Abstract
Alkaline phosphatase (ALP) enables intracellular targeting by peptide assemblies, but how the ALP substrates enter cells remains elusive. Here we show that nanoscale phosphopeptide assemblies cluster ALP to enable caveolae mediated endocytosis (CME) and endosomal escape. Specifically, fluorescent phosphopeptides undergo enzyme-catalyzed self-assembly to form nanofibers. Live cell imaging unveils that phosphopeptides nanoparticles, co-incubated with HEK293 cells overexpressing red fluorescent protein-tagged tissue-nonspecific ALP (TNAP-RFP), cluster TNAP-RFP in lipid rafts to enable CME. Further dephosphorylation of the phosphopeptides produce peptidic nanofibers for endosomal escape. Inhibiting TNAP, cleaving the membrane anchored TNAP, or disrupting lipid rafts abolishes the endocytosis. Decreasing the transformation to nanofibers prevents the endosomal escape. As the first study establishing a dynamic continuum of supramolecular assemblies for cellular uptake, this work illustrates an effective design for enzyme-responsive supramolecular therapeutics and provides mechanism insights for understanding the dynamics of cellular uptakes of proteins or exogenous peptide aggregates.
Keywords: Self-Assembly, Endocytosis, Endosomal Escape, Tissue Nonspecific Alkaline Phosphatase
INTRODUCTION
Because many drug targets identified by molecular cell biology are inside cells, considerable efforts have focused on engineering molecules for intracellular delivery of various cargo.1–3 Besides the use of cationic molecules for enhancing cellular uptake of therapeutics,4–6 one of the most explored approaches for intracellular delivery is to engineer molecules to be responsive to chemical or physical stimuli, such as redox,7–10 pH,11–13 enzymes,14–23 or light.24, 25 Among enzymatic approach, alkaline phosphatases (ALP) instructed self-assembly of peptides is particularly effective to facilitate the cellular uptake of the peptide assemblies.14–18, 26–28 However, the mechanism of this phenomenon is largely unknown. Coincidently, viruses also use responsive motif for cell entry. For example, the life cycle of virus (e.g., SARS-CoV-2)29 begins at the attachment to the plasma membrane of host cells30 to clustering plasma membrane-bound receptors, followed by viral proteolytic priming (VPP)31, 32 for endocytosis and endosomal escape. Prompted by the mechanism of viral cell entry, we hypothesize that the assemblies of phosphopeptides, as the substrate of ALP, act as a dynamic continuum33 to cluster the ALP to facilitate endocytosis and to undergo enzymatic morphological transition that enables the endosomal escape (Scheme 1). This hypothesis is also based on several additional rationales: (i) ALP is a GPI-anchor protein, and its clustering enables caveolae mediated endocytosis;30 (ii) ALP, locating in lipid rafts, plays a key role in the cellular uptake of aberrant protein aggregates; and (iii) ALP catalyzed dephosphorylation enables morphological transition (e.g., nanoparticles to nanofibers) and generates artificial intracellular filaments.17
Scheme 1.
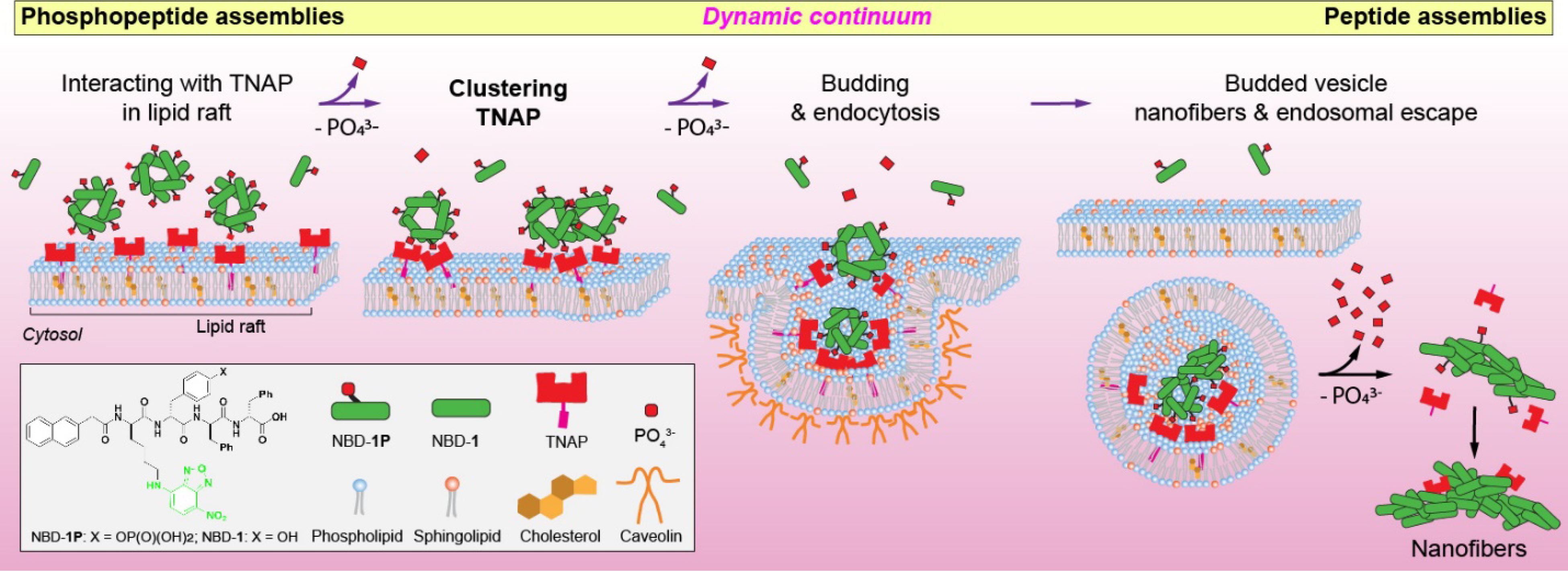
Illustration of a dynamic continuum of peptide assemblies for endocytosis and endosomal escape
To prove our hypothesis, we synthesize a phosphopeptide, which, above its critical micelle concentration (CMC), self-assembles to form nanoparticles, which transform into nanofibers upon the dephosphorylation catalyzed by ALP. The phosphopeptide molecules initially aggregate on the plasma membrane to cluster tissue nonspecific alkaline phosphatase (TNAP) in lipid rafts of HEK293 cells that overexpress TNAP, then enable caveolae-mediate endocytosis (CME). The assemblies of the phosphopeptides gradually changes their morphology from nanoparticles to nanofibers depending on the extent of dephosphorylation. Abolishing the interaction between the TNAP and the phosphopeptides significantly decrease the endocytosis. The TNAP, remained in endosome, catalyzes the further dephosphorylation of the phosphopeptides, generating peptide nanofibers to facilitate endosomal escape. As the first study that illustrates the intracellular peptide assemblies resulted from dynamic peptide assemblies rather than a monomeric peptide or static assemblies, this work not only provides insights for understanding the cellular uptakes of proteins or exogenous peptide aggregates, but also offers the guidance for designing enzyme responsive supramolecular assemblies as intracellular targeting therapeutics.34
RESULTS AND DISCUSSION
Scheme 1 shows the structure of the fluorescent phosphopeptide (NBD-1p). For studying the cellular uptake of the phosphopeptides, a nitrobenzoxadiazole (NBD), as the fluorophore, conjugates to the side chain of the D-lysine in a naphthyl capped D-phosphotetrapeptide (D-Lys-D-pTyr-D-Phe-D-Phe). Using Fmoc-based solid-phase peptide synthesis, we firstly produced this naphthyl capped D-phosphopeptide, then conjugated it to NBD. Then, the purification by high performance liquid chromatography (HPLC) produces the designed NBD-1p. Transmission electron microscopy (TEM) reveals that NBD-1p monomers primarily self-assembles into nanoparticles (22.4±7.2 nm in diameter, Figure 1A) at 200 μM (PBS, pH=7.4), which is above its critical micelle concentration (CMC, 159 μM, Figure S1). Upon the addition of ALP for catalytically dephosphorylating NBD-1p, the nanoparticles formed by NBD-1p transform into short nanofibers (21.9±3.7 nm in diameter and about 300–1000 nm in length) made of NBD-1 (Figure 1A and Scheme S1). This result confirms that NBD-1p undergoes enzyme-instructed self-assembly.35
Figure 1.
