Abstract
Background
Pediatric central nervous system (CNS) infections are potentially life-threatening and may incur significant morbidity. Identifying a pathogen is important, both in terms of guiding therapeutic management and in characterizing prognosis. Usual care testing by culture and polymerase chain reaction is often unable to identify a pathogen. We examined the systematic application of metagenomic next-generation sequencing (mNGS) for detecting organisms and transcriptomic analysis of cerebrospinal fluid (CSF) in children with central nervous system (CNS) infections.
Methods
We conducted a prospective multisite study that aimed to enroll all children with a CSF pleocytosis and suspected CNS infection admitted to 1 of 3 tertiary pediatric hospitals during the study timeframe. After usual care testing had been performed, the remaining CSF was sent for mNGS and transcriptomic analysis.
Results
We screened 221 and enrolled 70 subjects over a 12-month recruitment period. A putative organism was isolated from CSF in 25 (35.7%) subjects by any diagnostic modality. Metagenomic next-generation sequencing of the CSF samples identified a pathogen in 20 (28.6%) subjects, which were also all identified by usual care testing. The median time to result was 38 hours.
Conclusions
Metagenomic sequencing of CSF has the potential to rapidly identify pathogens in children with CNS infections.
Keywords: encephalitis, meningitis, metagenomics, next-generation sequencing, pediatric
We examined the utility of next-generation sequencing in comparison to usual care in detecting a pathogenic organism in children with central nervous system infections.
Pediatric central nervous system (CNS) infections are potentially life-threatening. Mortality and morbidity occur in up to 28% and 56% of patients, respectively [1, 2]. Central nervous system infections encompass a range of manifestations that vary based on pathogen [3]. Identifying a pathogen is vital, both in terms of guiding therapy and in characterizing prognosis. Culture and polymerase chain reaction (PCR) assays are frequently unable to identify a pathogen. For encephalitis, in more than 40% of patients, usual care testing is unable to identify a pathogen [3–5]. In bacterial meningitis, culture may be negative after antibiotic pretreatment, a common occurrence [2, 6]. Polymerase chain reaction offers rapid and sensitive testing, but it is restricted to the pathogens for which the platform is targeted. Given the limitations of available diagnostics, “Pan-omic” platforms offer promise [7, 8]. One such modality is metagenomic next-generation sequencing (mNGS) for pathogen deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) [9, 10]. This methodology poses the opportunity to broadly evaluate for pathogens by testing a single specimen. Turnaround times have shortened, making mNGS techniques more applicable as a method of achieving a timely diagnosis. We conducted a prospective, multisite, study to evaluate DNA sequencing (DNA-seq) and RNA sequencing (RNA-seq) of cerebrospinal fluid (CSF) for identification of pathogens in children with a suspected CNS infection.
METHODS
Trial Design and Oversight
Pediatric Infectious Disease Precision Medicine Using Sequencing Evaluation of CSF ([PIPSEC] ClinicalTrials.gov Identifier NCT03796546) was a prospective, multisite study to evaluate DNA and RNA sequencing of CSF for identification of pathogens in subjects who had undergone evaluation for a CNS infection. The members of the research team had final responsibility for the trial design and oversight.
Patient Consent Statement
Written consent was obtained from subjects or their guardian. The Western Institutional Review Board and the Children’s Hospital of Orange County Institutional Review Board provided human subject protection oversight.
Eligibility Criteria
Eligible subjects were less than 18 years of age undergoing evaluation for CNS infection and found to have a CSF pleocytosis. Suspicion for CNS infection was determined based on the clinical impression of the ordering provider. Cerebrospinal fluid pleocytosis was defined as a white blood cell count (WBC) >15 cells/µL with <5000 cells/µL red blood cells in the same sample [11]. A protocol deviation was approved for 2 subjects with 14 cells/µL WBCs in the CSF at the request of the study site principle investigator (PI).
