Abstract
Superovulation with gonadotropins alters the hormonal milieu during early embryo development and placentation, and may be responsible for fetal and placental changes observed after in vitro fertilization (IVF). We hypothesized that superovulation has differential effects depending on timing of exposure. To test our hypothesis, we isolated the effect of superovulation on pre- and peri-implantation mouse embryos. Blastocysts were obtained from either natural mating or following superovulation and mating, and were transferred into naturally mated or superovulated pseudopregnant recipient mice. Fetal weight was significantly lower after peri-implantation exposure to superovulation, regardless of preimplantation exposure (p = 0.006). Placentas derived from blastocysts exposed to superovulation pre- and peri-implantation were larger than placentas derived from natural blastocysts that are transferred into a natural or superovulated environment (p < 0.05). Fetal-to-placental weight ratio decreased following superovulation during the pre- or peri-implantation period (p = 0.05, 0.01, respectively) and these effects were additive. Peg3 DNA methylation levels were decreased in placentas derived from exposure to superovulation both pre- and peri-implantation compared with unexposed embryos and exposure of the preimplantation embryo only. Through RNA sequencing on placental tissue, changes were identified in genes involved in immune system regulation, specifically interferon signaling, which has been previously implicated in implantation and maintenance of early pregnancy in mice. Overall, we found that the timing of exposure to gonadotropin stimulation can have differential effects on fetal and placental growth. These findings could impact clinical practice and underscores the importance of dissecting the role of procedures utilized during IVF on pregnancy complications.
Keywords: in vitro fertilization (IVF), embryo, placenta, implantation, epigenetics
Preimplantation embryo exposure to superovulation affects placental growth, whereas peri-implantation exposure affects fetal growth. Placental effects occur via changes in immune-related gene expression and epigenetic changes in growth-related genes.
Introduction
Approximately one in eight couples in the United States suffer from infertility [1]. The most effective treatment for infertility is in vitro fertilization (IVF), which has led to the birth of more than 8 million babies worldwide and over 65 000 infants born in the United States annually [1]. IVF involves multiple interventions: the ovaries are stimulated with gonadotropins, or superovulated, leading to the development of multiple mature follicles; oocytes are surgically retrieved; oocytes are fertilized in vitro, and embryos are cultured in the laboratory for 3–5 days. Following in vitro culture, the embryos may be immediately transferred to the uterus and exposed to supraphysiological hormone levels resulting from the prior gonadotropin stimulation (fresh IVF), or vitrified for later use in a frozen embryo transfer (FET), where the hormonal environment of the recipient is more physiological [2]. All of these procedures could potentially impact the developing embryo and affect pregnancy outcomes.
Although the vast majority of infants born after IVF are healthy, epidemiologic data suggest that, compared with unassisted pregnancies, even singleton pregnancies conceived by IVF are associated with an increased risk of adverse perinatal outcomes. These include low birthweight, small for gestational age, and hypertensive disorders of pregnancy, such as preeclampsia [3–5]. Multiple studies have also demonstrated lower birthweight among infants born after fresh IVF compared with infants born following FET [6–10]. The specific intervention(s) and mechanisms responsible for these adverse outcomes are unclear, but studies indicate that superovulation leads to elevated serum estradiol levels, changes in serum/endometrial vascular endothelial growth factor (VEGF) expression, and placental/cord blood epigenetic changes, which may contribute to these adverse outcomes [11–16]. Mouse models of IVF have sought to further explore these mechanisms, as they allow for the independent assessment of individual interventions, which cannot be done in humans. Data from mouse models have shown that specific IVF procedures can lead to a decrease in fetal weight and placental abnormalities [17–19]. Embryos obtained after superovulation undergo DNA methylation changes in growth-related imprinted genes [20, 21]. Our laboratory has previously shown that transfer of embryos into the superovulated environment may affect placental vasculogenesis, which may be the result of altered VEGF levels during early implantation [17]. We have also previously shown that serum estradiol levels of superovulated mice are twice that of control mice during oocyte maturation and preimplantation but that these levels return to normal by the time of implantation, whereas the level of VEGF remains high after superovulation [17]. However, the critical window of exposure to gonadotropins and the resulting hormonal milieu, and the cumulative effect of these exposures remains unclear and extremely difficult to study in human IVF.
In this paper, we utilize a unique study design to isolate the effects of superovulation on pregnancy outcomes (Figure 1). This design allows us to transfer blastocysts obtained through superovulation or natural mating together into either natural or superovulated recipient mice, and identify each embryo’s individual exposure. In our model, we can separately examine pre- and peri-implantation effects of superovulation, as well as the combined effect. Preimplantation exposure involves exposure of the maturing oocyte and preimplantation embryo to the supraphysiological hormonal environment created though superovulation. Peri-implantation exposure includes exposure of both the uterus and implanting embryo to an altered environment during key events such as apposition, adhesion, and fetal trophoblast invasion [22]. A major advantage of our transgenic model is that natural and superovulated blastocysts can be transferred into a single recipient dam, and blastocyst origin can be ascertained through the use of the green fluorescent protein (GFP) tag (Figure 1A). This model generates four experimental groups: (1) natural blastocysts into natural recipients (Nat–Nat): a control group, allowing for comparable litter size and accounting for any effects from embryo transfer alone; (2) superovulated blastocysts into natural recipients (SO–Nat): examining preimplantation exposure and simulating the hormonal environment of a natural frozen-thawed embryo transfer cycle; (3) natural blastocysts into superovulated recipients (Nat–SO): examining the isolated effect of peri-implantation exposure, which cannot be examined in humans; and (4) superovulated blastocysts into superovulated recipients (SO–SO): examining the combined pre- and peri-implantation effect of superovulation, mimicking the hormonal environment of a human fresh IVF cycle (Figure 1B).
Figure 1.
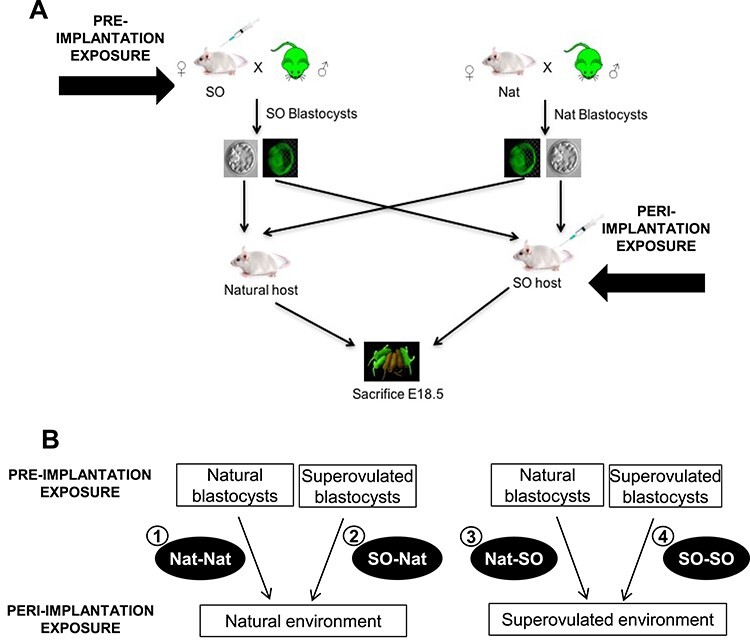
Experimental design: (A) Female mice were either superovulated and mated or naturally mated to males heterozygous for GFP. Blastocysts were obtained on postcoital day 3.5. Pseudopregnant females were generated through either natural mating or superovulation and mating to vasectomized males. Ten blastocysts (five from natural mating and five from superovulation and mating) were transferred into each recipient and GFP status was used to “label” the embryos (e.g., five GFP superovulated blastocysts were transferred along with five GFP-negative natural blastocysts into each pseudopregnant host). The reciprocal transfers were also performed. Pregnant mice were sacrificed at E18.5. (B) Four experimental groups were generated: (1) Nat–Nat; control group, allowing for comparable litter size and accounting for any affects from embryo transfer alone (n = 14); (2) SO–Nat; preimplantation exposure, simulating the hormonal environment(s) of a natural frozen-thawed embryo transfer cycle (n = 24); (3) Nat–SO; peri-implantation exposure that is not performed clinically (n = 12); (4) SO–SO; pre- and peri-implantation exposure, mimicking the hormonal milieu of a fresh IVF cycle (n = 14).
