Abstract
Sepsis is a series of clinical syndromes caused by the immunological response to infection. The clinical evidence for sepsis could typically attribute to bacterial infection or bacterial endotoxins, but infections due to viruses, fungi or parasites could also lead to sepsis. Regardless of the etiology, rapid clinical deterioration, prolonged stay in intensive care units and high risk for mortality correlate with the incidence of sepsis. Despite its prevalence and morbidity, improvement in sepsis outcomes has remained limited. In this comprehensive review, we summarize the current landscape of risk estimation, diagnosis, treatment and prognosis strategies in the setting of sepsis and discuss future challenges. We argue that the advent of modern technologies such as in-depth molecular profiling, biomedical big data and machine intelligence methods will augment the treatment and prevention of sepsis. The volume, variety, veracity and velocity of heterogeneous data generated as part of healthcare delivery and recent advances in biotechnology-driven therapeutics and companion diagnostics may provide a new wave of approaches to identify the most at-risk sepsis patients and reduce the symptom burden in patients within shorter turnaround times. Developing novel therapies by leveraging modern drug discovery strategies including computational drug repositioning, cell and gene-therapy, clustered regularly interspaced short palindromic repeats -based genetic editing systems, immunotherapy, microbiome restoration, nanomaterial-based therapy and phage therapy may help to develop treatments to target sepsis. We also provide empirical evidence for potential new sepsis targets including FER and STARD3NL. Implementing data-driven methods that use real-time collection and analysis of clinical variables to trace, track and treat sepsis-related adverse outcomes will be key. Understanding the root and route of sepsis and its comorbid conditions that complicate treatment outcomes and lead to organ dysfunction may help to facilitate identification of most at-risk patients and prevent further deterioration. To conclude, leveraging the advances in precision medicine, biomedical data science and translational bioinformatics approaches may help to develop better strategies to diagnose and treat sepsis in the next decade.
Keywords: sepsis, translational bioinformatics, computational medicine, precision medicine, genome informatics
Background
Sepsis is a persistently growing health concern in America, especially in light of the aging population demographics. More than 1.5 million people are affected by sepsis in the USA each year, leading to 250 000 deaths [1]. This results in an estimated mortality rate of 17%, which is staggering for such a prevalent disease. Sepsis is also a burden to the healthcare system financial state, accounting for $23.7 billion in 2013 alone (6.2% of all hospitalization costs) [2]. It is the primary cause of death of intensive care unit (ICU) patients, even as sepsis patients occupy only 10% of ICU beds [3]. Mortality rates attributed to sepsis remain disappointingly high at 15–20%, necessitating novel diagnostic and treatment strategies for earlier prevention and management to improve clinical outcomes [3, 4]. Traditional approaches, which assay single biomarkers, largely fail to identify most of the patients at risk for sepsis and instead are better suited to predict severity and mortality [5]. Hence, we need a systems medicine approach to diagnose, treat and estimate the prognoses of sepsis. With the introduction of longitudinal electronic health records (EHR) [6, 7], low-cost genomics and other molecular profiling technologies [8], novel machine intelligence algorithms [9, 10], modern drug discovery and companion diagnostics development strategies [11], we now are equipped with a plethora of powerful tools not previously available [10, 12]. In this review, we address some of the current challenges, and forecast how various aspects of precision medicine, biomedical data science and translational bioinformatics approaches will collaboratively help us to combat sepsis.
Defining sepsis
Difficulties in treating sepsis is partly due to challenges in understanding the mechanisms underlying the syndrome given its wide pathophysiological and clinical variability. Even a strict definition of sepsis has been elusive. In 1992, an international consensus panel first defined sepsis as a systemic inflammatory response syndrome (SIRS) [13]. In addition to establishing the SIRS criteria, the panel also defined sepsis, severe sepsis, septic shock, sepsis-induced hypotension and multiple organ dysfunction syndrome [14]. Severe sepsis includes acute organ dysfunction whereas septic shock results from significant reduction of tissue perfusion due to hypotension that is refractory to fluid resuscitation and with hyperlactatemia [14]. Sepsis was recently reevaluated by the task force of the European Society of Intensive Care Medicine (ESICM) and Society of Critical Care Medicine (SCCM) in their guidelines in 2014, in order to redefine sepsis and the criteria for diagnosis. In the Sepsis-3 paper, sepsis was re-defined as a ‘life-threatening organ dysfunction caused by a dysregulated host response to infection’ [15]. This definition was clinically supported by a source of infection with two or more quick sequential organ failure assessment (qSOFA) criteria. Sepsis-3 removed the severe sepsis classification entirely as no objective definition for end-organ dysfunction currently exists. It was also recognized by ESICM/SCCM guidelines that sepsis remains a poorly understood process. Therefore, no gold standard could yet be established to unequivocally identify a septic patient.
The wide variability in the clinical manifestations of sepsis contributes to the challenges in its definition. As a syndrome, the constellation of septic signs and symptoms is largely dependent on the site of infection, the causative pathogen, the pattern of end-organ dysfunction and the underlying healthy physiological profile of the patient [16]. Although the etiology of sepsis is unknown in one-half of cases, the majority of cases are caused by hospital or community acquired gram positive cocci Staphylococcus aureus, pathogenic Streptococcus spp. and gram negative bacilli such as Escherichia coli, Pseudomonas aeruginosa and Klebsiella [17]. In addition, fungi species such as Candida albicans, Histoplasma and (especially in the case of patients with AIDS) Pneumocystis jirovecii are known to cause sepsis in immunocompromised patients [18, 19]. Symptoms in severe sepsis and septic shock that indicate organ dysfunction include altered mental status, dyspnea, oliguria, jaundice and dysglycemia [16, 20]. Unfortunately, many of these symptoms are not highly specific to sepsis. Furthermore, the body’s physiologic response to increase systemic vascular resistance by endogenous catecholamines during the early stages of septic shock may offset the initial drop in blood pressure due to decreased effective intravascular volume from diffuse capillary leakage [21]. However, later stages of sepsis are characterized inability to effectively compensate and resulting systemic tissue hypoxia leading to acidosis and vasodilation [22]. A better understanding of the complex host response elicited during sepsis further suggests that the SIRS criteria of 1992 needed to be redefined in order to more accurately diagnose patients at risk for clinical deterioration. As the Sepsis-3 criteria has described, ‘sepsis is a multifaceted host response to an infecting pathogen that may be significantly amplified by endogenous factors’ [15]. Rather than a solely host pro-inflammatory response to infection, sepsis is now recognized as a complex interplay between both pro- and anti-inflammatory responses [23]. Inflammatory stimuli from the pathogens trigger host immune cells to release proinflammatory mediators such as TNF-a, IL-1, IFN-y, IL-12 and IL-18. These proinflammatory mediators interaction with prostaglandins, platelet activating factors, adhesion molecules and stress hormones results in vasodilation, increased vascular permeability and reduced perfusion. In addition, pro-inflammatory molecules activate opposing anti-inflammatory molecules, such as IL-10, that lead to periods of immunosuppression. These periods of immunosuppression can contribute to increased risk of nosocomial infections and reactivation of latent viruses [24]. This multifaceted host immune response with major nonimmunologic factors (cardiovascular, neuronal, autonomic, hormonal, metabolic and coagulation) may contribute to diminished oxygenation of the end-organs, leading to acute organ dysfunction.
Risk stratification and diagnoses
Sepsis is a syndromic condition that increases mortality in both pediatric and adult populations. Early identification may help set the course of treatment strategies and maximize therapeutic efficacy. The need to develop earlier preemptive identification and prophylactic treatment will benefit all ages but will clearly require personalized approaches based on different sub-populations (immunocompromised or non-immunocompromised; geriatric versus pediatric etc.). Development of new risk stratification algorithms, multianalyte diagnostics and biomarker panels are emerging to proactively identify patients at risk for sepsis. In this section, we propose the development of integrative approaches and multi-analyte-based diagnostic aids coupled with machine learning to enable better outcomes. We also discuss the possibilities of designing a polygenic risk score for sepsis and associated outcomes.