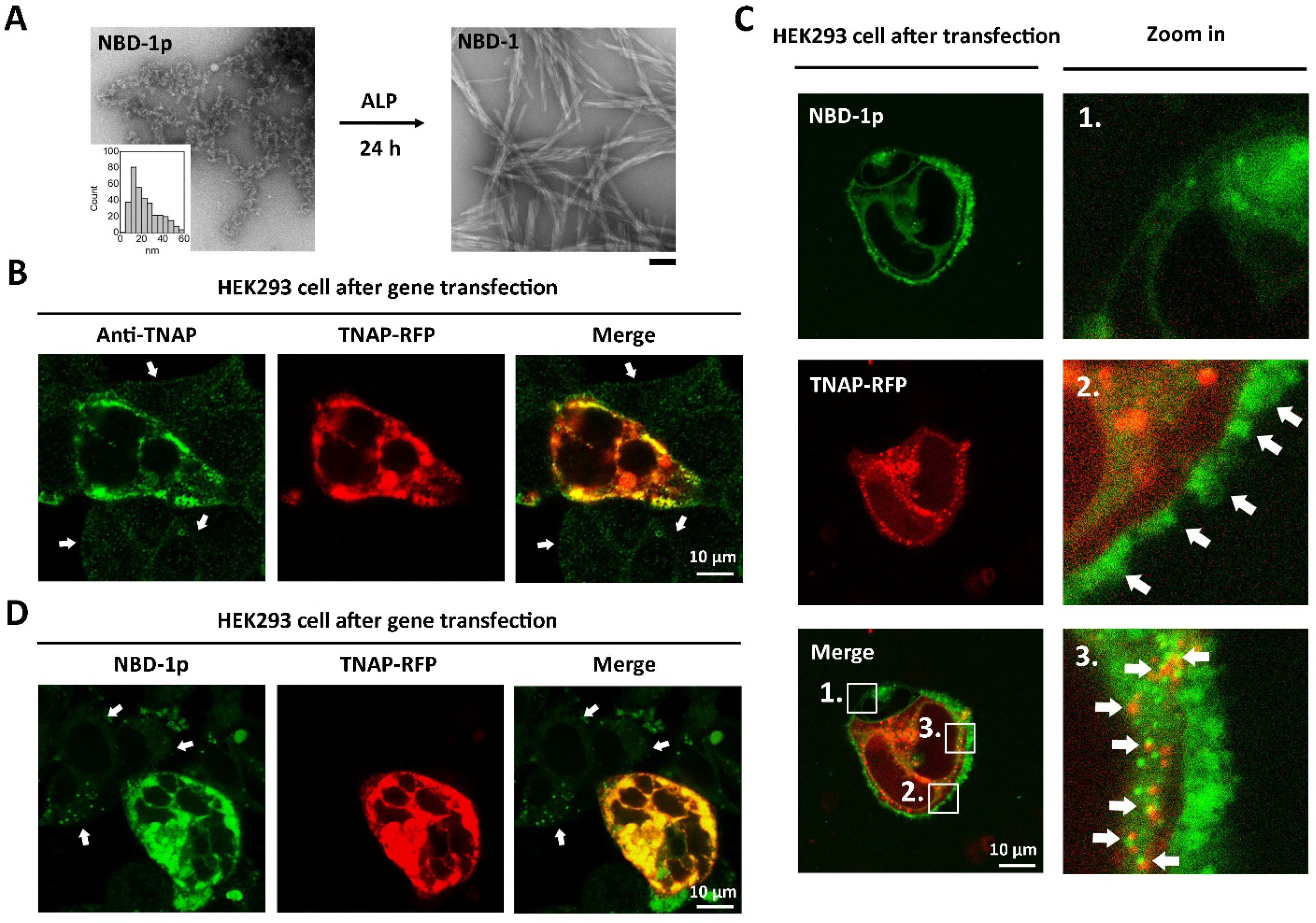
Upregulation of TNAP promotes the endocytosis of phosphopeptide assemblies. (A) TEM images of NBD-1p (200 μM) before and after the addition of ALP (0.1 U/mL, 37 °C) for 24 h. The insert is the diameter distribution of the nanoparticles formed by NBD-1p. Scale bar = 100 nm. (B) Immunofluorescence staining of TNAP in the transfected HEK293 cell. The non-transfected cells (pointed by arrows) show little TNAP. (C) Confocal fluorescence images of HEK293_TNAP-RFP cells co-incubated with NBD-1p (200 μM, 5 min, no washing). To visualize NBD-1p at early stage, the laser power here is 5 times stronger than that used in Figure 1D. (D) Confocal fluorescence imaging of TNAP-RFP transfected HEK293 cells treated by NBD-1p (200 μM, 12 h, washed). The non-transfected cells (indicated by arrows) show little intracellular NBD fluorescence.
