Abstract
A highly efficient and versatile method for construction of peptide macrocycles via palladium-catalyzed intramolecular S-arylation of alkyl and aryl thiols with aryl iodides under mild conditions is developed. The method exhibits a broad substrate scope for thiols, aryl iodides and amino acid units. Peptide macrocycles of a wide range of size and composition can be readily assembled in high yield from various easily accessible building blocks. This method has been successfully employed to prepare an 8-million-membered tetrameric cyclic peptide DNA-encoded library (DEL). Preliminary screening of the DEL library against protein p300 identified compounds with single digit micromolar inhibition activity.
A highly efficient and versatile method for construction of peptide macrocycles via palladium-catalyzed intramolecular S-arylation of alkyl and aryl thiols with aryl iodides under mild conditions is developed.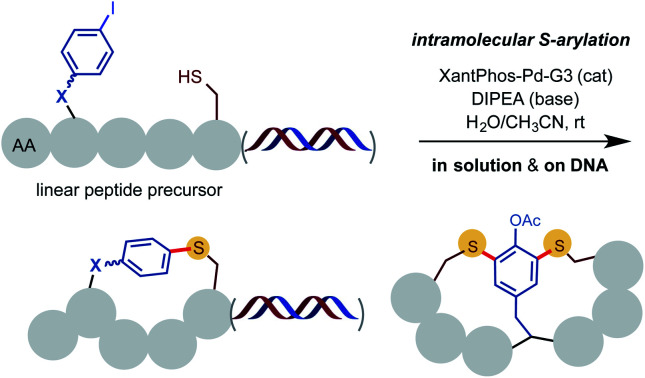
Introduction
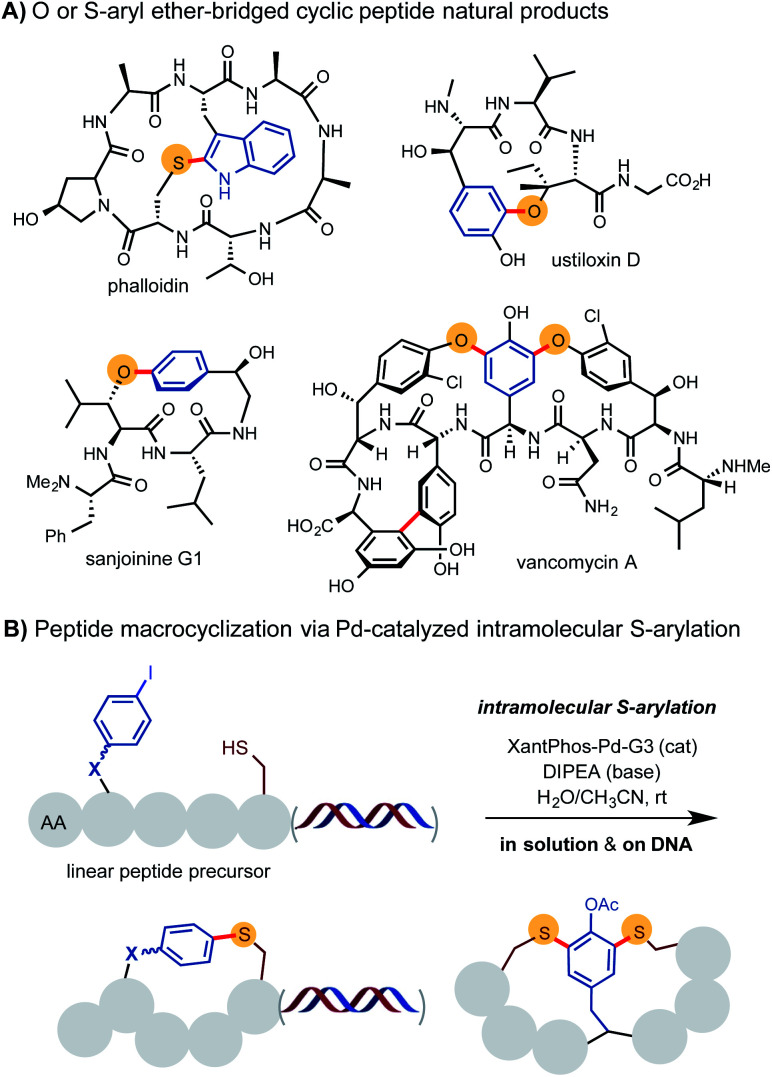
Peptide macrocycles have emerged as a powerful platform for drug discovery, especially for biological targets that are difficult to modulate by conventional small molecule drugs.1 To realize the full potential of this platform, new strategies are needed to streamline the synthesis of large libraries of peptide macrocycles with novel structures. Inspired by cyclophane-type peptide natural products such as vancomycin and celogentins,2 we have become interested in utilizing the aromatic side chains of amino acid building blocks as braces to construct cyclic peptides of rigid and constrained structures.3 The incorporation of these braces could potentially override the inherent conformational control of peptides by weak noncovalent interactions and give rise to unusual physiochemical properties.4 As shown by sanjoinine, ustiloxin, vancomycins and phalloidin, nature has frequently used aryl ether and even S-aryl ether motifs to crosslink the aromatic side chains to produce cyclic peptide natural products with interesting structures and biological activities (Scheme 1A).5,6 A range of intramolecular O-arylation methods including SNAr substitution and metal-catalyzed C–O coupling reactions have been successfully employed to facilitate the total synthesis of these complex natural products and prepare mimetic macrocyclic compounds.7 However, those methods typically require forcing reaction conditions and exhibit poor tolerance toward other nucleophilic functional groups such as amines and thiols. The associated protection/deprotection operations also can significantly complicate the synthetic route and diminish overall synthetic efficiency. In parallel with the development of O-arylation reactions, the chemistry of S-arylation of thiols has also been greatly advanced in the past few years.8–16 Notably, Pentelute and Buchwald reported the use of stoichiometric amount of aryl-palladium(ii) complexes carrying a RuPhos ligand for a highly efficient and chemoselective S-arylation of cysteine residue of unprotected peptides and proteins at room temperature (rt).10 Alami and Messaoudi reported a more economical and practical protocol for S-arylation of glycosyl thiols and cysteine in unprotected peptides and proteins with various aryl halides using Xantphos-Pd-G3 catalyst under biocompatible conditions.11b,c Use of this Pd-catalyzed S-arylation reaction in an intramolecular fashion could potentially provide a useful method for rapid synthesis of S-aryl ether bridged macrocyclic peptides.