An integrative approach for the accelerated diagnoses and personalized treatment
Much work has focused upon improving early diagnosis, treatment and prognosis of sepsis with diagnosis criteria, biomarkers of sepsis and host genomics. Identifying biomarkers associated with sepsis offers multiple clinical benefits. By expanding clinical biomarkers of sepsis, such as serum lactate and procalcitonin [25], with pro-inflammatory and anti-inflammatory cytokines, chemokines and acute phase proteins, we will be able to measure and create unique biochemical profiles for sepsis. Profiling of, for instance, 16 s ribosomal RNA via PCR can help identify a causative bacterial agent even when blood cultures are negative [26]. Current approaches for diagnosis and treatment include antimicrobial stewardship via microarray analysis [27] and point of care detection of bacterial DNA from whole blood [28], but combining new sequencing technologies with machine intelligence could improve the yield of these approaches. In addition, changes in biomarkers over the clinical course of sepsis can be used to monitor improvement or deterioration during management. Also, intensive biochemical profiling could help identify sepsis patients at higher risk for poorer outcomes such as with multiple organ dysfunction [25, 26]. Finally, the use of biomarkers in conjunction with a patient’s unique genetic susceptibility to infection will allow us to generate individualized prevention strategies and medical therapies for patients at risk with sepsis [29]. As a result, the clinical approach to sepsis will evolve to require a systems view with analysis of patient vitals, unique biomarker profile, metabolites, host genomic profile and his or her microbiome [30, 31]. Diagnostics aids can also be built by combining different omics modalities and multi-analyte technology platforms. For example, proteogenomics is a hybrid biotechnology approach that combines genomic sequencing or transcriptomic sequencing with proteomics profiling to provide a static-to-dynamic view of biological systems. A proteogenomics platform was recently developed to develop a liquid biopsy to detect multiple types of cancers [32]. By similarly using biomarker signatures from different analytes and combining predictive models using proteogenomics, metabo-proteomics and other hybrid-omics technologies can be built for diagnoses and prognoses (Figure 1) [31, 33, 34]. Furthermore, responders and non-responders can be identified using multi-modal diagnostic platforms. The refinement of these methods could be used to develop personalized, comorbidity-based drug discovery strategies.
Figure 1.
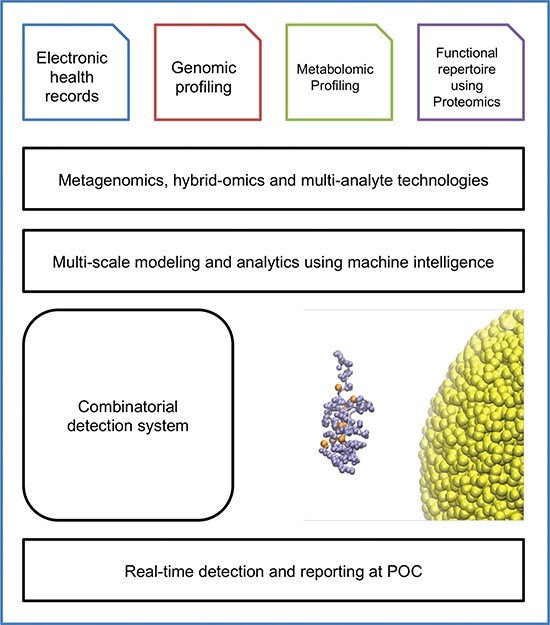
Translational bioinformatics framework for developing multi-analyte, heterogeneous, data-driven diagnostic aid for sepsis.
Predictive algorithms for accelerated diagnoses of sepsis
Designing predictive models may help stratify patients who may benefit with the end result of improved resource allocation and outcomes [35–41]. A variety of algorithms are already used to predict stages of clinical acuity in the setting of hospital admissions (See Table 1) [42]. Several risk stratification tools have also been reported that may potentially give an early indication of sepsis. Variants of algorithms like the modified early warning system, such as TREWScore [43] were recently proposed to help predict patients at risk. TREWScore is a targeted real-time early warning score that predicts which patients are most susceptible to septic shock within few hours. This prediction is achieved using a Cox proportional hazards model with L1 regularization as a supervised model with time until the onset of septic shock as the dependent variable. The model assumes that the onset of shock and the sepsis severity levels are critical. The baseline hazard function was fit using a multiple imputation method and estimated from a subset of 400 000 time-to-event and feature pairs from the development set. This has been used to repeatedly sample the event time for each interval-censored sample and generate 100 complete copies of the development dataset. A separate model was trained from each of the N copies of the development dataset. To predict on data from a new subject, predicted risk values were obtained from each of the N models; these values were combined using Rubin’s equations that compute the final risk value as the average of risk values outputted from each of the N models. The area under the curve obtained for the TREWScore was 0.83 (95% CI, 0.81–0.85). At a specificity of 0.67 [false-positive rate of 0.33], TREWScore achieved a sensitivity of 0.85. Patients were identified at a median of 28.2 h before shock onset.
Table 1.
Representative list of algorithms for predicting patient outcomes in the setting of sepsis
| Sepsis risk algorithms | Abbreviation | Benefits |
|---|---|---|
| BOMBARD | BOMBARD | BOMBARD was more accurate than SIRS and qSOFA at predicting severe sepsis/septic shock and sepsis mortality. |
| Hamilton early warnings system | HEWS | Good discriminative ability for predicting the occurrence of a critical event among septic patients. |
| Modified early warning system | MEWS | Provides great predictive value and accuracy for early clinical deterioration. |
| National early warning score | NEWS | NEWS has better performance than qSOFA and SIRS. |
| Quick version of sequential organ failure assessment | qSOFA | Ability to predict mortality based on organ dysfunction severity and easier to score clinically than SOFA. |
| Reasons for geographic and racial differences in stroke-severe sepsis risk score |
REGARDS-SSRS | Effective for predicting community-dwelling adults at high risk of sepsis. |
| Systemic inflammatory response syndrome | SIRS | Higher sensitivity to detect sepsis-related mortality than qSOFA |
| Sepsis ‘sniffer’ algorithm | SSA | SSA reduced the risk of incorrectly categorizing patients at low risk for sepsis, detected sepsis high risk in half the time, and reduced redundant NST screens. |
| Targeted real-time early warning score | TREWScore | Ability to detect at risk patients early using a learned algorithm model that takes into account many more factors. |
A polygenic risk score for sepsis and adverse outcomes
Risk stratification using panel of genetic variants to identify patients at risk for complex diseases like myocardial infarction, stroke and neuropsychiatric disorders is an emerging theme of precision medicine [44–46]. Emerging results from genome-wide association studies (GWAS) reveal several plausible risk alleles associated with sepsis and related clinical outcomes [47, 48]. A study that combines GWAS with deep sequencing identified association of several interesting genes with clinical outcomes like 28-day mortality after sepsis (VPS13A) and procalcitonin level in sepsis patients (CRISPLD2). Another GWAS investigation surveyed the genetic landscape to find the potential variants indicating survival from sepsis due to pneumonia. While such investigations address a critical knowledge gap, it remains elusive whether any common genomic markers could be predictive of the incidence of sepsis. In the absence of such direct genomic evidence for predictive sepsis risk, developing a genetic risk score for various detrimental outcomes associated with sepsis (heart failure, respiratory arrest, etc.) may help to find patients that need accelerated care in the context of an infection (Figure 2). While the health economic utility of whole-genome sequencing is still being debated, sequencing the genome and providing such information to compute risk scores may help to identify and accelerate treatments using genomic information [49].
Figure 2.
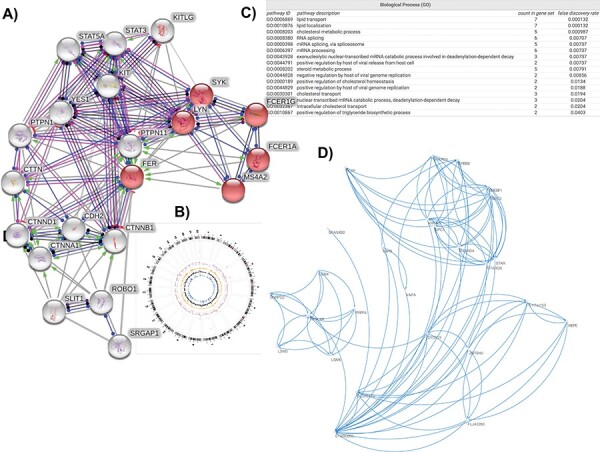
Emerging targets, pathways from genome-wide association studies of sepsis phenotypes. (A) Molecular neighborhood of FER protein; red nodes are proteins implicated in regulation of mast cell degranulation. (B) Genome-wide associations of variants implicated in response to sepsis therapy. (C) Biological processes mediated by the molecular neighborhood of STARD3NL. (D) First-degree interactome of STARD3NL.
Limitations of using singular biomarkers highlights the need for combinatorial markers of syndromic stages in sepsis
More than 100 biomarkers are listed in sepsis-related review articles and meta-analyses showing varying degree of clinical utilities for diagnoses. Many of these biomarkers are only effective after the infection reaches a stage where treatment is less effective or the beginning of major complications arise. Emerging advances in liquid biopsies [32, 50] and other molecular signature-based screening of diseases could help in designing a better diagnostic aid for various stages of sepsis. One such strategy could involve searching for a combination of existing biomarkers that could inform the preeminent stages of information. Leveraging data from EHR and using laboratory-based data points across different analytes and building predictive models may help in building such models preemptive diagnostic panels. Recently, we build a companion diagnostic aid to predict response of statin, a drug that reduces blood cholesterol, using a combination of data from gene expression profiling, biochemical assays and imaging data integrated using machine intelligence methods [51]. Similarly, models can be built by data from patients with sepsis using data already aggregated in EHR by integrating multi-analytes (genetic variation, protein level, metabolite signature, gene expression signature etc.) with machine learning for effective monitoring of sepsis [39, 52].