We choose HEK293 cells for testing our hypothesis because of their constitutively low expression of ALP, reliable growth, and propensity for transfection.36 Treating HEK293 cells with the polyethyleneimine (PEI)37 carrying the DNA plasmid encoding red fluorescent protein-tagged TNAP (TNAP-RFP), we generate a cell population consists of the HEK293 cells that overexpress TNAP-RFP (Figure 1B, the transfected cells are denoted as HEK293_TNAP-RFP) and the HEK293 cells without gene transfection (Figure 1B, indicated by arrows). To observe the fluorescence of NBD-1p at the early stage of endocytosis, we used high laser strength for imaging. Co-incubating NBD-1p (above CMC) with the HEK293 cells, after the transfection, reveals that, initially (5 min), NBD-1p mainly accumulates on the plasma membrane of HEK293_TNAP-RFP cells in a discrete pattern (Figure 1C, region 2, indicated by arrows), although the phosphopeptides barely interact with the plasma membrane of the non-transfected control (Figure 1C, region 1). The cell selective aggregation of NBD-1p on the plasma membrane of HEK293_TNAP-RFP cells indicates that the nanoparticles formed by NBD-1p associate with the plasma membrane-bound TNAP38, 39 for dephosphorylation. The interaction between TNAP and the nanoparticles (made of mainly NBD-1p and small amount of NBD-1) spatially induces the clustering of TNAP on plasma membrane (Scheme 1). Monomers of NBD-1p, being dephosphorylated rapidly, would likely interact with TNAP only briefly. Moreover, we observed not only the formation of more endocytic vesicles in HEK293_TNAP-RFP cells than in the non-transfected control (Figure 1C, region 1 and 3), but also some vesicles containing TNAP-RFP and NBD-1p in HEK293_TNAP-RFP cells (Figure 1C, region 3, and Movie S1). These results suggest that the TNAP-participating endocytosis of NBD-1p nanoparticles is enhanced in HEK293_TNAP-RFP cells. While some NBD fluorescence exists in some non-transfected HEK293 cells and the endosomal vesicles lacking TNAP in the transfected HEK293 cells, these NBD signals may originate from the cellular uptake of NBD-1p via other endocytosis pathways (e. g., macropinocytosis, a nonspecific endocytosis) and the enhanced intracellular NBD fluorescence or autofluorescence under high laser strength. Nonetheless, other endocytosis pathways, like macropinocytosis, less likely act as the main path for the phosphopeptide assemblies (Figure S2). Additionally, NBD-1p can still enter the non-transfected HEK293 cells via CME, although inefficiently due to low TNAP expression. The intracellular fluorescence from NBD increases significantly overtime in the HEK293_TNAP-RFP cells, while keeping mostly unchanged in the non-transfected HEK293 cells (Figure 1D, S3 and Movie S2). Trypan blue hardly stains the HEK293_TNAP-RFP cells with NBD-1p (Figure S4), confirming an intact cell membrane. Additionally, after the incubation with NBD-1p, the TNAP gene transfected HEK293 cells exhibit lower cell viability than that of the wild type (Figure S5). The cytotoxicity may originate from the formation of intracellular NBD-1 nanofibers after dephosphorylation. These results confirm that the overexpression of TNAP in HEK293 cell promotes the cellular uptake of the assemblies of NBD-1p.
Further study on the endocytosis of NBD-1p nanoparticles shows that pretreating the HEK293 cells (after TNAP transfection) with methyl-β-cyclodextrin (M-β-CD), a CME inhibitor via dissipating lipid rafts on plasma membrane,40, 41 at the cell compatible concentration (2.5 mM) drastically reduces the cellular uptake efficiency of NBD-1p (Figure 2A) compared to the control (Figure 1D). This result indicates that (i) M-β-CD decreases the CME efficiency at that working concentration, and (ii) CME contributes to the cell entry of NBD-1p which adheres on the plasma membrane of HEK293_TNAP-RFP cells (Figure 1C, region 2). Since TNAP is enriched in the lipid rafts on plasma membrane,42 this result also suggests that the assemblies of NBD-1p on plasma membrane (Figure 1C, region 2) associate with the TNAP in lipid rafts, and then cluster the TNAP, which signals the endocytosis of NBD-1p through CME30 (Scheme 1). The CME of NBD-1p nanoparticles triggered by TNAP aggregation agrees with the formation of endocytic vesicles containing TNAP-RFP and NBD-1p (Figure 1C, region 3). Using 2,5-dimethoxy-N-(quinolin-3-yl)-benzenesulfonamide (DQB), a noncompetitive inhibitor of TNAP with an allosteric inhibition mechanism,43 to inhibit the enzymatic reaction (i.e., TNAP catalyzing the dephosphorylation of NBD-1p) or the removal of plasma membrane-bound TNAP via phospholipase C (PLC) which cleaves the GPI anchors44 that link TNAP to plasma membrane also substantially decrease the cellular uptake of NBD-1p into HEK293_TNAP-RFP cells (Figure 2B and 2C). These results further support that the interaction between TNAP and the nanoparticles of NBD-1p is important for signaling CME. Below the CMC, the individual NBD-1p molecules neither aggregate on the plasma membrane of HEK293_TNAP-RFP cells (Figure S6 and Movie S3) nor enter the cells efficiently (Figure 2D), although the monomeric phosphopeptides may still associate with the TNAP on cell surface as the substrate of TNAP (Figure 2E). These results indicate that the clustering of TNAP in lipid rafts by the nanoparticles formed by the self-assembly of NBD-1p is essential for signaling the CME of the assemblies of NBD-1p (Figure 2E).
Figure 2.