Study Outcomes
The primary aim of this study was to investigate the diagnosis rate of mNGS for pathogen detection in CSF samples compared with usual care testing performed on CSF samples from the same subject [12]. Usual care testing consisted of all CSF diagnostic testing performed on the case in a CAP/CLIA certified laboratory (see Supplement Table 1). Usual care testing was done at the discretion of the clinical team and thus varied by subject. Data were extracted from the electronic medical record. Site PIs were additionally sent clinician surveys to assess clinical utility of mNGS results in relation to usual care. The treating physician, site PIs, and study PI then reviewed the CSF mNGS results in comparison to usual care testing from CSF and clinical presentation to determine the likelihood that the identified organism was indeed putative. Each mNGS result was identified as a true positive (TP), false positive (FP), true negative (TN), or false negative (FN) relative to usual care testing from CSF (composite of PCR, culture, and other CSF testing) as the reference standard. Determination of TP or TN was made when mNGS and usual care testing were concordant. In the event of discordant results, mNGS was compared with the reference standard composite of usual care testing. If mNGS identified an organism that was not identified by usual care testing, this was considered an FP. If mNGS failed to identify the organism identified by usual care testing, this was considered an FN. The P values for comparison of means were calculated using the Student t test.
Sequencing
Cerebrospinal fluid samples were collected from the laboratory after usual care testing had been performed. The minimum volume for mNGS testing was 0.5 mL. Samples for mNGS testing were stored at −80°C until they were shipped to the reference laboratory. Samples were shipped via express courier to achieve a less than 24-hour transit time. The CSF samples were tested with research use only next-generation shotgun DNA-seq and RNA-seq protocols; the resulting data were analyzed with the Explify Platform (IDbyDNA Inc., Salt Lake City, UT) as previously described [10, 13, 14]. The DNA (after host depletion) and RNA were extracted separately from residual samples. The DNA and RNA sequencing libraries were prepared with the Nextera DNA Flex Library Prep Kit (Illumina, San Diego, CA) and sequenced on a NextSeq550 instrument (Illumina) using mid-output kits. In this study, a range of 8–18 total libraries, including external controls, were sequenced per run for a median depth of 9.98 × 106 and 1.1 × 107 single-end, 150-base pair sequencing reads per sample-derived DNA and RNA library, respectively. Sequencing reads were adapter-trimmed and quality-filtered as part of the Explify analysis. The assay was quality controlled using internal control organisms spiked into each sample at the lysis step and the inclusion of 3 external controls (positive, negative, and blank) in each sample batch. Sample results were released after performance metrics for internal and external controls were evaluated for data quality and quantity. Sequencing data analysis was performed with the Explify platform. The RNA-seq and DNA-seq data were analyzed together, and the final result for a given sample was based on evidence from either.
Transcriptomic Analysis
Host transcriptomic counts were analyzed using Rosalind (OnRamp Bioinformatics, San Diego, CA). Sample transcript counts were generated with Kallisto (v.0.46.0) normalized by relative log expression using DESeq2 R library [15]. Heatmaps generation and clustering were performed using the Partitioning Around Medoids methods using the fpc R library [16]. Differential gene expression (DEG) was considered as a fold change of more than 1.5 and a false discovery rate cutoff of 0.05 [17]. Hypergeometric distribution was utilized to analyze pathway enrichment, gene ontology, domain structure, and other ontologies. The topGO R library was used to determine local similarities and dependencies between gene ontology terms to perform Elim pruning correction [18]. Several databases were utilized for enrichment analysis including REACTOME, HOMER, and molecular signatures database (MSigDB) [19–21]. Weighted correlation network analysis (WGCNA) was performed using filtered log2-transformed RNA-seq data [22]. This was applied to construct scale-free networks that specify gene modules [22]. To explore the modular structures of the coexpression network, the adjacency matrix was transformed into a topological overlap matrix [22]. The WGCNA parameters were as follows: minimum module size equal to 30 genes, tree-cut height set to 0.99. Significant modules were defined as those with a strong correlation (r > 0.4) and a P < .05. Interacting genes were visualized as a network using String version 11.0 (http://string-db.org). K-means clustering was used to determine gene clusters of interest and/or highly connected genes within this isolated M25 network [23]. The mNGS data were pseudo-aligned to the human genome (assembly hg38) using Kallisto to yield host response transcriptomic data and then filtered for ribosomal RNA reads before downstream analysis. The sequencing depth for mapped reads varied from 30 000 reads to 5 million reads (mean = 1.5 million).
