Abstract
Synthetic genomics aims to de novo synthesize a functional genome redesigned from natural sequences with custom features. Designed genomes provide new toolkits for better understanding organisms, evolution and the construction of cellular factories. Currently maintaining the fitness of cells with synthetic genomes is particularly challenging as defective designs and unanticipated assembly errors frequently occur. Mapping and correcting bugs that arise during the synthetic process are imperative for the successful construction of a synthetic genome that can sustain a desired cellular function. Here, we review recently developed methods used to map and fix various bugs which arise during yeast genome synthesis with the hope of providing guidance for putting the synthetic yeast chromosome to work.
This review summarizes strategies used to map and repair various bugs in synthetic genomic sequences and provides guidance for the construction of synthetic yeast chromosomes that are capable of maintaining cell fitness.
Introduction
The discovery of the chemical synthesis of urea and secretin opened the door to new organic chemicals and protein synthesis.1 The discovery of the DNA double helix structure along with more recent advances in DNA sequencing provided the impetus for the synthesis of genomes.2–4 Synthetic biologists are no longer satisfied with just copying a natural genome, and are ambitious to create new versions of genomes.5–19 Computer-aided simulation allows the redesign of genomes with specific functions and, in compliance with the most fundamental principle of genomic design, maintaining cell viability,7,11,12,20 custom genetic features can be introduced to increase the genome flexibility. For example, recoding, the introduction of recombination sites and watermark sequences,9 and the deletion of repetitive sequences and unstable elements may all be achieved.12,20 The newly designed genome sequence is hierarchically divided into oligonucleotides,7,9,21 and subsequently assembled into “short”,22,23 “medium”,24–26 and “long”13,20 DNA segments both in vivo and in vitro. Finally, the chemically synthesized DNA is transplanted into a bacterial or yeast cell replacing the native genetic material.11,27
Synthetic genomes have been made for a wide range of cell types, from unicellular prokaryotes7,11,12,18 and eukaryotes9,10,13–17 to multicellular plants and animals.28 However, generating a synthetic genome that is capable of maintaining cell fitness is still challenging.29 Although the genomes of Synechocystis PCC6803 and Mycoplasma genitalium have been synthesized and assembled in yeast cells, the step to replace the corresponding native genomes failed.5,19 Design flaws often occur due to limited biological knowledge or incomplete design principles,7,13,15 whilst spontaneous DNA variations will occur during cell division and genome assembly.7,14,16,17,30 For example, these variations include single-nucleotide variation (SNV), insertion/deletion (InDel), structural variation (SV) and copy number variation (CNV). Thus, researchers have to detect the bugs and debug following every assembly and sometimes even have to redesign the genome in order to maintain cell fitness (Fig. 1A).7,10 Genome synthesis is associated with debugging throughout the process,29 for example, about 15 years and US$40 million were spent on the synthesis and debugging of the Mycoplasma mycoides genome.29 Even the presence of a single base variant could have delayed the process for months.7 During our work on the synthetic yeast chromosome V,16 our group organized 5 sub-teams and spent more than 6 months debugging a lethal defect (unpublished data). Genome synthesis is tedious work and the most difficult task is to locate and debug errors. Here, we review recent progress in mapping and correcting bugs, revealing insights into genome synthesis.
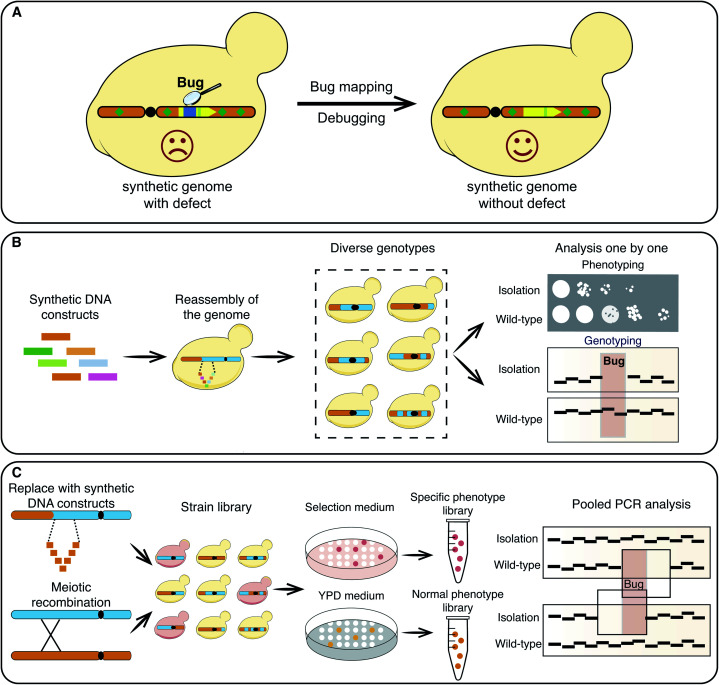
Fig. 1. Bug mapping of synthetic yeast chromosomes. (A) Mapping and debugging unanticipated design flaws or sequence variations on synthetic yeast chromosomes can recover cell growth fitness. (B) Bug mapping with a genome reassembly strategy. Synthetic DNA constructs are used to replace the corresponding native genomic sequences to generate colonies with diverse genotypes. Colonies with growth defects are isolated for genomic analysis to narrow down potential defective loci. (C) Bug mapping by PoPM strategy. A genome reassembly strategy or a meiotic recombination strategy are used to generate candidate colonies with diverse genotypes. Then, colonies are separated into two labraries based on their fitness, with one being robust and the other being defective in growth The two libraries undergo pooled PCR analysis to track potential loci that cause growth defects.