Scheme 1. Construction of S-aryl ether-linked peptide macrocycles.
Over the past two decades, DNA-encoded library (DEL) technologies for hit identification have gained substantial influence in drug discovery in both the pharmaceutical industry and academia.17 DEL combines the amplifiable DNA coding techniques with split-and-pool organic synthesis to allow the rapid and cost-effective synthesis and affinity-selection of ultra-large combinatorial libraries on miniaturized scales. However, the use of DNA-encoded synthesis platform also comes with a severe constraint that the libraries must be assembled in a DNA-compatible fashion.18 Despite the rapid expansion of toolbox for on-DNA reactions, the ability to generate large DEL library of high three-dimensional structural complexity and diversity is still very limited.19,20 Herein, we report the application of the Pd-catalyzed intramolecular S-arylation of thiols with aryl iodides for facile construction of S-aryl ether-bridged peptide macrocycles from unprotected linear peptide precursor under mild conditions.21 This method provides streamlined access to structural mimics of O-aryl ether-bridged cyclic peptide natural products and other related macrocyclic peptides of various ring size and composition. This chemistry has been successfully employed to synthesize 8-million-membered DNA-encoded library of tetrameric cyclic peptides. Screening the library against protein p300 revealed ligands with micromolar inhibition activity.
Results and discussion
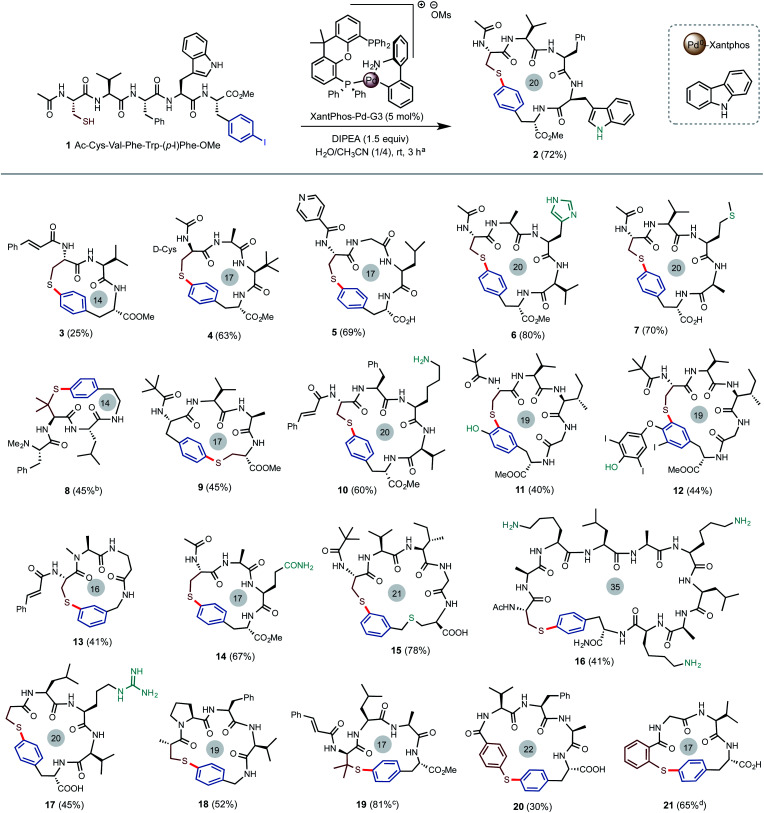
As shown in Scheme 2, our investigation commenced with the Pd-catalyzed intramolecular S-arylation of pentameric linear peptide 1 carrying a free cysteine and a para-iodinate phenylalanine (pI-Phe). To our delight, cyclization of 1 proceeded smoothly to give the desired cyclic product 2 in 72% isolated yield and exclusive chemoselectivity under the conditions of 5 mol% of Xantphos-Pd-G3 catalyst and 1.5 equiv. of diisopropyl ethylamine (DIPEA) base in the mixed solvents of CH3CN and H2O (4/1) at a concentration of 12.5 mM at rt for 3 hours. Only trace amount (<3%) of dimerization product were observed. No impurities arising from N-arylation of the indole side chain of Trp, oxidation of SH to disulfide, or deiodination of aryl iodide were observed. Use of triethylamine (TEA) base gave slightly decreased yield. Use of inorganic base such as K2CO3 gave lower yield. The use of H2O as co-solvent is critical to achieve high yield. The use of mixed solvents of THF/H2O gave comparable results (see ESI† for results under other reaction conditions). Previous studies indicated aminobiphenyl palladacycle precatalyst can readily undergo C–N reductive elimination to generate a carbazole and a 12-electron Xantphos-Pd0 species, which catalyzes the C–S cross coupling of SH and aryl iodides through a Pd0/II cycle.10,11
Scheme 2. Construction of peptide macrocycles via Pd-catalyzed intramolecular arylation of thiol groups with aryl iodides. (a) Isolated yield on a 0.1 mmol scale. (b) 16 h. (c) 45 °C (vs. 10% at rt). (d) 6 h.