RESULTS
We screened 221 subjects at 3 study sites from January 2019 to January 2020 and enrolled 70 subjects (Supplement Figure 1). See Table 1 for demographic information. The mNGS of the CSF samples identified a pathogen in 24 subjects. Twenty of these were deemed to be putative (TP) versus 25 by usual care testing of CSF (Table 2 and Table 3). Two subjects, one with acute disseminated encephalomyelitis and another with autoimmune encephalitis, received a noninfectious diagnosis. Nine subjects had enterovirus meningitis. Discordant results that the treating physician deemed to be FP results by mNGS were identified in 4 subjects. The organisms identified for these subjects included Cryptococcus spp, Staphylococcus warneri, Mucor circinelloides, and Streptococcus bovis. In these 4 cases, usual care testing, including gold standard CSF culture, did not identify an organism, and the treating clinician did not deem the mNGS result to be clinically relevant. In 5 subjects, CSF mNGS was deemed falsely negative with relation to usual care CSF testing. Two involved an infected ventriculoperitoneal shunt, one due to Staphylococcus epidermidis and the other to Staphylococcus aureus (both identified on culture). Upon chart review, the site of CSF sampling was not clear for either case (the sample may have been obtained proximally or distally from the shunt). Upon review of the mNGS data, both organisms were detected, but they did not meet prespecified thresholds for reporting. In the third case, PCR for Epstein-Barr virus (EBV) was positive from the CSF at just 151 IU/L. The subject improved without antiviral therapy. In the fourth case, CSF culture from a subject with chronic meningoencephalitis recovered a single colony of Cryptococcus neoformans and CSF cryptococcal antigen was positive. A prolonged antifungal course resulted in a full recovery. In the fifth case, human herpes virus 6 (HHV-6) was identified on a CSF multiplex PCR panel. This patient had full clinical recovery and antiviral therapy was not initiated. Of note, the HHV-6 target from the multiplex PCR panel was positive for 2 other subjects. In one subject, described above (Subject 2016), the CSF culture was also positive for S aureus and was accordingly adjudicated as an mNGS false negative. In the other subject (Subject 1004), a dedicated HHV-6 PCR was negative, and this subject was adjudicated as “concordant CSF mNGS and CSF usual care” and therefore an mNGS TN. Dedicated HHV-6 PCR was not sent for any other subjects.
Table 1.
Research Subject Demographic Characteristicsa
| Male Sex, n (%) | 40 (57.1) |
|---|---|
| Age (years), median (IQR) | 3.8 (0.2–11.8) |
| Rady Children’s Hospital San Diego, CA, n (%) | 31 (44.3) |
| Children’s Hopsital Orange County, CA, n (%) | 24 (34.3) |
| Nicklaus Children’s Hospital, FL, n (%) | 15 (21.4) |
| Immunocompromised, n (%) | 4 (5.7) |
| Hispanic ethnicity, n (%) | 41 (58.6) |
| Caucasian/White, n (%) | 46 (65.7) |
| African American/Black, n (%) | 2 (2.9) |
| Asian, n (%) | 2 (2.9) |
| Other, n (%) | 12 (28.6) |
| Presenting Symptoms | |
| Fever, n (%) | 51 (72.9) |
| Vomiting, n (%) | 28 (40.0) |
| Seizures, n (%) | 16 (22.9) |
| Lethargy, n (%) | 30 (42.9) |
| Altered Mental Status, n (%) | 26 (37.1) |
| VP shunt, n (%) | 6 (8.6) |
| Received antibitoics prior, n (%) | 28 (40.0) |
| CSF Parameters | |
| Nucleated cells cells/μL, median (IQR) | 109 (35.5–513.5) |
| Erythrocytes cells/μL, median (IQR) | 16 (6.0–368.0) |
| Protein mg/dL, median (IQR) | 97 (41.8–168.0) |
| Glucose mg/dL, median (IQR) | 47.5 (40.0–56.0) |
| Length of stay, median days (IQR) | 6 (3.0–18.5) |
| Admitted to ICU, n (%) | 37 (52.9) |
| Death, n (%) | 3 (4.3) |
| Total Patients | 70 |
Abbreviations: CSF, cerebrospinal fluid; ICU, intensive care unit; IQR, interquartile range; VP, ventriculoperitoneal.
aNOTE: Two unique tubes of residual CSF were received for 1027. Each was processed and analyzed separately by the reference laboratory; both samples produced the same result. The read counts listed in the table represent the mean sequencing reads for these 2 samples.
Table 2.