Bug mapping
Phenotypes, genotypes and omics can reflect the presence of bugs in synthetic genome sequences.13–17 Genome reassembly with semisynthetic DNA constructs,7 or iterative meiotic recombination and tetrad dissection have been used to narrow down possible defective regions (Fig. 1B).13,16 Using semisynthetic genome constructs, the failure to transplant a synthetic M. mycoides genome was attributed to the 811–900 construct.7 A yeast strain carrying a synthetic yeast chromosome VI (synVI) exhibited a respiratory deficient. In this case a meitotic recombination strategy and an “endoreduplication backcross” strategy were used to track the defect to the synthetic PCRTag closer to the PRE4 3′ end (Table 1).13
The pros and cons of various debugging methods.
| Methods | Pros | Cons |
|---|---|---|
| HR-based gene replacement | • The method is simple and based on two HR procedures. | • The selection process is laborious. |
| • The method can be used to repair SNVs,13,15 InDels14,15,17 and CNVs.16 | • False positive rates are relatively high. | |
| I-SceI endonuclease mediated method | • DSBs are introduced in specific genomic loci by I-SceI. | • An extra step to incorporate the I-SceI cassette is necessary. |
| • SNVs,48 SVs,14 and CNVs14 can be seamlessly modified with high efficiency. | ||
| Meiotic crossover | • This method can be used to repair extra copies of synthetic DNA that misintegrated on the synthetic chromosome.15 | • The strain carrying the synthetic chromosome must be mated with a native strain of the opposite mating-type. |
| • The insertion of selective markers can promote the selection efficiency. | • The chromosomal crossover randomly occurs throughout the whole chromosome, thus the selection process is laborious. | |
| Endoreduplication backcross | • This method can avoid crossover between synthetic chromosomes and corresponding native chromosomes during meiosis. | • The strain carrying the synthetic chromosome must be mated with a native strain of the opposite mating-type, in which the corresponding native chromosome needs to be engineered with a specific cassette of GAL1p-CEN::Kl.URA3. |
| • CNVs of long DNA sequences that misintegrated on other chromosome(s),16 2-micron plasmid deletions15 and mitochondria deficiencies13 can be efficiently repaired by using this method. | ||
| CRISPR/Cas9-mediated genomic editing | • DSBs are introduced in specific genomic loci by CRISPR/Cas9. | • The method relies on the presence and precise positioning of PAM site in the target sequence and therefore is not applicable to the entire genome. |
| • SNVs and InDels can be repaired in one step by subsequent homologous recombination of the donor DNA.16 | • Other problems of this method include off-target cleavage and variable efficiencies. | |
| Dual-labelling correction | • The PAM site and corresponding protospacer are inserted to make the target loci applicable to CRISPR/Cas9. | • It is necessary to incorporate a doubly selective cassette containing both an antibiotic gene and an auxotroph gene adjacently to a specific target for repair. |
| • DSBs are introduced in specific genomic loci by CRISPR/Cas9 to facilitate the repair of SNVs and InDels.16 | ||
| • The false positive rate is decreased by double selection of both an antibiotic and an auxotroph. | ||
| • Multiple targets can be repaired simultaneously. |
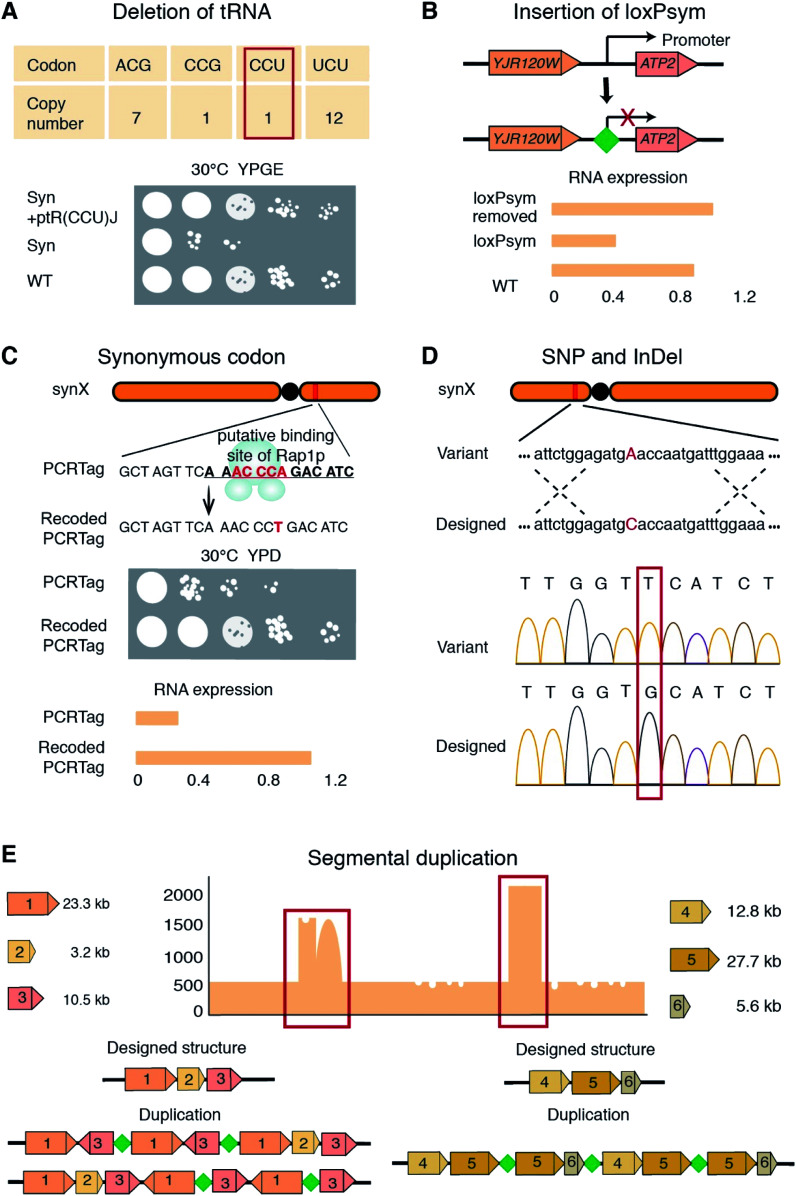
Both the aforementioned strategies require multiple steps that have to be carried out repeatedly over numerous rounds, and thus are extremely time consuming and labor-intensive. A pooled PCRTag mapping (PoPM) strategy was recently developed to locate bugs on the synthetic yeast chromosome X (synX).15 Utilizing this strategy one can rapidly locate bugs on a synthetic yeast chromosome (Fig. 1C).15 Yeast colonies were divided into two libraries based on their fitness, with one being normal and the other being defective in growth. Subsequently, PCR analysis was performed on both libraries using predesigned PCRTags based on synthetic and wild-type chromosomes. The specific regions only detected in the normal library but not in the defective library were considered potential defective loci. Using a PoPM strategy, the causes of growth defects were quickly pinpointed to the genomic loci including the deletion of tR(CCU)J, the inserted loxPsym site at the 3′UTR of YJR120W and a specific synonymous recoding of the essential gene FIP1 (Fig. 2A–C).15