As outlined in Scheme 2, the Pd-catalyzed intramolecular S-arylation reaction exhibited a very broad substrate scope under the optimized conditions. Linear peptide precursors were prepared with commercially available building blocks in high efficiency using the standard procedures of solid phase peptide synthesis (SPPS). Some substrates needed an additional amide or ester coupling step to modify the C-terminus of peptides. 4-Methoxytrityl (MMT) protecting group was used for most thiol building blocks and was removed under acidic conditions (trifluoroacetic acid/triethylsilane/dichloromethane). (1) All proteinogenic AAs were tolerated. Nucleophilic groups such as primary amine (Lys in 10, 16), carboxylic acid (free C-terminus in 5), primary amide (Gln in 14), guanidine (Arg in 17), imidazole (His in 6), indole (Trp in 2), phenol (11), and thioether (Met in 7) were untouched. A variety of N-terminal acyl capping units such as pivalic acid (Piv, 9), cinnamic acid (3) and isonicotinic acid (5) can be used. (2) Macrocycles of ring size between 17 (4-mer) and 20 (5-mer) can be generated in good to excellent yield. Trimeric cyclic peptides such as 3 and 8 of ring size 14 are formed in moderate yield. Notably, the thioether-linked para-cyclophane ring of 8 closely mimic the core structure of natural product sanjoinine. 16 with a 35-membered ring was obtained in 41% yield. As shown in 2 and 14, cyclization can proceed well without the assistance of turning unit such as Pro. (3) Beside iodinated Phe (4) and Tyr (11), aryl iodides with the iodo group on varied positions can be installed as a prosthetic group on the side chains (Cys of 15) or terminus (18) of peptides to facilitate macrocyclization. Product 12 was obtained as the main product via S-arylation of a thyroxine moiety. (4) Cysteine residue can be placed on various positions of the peptide (see 2vs.9). Besides Cys, other alkyl thiol units such as 3-mercaptopropionic acid (17), 3-mercaptoisobutyric acid (18), and penicillamine (8, 19) also work well. In comparison to alkyl thiols, arylthiols are slightly less reactive and form the corresponding diaryl thioethers in moderate to good yield (see 20, 21).
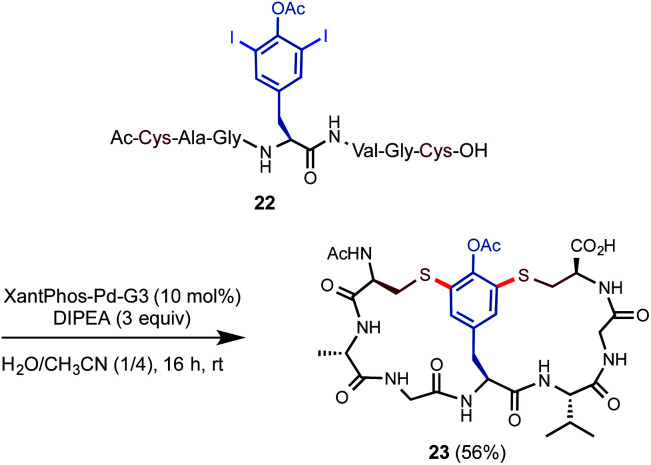
Inspired by the bis-diaryl ether scaffold of vancomycin, we went on to investigate whether double S-arylation of a center unit like 3,5-diiodotyrosine (DIT) could generate bicyclic structures in one operation (Scheme 3).22 DIT can be readily prepared from tyrosine and is commercially available at relatively low cost. A linear peptide precursor 22 containing one O–Ac–DIT and two free Cys units was prepared in high efficiency via standard SPPS. Subjecting 22 to slightly modified S-arylation conditions with 10 mol% of Pd catalyst and 3 equiv. of DIPEA in H2O/CH3CN (1/4) at rt gave the desired bicyclic product 23 in 56% isolated yield.
Scheme 3. Construction of bicyclic peptides via Pd-catalyzed double S-arylation. Isolated yield on a 0.1 mmol scale.
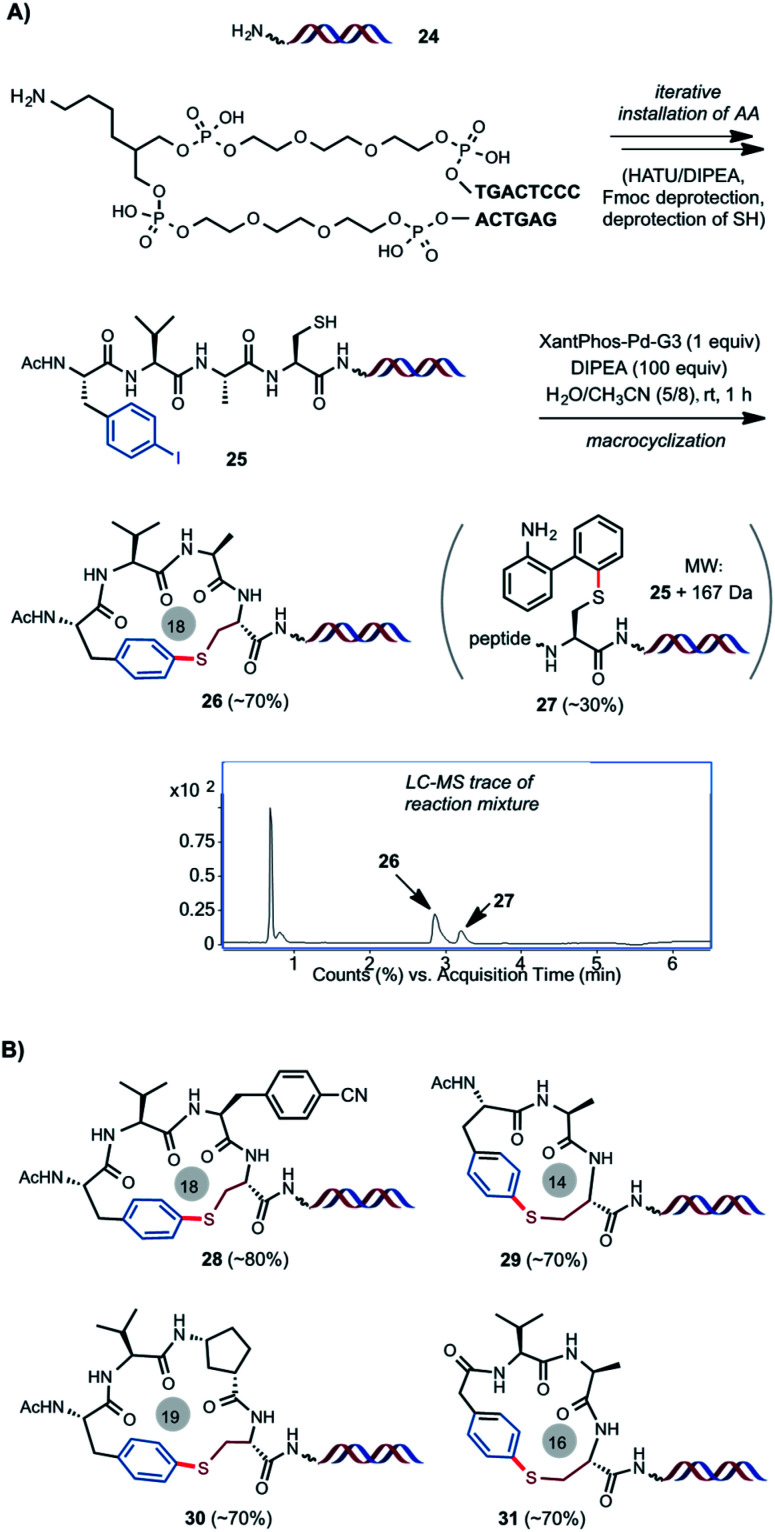