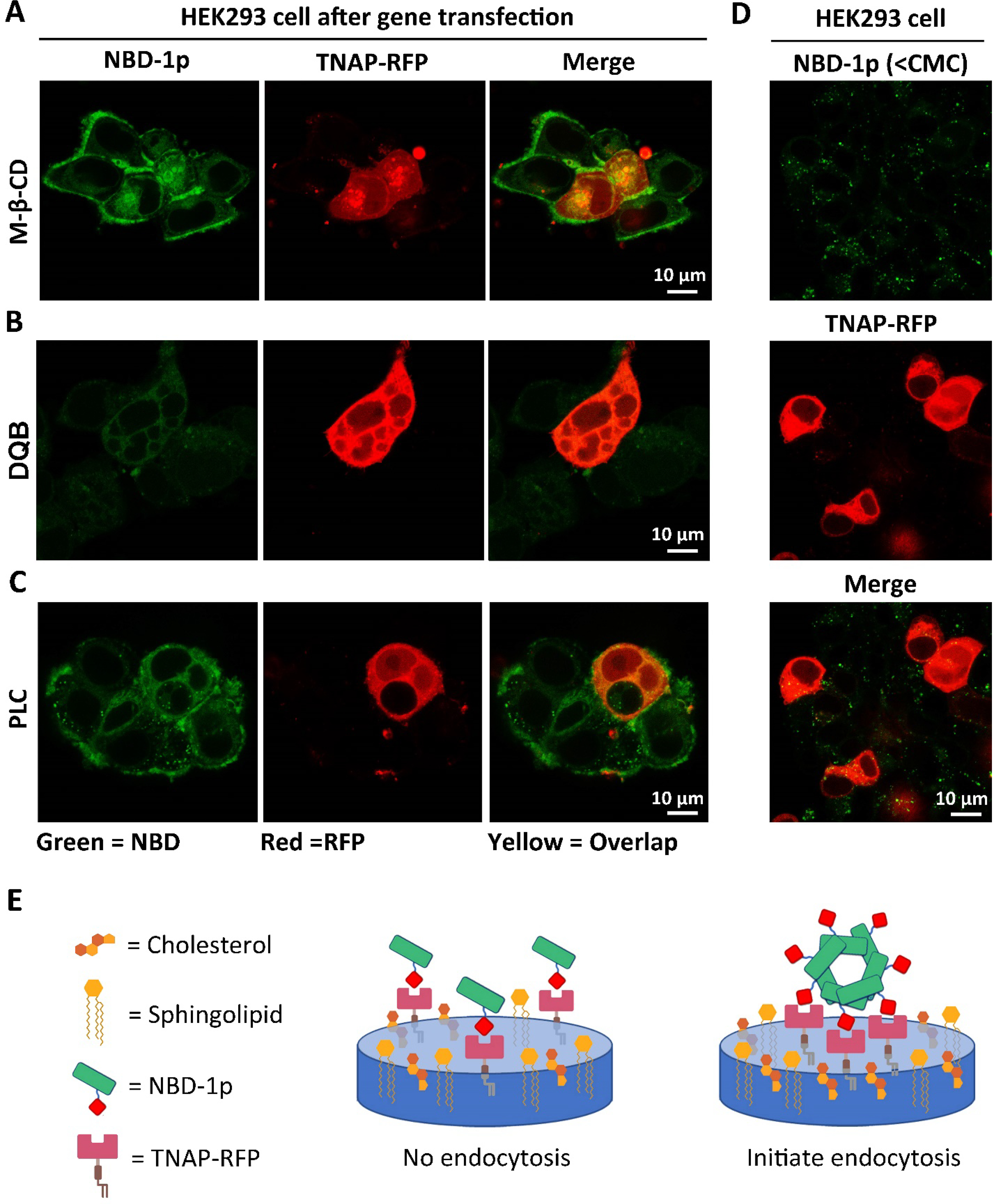
NBD-1p nanoparticles associate with the TNAP in lipid rafts followed by CME. (A-C) Confocal fluorescence images of HEK293_TNAP-RFP cells incubated with NBD-1p (200 μM, 12 h) after the pretreatment of (A) caveolin-dependent endocytosis inhibitors (M-β-CD), (B) TNAP inhibitor (DQB), and (C) phospholipase C which removes the GPI-anchored TNAP on plasma membrane. (D) Confocal fluorescence images of HEK293_TNAP-RFP cells incubated with NBD-1p (50 μM, 12 h) below CMC. (E) Illustration of that the assemblies of phosphopeptide rather than individual molecules initiate CME.
To further validate that the interactions between TNAP and the (D)-pTyr in the nanoparticles of NBD-1p initiates the CME, we incubated the HEK293_TNAP-RFP cells with NBD-1 (i.e., the dephosphorylated NBD-1p, Scheme S1). Clearly, the HEK293_TNAP-RFP cells treated by NBD-1 exhibit significantly less intracellular fluorescence intensity than those treated by NBD-1p (Figure 3A and 3B). This result suggests that the interaction between TNAP and the D-pTyr plays a role in the CME of the NBD-1p assemblies. Replacing the D-pTry by D-pSer, we generated a derivative of NBD-1p, named NBD-(D)Sp. The HEK293_TNAP-RFP cells incubated with NBD-(D)Sp show drastically reduced cellular uptake of the phosphopeptide compared to the incubation with NBD-1p (Figure 3C). Dephosphorylation assay reveals that ALP dephosphorylates NBD-1p much faster than NBD-(D)Sp at the same concentration (Figure 3D), indicating that ALP exhibits higher affinity for NBD-1p than NBD-(D)Sp. The results above suggest that the interaction between ALP and its substrate is necessary for the CME of the phosphopeptides. Moreover, a sequence-scrambled phosphopeptide derivative of NBD-1p, denoted as NBD-2p (with the sequence of D-Phe-D-Phe-D-Lys(ℇ-NBD)-D-pTyr, Scheme S1), also enters the HEK293_TNAP-RFP cells more efficiently than the non-transfected HEK293 cells (Figure S7). This result indicates that the cellular uptake of the phosphopeptide assemblies containing D-pTyr depends more on the interactions between TNAP and the enzyme triggers than on the binding between receptors and specific peptide sequences.45–47
Figure 3.
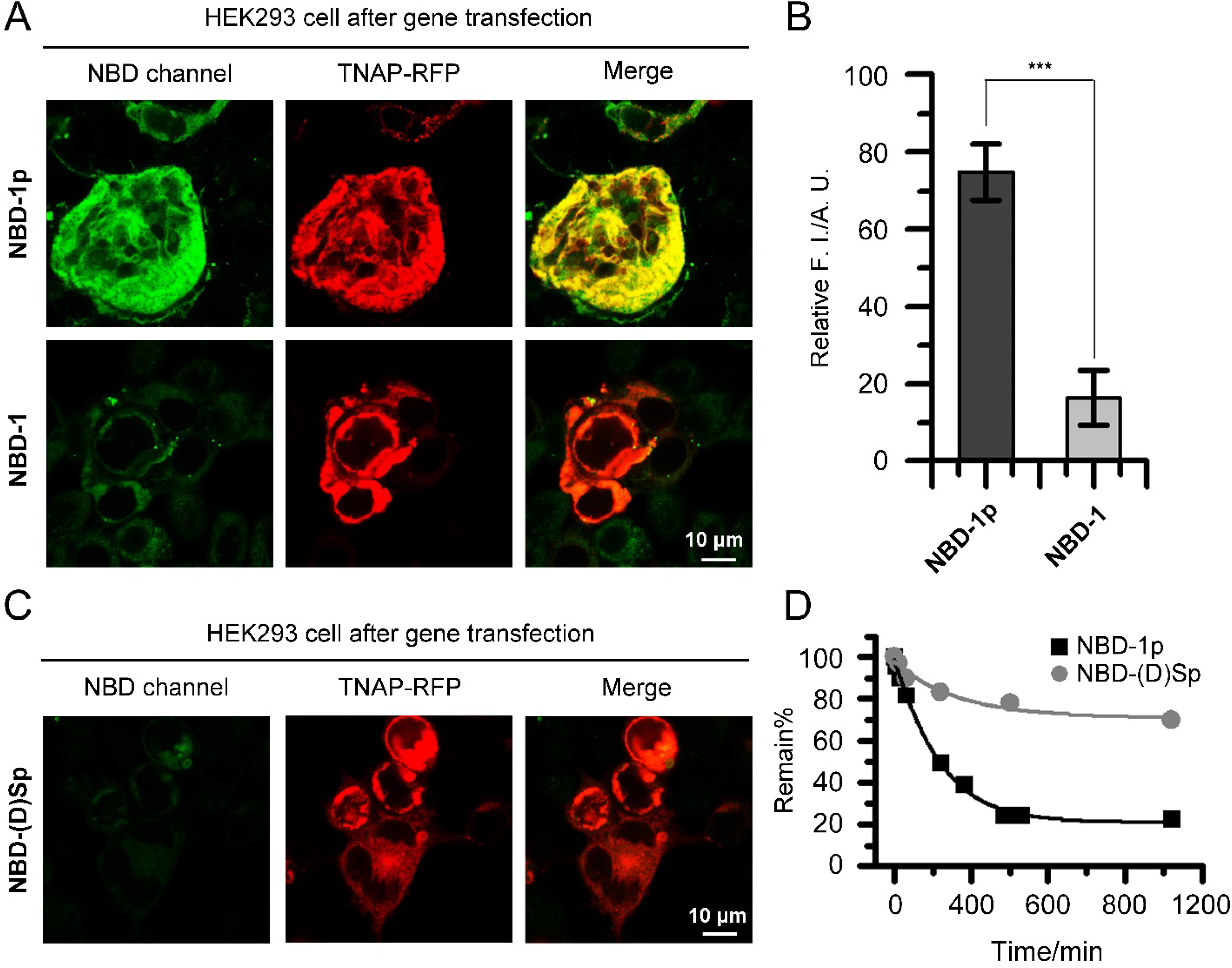
Phosphotyrosine is critical for rapid cellular uptake. (A) Confocal fluorescence images of HEK293_TNAP-RFP cells incubated with NBD-1p or NBD-1 (200 μM, 12 h, washed), respectively. (B) ImageJ analysis of intracellular fluorescence intensity of HEK293_TNAP-RFP cells in (A). Data is presented as mean ± standard deviation, *** means p<0.001. (C) Confocal fluorescence images of HEK293_TNAP-RFP cells incubated with NBD-(D)Sp (400 μM, 12 h, washed). (D) Dephosphorylation rate of NBD-1p and NBD-(D)Sp by ALP (0.1 U/mL, 37 °C).