Comparison of Subjects for Whom CSF Testing Was Positive by Any Method
| Subject | CSF Culture | HSV PCR | EV PCR | CSF Multiplex PCR | CSF Other Test (Positive Result) | CSF mNGS | Total Sequencing Reads | Number of Reads for Identified Organism |
|---|---|---|---|---|---|---|---|---|
| Concordant CSF mNGS and CSF Usual Care Results | ||||||||
| 1001 | Haemophilus influenzae | negative | not done | not done | H influenzae | 2.85E + 07 | 5.17E + 04 | |
| 1003 | no organisms | positive | not done | HSV | HSV | 2.38E + 07 | 9.80E + 01 | |
| 1007 | Enterococcus faecalis | not done | not done | not done | E faecalis | 2.63E + 07 | 2.05E + 04 | |
| 1017 | Streptococcus agalactiae | not done | not done | S agalactiae | S agalactiae | 1.81E + 07 | 5.61E + 05 | |
| 1019 | no organisms | not done | positive | not done | Enterovirus | 1.54E + 07 | 2.26E + 02 | |
| 1020 | no organisms | negative | positive | Enterovirus | Enterovirus | 1.84E + 07 | 1.43E + 03 | |
| 1023 | no organisms | negative | positive | not done | Enterovirus | 2.27E + 07 | 3.34E + 02 | |
| 1025 | no organisms | not done | positive | Enterovirus | Enterovirus | 1.60E + 07 | 9.90E + 01 | |
| 1026 | no organisms | negative | positive | Enterovirus | Enterovirus | 1.72E + 07 | 8.70E + 01 | |
| 1027 | no organisms | negative | positive | not done | Enterovirus | 1.60E + 07 | 1.27E + 04 | |
| 1028 | no organisms | not done | positive | not done | Enterovirus | 1.67E + 07 | 1.30E + 01 | |
| 1030 | Streptococcus pneumoniae | negative | not done | S pneumoniae | S pneumoniae | 2.22E + 07 | 5.53E + 03 | |
| 2001 | S agalactiae | not done | not done | not done | S agalactiae | 2.26E + 07 | 1.58E + 05 | |
| 2005 | S pneumoniae | not done | negative | S pneumoniae | S pneumoniae | 3.17E + 07 | 5.73E + 05 | |
| 2006 | no organisms | not done | not done | Enterovirus | Enterovirus | 2.41E + 07 | 1.98E + 03 | |
| 3001 | Neisseria meningitidis | not done | not done | N meningitidis | N meningitidis | 2.11E + 07 | 3.71E + 02 | |
| 3002 | no organisms | not done | not done | Enterovirus | Enterovirus | 1.48E + 07 | 3.40E + 01 | |
| 3012 | no organisms | not done | not done | negative | 16S PCR Streptococcus dysgalactiae | S dysgalactiae | 2.11E + 07 | 8.29E + 05 |
| 3013 | S agalactiae | not done | not done | negative | S agalactiae | 1.16E + 07 | 8.44E + 02 | |
| 3015 | S pneumoniae | not done | not done | S pneumoniae | S pneumoniae | 1.34E + 07 | 1.19E + 05 | |
| False-Positive CSF mNGS Results | ||||||||
| 1009 | no organisms | not done | not done | not done | Cryptococcus spp | 2.25E + 07 | 5.54E + 02 | |
| 2003 | no organisms | not done | not done | not done | Staphylococcus warneri | 3.06E + 07 | 1.17E + 03 | |
| 2011 | no organisms | not done | not done | not done | Mucor circinelloides | 2.27E + 07 | 6.77E + 04 | |
| 2022 | no organisms | not done | not done | negative | Streptococcus bovis | 2.20E + 07 | 1.41E + 02 | |
| False-Negative CSF mNGS Results | ||||||||
| 1016 | Cryptococcus neoformans | not done | not done | negative | no pathogens detected | 2.37E+07 | - | |
| 2015 | no organisms | not done | not done | HHV-6 | no pathogens detected | 1.33E + 07 | - | |
| 2016 | Staphylococcus aureus | not done | not done | HHV-6 | no pathogens detected | 1.83E + 07 | - | |
| 3010 | no organisms | not done | not done | negative | EBV (PCR) | no pathogens detected | 1.85E + 07 | - |
| 3014 | Staphylococcus epidermidis | not done | not done | Negative | no pathogens detected | 1.25E + 07 | - |
Abbreviations: CSF, cerebrospinal fluid; EBV, Epstein-Barr virus; EV, enterovirus; HHV-6, human herpes virus 6; HSV, herpes simplex virus; mNGS, metagenomic next-generation sequencing; PCR, polymerase chain reaction.