As outlined in Scheme 4A, we were pleased to find the Pd-catalyzed intramolecular S-arylation reaction can be readily adapted for the construction of peptide macrocycles on DNA. As a model experiment without attaching DNA coding tags, HATU-mediated iterative installation of four AA units and Fmoc deprotection on DNA template 24 gave DNA-linked tetrameric linear peptide 25 in good efficiency (see ESI† for details). Macrocyclization of 25 under the optimized conditions of 1 equiv. of Pd catalyst and 100 equiv. of DIPEA in the mixed solvents of CH3CN/H2O (8/5) at 0.38 mM concentration and rt for 1 h gave the desired cyclized product 26 in 70% conversion based on LC-MS estimation, along with 30% of byproduct 27. 27 has a molecular weight of 25 plus 167 Da, which matches with the intermolecular S-arylation by the aminobiphenyl ligand. Stoichiometric Pd catalyst is needed to achieve full conversion of starting material at rt. Use of larger excess of Pd catalyst can cause degradation of DNA. We suspect that binding of the SH group of 25 to the Pd(ii) center of Xantphos-Pd-G3 and subsequent S–C reductive elimination gave the intermolecular S-arylation byproduct. Little of similar impurities were observed in the off-DNA reactions using catalytic amount of Pd catalyst (Scheme 2). DNA-linked compounds 28–31 with varied ring size and composition were obtained in good efficiency (Scheme 4B). It is worth mentioning that the use of stoichiometric Pd catalyst is not an issue because of the tiny reaction scale for the eventual DEL library synthesis. Most of the reported methods involving metal-catalyzed reactions for DEL synthesis use large access of metal reagents (e.g. >5 equiv.).21
Scheme 4. (A) Construction of S-aryl ether-linked peptide macrocycle on uncoded DNA template. (B) Representative model synthesis on DNA. The side products sometimes overlap with the desired cyclized product on LC. Yields/conversion were based on the estimation of MS signals of the corresponding peaks on LC.
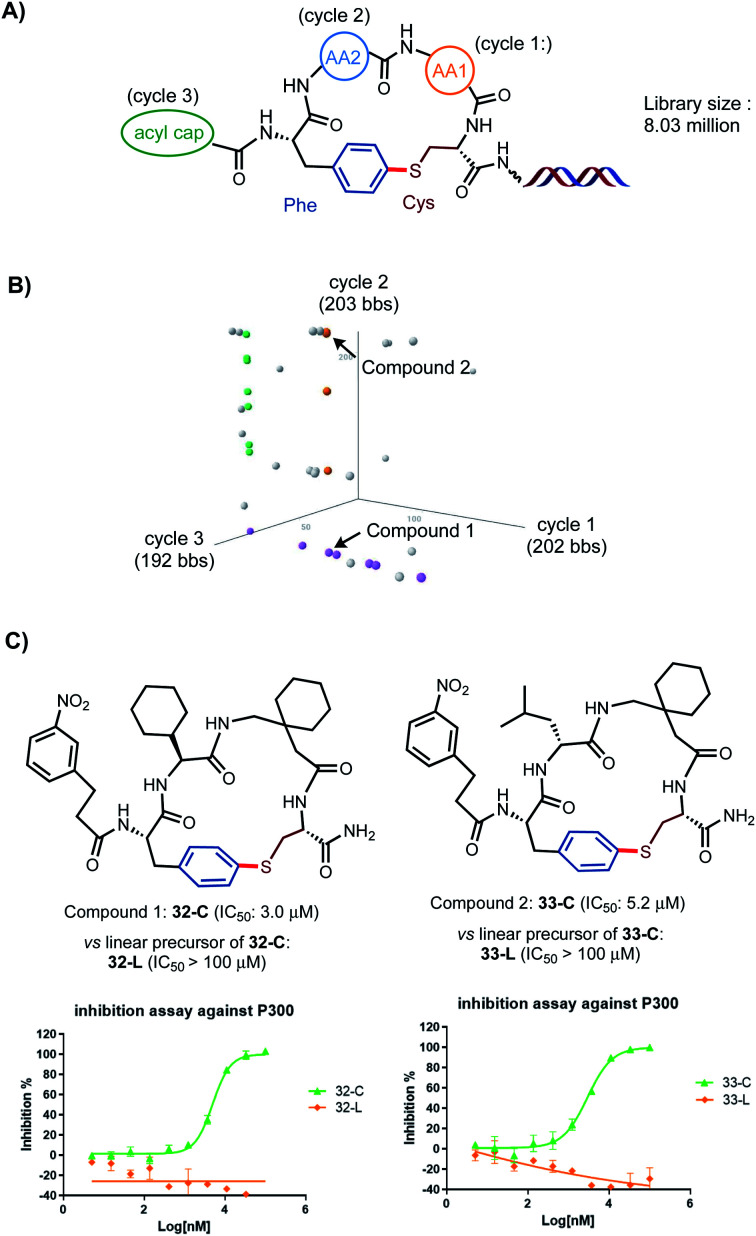
As outlined in Scheme 5A, an 8-million-membered DEL library of tetrameric peptide macrocycles consisting of two variable AA units (cycles 1, 2), one variable N-terminal acyl unit (cycle 3), and fixed Cys and pI-Phe were successfully constructed using the same procedures of peptide chain extension and macrocyclization optimized for the synthesis on the uncoded DNA template. 202 and 203 AA building blocks (bbs) and 192 acyl bbs were incorporated at cycles 1–3 respectively. Each cycle was encoded with 11 base pair of DNA via enzyme-mediated T4 ligation (see ESI† for details). The library was then screened against protein p300, which is a key transcriptional co-activator and has been implicated in a variety of pathological conditions including cancer.23 Protein binding assay was performed with a poly-His/bead (HisPur Ni-NTA Magnetic Beads) system. The binders were eluted, enriched, amplified by PCR, and sequenced using next generation high throughput sequencing. The affinity selection outcome was illustrated by a three-dimensional view of cubic scatter plot where each dot represented an encoded compound above the informatics-based cut-off filter (>500 enrichment value, Scheme 5B). Three lines of dot (orange, purple and green) were highlighted in the cube. Encoded compounds in the same line shared two common bbs. Compound 1 (32-C) at the cross point of orange and purple lines and compound 2 (33-C) randomly distributed on the orange line were selected for off-DNA resynthesis and validated by a radioactive acetyltransferase activity-based IC50 assay. To our delight, compounds 32-C and 33-C showed an IC50 of 3.0 and 5.2 μM respectively while the corresponding linear precursors 32-L and 33-L showed significantly lower inhibition activity (>100 μM), underscoring the importance of the conformational constrains in these S-aryl ether-braced cyclic scaffolds on their biological activities.