Fig. 2. Bugs on synthetic yeast chromosomes. (A) The deletion of tR(CCU)J leads to a growth defect in the synX strain under selective conditions. (B) The insertion of a loxPsym site at the 3′UTR of YJR120W disrupts the expression of ATP2. (C) The PCRTag in FIP1 is recoded to resemble the binding site of Rap1p, which down-regulates FIP1 expression. Spontaneously SNVs/InDels (D) and CNVs and SVs (E) are detected on synthetic yeast chromosomes. YPGE, yeast extract peptone glycerol ethanol. YPD, yeast extract peptone dextrose. Green diamond, loxPsym site.
SNVs, InDels, SVs and CNVs on synthetic yeast chromosomes have been detected by various methods including pulsed field gel electrophoresis (PFGE), whole genome sequencing (WGS), and PCR and qPCR analysis (Fig. 2D and E).9,10,13,14,16,17 CNVs on synthetic chromosomes are often associated with introduced loxPsym sites or cohesive termini resulting from restriction enzyme digestion during chromosome assembly.15,16 For example, a CNV on synX involved three fragments of 12.8, 27.7 and 5.6 kb in length, which were ligated by the loxPsym sequence (Fig. 2E).15 The quadruplication of a 2 kb sequence on synV was tandemly ligated by a short cohesive-end-like “GCGGCGC” sequence.16
Debugging
Reverting synthetic sequences at the defective loci to the corresponding native ones is a typical debugging method.13,17 For example, in the synthesis of synX, the PCRTag in FIP1, a synonymously recoded sequence, was designed and introduced to resemble the binding site of Rap1p. Consistent with Rap1p being a transcriptional repressor, the authors observed that FIP1 expression in the synX strain was down-regulated. Strikingly, merely reverting the base pairs in the putative Rap1p binding site was sufficient to completely repair the growth deficiency (Fig. 2C).15 Another debugging example from the synX project was at the ATP2 locus. In a qPCR screen, ATP2 expression in the synX strain was found to be significantly lower than the wild type level. An loxPsym site inserted downstream of the neighboring ORF YJR120W was hypothesized to alter the ATP2 promoter function. Deletion of this loxPsym site restored the ATP2 expression and recovered the fitness of the strain.15
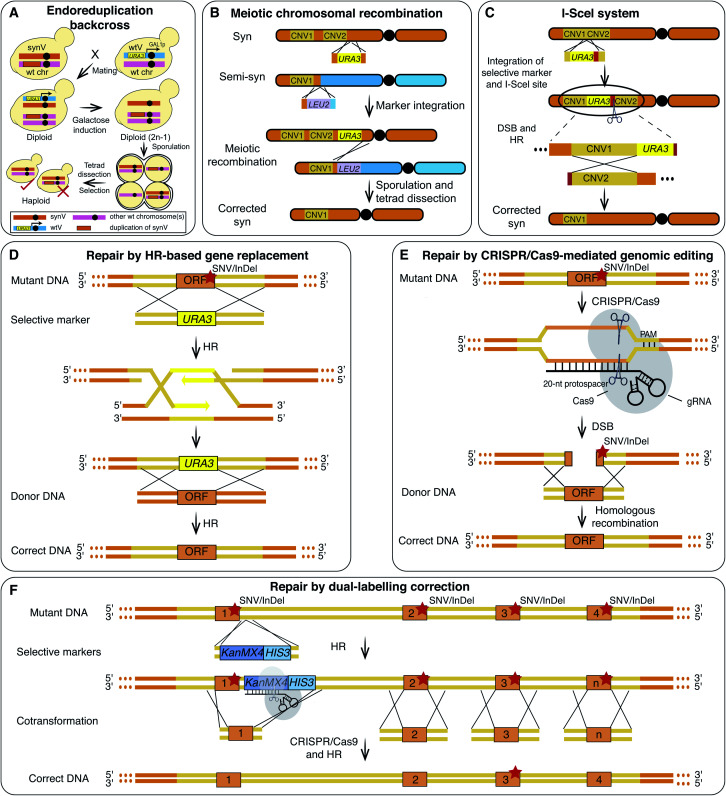
Chromosome abnormalities, SVs and CNVs, are not uncommon in synthetic yeast chromosomes and have been fixed to recover cell fitness. The “endoreduplication backcross” method was an efficient way to repair the CNVs of long sequences (Fig. 3A and Table 1).20,31 Using this method, an extra copy of two synV sequences (291 217–299 980 bp and 305 355–325 235 bp) on other chromosome(s) was removed, and the conditional growth defects of the strain were consequently repaired.16 Meiotic crossover is a frequently employed strategy to repair CNVs on the same chromosome (Fig. 3B). To remove the CNV on synX, haploids of opposite mating types carrying synX and semi-synX, with and without the CNV, respectively, were crossed. Following tetrad dissection, a haploid strain carrying a synX without the CNV was isolated using meiotic recombination.15 To facilitate the repair of SVs and CNVs, I-SceI endonuclease was employed to generate a double-strand break (DSB) at the target region and to promote homologous recombination (HR) (Fig. 3C).16,17
Fig. 3. Debugging of CNVs, SNVs and InDels. (A) An “endoreduplication backcross” strategy was used to repair large-scale duplications of synV that misintegrated on other chromosome(s). (B) A Meiotic crossover strategy was used to restore the duplication that misintegrated on the synthetic chromosome. (C) An I-SceI mediated repair strategy was used to correct SVs. The red segments denote I-Scel recognition sites. SNVs/InDels on synthetic yeast chromosomes are repaired using HR-based gene replacement (D), CRISPR/Cas9-mediated genomic editing (E) and dual-labelling correction (F) strategies. The orange rectangles denote the target regions that need to be corrected and the red stars represent the SNVs/InDels.