We hypothesize that superovulation has differential effects on fetal growth, placental development, DNA methylation, and vasculogenesis when the maturing oocyte and preimplantation embryo are exposed, compared to when the uterus and peri-implantation embryo are exposed, to a supraphysiological hormonal milieu. In this study, we aimed to isolate the critical windows of exposure responsible for the altered growth phenotype, and to determine potential mechanism(s) responsible for changes seen following superovulation.
Materials and methods
Embryo collection and transfer
All experiments and procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Six-week-old mice were obtained and housed in a temperature- and light-controlled environment with a 12-h dark/12-h light cycle and fed ad libitum. Female CF1 mice (Harlan Laboratories, Indianapolis, IN, USA) were either naturally mated or superovulated with intraperitoneal injections of 5 IU of pregnant mare serum gonadotropin (PMSG, EMD Millipore, Billerica, MA, USA) followed by 5 IU of human chorionic gonadotropin (hCG, Sigma-Aldrich, St. Louis, MO, USA) 48 h later and mated with male mice heterozygous for GFP (C57BL/6-Tg[CAG-EGFP]1Osb/J, Jackson Laboratory, Bar Harbor, ME, USA). Mating was confirmed on the morning following mating with the presence of a copulatory plug (postcoital day 0.5). On postcoital day 3.5, blastocysts were flushed from the uterine horns using standard techniques and separated by their GFP status.
Blastocysts were transferred into pseudopregnant CF1 female mice derived either through natural mating to vasectomized B6D2F1/J males (Jackson Laboratory) or superovulation (5 IU PMSG and 5 IU hCG), followed by mating to vasectomized B6D2F1/J males. The presence of a copulatory plug on postcoital day 0.5 confirmed mating. Ten blastocyst-stage embryos, five from superovulated dams and five from naturally mated dams, were non-surgically transferred into a single horn of each pseudopregnant female on postcoital day 3.5 using the non-surgical embryo transfer device (Paratechs, Lexington, KY, USA) per the manufacturer’s protocol. GFP status was used to distinguish whether the blastocysts were of superovulated or natural origin. Reciprocal transfers were performed to ensure observed changes were not due to the insertion of the GFP transgene.
Fetal evaluation and tissue collection
Pregnant recipient mice were sacrificed 15 days post-embryo transfer at E18.5. The number of implantation sites and resorption sites were counted for each recipient. The amniotic membranes were ruptured, and the conceptuses (fetus and placenta) were carefully dissected from the uterine horn. Fetus and placenta were separated and weighed. Each placenta was cut through the midplacental plane. Half the placenta was placed in 10% phosphate-buffered formalin for histological examination. The other half of the placenta as well as fetal liver, tail, and brain were snap frozen and stored at −80°C. GFP status of fetus and placenta was determined with the use of a fluorescent filter and confirmed with polymerase chain reaction (PCR) in order to determine whether the fetuses originated from embryos from superovulation or natural mating.
Histological analysis
Placentas were kept in formalin overnight at 4°C, dehydrated, and embedded in a paraffin block. Care was taken to orient the bivalved section vertically in order to obtain cross-sections of the placenta. Serial tissue sections of 4 μm thickness were cut and mounted on glass slides by the Abramson Cancer Center Histology Core (University of Pennsylvania). Tissues were deparaffinized using citrate buffer, pH 6.0 (Millipore) and stained with hematoxylin and eosin (H&E). Slides were digitized at the histology core at the Children’s Hospital of Philadelphia (CHOP, Philadelphia, PA, USA). Spectrum software (Leica Biosystems, Buffalo Grove, IL, USA) was used to view images, to outline each placental zone and calculate areas. All measurements were made through the midsagittal plane of the placenta and performed as previously described [23]. For the microvessel analysis, tissues were deparaffinized using citrate buffer and stained with monoclonal antibody to Plasmalemma vesicle-associated protein (PLVAP), clone MECA-32 (Bio-Rad, Raleigh, NC, USA). Slides were digitized at the histology core at the CHOP and quantification of vessel density was performed using Spectrum and ImageJ software [24]. Eight Nat–Nat placentas, 12 SO–Nat, 9 Nat-SO, and 7 SO–SO placentas were analyzed.
DNA/RNA extraction
Genomic DNA was extracted from frozen placenta, liver, and tail using the standard phenol chloroform method [25]. RNA extraction and isolation were performed using the Qiagen RNAeasy micro kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. RNA concentration and quality were determined with a NanoDrop spectrophotometer (Thermo Fischer Scientific, Waltham, MA, USA).
Sex determination
Sex determination of the fetus was performed using isolated tail DNA. PCR was performed using primers for the Sry gene: Sry forward primer: 5′- TTG TCT AGA GAG CAT GGA GGG CCA TGT CAA-3′, and reverse primer: 5’-CCA CTC CTC TGT GAC ACT TTA GCC CTC CGA-3′ as previously described [23].
LUminometric methylation assay (LUMA)
Samples of genomic DNA (2 μg) were digested with HpaII + EcoRI and MspI + EcoRI in parallel reactions containing 2 μL of Tango buffer (Fermentas), 5 U of HpaII or MspI, 5 U of EcoRI and dH2O to a final volume of 20 μL. Samples were incubated at 37°C for 4 h and stored overnight at 4°C. Samples were removed, briefly spun down, and 15 μL of pyrosequencing annealing buffer (Qiagen) was subsequently added to each sample. The reactions were mixed and 30 μL of the sample reaction containing the annealing buffer was transferred to a 96-well pyrosequencing plate. Nucleotides were dispensed in the following order: GTGTCACATGTGTG. Peak heights of nucleotide incorporation from the resulting pyrograms were used to calculate percent genomic DNA methylation using the formula: 1-[(HpaII(G)/EcoRI(T))/(MspI(G)/EcoRI(T))] × 100, where G and T are the peak heights for HpaII or MspI (methylation) and EcoRI (input DNA), respectively. All samples were run in triplicate.
Bisulfite mutagenesis and pyrosequencing
Bisulfite mutagenesis was performed with 2 μg of isolated genomic DNA from term embryonic liver and placenta, using a bisulfite kit (Qiagen) according to the manufacturer’s protocol. One microliter of bisulfite-treated DNA was used for PCR amplification using nested primers (Supplemental Table S1). The PyroMark PCR kit (Qiagen) was used in a 25 μL reaction. PCR conditions were: 95°C for 15 min followed by 45 cycles of 95°C for 15 s, 55°C for 30 s and 72°C for 15 s, 72°C for 10 min. Five microliters of the biotinylated PCR product was used for each sequencing assay with the respective sequencing primer. Pyrosequencing was done using the PyroMark Q96MD (Qiagen) system according to the manufacturer’s protocol and the PyroMark Gold 96 reagents kit (Qiagen). Methylation levels were analyzed using Qiagen’s Pyro Q-CpG software.
Statistical analysis
The primary outcome of the study was fetal weight. An a priori one-way analysis of variance (ANOVA) power calculation was performed and determined that nine fetuses per group would provide an 80% power to detect a 15% difference in fetal weight with an alpha of 0.05. One-way ANOVA followed by Tukey multiple comparisons testing were used to evaluate between-group differences when all four experimental groups were compared. Student t-test was used to compare differences between groups when comparing two individual groups. Two-way ANOVA with Sidak multiple comparisons was used to examine sex ratio. Multiple linear regression models were used to control for confounding factors, such as litter size. Linear mixed-effects modeling was used to account for any correlation between pups within a litter. Fisher exact test was used for categorical data. Results are presented as mean ± standard error of mean (SEM). A p-value < 0.05 was considered significant. Statistical analyses were performed with GraphPad Prism version 7.0 (San Diego, CA, USA) and STATA version 14 (StataCorp, College Station, TX, USA).