Scheme 5. Synthesis and affinity selection of an 8-million-membered library of tetrameric peptide macrocycles on DEL. (A) Design of library of tetrameric cyclic peptides with three cycles at AA1, AA2, acyl cap positions. (B) The selected library population is viewed in a cubic scatter plot generated by TIBCO Spotfire Analyst. In the cubic space, axis represents the bbs used in a given cycle of library synthesis and each dot represents an encoded compound above certain enrichment cut-off value (>500). Enrichment value is calculated by comparing the results from pre and post-selection counts. (C) Validation of two selected compounds 32-C and 33-C by off-DNA resynthesis and IC50 assays (see ESI† for details).
Conclusions
In summary, we have developed a highly efficient, flexible and robust strategy for construction of peptide macrocycles via palladium-catalyzed intramolecular S-arylation of alkyl and aryl thiols with aryl iodides under mild conditions. It enables rapid syntheses of a wide range of S-aryl ether-bridged peptide macrocycles using readily available building blocks in solution phase. It provides a powerful cross coupling tool to assemble complex peptide structures on DEL. The utility of this strategy for DEL has been successfully demonstrated by the identification of ligands of p300 of low micromolar inhibition activity from a proof-of-concept 8-million-membered library of tetrameric cyclic peptides. We expect this strategy can be readily expanded to prepare ultra-large peptide macrocycle libraries of different ring size, compositions and topological features for ligand identification for other important biological targets.
Author contributions
P. Y. carried out the reaction optimization and expanded the reaction scope. X. W. performed most of the DEL-related experiments. B. L. initiated the project and performed the preliminary method development. Y. Y., J. Y. and Y. S. contributed to DEL library synthesis and screening. H. T. helped with expanding the substrate scope. G. H. supervised part of the method development. X. L. supervised all the DEL-related work. G. C. oversaw the project and prepared most of the manuscript.
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
G. C. thanks NSFC-21672105, 21725204, 91753124, Natural Science Foundation of Tianjin 18JCZDJC32800, “111” project (B06005) of the Ministry of Education of China, and NCC2020FH02 for financial support of this work. G. C. dedicates this work to the 100th anniversary of Nankai University. X. L. thanks NSFC-21877117 and 91953203, National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program”, China (2018ZX09711002-005), and Shanghai Commission of Science and Technology (18431907100) for financial support of this work. X. W. thanks NSFC-for financial support of this work.
Electronic supplementary information (ESI) available: Detailed synthetic procedures, additional control experiments, compound characterization, LC-MS trace, NMR spectra and computational experiments. See DOI: 10.1039/d1sc00789k
Notes and references
- For selected reviews on peptide macrocycles: ; (a) Driggers E. M. Hale S. P. Lee J. Terrett N. K. Nat. Rev. Drug Discovery. 2008;7:608–624. doi: 10.1038/nrd2590. [DOI] [PubMed] [Google Scholar]; (b) Marsault E. Peterson M. L. J. Med. Chem. 2011;54:1961–2004. doi: 10.1021/jm1012374. [DOI] [PubMed] [Google Scholar]; (c) White C. J. Yudin A. K. Nat. Chem. 2011;3:509–524. doi: 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]; (d) Bockus A. T. McEwen C. M. Lokey R. S. Curr. Top. Med. Chem. 2013;13:821–836. doi: 10.2174/1568026611313070005. [DOI] [PubMed] [Google Scholar]; (e) Giordanetto F. Kihlberg J. J. Med. Chem. 2014;57:278–295. doi: 10.1021/jm400887j. [DOI] [PubMed] [Google Scholar]; (f) Hill T. A. Shepherd N. E. Diness F. Fairlie D. P. Angew. Chem., Int. Ed. 2014;53:2–24. doi: 10.1002/anie.201401058. [DOI] [PubMed] [Google Scholar]; (g) Lau Y. H. de Andrade P. Wu Y. Spring D. R. Chem. Soc. Rev. 2015;44:91–102. doi: 10.1039/c4cs00246f. [DOI] [PubMed] [Google Scholar]; (h) Vinogradov A. A. Yin Y. Suga H. J. Am. Chem. Soc. 2019;141:4167–4181. doi: 10.1021/jacs.8b13178. [DOI] [PubMed] [Google Scholar]; (i) Rivera D. G. Ojeda-Carralero G. M. Reguera L. Van der Eycken E. V. Chem. Soc. Rev. 2020;49:2039–2059. doi: 10.1039/c9cs00366e. [DOI] [PubMed] [Google Scholar]
- For selected reviews on cyclophane chemistry: ; (a) Cram D. J. Cram J. M. Acc. Chem. Res. 1970;4:204–213. [Google Scholar]; (b) Gulder T. Baran P. S. Nat. Prod. Rep. 2012;29:899–934. doi: 10.1039/c2np20034a. [DOI] [PubMed] [Google Scholar]