It is hard to predict the effects of each of the numerous SNVs and InDels that occur during genome synthesis. These may interfere with the evaluation of the design principles. Therefore, perfect matching of the synthesized sequence to the designed sequence is crucial and remains a major challenge in the field.16 Recently a combination of various methods including HR-based gene replacement, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated genomic editing and dual-labelling correction have been employed to fix SNVs and InDels. This resulted in the successful synthesis of a synV that “perfectly” matches the designed sequence (Table 1).16
For the initial synV, SNVs and InDels involving 3331 bp were revealed using WGS.16 The HR-based gene replacement method, requiring two steps of homologous recombination and selection (Fig. 3D), is laborious. A CRISPR/Cas9-mediated one-step method was then developed. It has been reported that the use of the CRISPR/Cas9 genomic editing tool to generate DSBs resulted in a 5–130 fold increase in the HR efficiency.32 Therefore, CRISPR/Cas9 was adapted in an effort to eliminate the SNVs and InDels during the construction of synV (Fig. 3E).16,33 However, this method relies on the presence and precise positioning of the NGG sequence, the protospacer adjacent motif (PAM) site in the target sequence and therefore is not applicable to the entire genome. Other problems with this method include off-target cleavage and variable efficiencies.
The problems associated with the aforementioned methods have hindered the complete removal of errors in synV. Therefore, a dual-labelling correction strategy (Fig. 3F) was developed to repair the remaining errors. A doubly selective cassette containing both an antibiotic gene (e.g. KanMX4) and an auxotroph gene (e.g. HIS3) was inserted adjacent to a specific target with the double selection minimizing false positive rates. The insertion of the cassette also introduced a protospacer and a PAM site. Subsequently, a pool of donor DNA fragments covering multiple target sites was co-transformed for multiplex correction. The predesigned artificial protospacer was cut using Cas9 and thus increasing the HR efficiency.16 The isolated candidate clone with the most correction events confirmed by sequencing was subjected to the next round of correction. After 24 rounds, all errors, including two new mutations, were corrected resulting in a synV with a sequence as designed and exhibiting wild-type fitness.16,30
Discussion and outlook
Advances in DNA assembly have enabled the synthesis of both prokaryotic and eukaryotic genomes. The “bottom-up” genome synthesis opens the door to many imaginative changes at the genomic scale. These include creating a functional genome of minimal size,11 data storage,34 recoding at the genomic scale,8,18,38–42 and directed genome evolution.10,35–38 However, design flaws and spontaneous variants often cause malfunctions of the synthetic chromosome, which then require bug finding and removal.39 We have summarized strategies that have been applied to the bug mapping and debugging process, which typically involve genotypically and phenotypically massive screening and are labor-intensive and time-consuming. Future strategies incorporating automation and high-throughput screenings are highly sought after. The endeavors by the Global Biofoundry Alliance (GBA) have made great advances in automation, which has been used widely for different purposes including genome assembly,40 directed enzyme evolution, and liquid-based high-throughput analysis.41–46 Furthermore, we expect that robotic systems and workflows will be automatically executed in the future via artificial intelligence and machine learning to facilitate the bug mapping and debugging process.47
Author contributions
Z. X. X. and Y. J. Y. conceived and designed the manuscript. Z. X. X., J. Z., J. F., and Y. J. Y. drafted the manuscript.
Conflicts of interest
There are no conflicts to declare.
Acknowledgments
This work was funded by the National Key Research and Development Program of China (2020YFC1316500), the National Natural Science Foundation of China (21907074) and the Natural Science Foundation of Tianjin (20JCQNJC02090).