The dispersive fluorescence of NBD-1p in HEK293_TNAP-RFP cells (Figure 1D and 3A) not only confirms the endocytosis of NBD-1p, but also the endosomal escape of the peptide assemblies. Considering the cell entry mechanism of SARS-CoV-2, in which the proteolytic cleavage of S proteins by endosomal proteases (e.g., cathepsin B and cathepsin L) promotes the release of viral genome from endosome to cytosol,48 we reckon that, after the CME of the assemblies of NBD-1p, further dephosphorylation catalyzed by the TNAP in endosomal compartments induces a phase transition on the peptide assemblies, which facilitates the endosomal escape of NBD-1 into cytosol. Liquid chromatography-mass spectrometry (LC-MS) analysis of the cell lysate of HEK293_TNAP-RFP cells that are preincubated with NBD-1p confirms the dephosphorylation of the peptide inside cells (Figure S8). Additionally, TEM images of liposomes encapsulating NBD-1p and ALP exhibit the generation of nanofibers that rupture the liposomes (Figure 4A). These results imply that the TNAP in endocytic vesicles convert the NBD-1p nanoparticles to nanofibers (NBD-1), which break the membrane of endocytic vesicles for escaping into cytosol. More TEM images reveal that the NBD-1p assemblies mixed with ALP initially remain as nanoparticles until certain dephosphorylation ratio that triggers the particle-to-fiber phase transition (Figure S9 and 3D). This result implies that although dephosphorylation begins once TNAP binds to NBD-1p assemblies on plasma membrane, the assemblies likely maintain as nanoparticles (which consist of mainly NBD-1p and small amount NBD-1) during internalization until being further dephosphorylated in endosomal vesicles where nanofibers form for endosome rupturing. This observation also agrees with the reports that filamentous nanostructures have much reduced/limited cellular uptake ability relative to their spherical counterparts.49, 50
Figure 4.
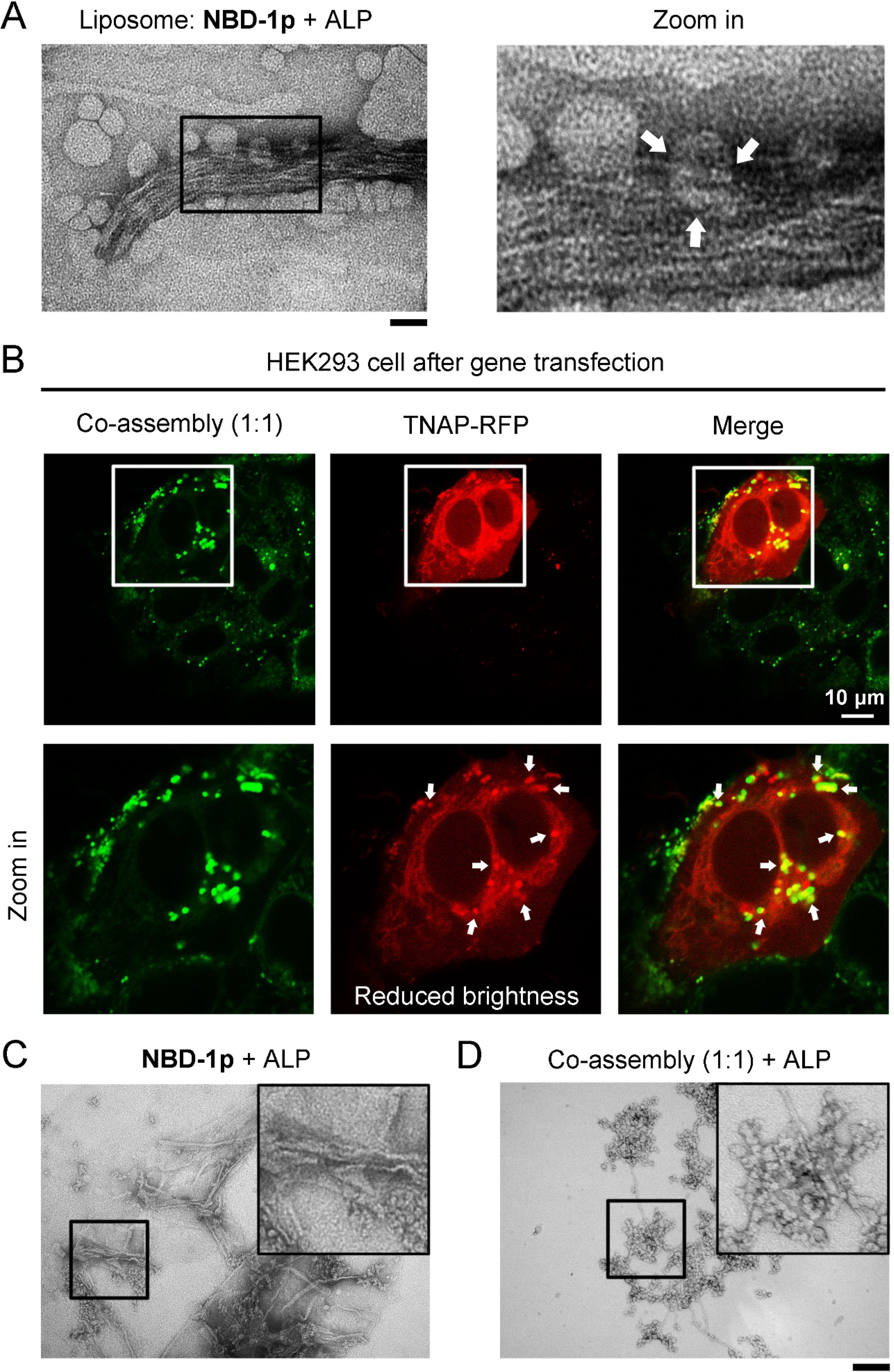
The co-assembly of NBD-1p and NBD-(D)Sp shows less efficient endosomal escape. (A) TEM image of liposomes carrying NBD-1p and ALP (1 U/mL, 37 °C, 6 h). Broken liposomes are indicated by arrows. Scale bar = 100 nm. (B) Confocal fluorescence images of HEK293_TNAP-RFP cells incubated with the mixture of NBD-1p (100 μM) and NBD-(D)Sp (100 μM) for 12 h. The peptides trapped in endosome are indicated by arrows. (C) TEM images of NBD-1p (200 μM) and (D) the co-assemblies of NBD-1p (100 μM) and NBD-(D)Sp (100 μM) after the addition of ALP (0.1 U/mL, 37 °C, 3 h) for partial dephosphorylation. ALP yields more nanofibers in (C) than in (D). Scale bar = 100 nm.