- (a) Feng Y. Chen G. Angew. Chem., Int. Ed. 2010;49:958–961. doi: 10.1002/anie.200905134. [DOI] [PubMed] [Google Scholar]; (b) He G. Zhang S.-Y. Nack W. A. Pearson R. Rabb-Lynch J. Chen G. Org. Lett. 2014;16:6488–6491. doi: 10.1021/ol503347d. [DOI] [PubMed] [Google Scholar]; (c) Zhang X. Lu G. Sun M. Mahankali M. Ma Y. Zhang M. Hua W. Hu Y. Wang Q. Chen J. He G. Qi X. Shen W. Liu P. Chen G. Nat. Chem. 2018;10:540–548. doi: 10.1038/s41557-018-0006-y. [DOI] [PubMed] [Google Scholar]; (d) Li B. Li X. Han B. Chen Z. Zhang X. He G. Chen G. J. Am. Chem. Soc. 2019;141:9401–9407. doi: 10.1021/jacs.9b04221. [DOI] [PubMed] [Google Scholar]
- For selected reports on cyclophane peptides: ; (a) Rogdan A. R. Jerome S. V. Houk K. N. James K. J. Am. Chem. Soc. 2010;13:2127–2138. doi: 10.1021/ja208503y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mendive-Tapia L. Preciado S. García J. Ramón R. Kielland N. Albericio F. Lavilla R. Nat. Commun. 2015;6:7160–7168. doi: 10.1038/ncomms8160. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Tang J. He Y. Chen H. Sheng W. Wang H. Chem. Sci. 2017;8:4565–4570. doi: 10.1039/c6sc05530c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Nguyen T. Q. N. Tooh Y. W. Sugiyama R. Nguyen T. P. D. Purushothaman M. Leow L. C. Hanif K. Yong R. H. S. Agatha I. Winnerdy F. R. Gugger M. Phan A. T. Morinaka B. I. Nat. Chem. 2020;12:1042–1053. doi: 10.1038/s41557-020-0519-z. [DOI] [PubMed] [Google Scholar]
- (a) Cao B. Park H. Joullie M. M. J. Am. Chem. Soc. 2002;124:520–521. doi: 10.1021/ja017277z. [DOI] [PubMed] [Google Scholar]; (b) Schuresko L. A. Lokey R. S. Angew. Chem., Int. Ed. 2007;46:3547–3549. doi: 10.1002/anie.200700017. [DOI] [PubMed] [Google Scholar]; (c) Temal-Laib T. Chastanet J. Zhu J. J. Am. Chem. Soc. 2002;124:583–590. doi: 10.1021/ja0170807. [DOI] [PubMed] [Google Scholar]
- For selected reviews on thioether motifs in natural products: ; (a) Dunbar K. L. Scharf D. H. Litomska A. Hertweck C. Chem. Rev. 2017;117:5521–5577. doi: 10.1021/acs.chemrev.6b00697. [DOI] [PubMed] [Google Scholar]; (b) Wang N. Saidhareddya P. Jiang X. Nat. Prod. Rep. 2020;37:246–275. doi: 10.1039/c8np00093j. [DOI] [PubMed] [Google Scholar]
- (a) Boger D. L. Zhou J. J. Am. Chem. Soc. 1993;115:11426–11433. [Google Scholar]; (b) Decicco C. P. Song Y. Evans D. A. Org. Lett. 2001;3:1029–1032. doi: 10.1021/ol015572i. [DOI] [PubMed] [Google Scholar]; (c) Cai Q. Zou B. Ma D. Angew. Chem., Int. Ed. 2006;45:1276–1279. doi: 10.1002/anie.200503538. [DOI] [PubMed] [Google Scholar]; (d) Lawson K. V. Rose T. E. Harran P. G. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E3753. doi: 10.1073/pnas.1311706110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Lee H. Boyer N. C. Deng Q. Kim H.-Y. Sawyer T. K. Sciammentta N. Chem. Sci. 2019;10:5073–5078. doi: 10.1039/c9sc00694j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For selected reviews on S-arylation: ; (a) Hartwig J. F. Acc. Chem. Res. 1998;31:852–860. [Google Scholar]; (b) Kondo T. Mitsudo T. A. Chem. Rev. 2000;100:3205–3220. doi: 10.1021/cr9902749. [DOI] [PubMed] [Google Scholar]; (c) Murata M. Buchwald S. L. Tetrahedron. 2004;60:7397–7403. [Google Scholar]; (d) Zhang C. Vinogradova E. V. Spokoyny A. M. Buchwald S. L. Pentelute B. L. Angew. Chem., Int. Ed. 2019;58:4810–4839. doi: 10.1002/anie.201806009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Migita T. Shimizu T. Asami Y. Shiobara J. Kato Y. Kosugi M. Bull. Chem. Soc. Jpn. 1980;53:1385–1389. [Google Scholar]; (b) Kosugi M. Ogata T. Terada M. Sano H. Migita T. Bull. Chem. Soc. Jpn. 1985;58:3657–3658. [Google Scholar]
- (a) Vinogradova E. V. Zhang C. Spokoyny A. M. Pentelute B. L. Buchwald S. L. Nature. 2015;526:687–691. doi: 10.1038/nature15739. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kubota K. Dai P. Pentelute B. L. Buchwald S. L. J. Am. Chem. Soc. 2018;140:3128–3133. doi: 10.1021/jacs.8b00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Brachet E. Brion J.-D. Messaoudi S. Alami M. Adv. Synth. Catal. 2013;355:477–490. [Google Scholar]; (b) Bruneau A. Roche M. Hamze A. Brion J.-D. Alami M. Messaoudi S. Chem.–Eur. J. 2015;21:8375–8379. doi: 10.1002/chem.201501050. [DOI] [PubMed] [Google Scholar]; (c) Shuaeeb R. A. A. Kolodych Al-S. Koniev O. Delacroix S. Erb S. Nicolay S. Cintrat J.-C. Brion J.-D. Cianforani S. Alami M. Wagner A. Messaoudi S. Chem.–Eur. J. 2016;22:11365–11370. doi: 10.1002/chem.201602277. [DOI] [PubMed] [Google Scholar]; (d) Probst N. Lartia R. Thory O. Alami M. Defrancq E. Messaoudi S. Chem.–Eur. J. 2018;24:1795–1800. doi: 10.1002/chem.201705371. [DOI] [PubMed] [Google Scholar]