RNA sequencing (RNA-seq)
A total of 16 placentas, 4 from each exposure, were selected for RNA-seq, with no more than one pup of each exposure per litter selected. RNA library preparations and sequencing reactions were conducted at GENEWIZ, LLC. (South Plainfield, NJ, USA). RNA samples received were quantified using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and RNA integrity was checked using Agilent TapeStation 4200 (Agilent Technologies, Palo Alto, CA, USA). RNA-seq libraries were prepared using the NEBNext Ultra RNA library prep kit for Illumina using manufacturer’s instructions (NEB, Ipswich, MA, USA). Briefly, mRNAs were initially enriched with Oligod(T) beads. Enriched mRNAs were fragmented for 15 min at 94°C. First- and second-strand cDNA were subsequently synthesized. Complementary DNA fragments were end repaired and adenylated at 3′ends, and universal adapters were ligated to cDNA fragments, followed by index addition and library enrichment by PCR with limited cycles. The sequencing library was validated on the Agilent TapeStation (Agilent Technologies) and quantified by using Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) as well as by quantitative PCR (KAPA Biosystems, Wilmington, MA, USA). The sequencing libraries were clustered on a single lane of a flow cell. After clustering, the flowcell was loaded on the Illumina HiSeq instrument (4000 or equivalent) according to the manufacturer’s instructions. The samples were sequenced using a 2 × 150-bp paired-end configuration. An average of 25 million reads were generated per sample. Image analysis and base calling were conducted by the HiSeq Control Software. Raw sequence data (.bcl files) generated from Illumina HiSeq were converted into fastq files and demultiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification.
After demultiplexing, sequence data were checked for overall quality and yield. Sequence reads were then trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were mapped to the reference genome GRCh38 available on ENSEMBL using the STAR aligner v.2.5.2b. The STAR aligner is a splice aware aligner that detects splice junctions and incorporates them to help align the entire read sequences. BAM files were generated as a result of this step. Unique gene hit counts were calculated by using featureCounts from the Subread package v.1.5.2. Only unique reads within exon regions were counted. After extraction of gene hit counts, the gene hit counts table was used for downstream differential expression analysis. Using DESeq2, a comparison of gene expression between the groups of samples was performed. The Wald test was used to generate p-values and Log2 fold changes. Genes with adjusted p-values < 0.1 and absolute log2 fold changes >1 were called as differentially expressed genes for each comparison. A gene ontology analysis was performed on the statistically significant set of genes by implementing the software GeneSCF v1.1. The GO list was used to cluster the set of genes based on their biological process and determine their statistical significance.
Gene expression analysis
Synthesis of cDNA was performed using the iScript Reverse Transcription Supermix (BioRad) using 1 μg of RNA. Real-time quantitative PCR (RT-qPCR) was performed with the QuantStudio 7 RT-qPCR system (Applied Biosystems). Each reaction was performed in 10 μL with 50 ng of cDNA template. Each sample was run in triplicate, and the reactions were carried out for 40 cycles. The Taqman probe used for the Ctss gene was: Mm01255859_m1. Gene expression was normalized to reference gene H2A and relative gene expression was calculated using the delta–delta Cycle threshold (Ct) method and then normalized to the Nat–Nat group.
Results
Superovulation may not affect implantation rate, pregnancy rate, litter size or fetal sex ratio
A total of 430 embryos (215 natural, 215 superovulated) were transferred into 43 pseudopregnant recipient mice obtained from mating to vasectomized male mice (22 naturally mated pseudopregnant female mice, 21 superovulated and mated pseudopregnant female mice). A total of 23 pregnancies were obtained, 15 in natural recipients and 8 in superovulated recipients, accounting for a pregnancy rate of 68% and 38% in natural recipients and superovulated recipients, respectively (p = 0.05) (Supplemental Table S2). Implantation rate was calculated as a percentage of the number of implantation sites by the number of embryos transferred. An implantation rate of 27% and 18% was observed in natural recipients and superovulated recipients, respectively (p = 0.24) (Supplemental Table S2, Supplemental Figure S2). Mean litter size was 2.5 (range 1–6) among natural recipients and 3.3 (range 1–6) among superovulated recipients, but the difference in litter size was not statistically significant (p = 0.29) (Supplemental Table S2, Supplemental Figure S2). We also examined implantation rate by GFP status and found no significant differences between the groups (p = 0.17) (Supplemental Table S2).
Overall, 64 fetuses were obtained from 15 natural recipients and 12 superovulated recipients. Among the natural recipients, there were 14 Nat–Nat fetuses and 24 SO–Nat fetuses. Among the superovulated recipients, there were 12 Nat–SO fetuses and 14 SO–SO fetuses. There were 29 male fetuses (5 Nat–Nat, 14 SO–Nat, 6 Nat-SO, and 4 SO–SO) and 35 female fetuses (9 Nat–Nat, 10 SO–Nat, 6 Nat–SO, and 10 SO–SO). The sex ratio of males to females in each group was also calculated to determine whether there was a difference between the groups. The ratio of males to females was 0.6 (5 males, 9 females) in the Nat–Nat group, 1.4 (14 males, 10 females) in the SO–Nat group, 1.0 (6 males, 6 females) in the Nat–SO group, and 0.4 (4 males, 10 females) in the SO–SO group. A two-way ANOVA with Sidak multiple comparisons determined that there was no significant difference in sex ratio between the groups.
Fetal weight decreases when embryos are exposed to a stimulated uterine environment peri-implantation
Fetal weights were assessed near term in the mouse at E18.5. Mean fetal weights were 1.49 ± 0.04 g in the Nat–Nat group, 1.55 ± 0.04 g in the SO–Nat group, 1.30 ± 0.05 g in the Nat–SO group, and 1.26 ± 0.07 g in the SO–SO group (Figure 2A). Fetal weight was significantly smaller in the SO–SO group compared with both the Nat–Nat (p < 0.05) and SO–Nat groups (p < 0.001); fetal weight in the Nat–SO group was also significantly smaller than the SO–Nat group (p < 0.01), and there was no difference in fetal weight between the SO–Nat group compared with the Nat–Nat control group (p = 0.85) (Figure 2A). These data suggest that the peri-implantation hormonal milieu created by superovulation affects fetal weight. When results were separated by sex, female mice had a significant decrease in fetal weight with superovulation, though there was insufficient power to detect these trends in male fetuses (Figure 2C). Female fetuses in the SO–SO group were significantly smaller than females in the Nat–Nat (p < 0.01) and SO–Nat groups (p < 0.01), respectively. Female fetuses in the Nat–SO group were also significantly smaller than females in the Nat–Nat (p < 0.05) and SO–Nat groups (p < 0.05) (Figure 2C). No impact of GFP status was seen on fetal weight (Supplemental Figure S1A). Multiple linear regression was performed to control for litter size and the decrease in fetal weight noted in embryos that were exposed to superovulation during peri-implantation, regardless of preimplantation exposure, remained statistically significant (p < 0.05). Mixed linear effects modeling showed that the superovulated peri-implantation environment results in a 0.24 g drop in fetal weight (p = 0.006), but that fetal weight was not affected by the origin of the blastocysts, i.e., whether blastocysts were obtained after superovulation and mating or natural mating (p = 0.541).
Figure 2.
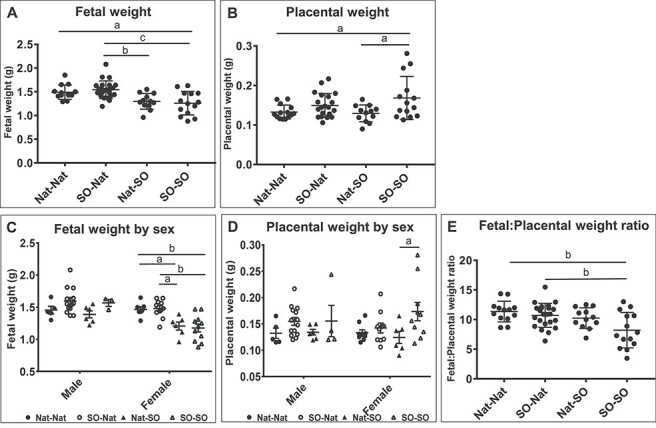
Fetal and placental weight. (A) Fetal weight at E18.5 in the four experimental groups. Fetal weight was significantly smaller in the superovulated peri-implantation groups compared with the natural peri-implantation groups, regardless of preimplantation exposure to superovulation. (B) Placental weight was significantly higher when embryos were exposed to a stimulated preimplantation environment compared with the natural peri-implantation environment. This difference was most pronounced when embryos from a stimulated preimplantation environment were transferred to a stimulated peri-implantation environment. (C) The differences in fetal weight between the superovulated and natural recipient dams persisted when fetal weights were compared for female fetuses but there was insufficient power to detect a difference among the male fetuses. (D) There were no significant differences in placental weight between the groups according to fetal sex. (E) Fetal:placental weight ratio was significantly lower for placentas derived from a stimulated peri-implantation environment. Data are presented as mean ± SEM. a = p < 0.05, b = p < 0.01, c = p < 0.001.