Recently, Sweeney et al. [53] have developed a diagnostic aid to classify the underlying infection as viral or bacterial using gene expression signatures. With a lower negative likelihood ratio than that of procalcitonin, their integrated antibiotics decision model (IADM) can be useful to rule out bacterial infections [36]. IADM has a lower negative likelihood ratio than procalcitonin. Thus, IADM may be more useful to rule out bacterial infections that require antibiotics compared to viral infections. While several statistical and machine learning algorithms are making progress in healthcare as decision aids, it should be noted that such methods have higher degree of false positive rates and the assessment of the patient’s post prediction—whether the patients flagged were eventually survived remains open. Hence, modeling the accelerated identification with progression of patients through various clinical pathways is critical. Modeling the end-to-end patient life cycle of the infection from probability, identification, treatment and recovery from sepsis using reinforcement learning or Bayesian deep learning methods may help to improve such models in the future [34, 36, 54].
Treatments and Prognosis
Emerging drug discovery strategies for developing treatments for sepsis
Current clinical pathways to target patients with sepsis include rapid response to septic patients and immediate initiation of antibiotic therapy. Depending on the clinical profile of the patient and time of sepsis detection, bona fide sepsis diagnosis can have a mortality rate as high as 30%. Furthermore, the evolution of drug-resistant forms of sepsis is also a major concern. Hence, leveraging modern drug discovery approaches may yield new therapies for sepsis. Here we briefly discuss some of the emerging themes (computational drug repositioning, cell and gene-therapy, clustered regularly interspaced short palindromic repeats (CRISPR)-based genetic editing systems, immunotherapy, microbiome restoration and nanomaterial-based therapy and phage therapy) in drug discovery in the context of sepsis.
New evidence from genomic and phenomic studies—druggable targets, mechanisms and therapeutic response in sepsis
As more research focuses on the host immunologic responses to infection, it has become increasingly clear that genomics may help broaden our understanding of sepsis’ pathogenesis. A landmark study in 1988 provided strong evidence for genetic susceptibility to infection. It demonstrated a 5.8-fold increase in mortality in adopted children who had at least one biological parent die from infection [55, 56]. This set off a gold rush in genomic research to identify the basis for genetic susceptibility to infection. The prime targets are genes involved in inflammatory pathways. Initial focus was directed primarily on single nucleotide polymorphisms (SNPs) in those genes [57]. Advances in sequencing technology and expression analysis led to the identification of candidate genes, genomic regions, structural variants, genetic variants and new molecular mechanisms implicated in sepsis susceptibility [48, 58–66].
A number of polymorphisms in genes for antigen recognition and inflammatory pathways have been implicated [23]. Recently, a GWAS has identified a single gene strongly associated with sepsis survival. Those who are homozygous for the C allele of the Tyrosine-protein kinase Fer (FER; P = 6.00e-6; dbSNP identifier: rs4723738) gene were shown to have a 44% reduction in sepsis mortality [47]. Given the high frequency of this allele, there could be a substantial population benefiting from this protective effect. The variant is encoded in chromosome 7 and has a compound role in mediating multiple autoimmune diseases including type 1 diabetes, rheumatoid arthritis, juvenile idiopathic arthritis, multiple sclerosis and Crohn’s disease. The precise role of FER or its interacting partners in sepsis response remains elusive and requires further biochemical discovery and translational research. Empirical protein structure results suggest that this gene encodes FCH, SH2, and tyrosine kinase domains. These different domains can be modeled using structural templates with sequence identity ranging from 40–100%. Thus, this could be a potential druggable target to modulate therapeutic responses in sepsis patients (Figure 3). We explored the molecular neighborhood of FER to assess its potential as a target and identified that within a network of 20 interactors, the protein–protein interaction network is highly enriched for relevant biological interactions. The interactors collectively mediate biological processes like regulation of mast cell degranulation, leukocyte activation and innate immune response. Molecular functions include IgE receptor activity, and signaling pathways mediated by tyrosine kinases, whereas cellular localization were enriched for various membrane locations. Proteins were also enriched across KEGG pathways including adherens junction, Fc epsilon RI pathway, cancer, asthma and leukocyte transendothelial migration (Figures 3 and 4).
Figure 3.
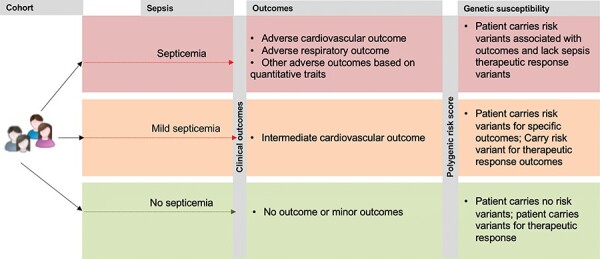
A personalized genomic framework for developing a polygenic risk for sepsis and associated outcomes.
Figure 4.
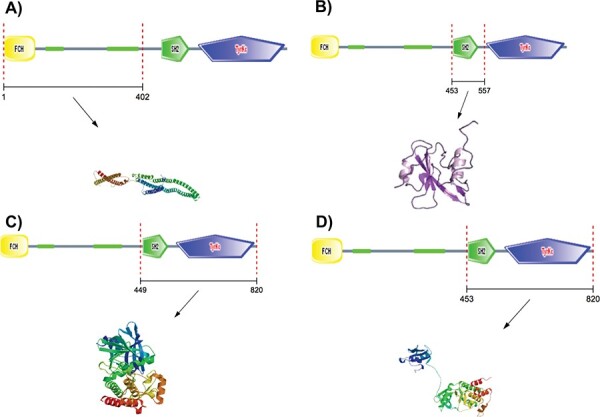
Homology modeling of FER protein, a putative target for therapeutic response in sepsis.
Phenome-wide association studies (PheWAS) enables the simultaneous identification of risk or protection for multiple phenotypes that are associated with genetic variants [67, 68]. We performed an empirical analysis of the data from PheWAS catalog (https://phewascatalog.org/phewas) for phenotype ‘Treatment response for severe sepsis’. We found that variants associated with sepsis response could mediate protective effects for 203 conditions including migraine, chondrocalcinosis and pelvic inflammatory disease among others (mapped genes includes KRT18P32, MKL1, TTC4P1, STARD3NL, NKX2-6; all observations P ≤ 0.05; OR ≤ 1.00). We further noted that conditions such as congenital anomalies of head and neck, congenital anomalies of intensive, hypertensive complications, jaundice, postinflammatory pulmonary fibrosis etc. were associated with mapped genes such as MKL1, NKX2-6, STARD3NL, KRT18P32, TTC4P1 (all observations P ≤ 0.05; OR ≤ 1.00) (Figures 3 and 4). Collectively these phenotypes are indicative of one or more complications in the setting of sepsis, and could be indicative of shared pathways driving common pathobiology. Hence, exploring the genomic and phenomic bases of these co-morbidity cascades may yield new druggable targets.
We explored the molecular neighborhood of one of these genes, STARD3NL, using predicted and experimental protein–protein interaction data from human proteome. The exact functional role of STARD3NL is yet to be elucidated beyond an indication as a component of cholesterol transport pathways, functional enrichment analyses of the molecular neighborhood (n = 20 genes) indicates that the protein could play a critical role in viral genome replication and positive regulation by host of viral release from host cell (Figure 4).
We also analyzed the current druggability status of the four emerging genes using the Illuminating Druggable Genome—PHAROS database ([69], see: https://pharos.nih.gov/idg): FER, STARD3NL, and VPS13A. Briefly, IDG-Pharos provides a four-level framework for simplifying the target development/druggability status. Tclin indicates the target have approved drugs with known mechanism of action; Tchem refers to the evidence targets with some level of chemical activity evidence; Tbio level suggests the availability of biological evidence and finally, Tdark applies to targets without any clinical, chemical or biological information. Interestingly, FER has nine compounds that could target the protein and have a Tchem level. Whereas both STARD3NL and VPS13A have a status of Tbio status that indicates the availability of biological knowledge about the target. However, both of these proteins are classified as ‘non-IDG’ candidates that illustrate the difficulty to target these using current approaches.