Table 3.
Evaluation of a Metagenomic Assay for Detection of CSF Pathogens Against a Composite Reference of Usual Care Testinga
| CSF mNGS % Positivity | 28.6% |
|---|---|
| CSF mNGS positive predictive agreement | 80.0 (95%, 59.3–93.2%) |
| CSF mNGS negative predictive agreement | 91.1 (95%, 78.8–97.5%) |
| % agreement (kappa) | 87.1 (0.80) |
Abbreviations: CSF, cerebrospinal fluid; mNGS, metagenomic next-generation sequencing.
aEach mNGS result was identified as a true positive, false positive, true negative, or false negative relative to usual care testing from CSF (composite of polymerase chain reaction [PCR], culture, and serologic testing for each subject) as the reference standard. A culture was sent for every subject. Dedicated enterovirus PCR was sent for 19 subjects, and dedicated herpes simplex virus PCR was sent for 13 subjects. A multiplex PCR was sent for 52 subjects. A dedicated Epstein-Barr virus PCR was sent for 1 subject. Broad-range universal PCR was sent for 1 subject.
Cerebrospinal fluid culture identified a putative organism in 12 cases (17.1%). A CSF PCR multiplex panel was utilized for 51 subjects, 15 (29.4%) of which were positive. In one subject with a negative culture and multiplex PCR panel, 16S broad-range PCR and mNGS both detected Streptococcus dysgalactiae. Median time to result from the time sample was received for CSF mNGS was 38.0 hours (interquartile range [IQR], 32.6–60.5 hours). The mean CSF nucleated cells was higher in the subjects for whom a pathogen was detected by mNGS in comparison to subjects for whom no organism was detected (1499.6 vs 410.7 cells/µL, P = .05). Mean CSF protein value (228.3 vs 184.4 mg/dL, P = .02) and mean glucose level (38.2 vs 52.6 mg/dL, P = .004) were also both higher in subjects for whom an organism was detected by mNGS versus subjects for whom no organism was detected. Based on survey results, clinicians indicated that CSF mNGS helped in the management in 17.1% of cases. In 10 of these cases, the negative mNGS results were used to buttress the clinical impression that a treatable infectious etiology was not missed by CSF testing. In one case, the clinician explicitly stated that the mNGS result was used to provide clearance for surgical intervention. In another case, the clinician was reassured by concordance between the positive mNGS and usual care result both identifying the same viral pathogen.
Transcriptome Analysis
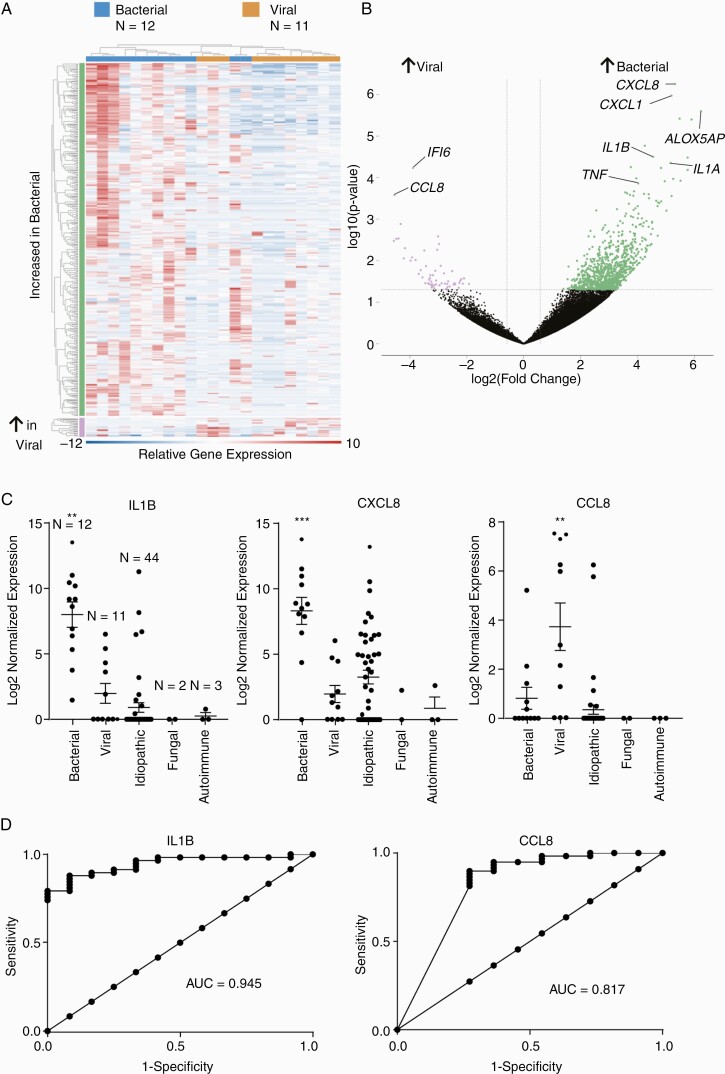
In addition to pathogen data, mNGS also generated incidental host transcriptomic data. Data were compared for all subjects using multidimensional scaling and did not globally cluster by diagnosis gender, age, CSF white blood cell count, or neutrophil predominance (Supplemental Figure 2A). A comparison of subjects diagnosed with viral (N = 12) compared with bacterial (N = 11) meningitis identified 409 differentially expressed genes (DEGs) (Figure 1) and overall clustered by infectious diagnosis. The DEGs included up-regulation of genes identified in host response to viral and bacterial infection, including interleukin 1A/B and interleukin 8 in bacterial meningitis. Gene ontology analysis identified pathways that included immune response and both leukocyte and neutrophil activation (Supplemental Figure 2). Interleukin 1B ([IL1B] P-adjusted value [p-adj] = .004) and CXCL8 (p-adj = .0002) were most associated with bacterial meningitis, and CCL8 (p-adj = .007) was most associated with viral meningitis compared with all other patient samples (Figure 1C). The WGCNA pathway analysis identified a statistically significant module of 68 coexpressed genes (Dark red module, Supplementary Figure 3). Quantitative trait analysis was not significant for any other patient features. Taken together, gene ontology and WGCNA analysis suggest that IL1B may be a key regulator of the proinflammatory response observed in bacterial meningitis (Supplement Figure 4). Diagnostic performance was assessed using receiver operator characteristic curves with a corresponding area under the curve of 0.95 (95% confidence interval, 0.89–0.998) for IL1B (Figure 1D). A subanalysis of the FN mNGS results suggested infection based on host response in 2 of these 4 patients.