References
- Rugnetta M., Synthetic biology, Encyclopedia Britannica, 2016, accessed 11 March 2021, https://www.britannica.com/science/synthetic-biology [Google Scholar]
- Watson J. D. Crick F. H. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Lander E. S. Linton L. M. Birren B. Nusbaum C. Zody M. C. Baldwin J. Devon K. Dewar K. Doyle M. FitzHugh W. Funke R. Gage D. Harris K. Heaford A. Howland J. Kann L. Lehoczky J. LeVine R. McEwan P. McKernan K. Meldrim J. Mesirov J. P. Miranda C. Morris W. Naylor J. Raymond C. Rosetti M. Santos R. Sheridan A. Sougnez C. Stange-Thomann Y. Stojanovic N. Subramanian A. Wyman D. Rogers J. Sulston J. Ainscough R. Beck S. Bentley D. Burton J. Clee C. Carter N. Coulson A. Deadman R. Deloukas P. Dunham A. Dunham I. Durbin R. French L. Grafham D. Gregory S. Hubbard T. Humphray S. Hunt A. Jones M. Lloyd C. McMurray A. Matthews L. Mercer S. Milne S. Mullikin J. C. Mungall A. Plumb R. Ross M. Shownkeen R. Sims S. Waterston R. H. Wilson R. K. Hillier L. W. McPherson J. D. Marra M. A. Mardis E. R. Fulton L. A. Chinwalla A. T. Pepin K. H. Gish W. R. Chissoe S. L. Wendl M. C. Delehaunty K. D. Miner T. L. Delehaunty A. Kramer J. B. Cook L. L. Fulton R. S. Johnson D. L. Minx P. J. Clifton S. W. Hawkins T. Branscomb E. Predki P. Richardson P. Wenning S. Slezak T. Doggett N. Cheng J. F. Olsen A. Lucas S. Elkin C. Uberbacher E. Frazier M. Gibbs R. A. Muzny D. M. Scherer S. E. Bouck J. B. Sodergren E. J. Worley K. C. Rives C. M. Gorrell J. H. Metzker M. L. Naylor S. L. Kucherlapati R. S. Nelson D. L. Weinstock G. M. Sakaki Y. Fujiyama A. Hattori M. Yada T. Toyoda A. Itoh T. Kawagoe C. Watanabe H. Totoki Y. Taylor T. Weissenbach J. Heilig R. Saurin W. Artiguenave F. Brottier P. Bruls T. Pelletier E. Robert C. Wincker P. Smith D. R. Doucette-Stamm L. Rubenfield M. Weinstock K. Lee H. M. Dubois J. Rosenthal A. Platzer M. Nyakatura G. Taudien S. Rump A. Yang H. Yu J. Wang J. Huang G. Gu J. Hood L. Rowen L. Madan A. Qin S. Davis R. W. Federspiel N. A. Abola A. P. Proctor M. J. Myers R. M. Schmutz J. Dickson M. Grimwood J. Cox D. R. Olson M. V. Kaul R. Raymond C. Shimizu N. Kawasaki K. Minoshima S. Evans G. A. Athanasiou M. Schultz R. Roe B. A. Chen F. Pan H. Ramser J. Lehrach H. Reinhardt R. McCombie W. R. de la Bastide M. Dedhia N. Blocker H. Hornischer K. Nordsiek G. Agarwala R. Aravind L. Bailey J. A. Bateman A. Batzoglou S. Birney E. Bork P. Brown D. G. Burge C. B. Cerutti L. Chen H. C. Church D. Clamp M. Copley R. R. Doerks T. Eddy S. R. Eichler E. E. Furey T. S. Galagan J. Gilbert J. G. Harmon C. Hayashizaki Y. Haussler D. Hermjakob H. Hokamp K. Jang W. Johnson L. S. Jones T. A. Kasif S. Kaspryzk A. Kennedy S. Kent W. J. Kitts P. Koonin E. V. Korf I. Kulp D. Lancet D. Lowe T. M. McLysaght A. Mikkelsen T. Moran J. V. Mulder N. Pollara V. J. Ponting C. P. Schuler G. Schultz J. Slater G. Smit A. F. Stupka E. Szustakowki J. Thierry-Mieg D. Thierry-Mieg J. Wagner L. Wallis J. Wheeler R. Williams A. Wolf Y. I. Wolfe K. H. Yang S. P. Yeh R. F. Collins F. Guyer M. S. Peterson J. Felsenfeld A. Wetterstrand K. A. Patrinos A. Morgan M. J. de Jong P. Catanese J. J. Osoegawa K. Shizuya H. Choi S. Chen Y. J. Szustakowki J. C. International Human Genome Sequencing Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Venter J. C. Adams M. D. Myers E. W. Li P. W. Mural R. J. Sutton G. G. Smith H. O. Yandell M. Evans C. A. Holt R. A. Gocayne J. D. Amanatides P. Ballew R. M. Huson D. H. Wortman J. R. Zhang Q. Kodira C. D. Zheng X. H. Chen L. Skupski M. Subramanian G. Thomas P. D. Zhang J. Gabor Miklos G. L. Nelson C. Broder S. Clark A. G. Nadeau J. McKusick V. A. Zinder N. Levine A. J. Roberts R. J. Simon M. Slayman C. Hunkapiller M. Bolanos R. Delcher A. Dew I. Fasulo D. Flanigan M. Florea L. Halpern A. Hannenhalli S. Kravitz S. Levy S. Mobarry C. Reinert K. Remington K. Abu-Threideh J. Beasley E. Biddick K. Bonazzi V. Brandon R. Cargill M. Chandramouliswaran I. Charlab R. Chaturvedi K. Deng Z. Di Francesco V. Dunn P. Eilbeck K. Evangelista C. Gabrielian A. E. Gan W. Ge W. Gong F. Gu Z. Guan P. Heiman T. J. Higgins M. E. Ji R. R. Ke Z. Ketchum K. A. Lai Z. Lei Y. Li Z. Li J. Liang Y. Lin X. Lu F. Merkulov G. V. Milshina N. Moore H. M. Naik A. K. Narayan V. A. Neelam B. Nusskern D. Rusch D. B. Salzberg S. Shao W. Shue B. Sun J. Wang Z. Wang A. Wang X. Wang J. Wei M. Wides R. Xiao C. Yan C. Yao A. Ye