Placental weight increases when preimplantation embryos are exposed to superovulation prior to transfer into a superovulated environment
Placental weight was significantly higher in the SO–SO group compared with the Nat–SO group (p < 0.05) and the Nat–Nat control group (p < 0.05), suggesting that exposure of the preimplantation embryo to superovulation has an effect on placental weight when the embryo is transferred into a superovulated environment (Figure 2B). Mean placental weights were 0.133 ± 0.005 g in the Nat–Nat group, 0.150 ± 0.006 g in the SO–Nat group, 0.129 ± 0.006 g in the Nat–SO group, and 0.168 ± 0.015 g in the SO–SO group. While there was a trend toward higher placental weight in the SO–Nat placentae compared with the Nat–Nat controls, this was not statistically significant (p = 0.53). This suggests that the stimulated preimplantation environment plays a role in placental size, but the effect is more profound when the peri-implantation embryos are transferred into a superovulated environment. This difference was significant in female fetuses, where the placental weight was higher in the SO–SO group compared with the Nat–SO group (p < 0.05), but not in male fetuses, potentially due to lack of power in this group (Figure 2D). GFP status did not have an independent effect on placental weight (Supplemental Figure S1B). Multiple linear regression was performed to control for litter size, and the increase in placental weight noted in preimplantation embryos exposed to superovulation remained significant (p < 0.05). Mixed linear effects modeling showed that placentas derived from superovulated blastocysts weighed 0.02 g higher on average than placentas derived from natural blastocysts (p = 0.006), but peri-implantation exposure to superovulation did not affect placental weight (p = 0.417).
Fetal-to-placental weight ratio is affected by both pre- and peri-implantation exposure to superovulation
Fetal-to-placental weight ratio was decreased in the SO–SO group compared with the Nat–Nat (p < 0.01) and SO–Nat groups (p < 0.01), but not the Nat–SO group (p = 0.10). Mixed linear effects modeling showed that the superovulated uterine environment encountered by the peri-implantation embryo has a negative impact on fetal-to-placental weight ratio (p = 0.01) and that superovulated preimplantation blastocysts have a significantly lower fetal-to-placental weight ratio (p = 0.05) (Figure 2E). Although the fetal-to-placental weight ratio is affected by both pre- and peri-implantation exposure, these effects are additive and do not interact (p = 0.20).
Placental architecture and microvessel density are not significantly affected by pre- or peri-implantation exposure to superovulation
We hypothesized that the changes in fetal and/or placental weight noted in the pups could be due to structural changes within the placenta. To investigate this, the cross-sectional areas of the junctional zone (JZ) and labyrinth zone (Lab) of each placenta were measured and the JZ:Lab ratios were determined. There was a trend toward an increase in JZ:Lab ratio after pre- and peri-implantation superovulation, but this was not statistically significant (Figure 3A and B). We further assessed whether the projections of the JZ into the Lab (JZ invaginations) were altered by the exposure to superovulation but found no differences between the experimental groups (Figure 3C). Previous work from our laboratory has shown differences in uterine arterial resistance and microvessel density after superovulation [23]. We therefore also examined placental microvessel density after staining histological sections from E18.5 placentas with PLVAP(Meca 32), but there was no significant difference between the groups in this model (Supplemental Figure S3).
Figure 3.
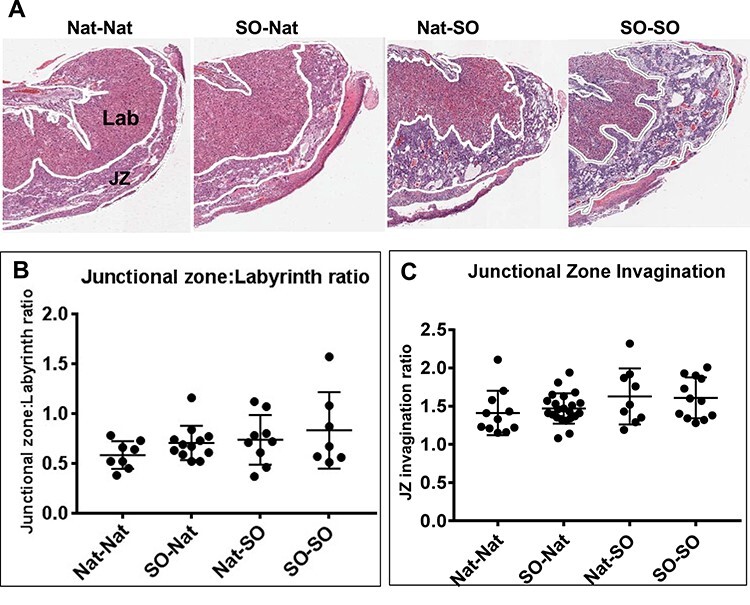
Histopathological examination of E18.5 placentae from the four experimental groups. (A) Representative sections of placentae stained with H&E showing the Lab and JZ. (B) The areas of the JZ and Lab were measured and the ratio of the JZ:Lab calculated. There was a trend toward increasing JZ:Lab ratio, but this was not statistically significant. (C) The invaginations of the JZ did not vary by experimental group.
Global DNA methylation levels in fetal and placental tissue are unaffected by pre- or peri-implantation superovulation
Assisted reproductive technologies, and specifically superovulation, affect DNA methylation in both the placenta and embryo [20, 21, 26, 27]. Global DNA methylation was assessed in E18.5 placenta and embryonic liver. Fetal liver was selected to study alterations in DNA methylation in embryonic tissue given that decreased in-utero fetal growth is associated with a risk of adverse metabolic outcomes [28–32]. Thirty-seven embryos were analyzed for DNA methylation studies: 9 embryos derived from Nat–Nat transfers, 10 embryos derived from SO–Nat transfers, 8 embryos derived from Nat–SO transfers, and 10 embryos derived from SO–SO transfers. Global DNA methylation levels were similar across experimental groups in both placenta and fetal liver (Figure 4A and B). Global DNA methylation remained similar in male fetuses and placentas across experimental groups. Global DNA methylation in the female Nat–SO placentas was significantly higher compared with female SO–Nat placentas (p < 0.05), but there was no difference in global DNA methylation in female livers across experimental groups.
Figure 4.
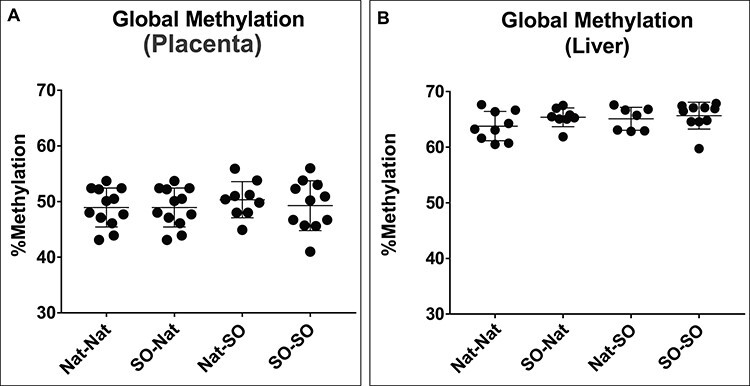
Global methylation at E18.5 as measured by LUMA. (A) Global DNA methylation is similar across groups in placentae. (B) Global DNA methylation is similar across groups in fetal tissue (liver).