To further prove the above assumption of endosomal escape, we incubate HEK293 cells, after TNAP-RFP gene transfection, with the mixture of NBD-1p and NBD-(D)Sp (total peptide concentration is 200 μM). The HEK293_TNAP-RFP cells incubated with the mixture of NBD-1p and NBD-(D)Sp at 1:1 or 1:3 ratio mainly generates fluorescent puncta colocalizing with TNAP-RFP inside the cells (Figure 4B and S10A), although the cells incubated with NBD-1p (100 or 50 μM) or NBD-(D)Sp (100 or 150 μM) ), respectively, below CMC exhibit little fluorescence (Figure S11). This result indicates that NBD-1p and NBD-(D)Sp form co-assemblies in solution, and the co-assemblies of NBD-1p and NBD-(D)Sp (1:1 or 1:3) primarily retain in the endocytic vesicles containing TNAP-RFP after the endocytosis. Our results also indicate that phosphopeptide assemblies exhibits emergent properties, which are absent in monomeric phosphopeptides. The nanoparticles formed by NBD-1p transform into nanofibers for endosomal rupturing after partial dephosphorylation by ALP (Figure 4C and 3D). Thus, the remained phosphopeptides, after releasing into cytosol, get further dephosphorylation by intracellular TNAP. This process produces networks of peptidic nanofibers that encapsulate intracellular TNAP (Figure 1D and 3A, TNAP colocalizes with NBD). However, the co-assemblies of NBD-1p and NBD-(D)Sp (1:1 or 1:3) mixed with ALP mostly remain as nanoparticles (Figure 4D and S10B) because NBD-(D)Sp is a poor substrate of ALP (Figure 3D). When the mixture has the ratio of NBD-1p and NBD-(D)Sp in 3 to 1, the co-assemblies behave similarly to the assemblies of NBD-1p (Figure S10). Moreover, a phosphopeptide derivative (NBD-3p) carrying D-pTry and pSer (Scheme S1) also ends up in the endosomal compartments of HEK293_TNAP-RFP cells after incubation (Figure S12). Like the co-assemblies of NBD-1p and NBD-(D)Sp with 1:1 or 1:3 ratio, NBD-3P, being treated by ALP, inefficiently undergoes the nanoparticle-to-nanofiber transition (Figure S13). This derivative also exhibits minimal cytotoxicity compared to other phosphopeptides (Figure S5). These results suggest that the particle-to-fiber transition of the phosphopeptide assemblies catalyzed by the ALP in endosome is critical for the endosomal escape of the peptide assemblies, likely via rupturing the endosome by the formation of nanofibers.
CONCLUSIONS
In conclusion, this work demonstrates the continuous morphological transformation of peptide assemblies, catalyzed by ALP, is responsible for the endocytosis, mainly through caveolae-mediated endocytic pathway, and endosomal escape of the peptide assemblies. Although other endocytosis pathways, such as macropinocytosis, may contribute to the cellular uptake of phosphopeptide assemblies, they are less likely act as the main path for the phosphopeptide assemblies. Here, TNAP catalytically controls a dynamic morphological transition, thus enables endocytosis and endosomal escape of peptide assemblies, in an analogy to the cell entry of virus. Because NBD-1p mainly exists as nanoparticles at the concentration above CMC, the effects of monomeric NBD-1p likely are insignificant. We suggest this type of cell uptake as “Enzyme Primed Endocytosis (EPE)”. While the contribution of VPP in the cell entry of virus has been extensively studied, little research elucidates how EPE controls the cellular uptake of other molecular assemblies. In analogy to the ligand-receptor mediated endocytosis that require tight binding, the clustering of enzymes in lipid rafts and the enzymatic reaction of supramolecular assemblies require rapid enzyme kinetics because NBD-(D)Sp hardly enter the cells. This study also provides a mechanistic understanding of the role of peptide assemblies for context-dependent signaling.51, 52 Although this work focuses on ALP and peptides, the mechanism revealed likely is applicable for the endocytosis of the substrates of esterase53 or proteases,54 including non-peptide substrates of enzymes.
Supplementary Material
ACKNOWLEDGMENT
The authors thank NIH (R01CA142746) and NSF (DMR-2011846) for their support.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at: https://pubs.acs.org/doi/10.1021/acs.nanolett.1c01029.
Experimental Procedures; (Scheme S1) Molecular structure; (Figure S1) Critical micelle concentration; (Figure S2) Time-dependent endocytosis; (Figure S3) Trypan blue staining of cells; (Figure S4) MTT cell viability assay; (Figure S5) Confocal fluorescence images of the HEK293 cells after TNAP-RFP gene transfection incubated with phosphopeptide below CMC; (Figure S6) Confocal fluorescence imaging of HEK293 cells after the transfection of TNAP-RFP gene incubated with sequence-scrambled phosphopeptide; (Figure S7) LC-MS analysis of cell lysate of HEK293 cells; (Figure S8) TEM images of phosphopeptide incubated with ALP for different time; (Figure S9) HEK293 cells, after TNAP-RFP gene transfection, incubated with the co-assemblies of NBD-1p and NBD-(D)Sp; (Figure S10) HEK293 cells, after TNAP-RFP gene transfection, incubated with NBD-1p or NBD-(D)Sp below CMC, respectively; (Figure S11) Confocal fluorescence imaging of HEK293 cells, after the transfection of TNAP-RFP gene, incubated with NBD-3p; (Figure S12) TEM images of NBD-3p incubated with ALP; (Figure S13) Confocal fluorescence images of HEK293_TNAP-RFP cells incubated with NBD-1p in the presence of macropinocytosis inhibitor; and (Figure S14) Mass spectrum.