Computational drug repositioning
Developing a new molecule or combination therapy to target a disease requires extensive funding, years of fundamental research to understand the mechanism, even before any clinical trials are initiated. However, computational drug positioning using Food and Drug Administration (FDA) approved compounds for new indications may reduce upfront costs and decrease time-to-market [41, 70]. Computational drug repositioning is a growing area of translational research with evidence for more than 200 compounds indicated across thousands of disease indications (See: RepurposeDB—Reference Database of Drug Repositioning Investigations, http://repurposedb.dudleylab.org/) [71]. Recently, Ghosh et al. [72] performed a randomized control trial to assess the efficacy of 650 compounds in Septic patients/C57Bl/6 mice and human endothelial cells. They identified compounds in the family of 3-hydroxy-3-methyl-glutaryl-CoA reductase inhibitor drug class (statins) as lead candidates. Mechanistically, they propose that −3-methyl-glutaryl-CoA reductase inhibitors may operate through a novel Foxo1-angiopoietin-2 mechanism to suppress de novo production of angiopoietin-2 to inhibit sepsis. Multiple studies have shown that inflammation plays an integral role in chronic cardiovascular diseases like coronary artery disease; see results from CANTOS (https://clinicaltrials.gov/ct2/show/NCT01327846) and Jupiter trials (https://clinicaltrials.gov/ct2/show/NCT00239681). Interestingly, it has been demonstrated that intensive statin therapy is most effective in those with high baseline levels of inflammation. This may be particularly relevant as patients die of cardiogenic shock in the setting of sepsis. Delineating the etiological routs of cardiogenic shock from bacterial toxins and life-style or genetic attributable cardiovascular disease is a challenge. Sepsis and cardiovascular diseases are potentially comorbid, given the role of chronic inflammation from endothelial dysfunction [73]. Identification of statins as a putative anti-sepsis agent is an interesting finding that needs further study. In another study, Kim et al. [74] used drug repositioning for identifying potential candidates for SIRS. Computational drug repositioning can also be used to identify combination of therapies and therapeutic opportunities especially for comorbid diseases [75–77]. For example, we recently reported a method to target molecular sub-network of genes shared by two diseases as plausible signature driving pathways associated with both diseases [78]. Such pathways could be targeted specifically using repurposed compounds for better outcomes.
Cell and gene therapy for sepsis
Cell therapy is typically defined as the administration of living whole cells to the patient for treatment of diseases. The use of mesenchymal stem cells as a potential therapy is being considered given the cells’ immunomodulatory properties on cytokine and chemokine synthesis in sepsis. MNCs are able to reduce tissue inflammation and produce antibacterial peptides that target offending pathogens, thereby reducing morbidity and mortality. However, there are effective dose uncertainties, high costs, production challenges and regulatory landscape that must be considered [79]. Gene therapy is a set of strategies that modify the expression of an individual’s genes or repair abnormal genes. This involves administering specific DNA or RNA molecules using viral or non-viral vectors (see https://www.asgct.org/education/gene-and-cell-therapy-defined and https://www.fda.gov/biologicsbloodvaccines/cellulargenetherapyproducts/default.htm). With the advent of single-cell RNA sequencing and other technologies, disease level cell profiles can help find population of cells increased or depleted in the setting of sepsis and its phenotypes. Kymriah (tisagenlecleucel) is a cell-based gene therapy chimeric antigen receptor-T cell that specifically induces an immune response against cancer cells for certain pediatric and young adult patients with a form of acute lymphoblastic leukemia. Similarly, developing cell-specific immune response to sepsis-associated cells may help to improve prognoses [80, 81].
Immunotherapy for sepsis
Application of immune therapy is an idea that is more than three decades old [82–85]. However, the availability of modern immunotherapy strategies combined with next-generation sequencing technologies [86–89] can be applicable to a variety of conditions including cancers, infectious diseases [90], respiratory diseases, [90] and cardiovascular diseases [91]. Several therapeutics that combine the concept of immunotherapy with cell and gene therapies are in the advanced stages of clinical trials for various cancers. Immunotherapies are currently being evaluated as a candidate for various infectious diseases including viral, bacterial and fungal infections [92–98]. Some of the most promising immunotherapies currently being studied for use in sepsis attempt to reduce T cell exhaustion and apoptosis or augment immune cell proliferation and activation. Recombinant interleukin-7 blocks apoptosis while enhances lymphocytic activation and proliferation (Figure 5). Programmed cell death 1 (PD1) or PDL1-specific antibodies inhibit PD1-PDL1 interaction to reduce apoptosis and augment T cell activation by macrophages as well. Recombinant interferon- ; (IFN
; (IFN ) and recombinant granulocyte/macrophage colony-stimulating factor (GM-CSF) primarily act on monocytes/macrophages to enhance activation of innate immunity. Both IFN
) and recombinant granulocyte/macrophage colony-stimulating factor (GM-CSF) primarily act on monocytes/macrophages to enhance activation of innate immunity. Both IFN
 and GM-CSF help increase expression of HLA-DR, enhancing antigen presenting capacity, and pro-inflammatory cytokine production.
and GM-CSF help increase expression of HLA-DR, enhancing antigen presenting capacity, and pro-inflammatory cytokine production.
Figure 5.
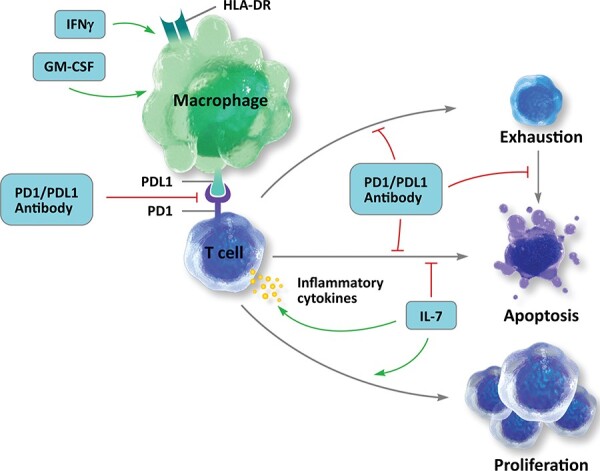
Emerging immunotherapy opportunities in the setting of sepsis.
Phage therapy to target sepsis
Phage therapy is an emerging therapeutic approach to leverage lytic bacteriophages to combat specific bacterial strains associated from infection. Phage therapy is currently being explored as a therapeutic strategy for sepsis [99–101]. An area of growing concern in the prevention of sepsis is the emergence of antimicrobial-resistant bacteria (AMR) [102]. AMR is defined as having either having a multi-drug resistance (MDR) or extensive drug resistance, formerly well-managed bacterial strains like Mycobacterium tuberculosis have emerged now as potentially lethal threats in cases where the bacterial load cannot be managed by traditional therapy. With the prolific use and misuse of antibiotics there is concern among the healthcare community that proper antibiotic stewardship may not be enough to prevent the increase of drug resistant bacteria involved in many sepsis cases [103]. Bacteria that have adapted to be resistant to modern forms of bacteriostatic medications are increasing in number and are beginning to present more often, so new approaches like bacteriophage therapy are emerging as a potential responses to bacteria-induced sepsis. Bacteriophages are viral bodies that have evolved to be highly specific in targeting specific bacterial strains for reproduction while typically ignoring human cells. Already MDR strains like P. aeruginosa have drastically low survival rates if they proliferate into extraintestinal spaces, but there is promising results that a combination of P. aeruginosa bacteriophage treatment and antibiotics can turn back the inflammatory response and control bacteria load to the point of recovery [104]. It should be noted that the optimal use of antibiotics is critical to ensuring the safety, efficacy and outcomes for such treatments.
Microbiome restoration using probiotics and synbiotics
Emerging evidence also suggests that gut microbiome may also play a critical role in sepsis and its acuity in the clinical setting [105–113]. For example, Bifidobacteria spp. were upregulated in controls compared to late onset sepsis samples and Escherichia spp. were upregulated in necrotizing enterocolitis and sepsis [106, 114]. Similarly, evidence for the role of microbial dysbiosis in the microbiome, pathobiome as well as microbiome restoration are emerging in sepsis. For example, a randomized symbiotic trial has shown that sepsis in rural areas of countries could be prevented using a symbiotic diet (mix of probiotic and prebiotic strains [115]) containing L. plantarum ATCC-202195 [116]. Further studies would need to understand whether such synbiotic treatment would be beneficial for adults and patients with multiple comorbidities.
Nanobiotechnology-based theranostic approaches for sepsis
Nanotechnology and its recent advances in the medicine offers a new class of treatment, diagnostic and theranostic opportunities for sepsis [117]. For example, sialic-acid decorated nanoparticles [118] biological microelectromechanical systems [119], nanoparticles (cerium oxide [120], antioxidant nanoparticles [121], biomimetic nanoparticles [122], sialic acid-coated nanoparticles [118]) are being explored as materials for developing diagnostic and therapeutic approaches to target sepsis.