J. Zhan M. Zhang W. Zhang H. Zhao Q. Zheng L. Zhong F. Zhong W. Zhu S. Zhao S. Gilbert D. Baumhueter S. Spier G. Carter C. Cravchik A. Woodage T. Ali F. An H. Awe A. Baldwin D. Baden H. Barnstead M. Barrow I. Beeson K. Busam D. Carver A. Center A. Cheng M. L. Curry L. Danaher S. Davenport L. Desilets R. Dietz S. Dodson K. Doup L. Ferriera S. Garg N. Gluecksmann A. Hart B. Haynes J. Haynes C. Heiner C. Hladun S. Hostin D. Houck J. Howland T. Ibegwam C. Johnson J. Kalush F. Kline L. Koduru S. Love A. Mann F. May D. McCawley S. McIntosh T. McMullen I. Moy M. Moy L. Murphy B. Nelson K. Pfannkoch C. Pratts E. Puri V. Qureshi H. Reardon M. Rodriguez R. Rogers Y. H. Romblad D. Ruhfel B. Scott R. Sitter C. Smallwood M. Stewart E. Strong R. Suh E. Thomas R. Tint N. N. Tse S. Vech C. Wang G. Wetter J. Williams S. Williams M. Windsor S. Winn-Deen E. Wolfe K. Zaveri J. Zaveri K. Abril J. F. Guigo R. Campbell M. J. Sjolander K. V. Karlak B. Kejariwal A. Mi H. Lazareva B. Hatton T. Narechania A. Diemer K. Muruganujan A. Guo N. Sato S. Bafna V. Istrail S. Lippert R. Schwartz R. Walenz B. Yooseph S. Allen D. Basu A. Baxendale J. Blick L. Caminha M. Carnes-Stine J. Caulk P. Chiang Y. H. Coyne M. Dahlke C. Mays A. Dombroski M. Donnelly M. Ely D. Esparham S. Fosler C. Gire H. Glanowski S. Glasser K. Glodek A. Gorokhov M. Graham K. Gropman B. Harris M. Heil J. Henderson S. Hoover J. Jennings D. Jordan C. Jordan J. Kasha J. Kagan L. Kraft C. Levitsky A. Lewis M. Liu X. Lopez J. Ma D. Majoros W. McDaniel J. Murphy S. Newman M. Nguyen T. Nguyen N. Nodell M. Pan S. Peck J. Peterson M. Rowe W. Sanders R. Scott J. Simpson M. Smith T. Sprague A. Stockwell T. Turner R. Venter E. Wang M. Wen M. Wu D. Wu M. Xia A. Zandieh A. Zhu X. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Gibson D. G. Benders G. A. Andrews-Pfannkoch C. Denisova E. A. Baden-Tillson H. Zaveri J. Stockwell T. B. Brownley A. Thomas D. W. Algire M. A. Merryman C. Young L. Noskov V. N. Glass J. I. Venter J. C. Hutchison III C. A. Smith H. O. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- Itaya M. Fujita K. Kuroki A. Tsuge K. Nat. Methods. 2008;5:41–43. doi: 10.1038/nmeth1143. [DOI] [PubMed] [Google Scholar]
- Gibson D. G. Glass J. I. Lartigue C. Noskov V. N. Chuang R. Y. Algire M. A. Benders G. A. Montague M. G. Ma L. Moodie M. M. Merryman C. Vashee S. Krishnakumar R. Assad-Garcia N. Andrews-Pfannkoch C. Denisova E. A. Young L. Qi Z. Q. Segall-Shapiro T. H. Calvey C. H. Parmar P. P. Hutchison III C. A. Smith H. O. Venter J. C. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- Gibson D. G. Smith H. O. Hutchison III C. A. Venter J. C. Merryman C. Nat. Methods. 2010;7:901–903. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- Dymond J. S. Richardson S. M. Coombes C. E. Babatz T. Muller H. Annaluru N. Blake W. J. Schwerzmann J. W. Dai J. Lindstrom D. L. Boeke A. C. Gottschling D. E. Chandrasegaran S. Bader J. S. Boeke J. D. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annaluru N. Muller H. Mitchell L. A. Ramalingam S. Stracquadanio G. Richardson S. M. Dymond J. S. Kuang Z. Scheifele L. Z. Cooper E. M. Cai Y. Zeller K. Agmon N. Han J. S. Hadjithomas M. Tullman J. Caravelli K. Cirelli K. Guo Z. London V. Yeluru A. Murugan S. Kandavelou K. Agier N. Fischer G. Yang K. Martin J. A. Bilgel M. Bohutski P. Boulier K. M. Capaldo B. J. Chang J. Charoen K. Choi W. J. Deng P. DiCarlo J. E. Doong J. Dunn J. Feinberg J. I. Fernandez C. Floria C. E. Gladowski D. Hadidi P. Ishizuka I. Jabbari J. Lau C. Y. Lee P. A. Li S. Lin D. Linder M. E. Ling J. Liu J. Liu J. London M. Ma H. Mao J. McDade J. E. McMillan A. Moore A. M. Oh W. C. Ouyang Y. Patel R. Paul M. Paulsen L. C. Qiu J. Rhee A. Rubashkin M. G. Soh I. Y. Sotuyo N. E. Srinivas V. Suarez A. Wong A. Wong R. Xie W. R. Xu Y. Yu A. T. Koszul R. Bader J. S. Boeke J. D. Chandrasegaran S. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison III C. A. Chuang R. Y. Noskov V. N. Assad-Garcia N. Deerinck T. J. Ellisman M. H. Gill J. Kannan K. Karas B. J. Ma L. Pelletier J. F. Qi Z. Q. Richter R. A. Strychalski E. A. Sun L. Suzuki Y. Tsvetanova B. Wise K. S. Smith H. O. Glass J. I. Merryman C. Gibson D. G. Venter J. C. Science. 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- Ostrov N. Landon M. Guell M. Kuznetsov G. Teramoto J. Cervantes N. Zhou M. Singh K. Napolitano M. G. Moosburner M. Shrock E. Pruitt B. W. Conway N. Goodman D. B. Gardner C. L. Tyree G. Gonzales A. Wanner B. L. Norville J. E. Lajoie M. J. Church G. M. Science. 