REFERENCES
- (1).Overchuk M; Zheng G Overcoming obstacles in the tumor microenvironment: Recent advancements in nanoparticle delivery for cancer theranostics. Biomaterials 2018, 156, 217–237. [DOI] [PubMed] [Google Scholar]
- (2).Goswami R; Jeon T; Nagaraj H; Zhai S; Rotello VM Accessing Intracellular Targets through Nanocarrier-Mediated Cytosolic Protein Delivery. Trends Pharmacol. Sci 2020, 41, (10), 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Glass Z; Lee M; Li Y; Xu Q Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 2018, 36, (2), 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Futaki S; Suzuki T; Ohashi W; Yagami T; Tanaka S; Ueda K; Sugiura Y Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2001, 276, (8), 5836–5840. [DOI] [PubMed] [Google Scholar]
- (5).MacEwan SR; Chilkoti A Controlled apoptosis by a thermally toggled nanoscale amplifier of cellular uptake. Nano Lett. 2014, 14, (4), 2058–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Lagerholm BC; Wang M; Ernst LA; Ly DH; Liu H; Bruchez MP; Waggoner AS Multicolor coding of cells with cationic peptide coated quantum dots. Nano Lett. 2004, 4, (10), 2019–2022. [Google Scholar]
- (7).Tang R; Wang M; Ray M; Jiang Y; Jiang Z; Xu Q; Rotello VM Active Targeting of the Nucleus Using Nonpeptidic Boronate Tags. J. Am. Chem. Soc 2017, 139, (25), 8547–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wu Y; Zhang L; Cui C; Cansiz S; Liang H; Wu C; Teng IT; Chen W; Liu Y; Hou W; Zhang X; Tan W Enhanced Targeted Gene Transduction: AAV2 Vectors Conjugated to Multiple Aptamers via Reducible Disulfide Linkages. J. Am. Chem. Soc 2018, 140, (1), 2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zeng H; Little HC; Tiambeng TN; Williams GA; Guan Z Multifunctional dendronized peptide polymer platform for safe and effective siRNA delivery. J. Am. Chem. Soc 2013, 135, (13), 4962–4965. [DOI] [PubMed] [Google Scholar]
- (10).Wang Y; Li Z; Shmidov Y; Carrazzone RJ; Bitton R; Matson JB Crescent-Shaped Supramolecular Tetrapeptide Nanostructures. J. Am. Chem. Soc 2020, 142, 20058–20065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Du JZ; Du XJ; Mao CQ; Wang J Tailor-Made dual pH-sensitive polymer-doxorubicin nanoparticles for efficient anticancer drug delivery. J. Am. Chem. Soc. 2011, 133, (44), 17560–17563. [DOI] [PubMed] [Google Scholar]
- (12).June RK; Gogoi K; Eguchi A; Cui XS; Dowdy SF Synthesis of a pH-sensitive nitrilotriacetic linker to peptide transduction domains to enable intracellular delivery of histidine imidazole ring-containing macromolecules. J. Am. Chem. Soc. 2010, 132, (31), 10680–10682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hu Y; Litwin T; Nagaraja AR; Kwong B; Katz J; Watson N; Irvine DJ Cytosolic delivery of membrane-impermeable molecules in dendritic cells using pH-responsive core-shell nanoparticles. Nano Lett. 2007, 7, (10), 3056–3064. [DOI] [PubMed] [Google Scholar]
- (14).He H; Liu S; Wu D; Xu B Enzymatically‐Formed Peptide Assemblies Sequestrate Proteins and Relocate Inhibitors for Selectively Killing Cancer Cells. Angew. Chem. Int. Ed 2020, 59, 16445–16450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Yan R; Hu Y; Liu F; Wei S; Fang D; Shuhendler AJ; Liu H; Chen HY; Ye D Activatable NIR Fluorescence/MRI Bimodal Probes for in Vivo Imaging by Enzyme-Mediated Fluorogenic Reaction and Self-Assembly. J. Am. Chem. Soc 2019, 141, (26), 10331–10341. [DOI] [PubMed] [Google Scholar]
- (16).He H; Tan W; Guo J; Yi M; Shy AN; Xu B Enzymatic Noncovalent Synthesis. Chem Rev 2020, 120, (18), 9994–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Feng Z; Wang H; Wang F; Oh Y; Berciu C; Cui Q; Egelman EH; Xu B Artificial Intracellular Filaments. Cell Rep Phys Sci 2020, 1, (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zhan J; Cai Y; He S; Wang L; Yang Z Tandem Molecular Self-Assembly in Liver Cancer Cells. Angew. Chem. Int. Ed 2018, 57, (7), 1813–1816. [DOI] [PubMed] [Google Scholar]
- (19).Chang J; Cai W; Liang C; Tang Q; Chen X; Jiang Y; Mao L; Wang M Enzyme-Instructed Activation of Pro-protein Therapeutics in Vivo. J. Am. Chem. Soc 2019, 141, (45), 18136–18141. [DOI] [PubMed] [Google Scholar]
- (20).Yuan Y; Wang L; Du W; Ding Z; Zhang J; Han T; An L; Zhang H; Liang G Intracellular self‐assembly of Taxol nanoparticles for overcoming multidrug resistance. Angew. Chem. Int. Ed 2015, 54, (33), 9700–9704. [DOI] [PubMed] [Google Scholar]
- (21).Wu C; Zhang R; Du W; Cheng L; Liang G Alkaline phosphatase-triggered self-assembly of near-infrared nanoparticles for the enhanced photoacoustic imaging of tumors. Nano Lett. 2018, 18, (12), 7749–7754. [DOI] [PubMed] [Google Scholar]
- (22).Hai Z; Ni Y; Saimi D; Yang H; Tong H; Zhong K; Liang G γ-glutamyltranspeptidase-triggered intracellular gadolinium nanoparticle formation enhances the T2-weighted MR contrast of tumor. Nano Lett. 2019, 19, (4), 2428–2433. [DOI] [PubMed] [Google Scholar]
- (23).Li G; Hu X; Nie P; Mang D; Jiao S; Zhang S; Roy SR; Yukawa S; Asahina S; Sugasawa H; William C; Zhaocai Z; Ye Z Lipid-Raft-Targeted Molecular Self-Assembly Inactivates YAP to Treat Ovarian Cancer. Nano Lett. 2021, 21, 747–755. [DOI] [PubMed] [Google Scholar]
- (24).Tan X; Li BB; Lu X; Jia F; Santori C; Menon P; Li H; Zhang B; Zhao JJ; Zhang K Light-triggered, self-immolative nucleic acid-drug nanostructures. J. Am. Chem. Soc 2015, 137, (19), 6112–6115. [DOI] [PubMed] [Google Scholar]
- (25).Wei T; Lu S; Sun J; Xu Z; Yang X; Wang F; Ma Y; Shi YS; Chen X Sanger’s Reagent Sensitized Photocleavage of Amide Bond for Constructing Photocages and Regulation of Biological Functions. J. Am. Chem. Soc 2020, 142, (8), 3806–3813. [DOI] [PubMed] [Google Scholar]