Figure 1.
(A) A heat map comparing Pediatric Infectious Disease Precision Medicine Using Sequencing Evaluation of CSF (PIPSEC) subjects with bacterial and viral diagnoses identified 409 differentially expressed genes with false discovery rate >0.05 and fold change ≥1.5. (B) Volcano plot identifying differentially expressed genes between bacterial and viral meningitis. (C) Scatter plot depicting mean, standard error of the mean, and individual values. **P-adjusted value (p-adj) < .01, ***p-adj < .0001 compared with all other groups with a fold change of 10.0 for interleukin (IL)1B and 18.0 for CXCL8 in bacterial meningitis, and 14.0 for CCL8 in viral meningitis compared with all other samples. (D) Receiver operator characteristic (ROC) for IL1B and CCL8. AUC, area under curve.
Post Hoc Cost Analysis
The median cost of usual care testing was $531.09 (IQR, $439.00–$721.09). An average of 3 usual care CSF tests (including culture) were sent per subject. Per-sample direct cost of CSF mNGS laboratory testing is dependent on the number of samples being run at a time and is estimated to range between $390.00 and $2000.00. In this study, no more than 5 CSF samples were sequenced per run to expedite the turnaround time. Per-sample direct cost for shotgun sequencing can be decreased by an order of magnitude through optimization of the balance between analytical sensitivity, test volume, and turnaround time considerations.
DISCUSSION
We describe application of metagenomics in the diagnosis of pediatric CNS infections that included all consenting subjects meeting enrollment criteria rather than only a referred subset. As described in previous studies, an infectious etiology was not identified for most subjects with suspected CNS infection [5]. A putative organism was isolated from CSF in 25 (35.7%) subjects, and, of these, mNGS of the CSF samples identified a pathogen in 20 (28.6%) subjects (Table 3). Using the above-described adjudication scheme, FP results by mNGS were identified in 4 subjects, and, in these subjects, usual care testing did not identify a pathogen. In 5 cases, a putative organism was recovered by usual care testing of the CSF, but not by CSF mNGS. Two of these cases involved hardware infection, and, in another subject with chronic meningoencephalitis, a single colony of C neoformans was isolated from CSF. In the fourth subject, although EBV was detected by PCR, the viral load was just 151 IU/L. The fifth case was a positive result for HHV-6 identified by CSF PCR multiplex panel, which is of unclear clinical significance (previous studies have described FP results for this target) [24]. In these cases, burden of organism was likely low even before initiation of antimicrobial therapy, which may indicate that, for some organisms and in certain circumstances, culture and PCR remain more sensitive [8]. Upon review of the subjects with hardware infections (Subjects 2016 and 3014) deemed as mNGS FNs, a signal for S aureus and S epidermidis was detected but below the reporting threshold (Supplement Figures 5 and 6). Limitations in the application of this study, including imperfect sampling, may have further degraded the sensitivity of mNGS.