2016;353:819–822. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- Mitchell L. A. Wang A. Stracquadanio G. Kuang Z. Wang X. Yang K. Richardson S. Martin J. A. Zhao Y. Walker R. Luo Y. Dai H. Dong K. Tang Z. Yang Y. Cai Y. Heguy A. Ueberheide B. Fenyo D. Dai J. Bader J. S. Boeke J. D. Science. 2017;355:eaaf4831. doi: 10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- Shen Y. Wang Y. Chen T. Gao F. Gong J. Abramczyk D. Walker R. Zhao H. Chen S. Liu W. Luo Y. Muller C. A. Paul-Dubois-Taine A. Alver B. Stracquadanio G. Mitchell L. A. Luo Z. Fan Y. Zhou B. Wen B. Tan F. Wang Y. Zi J. Xie Z. Li B. Yang K. Richardson S. M. Jiang H. French C. E. Nieduszynski C. A. Koszul R. Marston A. L. Yuan Y. Wang J. Bader J. S. Dai J. Boeke J. D. Xu X. Cai Y. Yang H. Science. 2017;355:eaaf4791. doi: 10.1126/science.aaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. Li B. Z. Zhao M. Mitchell L. A. Xie Z. X. Lin Q. H. Wang X. Xiao W. H. Wang Y. Zhou X. Liu H. Li X. Ding M. Z. Liu D. Zhang L. Liu B. L. Wu X. L. Li F. F. Dong X. T. Jia B. Zhang W. Z. Jiang G. Z. Liu Y. Bai X. Song T. Q. Chen Y. Zhou S. J. Zhu R. Y. Gao F. Kuang Z. Wang X. Shen M. Yang K. Stracquadanio G. Richardson S. M. Lin Y. Wang L. Walker R. Luo Y. Ma P. S. Yang H. Cai Y. Dai J. Bader J. S. Boeke J. D. Yuan Y. J. Science. 2017;355:eaaf4706. [Google Scholar]
- Xie Z. X. Li B. Z. Mitchell L. A. Wu Y. Qi X. Jin Z. Jia B. Wang X. Zeng B. X. Liu H. M. Wu X. L. Feng Q. Zhang W. Z. Liu W. Ding M. Z. Li X. Zhao G. R. Qiao J. J. Cheng J. S. Zhao M. Kuang Z. Wang X. Martin J. A. Stracquadanio G. Yang K. Bai X. Zhao J. Hu M. L. Lin Q. H. Zhang W. Q. Shen M. H. Chen S. Su W. Wang E. X. Guo R. Zhai F. Guo X. J. Du H. X. Zhu J. Q. Song T. Q. Dai J. J. Li F. F. Jiang G. Z. Han S. L. Liu S. Y. Yu Z. C. Yang X. N. Chen K. Hu C. Li D. S. Jia N. Liu Y. Wang L. T. Wang S. Wei X. T. Fu M. Q. Qu L. M. Xin S. Y. Liu T. Tian K. R. Li X. N. Zhang J. H. Song L. X. Liu J. G. Lv J. F. Xu H. Tao R. Wang Y. Zhang T. T. Deng Y. X. Wang Y. R. Li T. Ye G. X. Xu X. R. Xia Z. B. Zhang W. Yang S. L. Liu Y. L. Ding W. Q. Liu Z. N. Zhu J. Q. Liu N. Z. Walker R. Luo Y. Wang Y. Shen Y. Yang H. Cai Y. Ma P. S. Zhang C. T. Bader J. S. Boeke J. D. Yuan Y. J. Science. 2017;355:eaaf4704. [Google Scholar]
- Zhang W. Zhao G. Luo Z. Lin Y. Wang L. Guo Y. Wang A. Jiang S. Jiang Q. Gong J. Wang Y. Hou S. Huang J. Li T. Qin Y. Dong J. Qin Q. Zhang J. Zou X. He X. Zhao L. Xiao Y. Xu M. Cheng E. Huang N. Zhou T. Shen Y. Walker R. Luo Y. Kuang Z. Mitchell L. A. Yang K. Richardson S. M. Wu Y. Li B. Z. Yuan Y. J. Yang H. Lin J. Chen G. Q. Wu Q. Bader J. S. Cai Y. Boeke J. D. Dai J. Science. 2017;355:eaaf3981. doi: 10.1126/science.aaf3981. [DOI] [PubMed] [Google Scholar]
- Fredens J. Wang K. de la Torre D. Funke L. F. H. Robertson W. E. Christova Y. Chia T. Schmied W. H. Dunkelmann D. L. Beranek V. Uttamapinant C. Llamazares A. G. Elliott T. S. Chin J. W. Nature. 2019;569:514–518. doi: 10.1038/s41586-019-1192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya M. Tsuge K. Koizumi M. Fujita K. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15971–15976. doi: 10.1073/pnas.0503868102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S. M. Mitchell L. A. Stracquadanio G. Yang K. Dymond J. S. DiCarlo J. E. Lee D. Huang C. L. Chandrasegaran S. Cai Y. Boeke J. D. Bader J. S. Science. 2017;355:1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- Kannan K. Gibson D. G. Science. 2017;355:1024–1025. doi: 10.1126/science.aam9739. [DOI] [PubMed] [Google Scholar]
- Barnett R. W. Erfle H. Nucleic Acids Res. 1990;18:3094. doi: 10.1093/nar/18.10.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli R. B. Gunyuzlu P. Huang J. Scott C. Oakes F. T. Nucleic Acids Res. 1991;19:6007–6013. doi: 10.1093/nar/19.21.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymond J. S. Scheifele L. Z. Richardson S. Lee P. Chandrasegaran S. Bader J. S. Boeke J. D. Genetics. 2009;181:13–21. doi: 10.1534/genetics.108.096784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G. Young L. Chuang R. Y. Venter J. C. Hutchison III C. A. Smith H. O. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Lin Q. Jia B. Mitchell L. A. Luo J. Yang K. Zeller K. I. Zhang W. Xu Z. Stracquadanio G. Bader J. S. Boeke J. D. Yuan Y. J. ACS Synth. Biol. 2015;4:213–220. doi: 10.1021/sb500241e. [DOI] [PubMed] [Google Scholar]
