G. A. Croci D. O. Immunity. 2012;36:322–335. doi: 10.1016/j.immuni.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Dam T. K. Brewer F. C. Glycobiology. 2010;20:1061–1064. doi: 10.1093/glycob/cwq084. [DOI] [PubMed] [Google Scholar]
- Dennis J. W. Nabi I. R. Demetriou M. Cell. 2009;139:1229–1241. doi: 10.1016/j.cell.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaz L. Bonnyay D. Mahnke K. Rivera M. Nussenzweig M. C. Steinman R. M. J. Exp. Med. 2002;196:1627–1638. doi: 10.1084/jem.20021598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahnke K. Guo M. Lee S. Sepulveda H. Swain S. L. Nussenzweig M. Steinman R. M. J. Cell Biol. 2000;151:673–684. doi: 10.1083/jcb.151.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga H. Ueda H. Neurochem. Int. 2008;52:1076–1085. doi: 10.1016/j.neuint.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Kundranda M. N. Henderson M. Carter K. J. Gorden L. Binhazim A. Ray S. Baptiste T. Shokrani M. Leite-Browning M. L. Jahnen-Dechent W. Matrisian L. M. Ochieng J. Cancer Res. 2005;65:499. [PubMed] [Google Scholar]
- Morales F. C. Takahashi Y. Momin S. Adams H. Chen X. Georgescu M.-M. Mol. Cell. Biol. 2007;27:2527. doi: 10.1128/MCB.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingley E. Cell Commun. Signaling. 2012;10:21. doi: 10.1186/1478-811X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M. Roux A. Nat. Rev. Mol. Cell Biol. 2018;19:313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- Jiang Z. Liang Z. Shen B. Hu G. BioMed. Res. Int. 2015;2015:792904. doi: 10.1155/2015/792904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama F. Thevapala S. Ridley A. J. J. Cell Sci. 2012;125:3310. doi: 10.1242/jcs.094383. [DOI] [PubMed] [Google Scholar]
- Seifried A. Schultz J. Gohla A. FEBS J. 2013;280:549–571. doi: 10.1111/j.1742-4658.2012.08633.x. [DOI] [PubMed] [Google Scholar]
- Shoemaker B. A. Panchenko A. R. PLoS Comput. Biol. 2007;3:e42. doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingström T. Plewczynski D. Briefings Bioinf. 2010;12:702–713. doi: 10.1093/bib/bbq064. [DOI] [PubMed] [Google Scholar]
- Stelzl U. Worm U. Lalowski M. Haenig C. Brembeck F. H. Goehler H. Stroedicke M. Zenkner M. Schoenherr A. Koeppen S. Timm J. Mintzlaff S. Abraham C. Bock N. Kietzmann S. Goedde A. Toksöz E. Droege A. Krobitsch S. Korn B. Birchmeier W. Lehrach H. Wanker E. E. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Klapa M. I. Tsafou K. Theodoridis E. Tsakalidis A. Moschonas N. K. BMC Syst. Biol. 2013;7:96. doi: 10.1186/1752-0509-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nastou K. C. Tsaousis G. N. Kremizas K. E. Litou Z. I. Hamodrakas S. J. BioMed Res. Int. 2014;2014:397145. doi: 10.1155/2014/397145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A.-L. Oltvai Z. N. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Lima-Mendez G. van Helden J. Mol. BioSyst. 2009;5:1482–1493. doi: 10.1039/B908681A. [DOI] [PubMed] [Google Scholar]
- Little K. D. Hemler M. E. Stipp C. S. Mol. Biol. Cell. 2004;15:2375–2387. doi: 10.1091/mbc.e03-12-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draber P. Vonkova I. Stepanek O. Hrdinka M. Kucova M. Skopcova T. Otahal P. Angelisova P. Horejsi V. Yeung M. Weiss A. Brdicka T. Mol. Cell. Biol. 2011;31:4550. doi: 10.1128/MCB.05817-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vences-Catalán F. Duault C. Kuo C.-C. Rajapaksa R. Levy R. Levy S. Biochem. Soc. Trans. 2017;45:531–535. doi: 10.1042/BST20160478. [DOI] [PubMed] [Google Scholar]
- O'Reilly F. J. Rappsilber J. Nat. Struct. Mol. Biol. 2018;25:1000–1008. doi: 10.1038/s41594-018-0147-0. [DOI] [PubMed] [Google Scholar]
- Fürsch J. Kammer K.-M. Kreft S. G. Beck M. Stengel F. Anal. Chem. 2020;92:4016–4022. doi: 10.1021/acs.analchem.9b05559. [DOI] [PubMed] [Google Scholar]
- Steigenberger B. Pieters R. J. Heck A. J. R. Scheltema R. A. ACS Cent. Sci. 2019;5:1514–1522. doi: 10.1021/acscentsci.9b00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez J. D. Tang X. Campbell M. D. Reyes G. Kramer P. A. Stuppard R. Keller A. Zhang H. Rabinovitch P. S. Marcinek D. J. Bruce J. E. Proc. Natl. Acad. Sci. U. S. A. 2020;117:15363–15373. doi: 10.1073/pnas.2002250117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D. Li Q. Zhang M. J. Liu C. Ma C. Zhang P. Ding Y. H. Fan S. B. Tao L. Yang B. Li X. Ma S. Liu J. Feng B. Liu X. Wang H. W. He S. M. Gao N. Ye K. Dong M. Q. Lei X. Elife. 2016;5:e12509. doi: 10.7554/eLife.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigenberger B. Albanese P. Heck A. J. R. Scheltema R. A. J. Am. Soc. Mass Spectrom. 2020;31:196–206. doi: 10.1021/jasms.9b00085. [DOI] [PubMed] [Google Scholar]
- Liu F. Lössl P. Scheltema R. Viner R. Heck A. J. R. Nat. Commun. 2017;8:15473. doi: 10.1038/ncomms15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J. K. McCormack A. L. Yates J. R. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Elias J. E. Gygi S. P. Nat. Methods. 2007;4:207. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Kall L. Canterbury J. D. Weston J. Noble W. S. MacCoss M. J. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Chen Z.-L. Meng J.-M. Cao Y. Yin J.-L. Fang R.-Q. Fan S.-B. Liu C. Zeng W.-F. Ding Y.-H. Tan D. Wu L. Zhou W.-J. Chi H. Sun R.-X. Dong M.-Q. He S.-M. Nat. Commun. 2019;10:3404. doi: 10.1038/s41467-019-11337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W. Sherman B. T. Lempicki R. A. Nat. Protoc. 2008;4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Tyanova S. Temu T. Sinitcyn P. Carlson A. Hein M. Y. Geiger T. Mann M. Cox J. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Consortium T. U. Nucleic Acids Res. 2018;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S. Ammari M. Aranda B. Breuza L. Briganti L. Broackes-Carter F. Campbell N. H. Chavali G. Chen C. del-Toro N. Duesbury M. Dumousseau M. Galeota E. Hinz U. Iannuccelli M. Jagannathan S. Jimenez R. Khadake J. Lagreid A. Licata L. Lovering R. C. Meldal B. Melidoni A. N. Milagros M. Peluso D. Perfetto L. Porras P. Raghunath A. Ricard-Blum S. Roechert B. Stutz A. Tognolli M. van Roey K. Cesareni G. Hermjakob H. Nucleic Acids Res. 2013;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. Markiel A. Ozier O. Baliga N. S. Wang J. T. Ramage D. Amin N. Schwikowski B. Ideker T. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.