Peri-implantation exposure of the embryo to superovulation results in hypomethylation of the Peg3 locus
DNA methylation profiles were analyzed using bisulfite pyrosequencing assays for Peg3, KvDMR1, H19/Igf2 ICR, and Igf2 DMR (Figure 5). Previous studies have shown that many IVF-derived samples exhibit variability in DNA methylation levels within different litters; therefore, single placentae were used as statistical units [33]. There was a significant decrease in average DNA methylation at the Peg3 locus when comparing Nat–Nat and SO–SO placenta and SO–Nat and SO–SO placenta (p < 0.01) (Figure 5A). There was no significant difference in average DNA methylation at KvDMR1, the H19/Igf2 ICR, or Igf2 DMR in placentas (Figure 5B–D).
Figure 5.
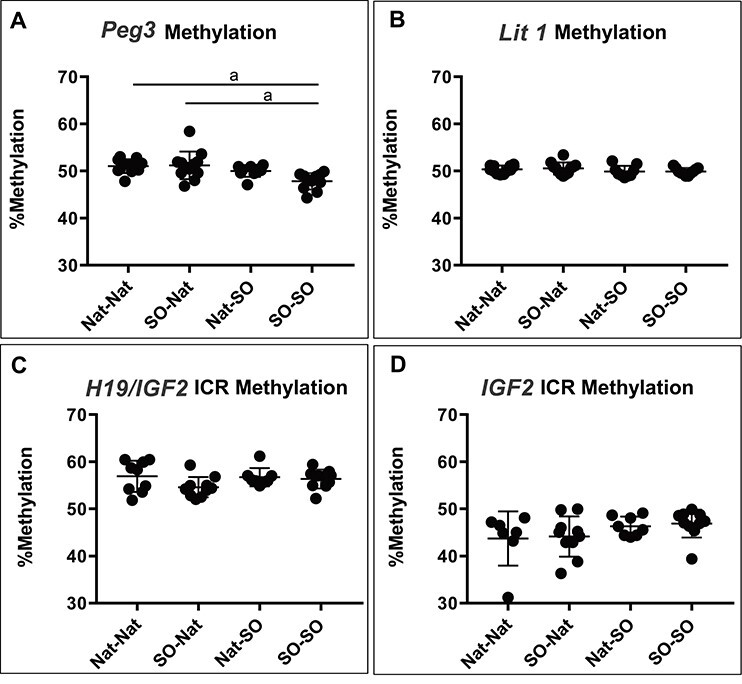
DNA methylation in the placenta. DNA methylation profiles were analyzed using bisulfite pyrosequencing assays for Peg3, KvDMR1, the H19/Igf2 ICR, and Igf2 DMR in placenta. (A) There was a significant decrease in average DNA methylation at the Peg3 locus when comparing the Nat–Nat and the SO–SO placenta and SO–NAT and SO–SO (p < 0.01). (B) There was no significant difference in average DNA methylation at KvDMR1, (C) the H19/Igf2 ICR, or (D) Igf2 DMR in placentas. a = p < 0.01.
Superovulation results in changes of gene expression in genes associated with immune response
To further understand the mechanism(s) responsible for the phenotypic changes seen, we carried out RNA-seq on placental tissue obtained from all four experimental groups. We found gene expression changes in pairwise comparisons between individual groups (Supplemental Table S3). Comparing Nat–Nat vs. Nat–SO and Nat–Nat vs. SO–Nat groups did not show statistically significant phenotypic changes, but led to gene expression changes in 11 and 36 genes, respectively. The most significant changes in gene expression were noted when both the pre- and peri-implantation environments were altered by superovulation, i.e., when SO–SO and Nat–Nat groups were compared. There were a total of 93 genes differentially expressed between these groups. When the effect of superovulation on the peri-implantation embryo was examined (SO–Nat vs. SO–SO), 25 genes were changed. When comparing SO–Nat vs. Nat–SO groups, we found that 24 genes were differentially expressed. While isolated exposure of the preimplantation embryo to superovulation appeared to lead to significant placental weight differences, we did not find any genes changed when we compared the Nat–SO and the SO–SO groups (Figure 6A). We also examined overlap between groups to discern genes commonly changed by exposure to superovulation. We found the largest overlap when the combined pre- and peri-implantation exposure to superovulation was compared with peri-implantation exposure to superovulation only (nine of the same genes were differentially affected in both groups) (Figure 6A). When these results were represented as a Venn diagram [34] to visually examine extent of overlap, we found that each exposure primarily led to unique gene expression changes (Figure 6B). We also looked for common genes that were differentially expressed and found that the gene H2-Q6 was changed in four pairwise comparisons and Naip5 was changed in three pairwise comparisons (Figure 6C, Supplemental Table S4).
Figure 6.
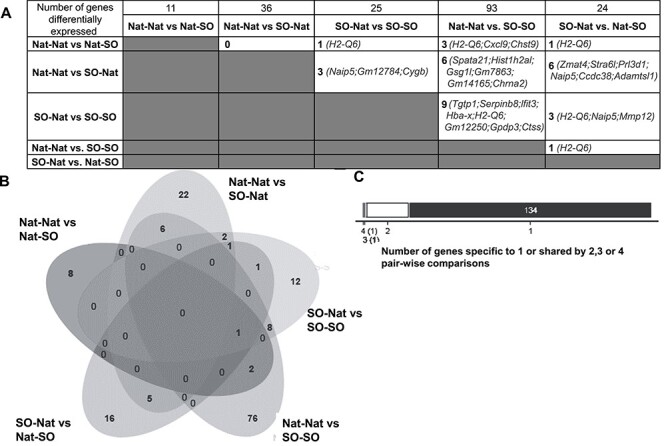
Gene expression changes (A) Table showing pairwise changes in gene expression noted between experimental groups. Total numbers are displayed in the top row: comparing Nat–Nat vs. Nat–SO groups resulted in 11 genes changed, Nat–Nat vs. SO–Nat groups resulted in 36 genes changed, SO–Nat vs. SO–SO groups resulted in 25 genes changed, Nat–Nat vs. SO–SO groups resulted in 93 genes changed and SO–Nat vs. Nat–SO groups resulted in 24 genes changed. Overlap between each pairwise comparison was also examined (numbers and specific genes). (B) Venn diagram visually examining overlap between each pairwise comparison shows that unique genes were differentially expressed when each pairwise comparison was examined. (C) Number of genes shared by four, three, two, or one pairwise comparison was examined.
To identify functional changes that could result from the gene expression changes found, we carried out ingenuity pathway analysis (IPA) on gene sets obtained after comparing Nat–Nat vs. SO–SO, SO–Nat vs. SO–SO, and SO–Nat vs. Nat–SO, all of which showed phenotypic effects on fetal and placental size. Our pathway analysis for all comparisons commonly highlighted canonical pathways, upstream regulators, and diseases or physiological functions associated with the immune response. Genes involved in interferon signaling were specifically highlighted, such as Ifn α/β, Ifn B1, Tmem173, Ifnar1, IfI30, and Irf1 among others. We also attempted to validate several genes that were differentially affected in the SO–SO vs. Nat–Nat and SO–SO vs. SO–Nat groups, since they were most likely to play a role in the phenotypic changes observed between these groups (Supplemental Figure S4A). Many of these genes were virtually undetectable by RT-qPCR with extremely high Ct values when utilizing RNA isolated from whole placenta. Only Ctss demonstrated significant differences in expression in females between Nat–Nat vs. SO–Nat groups, Nat–Nat vs. Nat–SO groups, and Nat-Nat vs. SO–SO groups (Supplemental Figure S4B).
Discussion
IVF is an effective technology for the treatment of infertility and has helped millions of people worldwide to conceive. However, IVF has been associated with an increased risk of adverse obstetric and perinatal outcomes including low birthweight and hypertensive disorders of pregnancy that may be related to changes in placentation [35, 36]. It is not possible to separate out the multiple parts of IVF in humans in order to determine which steps in the process may contribute to adverse outcomes. Therefore, the mouse has been instrumental in isolating which modifiable factor could be contributing to these outcomes.