Genetic editing to target sepsis
Genetic editing is a modern approach for precise genetic engineering to edit (insert, delete or modify) genomic regions [123–125]. New methods including CRISPR and CRISPR-associated genes based systems are particularly precise in their ability to edit genomic regions with varying degree off-target effects [126, 127]. While the use of genetic editing is an active area of research, genetic editing could be used to diagnose or target the infectious agents in the context sepsis [128–132].
Prognosis of sepsis therapies
Compared to risk stratification diagnoses and treatment, sepsis prognoses may be the least studied research theme in the context of the disease. Long-term prognoses of sepsis patients are poor. Patients with no organ damage have 15–30% mortality rates, whereas patients with severe sepsis or septic shock have a mortality rate of 40–60% [133–136]. While there are several studies reporting advances in the early detection of sepsis, factors that drive successful, long-term prognoses remain elusive. As a range that involves ~20% differences, precisely subtyping sepsis and modeling care pathways may help to improve prognoses. Care pathway modeling is an emerging theme in biomedical and healthcare data science [137–139]. Briefly, care pathway modeling estimates the disease or syndromic trajectory for a given patient and how perturbations via medications or other clinical interventions could help to lead to a positive outcomes and better prognostic outlooks [132, 140, 141]. Modeling various care pathways for different age groups and patient subtypes may lead to the development of intelligent clinical decision systems. Combing various biomedical and healthcare data sources and building a national and international case repository with extensive clinical history and patient reported information may help to tackle this challenging problem [38].
Discussion
Sepsis is a complex infectious disease with diverse clinical manifestations. Viral, bacterial and fungal agents as well as metagenomic interactions are implicated in sepsis and differential phenotypes. Although not well understood, there is an association between pneumococcal pneumonia and septic shock. In a recent study, 114 of 1041 patients with pneumonia had a septic shock upon admission [142]. However, independent risk factors could be involved such as tobacco smoking and chronic corticosteroid treatment. As well as pneumonia, bloodstream infection has a higher correlation to triggering sepsis. Septicemia, a bloodstream infection, which involves dangerous bacteria and toxins transporting through the human body, that can eventually lead to sepsis if left untreated since after all sepsis is a severe complication of septicemia. Analyzing single-modality data would only lead to the limited understanding of the pathobiology. However, leveraging the combination of biomarkers, data from EHR and combining with in-depth molecular profiling and modeling using multi-scale modeling may yield new insights. Sepsis is a syndrome with a high risk of mortality, ICU visits and hospital readmissions. Sepsis is evolving as a significant clinical, scientific and operational encounter that influences healthcare outcomes. Precise diagnoses with ample time for clinical interventions, treatment with higher response rates and progressive outcomes in sepsis remain as significant challenges. In this article, we provided an overview of emerging technologies including the role of computational medicine, novel therapeutic opportunities and application of machine intelligence methods that may help improve them. As a primary cause for mortality rates associated with hospitalization, developing informatics solutions, predictive models and personalizing risk estimates would have significant implications in quality of healthcare delivery and influence patient outcomes. However, mere data-driven methods or informatics approaches may not provide results without constant feedback from clinical pathways and patient trajectories.
Emerging role of translational bioinformatics approaches in sepsis
Translational bioinformatics approaches are collectively accelerating the discovery of therapeutic strategies for rare and common diseases. Recent progress in cardiology, oncology, immuno-oncology and autoimmune diseases was the net result of growth in computing, artificial intelligence, biomedical technologies and translational bioinformatics. However, the impact of translational bioinformatics approaches in the area of sepsis is very limited. For example, PubMed search retrieves around 700 papers for a query with ‘sepsis + bioinformatics’ compared to 10× times publications that discuss bioinformatics approaches in the setting of cancer and cardiovascular diseases. The apparent lack of translational bioinformatics research could be attributed to various reasons; one reason could be the lack of centralized informatics resources that compile a variety of biomedical and clinical data in the setting of sepsis. Organizing the biomedical data and combining clinical data including patient trajectories, clinical history and therapeutic responses may help to build a community that could ask broad research questions to understand various molecular etiologies of sepsis and target them using individualized approaches.
Future prospects
Despite the availability of care guidelines that reliably improve outcomes, overall sepsis mortality has increased in the past decade. Most healthcare organizations are struggling with mortality rates between 19–30% for bona fide sepsis patients. This high morbidity and mortality adversely influence quality of healthcare delivery, revenue-cycle, and reduce patient recovery rates. The clinical implications from different sepsis cases must be considered for data analysis. Understanding root and route of sepsis incidents from heterogeneous biomedical and health care would help to find identify drivers that prevent infection. Designing translational bioinformatics resource that aid in developing predictive models, multianalyte diagnostic aids, targeted drug discovery and repositioning strategies would help to stratify and treat patients at risk and improve prognoses.
Competing Interests
KS has received consulting fees or honoraria from McKinsey & Company, Alphabet, LEK Consulting, Parthenon-EY, Philips Healthcare, OccamzRazor and Kencore Health. JTD has received consulting fees or honoraria from Janssen Pharmaceuticals, GlaxoSmithKline, AstraZeneca, and Hoffman-La Roche; is a scientific advisor to LAM Therapeutics, NuMedii, and Ayasdi; and holds equity in NuMedii, Ayasdi, and Ontomics. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Key Points
Sepsis is a life-threatening condition that affects more than 30 million people worldwide every year with high mortality rates (15–20%).
Irrespective of advances in translational bioinformatics, sepsis remains to be an understudied theme compared to cancer and cardiovascular diseases.
Understanding the clinical trajectory that leads to sepsis incidents from real-world data streams would help to develop diagnostics and therapeutic approaches to target sepsis and reduce its global disease burden.
Developing predictive models using molecular data combined with clinical data would help to stratify patients at risk for improved care and design healthcare strategies that improve outcome and reduce mortality.
Combining biomedical big data with translational bioinformatics approaches and machine intelligence could help to develop novel strategies to combat sepsis.
Acknowledgements
The authors would like to thank members of the Mount Sinai Health System—Hospital Big Data initiative, Mount Sinai Data Warehouse for facilitating data accessibility and the Mount Sinai Scientific Computing team for infrastructural support. KS and JTD would like to acknowledge the National Institutes of Health, National Center for Advancing Translational Sciences (NCATS), Clinical and Translational Science Awards (UL1TR001433). KS would like to acknowledge Drs Jamie S. Hirsch, John Chelico, Michael Oppenheim, Kevin Bock (Northwell Health, Northwell Health, New York) and Chaitanya Gupta, Emmanuel Query (ProbiusDX Inc, El Cerrito, CA), organizers and attendees of Friend or Foe Proposers Day organized by Defense Advanced Research Projects Agency (DARPA, Arlington County, VA) who gave feedback on the multi-analyte, heterogenous, data-driven diagnostic aid for sepsis.
Andrew C. Liu is a medical student at Donald and Barbara School of Medicine at Hofstra/Northwell, Northwell Health and worked as a health informatics intern at Department of Information Services, Northwell Health, New Hyde Park.
Krishna Patel is a medical student at Donald and Barbara School of Medicine at Hofstra/Northwell, Northwell Health and worked as a health informatics intern at Department of Information Services, Northwell Health, New Hyde Park.
Ramya Dhatri Vunikili is a Master’s student at Courant Institute of Mathematical Sciences, New York University, New York and a summer intern at Center for Research Informatics and Innovation, Northwell Health, New Hyde Park.
Kipp W. Johnson is an MD–PhD student in the Dudley Laboratory at the Icahn School of Medicine at Mount Sinai, Mount Sinai Health System, New York, NY.
Fahad Abdu is a pre-med student and undergraduate at Stonybrook University and a summer intern at Center for Research Informatics and Innovation, Northwell Health, New Hyde Park.
Shivani Kamath Belman is a Bioinformatics Master’s student at Department of Bioengineering, the University of Illinois at Chicago and a summer intern at Center for Research Informatics and Innovation, Northwell Health, New Hyde Park.
Benjamin S. Glicksberg was a PhD candidate in the Dudley and Chen Laboratories, Icahn School of Medicine at Mount Sinai, Mount Sinai Health System, New York, NY. He is currently working as a post-doctoral fellow at the University of California, San Francisco (UCSF).
Pratyush Tandale is a bioinformatics undergraduate student at School of Biotechnology and Bioinformatics, D Y Patil University, Navi Mumbai, India, and a thesis student Center for Research Informatics and Innovation, Northwell Health, New Hyde Park.
Roberto Fontanez is a summer intern at the Center for Research Informatics and Innovation, Northwell Health, New Hyde Park.
Oommen K. Mathew is a computational biologist with a PhD specialization in computational biology and algorithm development.
Andrew Kasarskis is the Chief Data Officer of Mount Sinai Health System, New York, NY.