- For selected reports on S-arylation under the catalysis of Ni, Cu, Fe and Co: ; (a) Rout L. Jammi S. Punniyamurthy T. Barua P. Saha P. Tetrahedron Lett. 2008;49:1484–1487. [Google Scholar]; (b) Oderinde M. S. Frenette M. Robbins D. W. Aquila B. Johannes J. W. J. Am. Chem. Soc. 2016;138:1760–1763. doi: 10.1021/jacs.5b11244. [DOI] [PubMed] [Google Scholar]; (c) Jones K. D. Power D. J. Bierer D. Gericke K. M. Stewart S. G. Org. Lett. 2018;20:208–211. doi: 10.1021/acs.orglett.7b03560. [DOI] [PubMed] [Google Scholar]; (d) Kwong F. Y. Buchwald S. L. Org. Lett. 2002;4:3517–3520. doi: 10.1021/ol0266673. [DOI] [PubMed] [Google Scholar]; (e) Uyeda C. Tan Y. Fu G. C. Peters J. C. J. Am. Chem. Soc. 2013;135:9548–9552. doi: 10.1021/ja404050f. [DOI] [PubMed] [Google Scholar]; (f) Rout L. Sen T. K. Punniyamurthy T. Angew. Chem., Int. Ed. 2007;46:5583–5586. doi: 10.1002/anie.200701282. [DOI] [PubMed] [Google Scholar]; (g) Correa A. Carril M. Bolm C. Angew. Chem., Int. Ed. 2008;47:2880–2883. doi: 10.1002/anie.200705668. [DOI] [PubMed] [Google Scholar]; (h) Wong Y.-C. Thatai T. Cheng C.-H. Org. Lett. 2006;8:5613–5616. doi: 10.1021/ol062344l. [DOI] [PubMed] [Google Scholar]
- For selected reports of metal-catalyzed S-arylation of peptides and proteins: ; (a) Forbes C. R. Zondlo N. J. Org. Lett. 2012;14:464–467. doi: 10.1021/ol202947f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Willwacher J. Raj R. Mohammed S. Davis B. G. J. Am. Chem. Soc. 2016;138:8678–8681. doi: 10.1021/jacs.6b04043. [DOI] [PubMed] [Google Scholar]; (c) Cheng W.-M. Lu X. Shi J. Liu L. Org. Chem. Front. 2018;5:3186–3193. [Google Scholar]; (d) Messina M. S. Stauber J. M. Waddington M. A. Rheingold A. L. Maynard H. D. Spokoyny A. M. J. Am. Chem. Soc. 2018;140:7065–7069. doi: 10.1021/jacs.8b04115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For selected reports on SNAr-mediated S-arylation for peptide macrocyclization: ; (a) Spokoyny A. M. Zou Y. Ling J. J. Yu H. Lin Y.-S. Pentelute B. L. J. Am. Chem. Soc. 2013;135:5946–5949. doi: 10.1021/ja400119t. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Brown S. P. Smith A. B. J. Am. Chem. Soc. 2015;137:4034–4037. doi: 10.1021/ja512880g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kalhor-Monfared S. M. Jafari R. Patterson J. T. Kitov P. I. Dwyer J. J. Nuss J. M. Derda R. Chem. Sci. 2016;7:3785–3790. doi: 10.1039/c5sc03856a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Liu W. Zheng Y. Kong X. Heinis C. Zhao Y. Wu C. Angew. Chem., Int. Ed. 2017;56:4458–4463. doi: 10.1002/anie.201610942. [DOI] [PubMed] [Google Scholar]; (e) Hanaya K. Ohata J. Miller M. K. Mangubat-Medina A. E. Swierczynski M. J. Yang D. C. Rosenthal R. M. Popp B. V. Ball Z. T. Chem. Commun. 2019;55:2841–2844. doi: 10.1039/c9cc00159j. [DOI] [PubMed] [Google Scholar]
- Rojas A. J. Zhang C. Vinogradova E. V. Buchwald N. Reilly J. Pentelute B. L. Buchwald S. L. Chem. Sci. 2017;8:4157–4263. doi: 10.1039/c6sc05454d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For selected reports on peptide macrocyclization mediated by S-alkylation: ; (a) Timmerman P. Beld J. Puijk W. C. Meloen R. H. ChemBioChem. 2005;6:821–824. doi: 10.1002/cbic.200400374. [DOI] [PubMed] [Google Scholar]; (b) Goto Y. Ohta A. Sako Y. Yamagishi Y. Murakami H. Suga H. ACS Chem. Biol. 2008;3:120–129. doi: 10.1021/cb700233t. [DOI] [PubMed] [Google Scholar]; (c) Muppidi A. Wang Z. Li X. Chen J. Lin Q. Chem. Commun. 2011;47:9396–9398. doi: 10.1039/c1cc13320a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Chen S. Y. Bertoldo D. Angelini A. Pojer F. Heinis C. Angew. Chem., Int. Ed. 2014;53:1602–1606. doi: 10.1002/anie.201309459. [DOI] [PubMed] [Google Scholar]; (e) Wang Y. Chou D. H. Angew. Chem., Int. Ed. 2015;54:10931–10934. doi: 10.1002/anie.201503975. [DOI] [PubMed] [Google Scholar]
- For selected reviews of DEL: ; (a) Brenner S. Lerner R. A. Proc. Natl. Acad. Sci. U. S. A. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Needels M. C. Jones D. G. Tate E. H. Heinkel G. L. Kochersperger L. M. Dower W. J. Barrett R. W. Gallop M. A. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10700–10704. doi: 10.1073/pnas.90.22.10700. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Clark M. A. Curr. Opin. Chem. Biol. 2010;14:396–403. doi: 10.1016/j.cbpa.2010.02.017. [DOI] [PubMed] [Google Scholar]; (d) Kleiner R. E. Dumelin C. E. Liu D. R. Chem. Soc. Rev. 2011;40:5707–5717. doi: 10.1039/c1cs15076f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Mannocci L. Leimbacher M. Wichert M. Scheuermann J. Neri D. Chem. Commun. 2011;47:12747–12753. doi: 10.1039/c1cc15634a. [DOI] [PubMed] [Google Scholar]; (f) Goodnow Jr R. A. Dumelin C. E. Keefe A. D. Nat. Rev. Drug Discovery. 2017;16:131–147. doi: 10.1038/nrd.2016.213. [DOI] [PubMed] [Google Scholar]