- Gibson D. G. Glass J. I. Lartigue C. Noskov V. N. Chuang R. Y. Algire M. A. Benders G. A. Montague M. G. Ma L. Moodie M. M. Merryman C. Vashee S. Krishnakumar R. Assad-Garcia N. Andrews-Pfannkoch C. Denisova E. A. Young L. Qi Z. Q. Segall-Shapiro T. H. Calvey C. H. Parmar P. P. Hutchison III C. A. Smith H. O. Venter J. C. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- Logsdon G. A. Gambogi C. W. Liskovykh M. A. Barrey E. J. Larionov V. Miga K. H. Heun P. Black B. E. Cell. 2019;178:624–639. doi: 10.1016/j.cell.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. Nature. 2011;473:403–408. doi: 10.1038/473403a. [DOI] [PubMed] [Google Scholar]
- Xie Z. X. Liu D. Li B. Z. Zhao M. Zeng B. X. Wu Y. Shen Y. Lin T. Yang P. Dai J. Cai Y. Yang H. Yuan Y. J. Chem. Soc. Rev. 2017;46:7191–7207. doi: 10.1039/c7cs00208d. [DOI] [PubMed] [Google Scholar]
- Xie Z. X. Mitchell L. A. Liu H. M. Li B. Z. Liu D. Agmon N. Wu Y. Li X. Zhou X. Li B. Xiao W. H. Ding M. Z. Wang Y. Yuan Y. J. Boeke J. D. G3: Genes, Genomes, Genet. 2018;8:173–183. [Google Scholar]
- DiCarlo J. E. Norville J. E. Mali P. Rios X. Aach J. Church G. M. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L. Ran F. A. Cox D. Lin S. Barretto R. Habib N. Hsu P. D. Wu X. Jiang W. Marraffini L. A. Zhang F. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. Han M. Zhou J. Ge Q. Wang P. Zhang X. Zhu S. Song L. Yuan Y. Natl. Sci. Rev. 2021 doi: 10.1093/nsr/nwab028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. Luo Z. Wang Y. Pham N. T. Tuck L. Perez-Pi I. Liu L. Shen Y. French C. Auer M. Marles-Wright J. Dai J. Cai Y. Nat. Commun. 2018;9:1936. doi: 10.1038/s41467-018-04254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B. Wu Y. Li B. Z. Mitchell L. A. Liu H. Pan S. Wang J. Zhang H. R. Jia N. Li B. Shen M. Xie Z. X. Liu D. Cao Y. X. Li X. Zhou X. Qi H. Boeke J. D. Yuan Y. J. Nat. Commun. 2018;9:1933. doi: 10.1038/s41467-018-03084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Jia B. Xie Z. X. Li Y. X. Yuan Y. J. Front. Chem. Sci. Eng. 2018;12:806–814. [Google Scholar]
- Wang J. Xie Z. X. Ma Y. Chen X. R. Huang Y. Q. He B. Jia B. Li B. Z. Yuan Y. J. Nat. Commun. 2018;9:3783. doi: 10.1038/s41467-018-06216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Xie Z. X. Yuan Y. J. FEMS Yeast Res. 2020;20:foaa012. doi: 10.1093/femsyr/foaa012. [DOI] [PubMed] [Google Scholar]
- Hillson N. Caddick M. Cai Y. Carrasco J. A. Chang M. W. Curach N. C. Bell D. J. Le Feuvre R. Friedman D. C. Fu X. Gold N. D. Herrgard M. J. Holowko M. B. Johnson J. R. Johnson R. A. Keasling J. D. Kitney R. I. Kondo A. Liu C. Martin V. J. J. Menolascina F. Ogino C. Patron N. J. Pavan M. Poh C. L. Pretorius I. S. Rosser S. J. Scrutton N. S. Storch M. Tekotte H. Travnik E. Vickers C. E. Yew W. S. Yuan Y. Zhao H. Freemont P. S. Nat. Commun. 2019;10:2040. doi: 10.1038/s41467-019-10079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao R. Liang J. Tasan I. Si T. Ju L. Y. Zhao H. M. ACS Synth. Biol. 2017;6:678–685. doi: 10.1021/acssynbio.6b00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si T. Chao R. Min Y. Wu Y. Ren W. Zhao H. Nat. Commun. 2017;8:15187. doi: 10.1038/ncomms15187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F. Chung M. T. Yao Y. Nidetz R. Lee L. M. Liu A. P. Feng Y. Kurabayashi K. Yang G. Y. Nat. Commun. 2018;9:1030. doi: 10.1038/s41467-018-03492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers G. O. F. Chee S. M. Bell D. Suckling L. Kern M. Tew D. McClymont D. W. Ellis T. Nat. Commun. 2020;11:868. doi: 10.1038/s41467-020-14708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abate A. R. Hung T. Sperling R. A. Mary P. Rotem A. Agresti J. J. Weiner M. A. Weitz D. A. Lab Chip. 2013;13:4864–4869. doi: 10.1039/c3lc50905b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H. S. Je K. Min J. W. Park D. Han K. Y. Shin S. H. Park W. Y. Yoo C. E. Kim S. H. Lab Chip. 2018;18:775–784. doi: 10.1039/c7lc01284e. [DOI] [PubMed] [Google Scholar]
- Mehr S. H. M. Craven M. Leonov A. I. Keenan G. Cronin L. Science. 2020;370:101–108. doi: 10.1126/science.abc2986. [DOI] [PubMed] [Google Scholar]
- Noskov V. N. Segall-Shapiro T. H. Chuang R. Y. Nucleic Acids Res. 2010;38:2570–2576. doi: 10.1093/nar/gkq099. [DOI] [PMC free article] [PubMed] [Google Scholar]