Priyabrata Mukherjee is a Professor in the Department of Pathology and Experimental Pathology and Peggy and Charles Stephenson Endowed Chair in Cancer Laboratory Research, College of Medicine and Stephenson Cancer Center, The University of Oklahoma Health Sciences Center.
Lakshminarayanan Subramanian is a professor in the Courant Institute of Mathematical Sciences at New York University. His research interests are in the areas of networked systems and data science with applications in computing for development.
Joel T. Dudley is an associate professor of Genetics and Genomic Sciences and founding Director of the Institute for Next Generation Healthcare at the Icahn School of Medicine at Mount Sinai.
Khader Shameer is the Director of Bioinformatics and Data Science Programs at Northwell Health, New Hyde Park. He was a senior biomedical and health care data scientist in the Dudley Laboratory and senior scientist at the Institute of Next Generation Healthcare, Mount Sinai Health System, New York, NY.
Reference
- 1. Center for Disease Control . CDC Urges Early Recognition, Prompt Treatment of Sepsis. https://www.cdc.gov/media/releases/2017/p0831-sepsis-recognition-treatment.html (9 February 2019, date last accessed).
- 2. Torio CM, Moore BJ. National Inpatient Hospital Costs: The Most Expensive Conditions by Payer, 2013: Statistical Brief #204. Healthcare Cost and Utilization Project (HCUP) Statistical Briefs. Rockville (MD), Agency for Healthcare Research and Quality (US), 2006. [PubMed] [Google Scholar]
- 3. Angus DC, Linde-Zwirble WT, Lidicker J, et al. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 2001;29:1303–10. [DOI] [PubMed] [Google Scholar]
- 4. Lagu T, Rothberg MB, Shieh MS, et al. Hospitalizations, costs, and outcomes of severe sepsis in the United States 2003 to 2007. Crit Care Med 2012;40:754–61. [DOI] [PubMed] [Google Scholar]
- 5. Dolin HH, Papadimos TJ, Stepkowski S, et al. A novel combination of biomarkers to herald the onset of Sepsis prior to the manifestation of symptoms. Shock 2018;49:364–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012;13:395–405. [DOI] [PubMed] [Google Scholar]
- 7. Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet 2011;12:417–28. [DOI] [PubMed] [Google Scholar]
- 8. Payne K, Gavan SP, Wright SJ, et al. Cost-effectiveness analyses of genetic and genomic diagnostic tests. Nat Rev Genet 2018;19:235–46. [DOI] [PubMed] [Google Scholar]
- 9. Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019;25:44–56. [DOI] [PubMed] [Google Scholar]
- 10. Norgeot B, Glicksberg BS, Butte AJ. A call for deep-learning healthcare. Nat Med 2019;25:14–5. [DOI] [PubMed] [Google Scholar]
- 11. Dugger SA, Platt A, Goldstein DB. Drug development in the era of precision medicine. Nat Rev Drug Discov 2018;17:183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glicksberg BS, Johnson KW, Dudley JT. The next generation of precision medicine: observational studies, electronic health records, biobanks and continuous monitoring. Hum Mol Genet 2018;27:R56–62. [DOI] [PubMed] [Google Scholar]
- 13. Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM consensus conference committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 1992;101:1644–55. [DOI] [PubMed] [Google Scholar]
- 14. Kalantari A, Mallemat H, Weingart SD. Sepsis definitions: the search for gold and what CMS got wrong. West J Emerg Med 2017;18:951–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016;315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Angus DC, van der Poll T. Severe sepsis and septic shock. New England Journal of Medicine 2013;369:840–51. [DOI] [PubMed] [Google Scholar]
- 17. Opal SM, Garber GE, LaRosa SP, et al. Systemic host responses in severe sepsis analyzed by causative microorganism and treatment effects of drotrecogin alfa (activated). Clin Infect Dis 2003;37:50–8. [DOI] [PubMed] [Google Scholar]
- 18. Lepak A, Andes D. Fungal sepsis: optimizing antifungal therapy in the critical care setting. Crit Care Clin 2011;27:123–47. [DOI] [PubMed] [Google Scholar]
- 19. Duggan S, Leonhardt I, Hunniger K, et al. Host response to Candida albicans bloodstream infection and sepsis. Virulence 2015;6:316–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cawcutt KA, Peters SG. Severe sepsis and septic shock: clinical overview and update on management. Mayo Clin Proc 2014;89:1572–8. [DOI] [PubMed] [Google Scholar]
- 21. Harrison TR, Kasper DL, Fauci AS. Harrison's Principles of Internal Medicine, 19th edition. New York: McGraw-Hill, 2015.
- 22. Landry DW, Oliver JA. The pathogenesis of vasodilatory shock. N Engl J Med 2001;345:588–95. [DOI] [PubMed] [Google Scholar]
- 23. Chung LP, Waterer GW. Genetic predisposition to respiratory infection and sepsis. Crit Rev Clin Lab Sci 2011;48:250–68. [DOI] [PubMed] [Google Scholar]
- 24. Boomer JS, To K, Chang KC, et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA 2011;306:2594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wanner GA, Keel M, Steckholzer U, et al. Relationship between procalcitonin plasma levels and severity of injury, sepsis, organ failure, and mortality in injured patients. Crit Care Med 2000;28:950–7. [DOI] [PubMed] [Google Scholar]
- 26. Faix JD. Biomarkers of sepsis. Crit Rev Clin Lab Sci 2013;50:23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aitken SL, Hemmige VS, Koo HL, et al. Real-world performance of a microarray-based rapid diagnostic for gram-positive bloodstream infections and potential utility for antimicrobial stewardship. Diagn Microbiol Infect Dis 2015;81:4–8. [DOI] [PubMed] [Google Scholar]
- 28. Hassan MM, Ranzoni A, Cooper MA. A nanoparticle-based method for culture-free bacterial DNA enrichment from whole blood. Biosens Bioelectron 2018;99:150–5. [DOI] [PubMed] [Google Scholar]
- 29. Russell JA. Genomics and pharmacogenomics of sepsis: so close and yet so far. Crit Care 2016;20:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sweeney TE, Shidham A, Wong HR, et al. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med 2015;7:287ra271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Monte AA, Brocker C, Nebert DW, et al. Improved drug therapy: triangulating phenomics with genomics and metabolomics. Hum Genomics 2014;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sweeney TE, Perumal TM, Henao R, et al. A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun 2018;9:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langley RJ, Wong HR. Early diagnosis of sepsis: is an integrated Omics approach the way forward? Mol Diagn Ther 2017;21:525–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shameer K, Johnson KW, Glicksberg BS, et al. Machine learning in cardiovascular medicine: are we there yet? Heart 2018;104:1156–64. [DOI] [PubMed] [Google Scholar]
- 36. Johnson KW, Torres Soto J, Glicksberg BS, et al. Artificial intelligence in cardiology. J Am Coll Cardiol 2018;71:2668–79. [DOI] [PubMed] [Google Scholar]
- 37. Shameer K, Johnson KW, Yahi A, et al. Predictive modeling of hospital readmission rates using electronic medical record-wide machine learning: a case-study using Mount Sinai heart failure cohort. Pac Symp Biocomput 2017;22:276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shameer K, Badgeley MA, Miotto R, et al. Translational bioinformatics in the era of real-time biomedical, health care and wellness data streams. Brief Bioinform 2017;18:105–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peters LA, Perrigoue J, Mortha A, et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet 2017;49:1437–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yadav KK, Shameer K, Readhead B, et al. Systems medicine approaches to improving understanding, treatment, and clinical Management of Neuroendocrine Prostate Cancer. Curr Pharm Des 2016;22:5234–48. [DOI] [PubMed] [Google Scholar]
- 41. Hodos RA, Kidd BA, Shameer K, et al. In silico methods for drug repurposing and pharmacology. Wiley Interdiscip Rev Syst Biol Med 2016;8:186–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Badgeley MA, Shameer K, Glicksberg BS, et al. EHDViz: clinical dashboard development using open-source technologies. BMJ Open 2016;6:e010579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Henry KE, Hager DN, Pronovost PJ, et al. A targeted real-time early warning score (TREWScore) for septic shock. Sci Transl Med 2015;7:299ra122. [DOI] [PubMed] [Google Scholar]
- 44. Kullo IJ, Jouni H, Austin EE, et al. Incorporating a genetic risk score into coronary heart disease risk estimates: effect on low-density lipoprotein cholesterol levels (the MI-GENES clinical trial). Circulation 2016;133:1181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nature Reviews Genetics, 2018;19:9–581. [DOI] [PubMed] [Google Scholar]