Cerebrospinal fluid sampling is important for management of CNS infections. Sensitivity of culture in bacterial meningitis is high, but it is diminished if CSF is obtained after initiation of antibiotics [6]. Culture is less sensitive for fungi and mycobacteria. Polymerase chain reaction may demonstrate high sensitivity for certain targets, but it requires clinicians to suspect the organism before testing, which may lead to missed diagnoses and to broad and potentially unnecessary testing. Metagenomic next-generation sequencing may be used to evaluate for the presence of many pathogens with a single test. Wilson et al [9] examined the use of mNGS in 204 subjects with suspected CNS infections. Among 58 subjects in whom an organism was identified, mNGS identified 13 (22%) that were not identified by usual care testing. A notable difference in this cohort from our study is the older study population, many of whom were immunocompromised. In addition, many of these subjects were recruited after an extensive evaluation, which increased the likelihood that mNGS would recover an uncommon pathogen where usual care had already failed to discover an etiology. The subjects recruited in our study were more often previously healthy, and therefore they were more likely to have infection with common pathogens. Metagenomic next-generation sequencing was additionally used as a first-tier test in conjunction with usual care. This decreased the likelihood that mNGS would detect an organism not identified by usual care testing. In a cohort more similar to our study, Hong et al [25] evaluated mNGS for CNS infections in a resource-limited setting, and they found that mNGS of the CSF identified a pathogen in 14 of 19 CSF samples that were positive by PCR. Using PCR as the reference assay, they calculated a sensitivity and specificity of 74% and 66%, respectively. Our data show that mNGS offers similar diagnostic yield, although to fully understand the limit of detection and the importance for clinical care, larger studies will be needed, especially as mNGS techniques involving enrichment and depletion technologies are refined. Metagenomic next-generation sequencing may be better utilized as an adjunct to current usual care testing, especially when usual care testing has already failed to recover a pathogen. In addition, although cost has been a limiting factor in the adoption of mNGS, if mNGS is ultimately shown to provide similar results to usual care in a shorter time at a similar price point, mNGS may prove to be more attractive [26]. Turnaround time can additionally be improved by local implementation of mNGS testing, a model that has become more common in tertiary care centers.
Transcriptome analysis was used to evaluate host expression of genes associated with the immunologic response. Subjects with bacterial meningitis were found to have a distinct pattern of gene expression compared with those with a viral etiology. It is notable that CXCL8, CXCL1, tumor necrosis factor (TNF), and interleukin 1A/B have been found to be expressed in bacterial meningitis [27–29]. In contrast, CCL8 has been found to be associated with viral infections [27, 30, 31]. The results of our analysis show increased expression of CXCL8, CXCL1, and TNF in the subjects with a bacterial infection and increased expression of both CCL8 and IFI6 in subjects with viral infection (Figure 1). The WGCNA independently identified interleukin 1B as a hub gene in the host response to bacterial meningitis, which suggests that it may regulate proinflammatory gene expression. Interleukin 1 genotype has been implicated as contributing to the risk of fatal meningococcal meningitis [32]. The clinical utility of these findings needs to be clarified with further studies, but, potentially, host response transcriptome analysis may prove useful in characterizing inflammatory response associated with specific pathogens and in attributing pathogenicity when an organism is identified [28, 33–37]. Furthermore, characterizing host response may additionally inform the differential diagnosis when an organism is not found.