- (26).Feng Z; Han X; Wang H; Tang T; Xu B Enzyme-instructed peptide assemblies selectively inhibit bone tumors. Chem 2019, 5, (9), 2442–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Zhou J; Du X; Chen X; Wang J; Zhou N; Wu D; Xu B Enzymatic self-assembly confers exceptionally strong synergism with NF-κB targeting for selective necroptosis of cancer cells. J. Am. Chem. Soc 2018, 140, (6), 2301–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zhan J; Cai Y; He S; Wang L; Yang Z Tandem molecular self‐assembly in liver cancer cells. Angew. Chem. Int. Ed 2018, 130, (7), 1831–1834. [DOI] [PubMed] [Google Scholar]
- (29).Ou X; Liu Y; Lei X; Li P; Mi D; Ren L; Guo L; Guo R; Chen T; Hu J Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun 2020, 11, (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Parton RG; Joggerst B; Simons K Regulated internalization of caveolae. J. Cell Biol 1994, 127, (5), 1199–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Dube D; Brecher MB; Delos SE; Rose SC; Park EW; Schornberg KL; Kuhn JH; White JM The primed ebolavirus glycoprotein (19-Kilodalton GP 1,2): Sequence and residues critical for host cell binding. J. Virol 2009, 83, (7), 2883–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Hoffmann M; Kleine-Weber H; Schroeder S; Kruger N; Herrler T; Erichsen S; Schiergens TS; Herrler G; Wu NH; Nitsche A; Muller MA; Drosten C; Pohlmann S SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, (2), 271–280 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wu H; Fuxreiter M The structure and dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell 2016, 165, (5), 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wang Q; Jiang N; Fu B; Huang F; Liu J Self-assembling peptide-based nanodrug delivery systems. Biomater. Sci 2019, 7, (12), 4888–4911. [DOI] [PubMed] [Google Scholar]
- (35).Li J; Kuang Y; Shi J; Zhou J; Medina JE; Zhou R; Yuan D; Yang C; Wang H; Yang Z Enzyme‐instructed intracellular molecular self‐assembly to boost activity of cisplatin against drug‐resistant ovarian cancer cells. Angew. Chem. Int. Ed 2015, 54, (45), 13307–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Thomas P; Smart TG HEK293 cell line: a vehicle for the expression of recombinant proteins. J Pharmacol Toxicol 2005, 51, (3), 187–200. [DOI] [PubMed] [Google Scholar]
- (37).Longo PA; Kavran JM; Kim M-S; Leahy DJ, Transient mammalian cell transfection with polyethylenimine (PEI). In Method Enzymol, Elsevier: 2013; Vol. 529, pp 227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Lin C; Sasaki M; Orcutt M; Miyayama H; Singer R Plasma membrane localization of alkaline phosphatase in HeLa cells. J Histochem Cytochem 1976, 24, (5), 659–667. [DOI] [PubMed] [Google Scholar]
- (39).De Broe ME; Roels F; Nouwen EJ; Claeys L; Wieme RJ Liver plasma membrane: the source of high molecular weight alkaline phosphatase in human serum. Hepatology 1985, 5, (1), 118–128. [DOI] [PubMed] [Google Scholar]
- (40).Le PU; Guay G; Altschuler Y; Nabi IR Caveolin-1 is a negative regulator of caveolae-mediated endocytosis to the endoplasmic reticulum. J Biol Chem 2002, 277, (5), 3371–3379. [DOI] [PubMed] [Google Scholar]
- (41).Mahammad S; Parmryd I, Cholesterol depletion using methyl-β-cyclodextrin. In Methods in membrane lipids, Springer: 2015; pp 91–102. [DOI] [PubMed] [Google Scholar]
- (42).Milhiet PE; Giocondi MC; Baghdadi O; Ronzon F; Roux B; Le Grimellec C Spontaneous insertion and partitioning of alkaline phosphatase into model lipid rafts. Embo Rep 2002, 3, (5), 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Dahl R; Sergienko EA; Su Y; Mostofi YS; Yang L; Simao AM; Narisawa S; Brown B; Mangravita-Novo A; Vicchiarelli M Discovery and validation of a series of aryl sulfonamides as selective inhibitors of tissue-nonspecific alkaline phosphatase (TNAP). J. Med. Chem 2009, 52, (21), 6919–6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Müller A; Klöppel C; Smith-Valentine M; Van Houten J; Simon M Selective and programmed cleavage of GPI-anchored proteins from the surface membrane by phospholipase C. Biochim. Biophys. Acta, Biomembr 2012, 1818, (1), 117–124. [DOI] [PubMed] [Google Scholar]
- (45).Vasile E; Simionescu M; Simionescu N Visualization of the binding, endocytosis, and transcytosis of low-density lipoprotein in the arterial endothelium in situ. J. Cell Biol 1983, 96, (6), 1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Goldstein JL; Anderson R; Brown MS In Receptor-mediated endocytosis and the cellular uptake of low density lipoprotein, Ciba Foundation symposium, 1982; pp 77–95. [DOI] [PubMed] [Google Scholar]
- (47).Goldstein JL; Brown MS; Anderson RG; Russell DW; Schneider WJ Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annu. Rev. Cell. Biol 1985, 1, (1), 1–39. [DOI] [PubMed] [Google Scholar]
- (48).V’kovski P; Kratzel A; Steiner S; Stalder H; Thiel V Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol 2020, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lock LL; Reyes CD; Zhang P; Cui H Tuning cellular uptake of molecular probes by rational design of their assembly into supramolecular nanoprobes. J. Am. Chem. Soc 2016, 138, (10), 3533–3540. [DOI] [PubMed] [Google Scholar]
- (50).Nishiyama N Nanocarriers shape up for long life. Nat. Nanotechnol 2007, 2, (4), 203–204. [DOI] [PubMed] [Google Scholar]
- (51).Wang H; Feng Z; Xu B Instructed Assembly as Context‐Dependent Signaling for the Death and Morphogenesis of Cells. Angew. Chem. Int. Ed 2019, 58, (17), 5567–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Shang Y; Zhi D; Feng G; Wang Z; Mao D; Guo S; Liu R; Liu L; Zhang S; Sun S Supramolecular nanofibers with superior bioactivity to insulin-like growth factor-I. Nano Lett. 2019, 19, (3), 1560–1569. [DOI] [PubMed] [Google Scholar]
- (53).Mix KA; Lomax JE; Raines RT Cytosolic delivery of proteins by bioreversible esterification. J. Am. Chem. Soc 2017, 139, (41), 14396–14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Tanaka A; Fukuoka Y; Morimoto Y; Honjo T; Koda D; Goto M; Maruyama T Cancer cell death induced by the intracellular self-assembly of an enzyme-responsive supramolecular gelator. J. Am. Chem. Soc 2015, 137, (2), 770–775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