Prior studies have shown that manipulations involved in IVF, including superovulation, the fertilization of oocytes in vitro, embryo culture, and embryo transfer, can affect fetal and placental development and DNA methylation in mice [17, 18, 20, 21, 23, 26, 27, 33]. We designed the current study to isolate the potential impact of superovulation on the developing oocyte and preimplantation embryo from its effects on the peri-implantation embryo and uterus. In our model, the use of flushed blastocysts allowed us to eliminate the effects of IVF and in vitro embryo culture, as studies show that these manipulations alone may affect fetal and placental development [37, 38]. In this study, we transferred naturally derived blastocysts into pseudopregnant recipient mice in order to control for any effect from embryo transfer alone, another manipulation shown to have isolated effects, which is clearly impossible in humans. [18] Our unique study design, using GFP as a marker, allowed us to transfer both superovulated and natural blastocysts into a single pseudopregnant recipient mouse. With this model, we determined that exposure of peri-implantation embryos during early development to a superovulated uterine environment contributes most significantly to the reduction in fetal weight seen during assisted reproductive technologies, whereas the exposure of the gamete and preimplantation embryo to superovulation appeared to predominantly affect placental weight.
In the current study, we demonstrate that exposure to superovulation in the peri-implantation period leads to lower fetal weight, which parallels the increased incidence of low birthweight phenotype that is seen after fresh IVF cycles [35]. Consistent with the current study, a previous study in our laboratory assessing fetal weight in mice at E18.5 after naturally derived blastocysts were flushed at E3.5 and transferred to either superovulated or natural pseudopregnant mice found that pups were smaller if the blastocysts were transferred into a superovulated uterine environment [17]. We have also previously shown that the superovulated uterine environment leads to a decrease in fetal weight, independent of the process of cryopreservation [23]. This confirmed findings of an older study that showed, compared with transfer into a natural recipient, superovulated preimplantation embryos transferred into a stimulated uterine environment had a higher resorption rate (69% vs. 36%; p = 0.01) and lower fetal weight (0.51 g vs. 0.72 g; p = 0.006) [39]. This latter study, however, involved embryo culture as well as surgical embryo transfer, which could have independent effects on fetal and placental weight. In this study, we attempt to isolate gonadotropin exposure and control for the hormonal environment of the recipient by utilizing the GFP transgene, which allows us to transfer both exposed and unexposed embryos in a single recipient. Our findings demonstrate that, regardless of perimplantation exposure, the superovulated hormonal milieu affects fetal weight near term.
The current study showed an increase in placental weight in blastocysts exposed to superovulation both pre- and peri-implantation as compared with natural blastocysts that were transferred into a natural or superovulated environment. An increase in placental weight was not seen when superovulated preimplantation embryos were transferred into a natural environment suggesting that it is the combined effect of superovulation pre-and peri-implantation that leads to the changes in placental weight. The study was not powered to detect a difference in placental weight; therefore, it is possible that preimplantation exposure alone may lead to alterations in placental weight, but this was not demonstrated in our study. Prior mouse studies have also shown that blastocysts generated following superovulation, IVF, embryo culture, and embryo transfer have increased placental weight and decreased fetal-to-placental weight ratio at late gestation (E18.5) compared with blastocysts obtained following superovulation and embryo transfer alone. These data suggest that other aspects of assisted reproduction, such as method of fertilization and embryo culture, may also have an effect on placental size [40]. In this study, gross morphology of placentae was not affected. A previous study has shown that gene expression of the Glut3 glucose transporter, amino acid transporters, and imprinted genes were downregulated in IVF placentae compared with superovulated flushed blastocysts suggesting that placental changes are present, but may not be physically evident [40]. Here, we show that exposure of the preimplantation embryo may lead to placental changes, though these may be differentially affected by the peri-implantation environment. Interestingly, in a prior study in our laboratory in which naturally derived blastocysts were transferred into pseudopregnant recipients, we found a decrease in placental weight after exposure to a superovulated uterine environment compared with the natural uterine environment [17]. The prior study had similar numbers of pups (16 Nat–Nat and 23 Nat–SO) but was not performed with the same transgenic mouse strain used in this study, suggesting possible strain-specific effects. Moreover, it is also possible that there may be a negative effect of the peri-implantation uterine environment on placental weight that is masked by the larger effect of preimplantation blastocyst exposure seen in this study.
Placental histopathological study was performed to determine whether structural changes in the placenta were responsible for the placental enlargement. There was a trend toward an increase in the JZ:Lab, although this did not reach statistical significance. Increased JZ:Lab has been noted in term placentas derived from in vitro fertilized embryos that are transferred into recipient mice that were not superovulated [18]. The current study demonstrates that superovulation alone without in vitro manipulation of gametes may lead to placental enlargement, but structural change may only be partially responsible.
In our study, we noted a sex difference in fetal and placental weight trends. The reasons for this are not completely clear as the sex ratio among the groups were not significantly different. However, the study was powered to detect overall difference in fetal weight without regard to sex. It is likely that we did not have sufficient power to detect a difference among the male pups. The number of females largely met the calculated sample size with 9 Nat–Nat, 10 SO–Nat, 6 Nat–SO, and 10 SO–SO females compared with the relatively smaller sample size of males 5 Nat–Nat, 14 SO–Nat, 6 Nat–SO, and 4 SO–SO males. Future study is necessary to determine whether a true sex difference exists with respect to fetal and placental weight.
Superovulation and embryo transfer occurs during critical periods of global DNA demethylation and remethylation, where imprinted genes must escape these processes to preserve germline imprints [41]. Multiple studies have shown that procedures utilized during IVF may disrupt the establishment or maintenance of these imprints [18, 20, 21, 27, 42]. In the current study, SO–SO-derived fetuses were smaller and had larger placentas compared with the fetuses that were not exposed to superovulation at all or those in which only the preimplantation embryo was exposed. Given these alterations in growth, methylation changes in the imprinted growth-related Peg3, KvDMR1, H19/Igf2 ICR, and the Igf2 DMR loci were investigated. We found that the Peg3 gene was hypomethylated in placentas exposed to hormonal stimulation pre- and peri-implantation as well as after peri-implantation exposure alone. Peg3 is a paternally expressed, maternally methylated gene that has been implicated in fetal growth through a reduction in muscle mass and myofiber number [43]. We found hypomethylation in Peg3 methylation in the smaller pups, which potentially could lead to biallellic expression of PEG3. Further studies are needed to examine how these changes in expression affect prenatal skeletal muscle development.
Gene expression analyses performed on placental tissue showed gene changes when the group with exposure to superovulation during both pre- and peri-implantation was compared with the unexposed group or with the group with exposure to the preimplantation embryo. IPAs on the differentially expressed gene list specifically identified physiological functions associated with the immune response and interferon signaling. Interferon signaling is complex, and involves several factors, of which interferon gamma (Ifng) is the most well-studied with relevance to the maternal–fetal interface. IFNG has been shown to be essential to normal implantation and maintenance of early pregnancy in the mouse [44]. The majority of IFNG appears to be derived from uterine natural killer cells (uNK) that, in the mouse, appear in the decidua at E5.5 and proliferate from E8 to 11 in the mesometrial lymphoid aggregate of pregnancy after which they move into the decidua [45]. Murine uNK cells decline in number starting midgestation [44, 45]. Mice with deficient IFNG signaling (Ifn-γ−/− and Ifn-γRα−/−) have increased rates of fetal loss in their first pregnancy and are noted to have an increase in uNKs later in gestation, edematous decidua, and abnormal decidual vasculature [44]. Thus, the changes seen in the placenta after superovulation may in part be due to impairment in the Ifng signaling pathway. In humans, uNK cells appear in the late secretory phase of the menstrual cycle and increase until midgestation when they begin to decline [45]. As in mice, human uNK cells secrete IFNG and the uterine epithelium and vascular smooth muscle cells appear to be targets [45]. The interferon pathway is therefore likely important in early pregnancy for placentation and perturbation of the hormonal environment created by superovulation may alter this process, ultimately leading to a growth-restricted phenotype seen in some infants after fresh embryo transfer.