- 46. Lewis CM, Vassos E. Prospects for using risk scores in polygenic medicine. Genome Med 2017;9:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rautanen A, Mills TC, Gordon AC, et al. Genome-wide association study of survival from sepsis due to pneumonia: an observational cohort study. Lancet Respir Med 2015;3:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scherag A, Schoneweck F, Kesselmeier M, et al. Genetic factors of the disease course after sepsis: a genome-wide study for 28Day mortality. EBioMedicine 2016;12:239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schwarze K, Buchanan J, Taylor JC, et al. Are whole-exome and whole-genome sequencing approaches cost-effective A systematic review of the literature? Genetics in Medicine, 2018;20:1122–1130. [DOI] [PubMed] [Google Scholar]
- 50. Aro K, Wei F, Wong DT, et al. Saliva liquid biopsy for point-of-care applications. Front Public Health 2017;5:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Johnson K, Khader S, Glicksberg BS, et al. A machine learning model predicts individuals who improve coronary artery plaque fibrous cap thickness following high-intensity statin therapy. J Am Coll Cardiol 2018;71:A1348. [Google Scholar]
- 52. Clancy CE, An G, Cannon WR, et al. Multiscale modeling in the clinic: drug design and development. Ann Biomed Eng 2016;44:2591–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sweeney TE, Wong HR, Khatri P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci Transl Med 2016;8:346ra391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shameer K, Johnson KW, Glicksberg BS, et al. The whole is greater than the sum of its parts: combining classical statistical and machine intelligence methods in medicine. Heart 2018;104:1228. [DOI] [PubMed] [Google Scholar]
- 55. Sørensen TIA, Nielsen GG, Andersen PK, et al. Genetic and environmental influences on premature death in adult adoptees. New England Journal of Medicine 1988;318:727–32. [DOI] [PubMed] [Google Scholar]
- 56. Giamarellos-Bourboulis EJ, Opal SM. The role of genetics and antibodies in sepsis. Ann Transl Med 2016;4:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Namath A, Patterson AJ. Genetic polymorphisms in sepsis. Crit Care Nurs Clin North Am 2011;23:181–202. [DOI] [PubMed] [Google Scholar]
- 58. Tunjungputri RN, Mobegi FM, Cremers AJ, et al. Phage-derived protein induces increased platelet activation and is associated with mortality in patients with invasive pneumococcal disease. MBio 2017;8:e01984-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dolgachev VA, Goldberg R, Suresh MV, et al. Electroporation-mediated delivery of the FER gene in the resolution of trauma-related fatal pneumonia. Gene Ther 2016;23:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ferguson JF, Meyer NJ, Qu L, et al. Integrative genomics identifies 7p11.2 as a novel locus for fever and clinical stress response in humans. Hum Mol Genet 2015;24:1801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Salipante SJ, Roach DJ, Kitzman JO, et al. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 2015;25:119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mikacenic C, Reiner AP, Holden TD, et al. Variation in the TLR10/TLR1/TLR6 locus is the major genetic determinant of interindividual difference in TLR1/2-mediated responses. Genes Immun 2013;14:52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rowell JL, Dowling NF, Yu W, et al. Trends in population-based studies of human genetics in infectious diseases. PLoS One 2012;7:e25431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kayagaki N, Stowe IB, Lee BL, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015;526:666–71. [DOI] [PubMed] [Google Scholar]
- 65. Rittirsch D, Flierl MA, Ward PA. Harmful molecular mechanisms in sepsis. Nat Rev Immunol 2008;8:776–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hotchkiss RS, Moldawer LL, Opal SM, et al. Sepsis and septic shock. Nat Rev Dis Primers 2016;2:16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shameer K, Denny JC, Ding K, et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet 2014;133:95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 2013;31:1102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nguyen DT, Mathias S, Bologa C, et al. Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res 2017;45:D995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shameer K, Readhead B, Dudley JT. Computational and experimental advances in drug repositioning for accelerated therapeutic stratification. Curr Top Med Chem 2015;15:5–20. [DOI] [PubMed] [Google Scholar]
- 71. Shameer K, Glicksberg BS, Hodos R, et al. Systematic analyses of drugs and disease indications in RepurposeDB reveal pharmacological, biological and epidemiological factors influencing drug repositioning. Brief Bioinform 2018;19:656–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ghosh CC, Thamm K, Berghelli AV, et al. Drug repurposing screen identifies Foxo1-dependent Angiopoietin-2 regulation in sepsis. Crit Care Med 2015;43:e230–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang HE, Shapiro NI, Griffin R, et al. Chronic medical conditions and risk of sepsis. PLoS One 2012;7:e48307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim JH, Lee J, Park KS, et al. Drug repositioning to alleviate systemic inflammatory response syndrome caused by gram-negative bacterial outer membrane vesicles. Adv Healthc Mater 2018;7:e1701476. [DOI] [PubMed] [Google Scholar]
- 75. Gayvert KM, Aly O, Platt J, et al. A computational approach for identifying synergistic drug combinations. PLoS Comput Biol 2017;13:e1005308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sun W, Sanderson PE, Zheng W. Drug combination therapy increases successful drug repositioning. Drug Discov Today 2016;21:1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Clouser CL, Patterson SE, Mansky LM. Exploiting drug repositioning for discovery of a novel HIV combination therapy. J Virol 2010;84:9301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shameer K, Dow G, Glicksberg BS, et al. A network-biology informed computational drug repositioning strategy to target disease risk trajectories and comorbidities of peripheral artery disease. AMIA Jt Summits Transl Sci Proc 2017;2018:108–17. [PMC free article] [PubMed] [Google Scholar]
- 79. Laroye C, Gibot S, Reppel L, et al. Concise review: Mesenchymal stromal/stem cells: a new treatment for sepsis and septic shock? Stem Cells 2017;35:2331–9. [DOI] [PubMed] [Google Scholar]
- 80. Avraham R, Haseley N, Brown D, et al. Pathogen cell-to-cell variability drives heterogeneity in host immune responses. Cell 2015;162:1309–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rimmele T, Payen D, Cantaluppi V, et al. Immune cell phenotype and function in sepsis. Shock 2016;45:282–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Durant JR. Immunotherapy of cancer: the end of the beginning? N Engl J Med 1987;316:939–41. [DOI] [PubMed] [Google Scholar]
- 83. Farkona S, Diamandis EP, Blasutig IM. Cancer immunotherapy: the beginning of the end of cancer? BMC Med 2016;14:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lazzari C, Karachaliou N, Bulotta A, et al. Combination of immunotherapy with chemotherapy and radiotherapy in lung cancer: is this the beginning of the end for cancer? Ther Adv Med Oncol 2018;10:1758835918762094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Toh HC. Cancer immunotherapy-the end of the beginning. Chin Clin Oncol 2018;7:12. [DOI] [PubMed] [Google Scholar]
- 86. Leentjens J, Kox M, van der Hoeven JG, et al. Immunotherapy for the adjunctive treatment of sepsis: from immunosuppression to immunostimulation. Time for a paradigm change? Am J Respir Crit Care Med 2013;187:1287–93. [DOI] [PubMed] [Google Scholar]
- 87. van der Poll T. Immunotherapy of sepsis. Lancet Infect Dis 2001;1:165–74. [DOI] [PubMed] [Google Scholar]
- 88. Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol 2013;13:862–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hotchkiss RS, Opal S. Immunotherapy for sepsis—a new approach against an ancient foe. N Engl J Med 2010;363:87–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Manohar A, Ahuja J, Crane JK. Immunotherapy for infectious diseases: past, present, and future. Immunol Invest 2015;44:731–7. [DOI] [PubMed] [Google Scholar]
- 91. Martini E, Stirparo GG, Kallikourdis M. Immunotherapy for cardiovascular disease. J Leukoc Biol 2018;103:493–500. [DOI] [PubMed] [Google Scholar]
- 92. Kullberg BJ. Trends in immunotherapy of fungal infections. Eur J Clin Microbiol Infect Dis 1997;16:51–5. [DOI] [PubMed] [Google Scholar]
- 93. Hamad M. Antifungal immunotherapy and immunomodulation: a double-hitter approach to deal with invasive fungal infections. Scand J Immunol 2008;67:533–43. [DOI] [PubMed] [Google Scholar]
- 94. Hirsch HH, Hartmann A. Immunotherapy for invasive fungal infections in transplant patients: back to the future? Am J Transplant 2010;10:1719–20. [DOI] [PubMed] [Google Scholar]
- 95. Delsing CE, Gresnigt MS, Leentjens J, et al. Interferon-gamma as adjunctive immunotherapy for invasive fungal infections: a case series. BMC Infect Dis 2014;14:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Einsele H, Loffler J, Kapp M, et al. Immunotherapy for viral and fungal infections. Bone Marrow Transplant 2015;50(Suppl 2):S51–4. [DOI] [PubMed] [Google Scholar]