- Selected reports on DNA-compatible chemistry: ; (a) Luk K.-C. and Satz A. L., DNA-Compatible Chemistry. Handbook for DNA Encoded Chemistry: Theory and Applications for Exploring Chemical Space and Drug Discovery, ed. R. A. Goodnow Jr, John Willey & Sons, Inc., Hoboken, NJ, 1st edn, 2014, ch. 4, pp. 67−98 [Google Scholar]; (b) Satz A. L. Cai J. Yi C. Goodnow R. Gruber F. Kowalczyk A. Petersen A. Naderi-Oboodi G. Orzechowski L. Strebel Q. Bioconjugate Chem. 2015;26:1623–1632. doi: 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]
- (a) Phelan J. P. Lang S. B. Sim J. Berritt S. Peat A. J. Billings K. Fan L. Molander G. A. J. Am. Chem. Soc. 2019;141:3723–3732. doi: 10.1021/jacs.9b00669. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gerry C. J. Wawer M. J. Clemons P. A. Schreiber S. L. J. Am. Chem. Soc. 2019;141:10225–10235. doi: 10.1021/jacs.9b01203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For selected reports on macrocycle synthesis on DEL: ; (a) Gartner Z. J. Tse B. N. Grubina R. Doyon J. B. Snyder T. M. Liu D. R. Science. 2004;305:1601–1605. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tse B. N. Snyder T. M. Shen Y. Liu D. R. J. Am. Chem. Soc. 2008;130:15611–15626. doi: 10.1021/ja805649f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lu X. Fan L. Phelps C. B. Davie C. P. Donahue C. P. Bioconjugate Chem. 2017;28:1625–1629. doi: 10.1021/acs.bioconjchem.7b00292. [DOI] [PubMed] [Google Scholar]; (d) Li Y. De Luca R. Cazzamalli S. Pretto F. Bajic D. Scheuermann J. Neri D. Nat. Chem. 2018;10:441–448. doi: 10.1038/s41557-018-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Usanov D. L. Chan A. I. Maianti J. P. Liu D. R. Nat. Chem. 2018;10:704–714. doi: 10.1038/s41557-018-0033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Zhu Z. Shaginian A. Grady L. C. O'Keeffe T. Shi X. E. Davie C. P. Simpson G. L. Messer J. A. Evindar G. Bream R. N. Thansandote P. P. Prentice N. R. Mason A. M. Pal S. ACS Chem. Biol. 2018;13:53–59. doi: 10.1021/acschembio.7b00852. [DOI] [PubMed] [Google Scholar]; (g) Stress C. J. Sauter B. Schneider L. A. Sharpe T. Gillingham D. Angew. Chem., Int. Ed. 2019;58:9570–9674. doi: 10.1002/anie.201902513. [DOI] [PubMed] [Google Scholar]; (h) Pham M. V. Bergeron-Brlek M. Heinis C. ChemBioChem. 2020;21:543–549. doi: 10.1002/cbic.201900390. [DOI] [PubMed] [Google Scholar]
- For selected reports on Pd-mediated reactions on DNA: ; (a) Omumi A. Beach D. G. Baker M. Gabryelski W. Manderville R. A. J. Am. Chem. Soc. 2011;133:42–50. doi: 10.1021/ja106158b. [DOI] [PubMed] [Google Scholar]; (b) Lercher L. McGouran J. F. Kessler B. M. Schofield C. J. Davis B. G. Angew. Chem., Int. Ed. 2013;52:10553–10558. doi: 10.1002/anie.201304038. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Krause A. Hertl A. Muttach F. Jäschke A. Chem.–Eur. J. 2014;20:16613–16619. doi: 10.1002/chem.201404843. [DOI] [PubMed] [Google Scholar]; (d) Ding Y. Clark M. A. ACS Comb. Sci. 2015;17:1–4. doi: 10.1021/co5001037. [DOI] [PubMed] [Google Scholar]; (e) Malone M. L. Paegel B. M. ACS Comb. Sci. 2016;18:182–187. doi: 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Lu X. Roberts S. E. Franklin G. J. Davie C. P. MedChemComm. 2017;8:1614–1617. doi: 10.1039/c7md00289k. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li J.-Y. Huang H. Bioconjugate Chem. 2018;29:3841–3846. doi: 10.1021/acs.bioconjchem.8b00676. [DOI] [PubMed] [Google Scholar]; Wang X. Hui Sun H. Liu J. Zhong W. Zhang M. Zhou H. Dai D. Lu X. Org. Lett. 2019;21:719–723. doi: 10.1021/acs.orglett.8b03926. [DOI] [PubMed] [Google Scholar]; Favalli N. Bassi G. Zanetti T. Scheuermann J. Neri D. Helv. Chim. Acta. 2019;102:e1900033. doi: 10.1002/hlca.201900033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For selected reports on synthesis of bicyclic peptides: ; (a) Heinis C. Rutherford T. Freund S. Winter G. Nat. Chem. Biol. 2009;7:502–507. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]; (b) Assem N. Ferreira D. J. Wolan D. W. Dawson P. E. Angew. Chem., Int. Ed. 2015;54:8665–8668. doi: 10.1002/anie.201502607. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lian W. Upadhyaya P. Rhodes C. A. Liu Y. Pei D. J. Am. Chem. Soc. 2013;135:11990–11995. doi: 10.1021/ja405106u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Richelle G. J. J. Ori S. Hiemstra H. Maarseveen van J. H. Timmerman P. Angew. Chem., Int. Ed. 2017;56:501–505. doi: 10.1002/anie.201709127. [DOI] [PubMed] [Google Scholar]; (e) Yin Y. Fei Q. Liu W. Li Z. Suga H. Wu C. Angew. Chem., Int. Ed. 2019;58:4880–4885. doi: 10.1002/anie.201813827. [DOI] [PubMed] [Google Scholar]
- (a) Iyer N. G. Ozdag H. Caldas C. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]; (b) Lasko L. M. et. al. . Nature. 2017;550:128–132. doi: 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.