Our current study focused on the effects of superovulation on the fetus and placenta. Maternal cells of the endometrium, however, also play a significant role in ensuring normal embryo implantation, placentation, and healthy pregnancy. Indeed, superovulation results in changes in gene expression in human endometrium [46], and work from our laboratory shows changes in endometrial gene expression specifically in genes important for early placentation [47]. Work by Segal et al. [48] has also shown that superovulation alters the expression of angiogenic factors in the murine uterus, which may result in altered blood flow to fetuses, thereby adversely impacting fetal growth. It is, therefore, likely that at least some of the changes we see in our study are secondary to changes in the uterus following superovulation.
In summary, our study has shown that peri-implantation exposure of the developing embryo to superovulation at the level of the uterus most significantly affects birthweight independent of preimplantation exposure and of other IVF procedures. Furthermore, it appears that placental weight is impacted more significantly by exposure to superovulation preimplantation than during peri-implantation development. These data suggest that exposure to superovulation at each time point has differential effects on fetal and placental weight and these exposures at least partly contribute to the adverse outcomes seen following IVF. The superovulated blastocysts transferred into a physiological uterine environment did not have growth-restricted fetuses compared with the Nat–Nat group, reinforcing clinical data that growth disturbances are less likely following FETs [49, 50]. However, the placenta itself is an important mediator of fetal cardiometabolic health and the enlarged placentae seen with superovulated blastocysts may have long-term negative effects on cardiovascular health at a range of fetal birthweights [51, 52]. Though gonadotropin treatment is necessary for efficient IVF, further investigation is necessary to determine whether milder stimulation protocols and lower gonadotropin exposure can ameliorate some of the changes seen in this study.
Understanding the mechanisms by which superovulation leads to adverse perinatal outcomes in fresh IVF can also provide valuable information regarding the pathogenesis of pregnancy disorders unrelated to assisted reproductive technologies. Our expression data suggest potentially altered immune cell function following superovulation. Further research is necessary to gain understanding of the role of immune cells during implantation. Our current studies are investigating the function of both uNK cells and macrophages on trophoblast invasion and placentation and the effect of superovulation on these cell types. In addition, we are examining earlier time points during gestation to determine when fetal and placental changes are first evident and whether changes in one precede the other. Most importantly, long-term studies are needed, in both animal models and humans, to assess the potential downstream effects on health that may result from these early exposures.
Supplementary Material
Acknowledgments
The authors would like to thank Mary Sammel, ScD, Ji Young Kim, PhD, and Andrew Cucchiara, PhD for statistical advice and Christopher Krapp for his assistance with LUMA and pyrosequencing experiments.
Footnotes
† Grant Support: Research reported in the publication was supported by grants HD068157 (CC, MB, MM), T32-HD040135-15 (CSP), the Jones Foundation for Reproductive Medicine (MM), and by the Institute for Translational Medicine and Therapeutics of the Perelman School of Medicine at the University of Pennsylvania (CSP).
Conference Presentation: Presented in part at the 74th Annual Meeting of the American Society for Reproductive Medicine, 6–10 October 2018, Denver, Colorado.
Conflict of Interest
The authors have declared that no conflict of interest exists.
Author’s contributions
C.S.P. was involved in study concept and design, performing experiments, statistical analysis, drafting, and revising the manuscript. S.M. was involved in performing experiments, drafting, and revising the manuscript. E.R.C. and T.O. were involved in performing experiments and collecting data. C.C. and M.B. were involved in study concept and design. M.B. provided the laboratory space and equipment for epigenetic analyses and was involved in the data interpretation. M.M. was involved in study concept and design, data interpretation, and critical revision of the manuscript and figures. All authors were involved in critical revision of the manuscript and the approval of the final manuscript.
References
- 1. Sunderam S, Kissin DM, Zhang Y, Folger SG, Boulet SL, Warner L, Callaghan WM, Barfield WD. Assisted reproductive technology surveillance - United States, 2016. MMWR Surveill Summ 2019; 68:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Voorhis BJ. In vitro fertilization. N Engl J Med 2007; 356:379–386. [DOI] [PubMed] [Google Scholar]
- 3. Pandey S, Shetty A, Hamilton M, Bhattacharya S, Maheshwari A. Obstetric and perinatal outcomes in singleton pregnancies resulting from IVF/ICSI: a systematic review and meta-analysis. Hum Reprod Update 2012; 18:485–503. [DOI] [PubMed] [Google Scholar]
- 4. Qin J, Sheng X, Wu D, Gao S, You Y, Yang T, Wang H. Adverse obstetric outcomes associated with in vitro fertilization in singleton pregnancies. Reprod Sci 2017; 24:595–608. [DOI] [PubMed] [Google Scholar]
- 5. Silberstein T, Levy A, Harlev A, Saphier O, Sheiner E. Perinatal outcome of pregnancies following in vitro fertilization and ovulation induction. J Matern Fetal Neonatal Med 2014; 27:1316–1319. [DOI] [PubMed] [Google Scholar]
- 6. Kalra SK, Ratcliffe SJ, Coutifaris C, Molinaro T, Barnhart KT. Ovarian stimulation and low birth weight in newborns conceived through in vitro fertilization. Obstet Gynecol 2011; 118:863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pelkonen S, Koivunen R, Gissler M, Nuojua-Huttunen S, Suikkari AM, Hyden-Granskog C, Martikainen H, Tiitinen A, Hartikainen AL. Perinatal outcome of children born after frozen and fresh embryo transfer: the Finnish cohort study 1995-2006. Hum Reprod 2010; 25:914–923. [DOI] [PubMed] [Google Scholar]
- 8. Maheshwari A, Raja EA, Bhattacharya S. Obstetric and perinatal outcomes after either fresh or thawed frozen embryo transfer: an analysis of 112,432 singleton pregnancies recorded in the human fertilisation and embryology authority anonymized dataset. Fertil Steril 2016; 106:1703–1708. [DOI] [PubMed] [Google Scholar]
- 9. Vidal M, Vellve K, Gonzalez-Comadran M, Robles A, Prat M, Torne M, Carreras R, Checa MA. Perinatal outcomes in children born after fresh or frozen embryo transfer: a Catalan cohort study based on 14,262 newborns. Fertil Steril 2017; 107:940–947. [DOI] [PubMed] [Google Scholar]
- 10. Shapiro BS, Daneshmand ST, Bedient CE, Garner FC. Comparison of birth weights in patients randomly assigned to fresh or frozen-thawed embryo transfer. Fertil Steril 2016; 106:317–321. [DOI] [PubMed] [Google Scholar]
- 11. Chung K, Coutifaris C, Chalian R, Lin K, Ratcliffe SJ, Castelbaum AJ, Freedman MF, Barnhart KT. Factors influencing adverse perinatal outcomes in pregnancies achieved through use of in vitro fertilization. Fertil Steril 2006; 86:1634–1641. [DOI] [PubMed] [Google Scholar]
- 12. Imudia AN, Awonuga AO, Doyle JO, Kaimal AJ, Wright DL, Toth TL, Styer AK. Peak serum estradiol level during controlled ovarian hyperstimulation is associated with increased risk of small for gestational age and preeclampsia in singleton pregnancies after in vitro fertilization. Fertil Steril 2012; 97:1374–1379. [DOI] [PubMed] [Google Scholar]
- 13. Pinborg A, Wennerholm UB, Romundstad LB, Loft A, Aittomaki K, Söderström-Anttila V, Nygren KG, Hazekamp J, Bergh C. Why do singletons conceived after assisted reproduction technology have adverse perinatal outcome? Systematic review and meta-analysis. Hum Reprod Update 2013; 19:87–104. [DOI] [PubMed] [Google Scholar]