- 97. Cutino-Moguel MT, Eades C, Rezvani K, et al. Immunotherapy for infectious diseases in haematological immunocompromise. Br J Haematol 2017;177:348–56. [DOI] [PubMed] [Google Scholar]
- 98. Hill JA, Li D, Hay KA, et al. Infectious complications of CD19-targeted chimeric antigen receptor-modified T-cell immunotherapy. Blood 2018;131:121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gorski A, Jonczyk-Matysiak E, Lusiak-Szelachowska M, et al. The potential of phage therapy in sepsis. Front Immunol 2017;8:1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pouillot F, Chomton M, Blois H, et al. Efficacy of bacteriophage therapy in experimental sepsis and meningitis caused by a clone O25b:H4-ST131 Escherichia coli strain producing CTX-M-15. Antimicrob Agents Chemother 2012;56:3568–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Watanabe R, Matsumoto T, Sano G, et al. Efficacy of bacteriophage therapy against gut-derived sepsis caused by Pseudomonas aeruginosa in mice. Antimicrob Agents Chemother 2007;51:446–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Nazir T, Abraham S, Islam A. Emergence of potential superbug mycobacterium tuberculosis, lessons from New Delhi mutant-1 bacterial strains. Int J Health Sci (Qassim) 2012;6:87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Leeds IL, Fabrizio A, Cosgrove SE, et al. Treating wisely: the surgeon's role in antibiotic stewardship. Ann Surg 2017;265:871–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Can K, Aksu U, Yenen OS. Investigation of PhiKZ phage therapy against Pseudomonas aeruginosa in mouse pneumonia model. Turk J Med Sci 2018;48:670–8. [DOI] [PubMed] [Google Scholar]
- 105. Wandro S, Osborne S, Enriquez C, et al. The microbiome and Metabolome of preterm infant stool are personalized and not driven by health outcomes, including necrotizing enterocolitis and late-onset sepsis. mSphere 2018;3:e00104-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Stewart CJ, Embleton ND, Marrs ECL, et al. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome 2017;5:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Alverdy JC, Krezalek MA. Collapse of the microbiome, emergence of the Pathobiome, and the immunopathology of sepsis. Crit Care Med 2017;45:337–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lachar J, Bajaj JS. Changes in the microbiome in cirrhosis and relationship to complications: hepatic encephalopathy, spontaneous bacterial peritonitis, and sepsis. Semin Liver Dis 2016;36:327–30. [DOI] [PubMed] [Google Scholar]
- 109. Dickson RP, Singer BH, Newstead MW, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol 2016;1:16113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cabrera-Perez J, Badovinac VP, Griffith TS. Enteric immunity, the gut microbiome, and sepsis: rethinking the germ theory of disease. Exp Biol Med (Maywood) 2017;242:127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kumar PS. From focal sepsis to periodontal medicine: a century of exploring the role of the oral microbiome in systemic disease. J Physiol 2017;595:465–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Krezalek MA, DeFazio J, Zaborina O, et al. The shift of an intestinal "microbiome" to a "Pathobiome" governs the course and outcome of sepsis following surgical injury. Shock 2016;45:475–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Stewart CJ, Marrs EC, Nelson A, et al. Development of the preterm gut microbiome in twins at risk of necrotising enterocolitis and sepsis. PLoS One 2013;8:e73465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Denning N-L, Prince JM. Neonatal intestinal dysbiosis in necrotizing enterocolitis. Mol Med 2018;24:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Pandey KR, Naik SR, Vakil BV. Probiotics, prebiotics and synbiotics- a review. J Food Sci Technol 2015;52:7577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Panigrahi P, Parida S, Nanda NC, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 2017;548:407–12. [DOI] [PubMed] [Google Scholar]
- 117. Buxton DB. Nanomedicine for the management of lung and blood diseases. Nanomedicine (Lond) 2009;4:331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Spence S, Greene MK, Fay F, et al. Targeting Siglecs with a sialic acid-decorated nanoparticle abrogates inflammation. Sci Transl Med 2015;7:303ra140. [DOI] [PubMed] [Google Scholar]
- 119. Hassan U, Ghonge T, Reddy B Jr, et al. A point-of-care microfluidic biochip for quantification of CD64 expression from whole blood for sepsis stratification. Nat Commun 2017;8:15949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Selvaraj V, Manne ND, Arvapalli R, et al. Effect of cerium oxide nanoparticles on sepsis induced mortality and NF-kappaB signaling in cultured macrophages. Nanomedicine (Lond) 2015;10:1275–88. [DOI] [PubMed] [Google Scholar]
- 121. Soh M, Kang DW, Jeong HG, et al. Ceria-zirconia nanoparticles as an enhanced multi-antioxidant for sepsis treatment. Angew Chem Int Ed Engl 2017;56:11399–403. [DOI] [PubMed] [Google Scholar]
- 122. Thamphiwatana S, Angsantikul P, Escajadillo T, et al. Macrophage-like nanoparticles concurrently absorbing endotoxins and proinflammatory cytokines for sepsis management. Proc Natl Acad Sci U S A 2017;114:11488–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet 2014;23:R40–6. [DOI] [PubMed] [Google Scholar]
- 125. Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of a*T to G*C in genomic DNA without DNA cleavage. Nature 2017;551:464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. White MK, Kaminski R, Wollebo H, et al. Gene editing for treatment of neurological infections. Neurotherapeutics 2016;13:547–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Narula S, Shameer K, Salem Omar AM, et al. Reply: deep learning with unsupervised feature in echocardiographic imaging. J Am Coll Cardiol 2017;69:2101–2. [DOI] [PubMed] [Google Scholar]
- 128. Tang L, Zeng Y, Du H, et al. CRISPR/Cas9-mediated gene editing in human zygotes using Cas9 protein. Mol Genet Genomics 2017;292:525–33. [DOI] [PubMed] [Google Scholar]
- 129. Ris MM, Deitrich RA, Von Wartburg JP. Inhibition of aldehyde reductase isoenzymes in human and rat brain. Biochem Pharmacol 1975;24:1865–9. [DOI] [PubMed] [Google Scholar]
- 130. Suzuki T, Asami M, Perry AC. Asymmetric parental genome engineering by Cas9 during mouse meiotic exit. Sci Rep 2014;4:7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Bakhrebah MA, Nassar MS, Alsuabeyl MS, et al. CRISPR technology: new paradigm to target the infectious disease pathogens. Eur Rev Med Pharmacol Sci 2018;22:3448–52. [DOI] [PubMed] [Google Scholar]
- 132. Caliendo AM, Hodinka RL. A CRISPR way to diagnose infectious diseases. N Engl J Med 2017;377:1685–7. [DOI] [PubMed] [Google Scholar]
- 133. Schoenberg MH, Weiss M, Radermacher P. Outcome of patients with sepsis and septic shock after ICU treatment. Langenbecks Arch Surg 1998;383:44–8. [DOI] [PubMed] [Google Scholar]
- 134. Cuthbertson BH, Elders A, Hall S, et al. Mortality and quality of life in the five years after severe sepsis. Crit Care 2013;17:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Kibe S, Adams K, Barlow G. Diagnostic and prognostic biomarkers of sepsis in critical care. J Antimicrob Chemother 2011;66(Suppl 2):ii33–40. [DOI] [PubMed] [Google Scholar]
- 136. Garcia-Simon M, Morales JM, Modesto-Alapont V, et al. Prognosis biomarkers of severe sepsis and septic shock by 1H NMR urine metabolomics in the intensive care unit. PLoS One 2015;10:e0140993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Augusto V, Xie X, Prodel M et al. Evaluation of discovered clinical pathways using process mining and joint agent-based discrete-event simulation. In: 2016 Winter Simulation Conference (WSC), 2016, pp. 2135–2146. IEEE Press.
- 138. Yoo S, Cho M, Kim S, et al. Conformance analysis of clinical pathway using electronic health record data. Healthc Inform Res 2015;21:161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Weerdt JD, Caron F, Vanthienen J et al. Getting a grasp on clinical pathway data: an approach based on process mining. Proceedings of the 2012 Pacific-Asia conference on Emerging Trends in Knowledge Discovery and Data Mining. Kuala Lumpur, Malaysia: Springer-Verlag, 2013, 22–35. [Google Scholar]
- 140. Miotto R, Li L, Kidd BA, et al. Deep patient: An unsupervised representation to predict the future of patients from the electronic health records. Sci Rep 2016;6:26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Yang W, Su Q. Process mining for clinical pathway: literature review and future directions. In: 2014 11th International Conference on Service Systems and Service Management (ICSSSM). 2014, pp. 1–5.
- 142. Garcia-Vidal C, Ardanuy C, Tubau F, et al. Pneumococcal pneumonia presenting with septic shock: host- and pathogen-related factors and outcomes. Thorax 2010;65:77–81. [DOI] [PubMed] [Google Scholar]