- 14. Pereira N, Elias RT, Christos PJ, Petrini AC, Hancock K, Lekovich JP, Rosenwaks Z. Supraphysiologic estradiol is an independent predictor of low birth weight in full-term singletons born after fresh embryo transfer. Hum Reprod 2017; 32:1410–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, Gaughan JP, Coutifaris C, Sapienza C. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet 2009; 18:3769–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Turan N, Ghalwash MF, Katari S, Coutifaris C, Obradovic Z, Sapienza C. DNA methylation differences at growth related genes correlate with birth weight: a molecular signature linked to developmental origins of adult disease? BMC Med Genomics 2012; 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mainigi MA, Olalere D, Burd I, Sapienza C, Bartolomei M, Coutifaris C. Peri-implantation hormonal milieu: elucidating mechanisms of abnormal placentation and fetal growth. Biol Reprod 2014; 90:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Waal E, Vrooman LA, Fischer E, Ord T, Mainigi MA, Coutifaris C, Schultz RM, Bartolomei MS. The cumulative effect of assisted reproduction procedures on placental development and epigenetic perturbations in a mouse model. Hum Mol Genet 2015; 24:6975–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Van der Auwera I, D'Hooghe T. Superovulation of female mice delays embryonic and fetal development. Hum Reprod 2001; 16:1237–1243. [DOI] [PubMed] [Google Scholar]
- 20. Fortier AL, Lopes FL, Darricarrere N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet 2008; 17:1653–1665. [DOI] [PubMed] [Google Scholar]
- 21. Fortier AL, McGraw S, Lopes FL, Niles KM, Landry M, Trasler JM. Modulation of imprinted gene expression following superovulation. Mol Cell Endocrinol 2014; 388:51–57. [DOI] [PubMed] [Google Scholar]
- 22. Kim SM, Kim JS. A review of mechanisms of implantation. Dev Reprod 2017; 21:351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weinerman R, Ord T, Bartolomei MS, Coutifaris C, Mainigi M. The superovulated environment, independent of embryo vitrification, results in low birthweight in a mouse model. Biol Reprod 2017; 97:133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Meth 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc 2006; 2006. [DOI] [PubMed] [Google Scholar]
- 26. Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MRW. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet 2010; 19:36–51. [DOI] [PubMed] [Google Scholar]
- 27. Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod 2007; 22:26–35. [DOI] [PubMed] [Google Scholar]
- 28. Jaquet D, Gaboriau A, Czernichow P, Levy-Marchal C. Insulin resistance early in adulthood in subjects born with intrauterine growth retardation. J Clin Endocrinol Metab 2000; 85:1401–1406. [DOI] [PubMed] [Google Scholar]
- 29. Jaquet D, Leger J, Czernichow P, Levy-Marchal C. The effect of in-utero undernutrition on the insulin resistance syndrome. Curr Diab Rep 2002; 2:77–82. [DOI] [PubMed] [Google Scholar]
- 30. Jaquet D, Léger J, Lévy-Marchal C, Czernichow P. Low birth weight: effect on insulin sensitivity and lipid metabolism. Horm Res Paediatr 2003; 59:1–6. [DOI] [PubMed] [Google Scholar]
- 31. Arends NJ, Boonstra VH, Duivenvoorden HJ, Hofman PL, Cutfield WS, Hokken-Koelega AC. Reduced insulin sensitivity and the presence of cardiovascular risk factors in short prepubertal children born small for gestational age (SGA). Clin Endocrinol (Oxf) 2005; 62:44–50. [DOI] [PubMed] [Google Scholar]
- 32. Ong KK, Ahmed ML, Emmett PM, Preece MA, Dunger DB. Association between postnatal catch-up growth and obesity in childhood: prospective cohort study. BMJ 2000; 320:967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Waal E, Mak W, Calhoun S, Stein P, Ord T, Krapp C, Coutifaris C, Schultz RM, Bartolomei MS. In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies. Biol Reprod 2014; 90:21–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bardou P, Mariette J, Escudie F, Djemiel C, Klopp C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform 2014; 15:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qin J, Sheng X, Wu D, Gao S, You Y, Yang T, Wang H. Adverse obstetric outcomes associated with in vitro fertilization in singleton pregnancies. Reprod Sci 2016. doi: 10.1177/1933719116667229. [DOI] [PubMed] [Google Scholar]
- 36. Qin J-B, Sheng X-Q, Wu D, Gao S-Y, You Y-P, Yang T-B, Wang H. Worldwide prevalence of adverse pregnancy outcomes among singleton pregnancies after in vitro fertilization/intracytoplasmic sperm injection: a systematic review and meta-analysis. Arch Gynecol Obstet 2016; 295:1–17. [DOI] [PubMed] [Google Scholar]
- 38. Hemkemeyer SA, Schwarzer C, Boiani M, Ehmcke J, Le Gac S, Schlatt S, Nordhoff V. Effects of embryo culture media do not persist after implantation: a histological study in mice. Hum Reprod 2014; 29:220–233. [DOI] [PubMed] [Google Scholar]
- 39. Ertzeid G, Storeng R. The impact of ovarian stimulation on implantation and fetal development in mice. Hum Reprod 2001; 16:221–225. [DOI] [PubMed] [Google Scholar]
- 40. Bloise E, Lin W, Liu X, Simbulan R, Kolahi KS, Petraglia F, Maltepe E, Donjacour A, Rinaudo P. Impaired placental nutrient transport in mice generated by in vitro fertilization. Endocrinology 2012; 153:3457–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smallwood SA, Kelsey G. De novo DNA methylation: a germ cell perspective. Trends Genet 2012; 28:33–42. [DOI] [PubMed] [Google Scholar]
- 42. Song S, Ghosh J, Mainigi M, Turan N, Weinerman R, Truongcao M, Coutifaris C, Sapienza C. DNA methylation differences between in vitro- and in vivo-conceived children are associated with ART procedures rather than infertility. Clin Epigenetics 2015; 7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Correra RM, Ollitrault D, Valente M, Mazzola A, Adalsteinsson BT, Ferguson-Smith AC, Marazzi G, Sassoon DA. The imprinted gene Pw1/Peg3 regulates skeletal muscle growth, satellite cell metabolic state, and self-renewal. Sci Rep 2018; 8:14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ashkar AA, Croy BA. Interferon-γ contributes to the normalcy of murine pregnancy. Biol Reprod 1999; 61:493–502. [DOI] [PubMed] [Google Scholar]
- 45. Murphy SP, Tayade C, Ashkar AA, Hatta K, Zhang J, Croy BA. Interferon gamma in successful pregnancies. Biol Reprod 2009; 80:848–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Horcajadas JA, Pellicer A, Simon C. Wide genomic analysis of human endometrial receptivity: new times, new opportunities. Hum Reprod Update 2007; 13:77–86. [DOI] [PubMed] [Google Scholar]
- 47. Senapati S, Wang F, Ord T, Coutifaris C, Feng R, Mainigi M. Superovulation alters the expression of endometrial genes critical to tissue remodeling and placentation. J Assist Reprod Genet 2018; 35:1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Segal TR, Amini P, Wang J, Peters G, Skomorovska-Prokvolit Y, Mainigi MA, Goldfarb JM, Mesiano S, Weinerman R. Superovulation with different trigger agents differentially alter essential angiogenic factors in the endometrium in a mouse model. Biol Reprod 2020;102:1122–1133. [DOI] [PubMed] [Google Scholar]
- 49. Kansal Kalra S, Ratcliffe SJ, Milman L, Gracia CR, Coutifaris C, Barnhart KT. Perinatal morbidity after in vitro fertilization is lower with frozen embryo transfer. Fertil Steril 2011; 95:548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kalra SK. Adverse perinatal outcome and in vitro fertilization singleton pregnancies: what lies beneath? Further evidence to support an underlying role of the modifiable hormonal milieu in in vitro fertilization stimulation. Fertil Steril 2012; 97:1295–1296. [DOI] [PubMed] [Google Scholar]
- 51. Risnes KR, Romundstad PR, Nilsen TI, Eskild A, Vatten LJ. Placental weight relative to birth weight and long-term cardiovascular mortality: findings from a cohort of 31,307 men and women. Am J Epidemiol 2009; 170:622–631. [DOI] [PubMed] [Google Scholar]
- 52. Lewis RM, Cleal JK, Hanson MA. Review: placenta, evolution and lifelong health. Placenta 2012; 33Suppl:S28–S32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