This study has several limitations. The epidemiology of CNS infections may reflect geographic distribution, limiting generalizability. The number of enrollees is also likely too small to adequately capture rarer pathogens associated with CNS infections. Adjudication of results as putative or not with respect to a composite of usual care tests used to create a reference standard was hampered by the variability in the types of testing sent. Although every subject had CSF culture sent, the choice to use serologic- or molecular-based assays was purely at the discretion of the clinical team, and the validity of comparing mNGS results to a standard reference that is not uniform throughout the cohort is limited. In addition, characterization of mNGS results as FP are based on published epidemiology of CNS infection in children, which are biased toward historic norms and limited by the sensitivity of usual care testing modalities (including CSF culture). Furthermore, we were unable to utilize adjunctive orthogonal testing such as targeted or broad-range PCR due to unavailability of additional CSF sample. False-positve mNGS results may be attributable to sample misidentification, contamination introduced during sample collection or processing, biases within reference databases, or the analysis tools used. For the determination of a TN specifically, by using the criteria of no organism detected by any testing method, we may have incorrectly adjudicated subjects with organisms that were either in low abundance or difficult to isolate. In addition, subjects with suspected infection may instead have autoinflammatory or autoimmune phenomenon that may be challenging to differentiate from an infection [3, 9, 38, 39]. Cerebrospinal fluid parameters vary based on age, and the low cutoff used in the inclusion criteria may have allowed for enrollment of some subjects with physiologically normal CSF [11, 40]. In contrast, more indolent CNS infections may not generate a significant CSF pleocytosis. Finally, the protocol stipulated that only residual CSF be used for mNGS, which likely limited diagnostic yield, especially in cases in which burden of organism was lower [8].
CONCLUSIONS
In this prospective study of pediatric CNS infections, mNGS of the CSF identified an organism in 28.6% of subjects. Although certain usual care tests remain more sensitive for some targets, mNGS may have value as an adjunctive diagnostic tool, especially in situations in which standard testing is known to have limited yield. Furthermore, this is specific to the sequencing and analysis methods used in this study, because sensitivity and specificity of mNGS vary depending on choice of wet laboratory methodology and analysis tools. Further studies are required to clarify the best use of mNGS in the evaluation of pediatric CNS infections.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We acknowledge the contributions of Drs. Magaly Diaz-Barbosa and Fabian Castillo from Nicklaus Children’s Hospital in Miami and Drs. Stephen Kingsmore and Charlotte Hobbs of Rady Children’s Institute for Genomic Medicine in support of this study.
Financial support. This work was funded by IDbyDNA (Salt Lake City, UT) (Grant NCT03796546).
Potential conflicts of interest. R. Sc. is the Chief Medical Officer of IDbyDNA. L. F. is the Director of Medical and Scientific Affairs at IDbyDNA. B. B. is the Associate Director of Medical and Scientific Affairs at IDbyDNA. R. St. is the Associate Director of Clinical Studies at IDbyDNA. T. S. is a scientist with IDbyDNA’s Medical and Scientific Affairs Department. H. X. is the Associate Director of Laboratory Technology at IDbyDNA. IDbyDNA sponsored this study. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Okike IO, Ladhani SN, Johnson AP, et al. ; neoMen Study Group . Clinical characteristics and risk factors for poor outcome in infants less than 90 days of age with bacterial meningitis in the United Kingdom and Ireland. Pediatr Infect Dis J 2018; 37:837–43. [DOI] [PubMed] [Google Scholar]
- 2. Tunkel AR, Hartman BJ, Kaplan SL, et al. . Practice guidelines for the management of bacterial meningitis. Clin Infect Dis 2004; 39:1267–84. [DOI] [PubMed] [Google Scholar]
- 3. Britton PN, Dale RC, Blyth CC, et al. . Causes and clinical features of childhood encephalitis: a multicenter, prospective cohort study. Clin Infect Dis 2020; 70:2517–26. [DOI] [PubMed] [Google Scholar]
- 4. Tunkel AR, Glaser CA, Bloch KC, et al. ; Infectious Diseases Society of America . The management of encephalitis: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis 2008; 47:303–27. [DOI] [PubMed] [Google Scholar]
- 5. Erickson TA, Muscal E, Munoz FM, et al. . Infectious and autoimmune causes of encephalitis in children. Pediatrics 2020; 145:e20192543. [DOI] [PubMed] [Google Scholar]
- 6. Kanegaye JT, Soliemanzadeh P, Bradley JS. Lumbar puncture in pediatric bacterial meningitis: defining the time interval for recovery of cerebrospinal fluid pathogens after parenteral antibiotic pretreatment. Pediatrics 2001; 108:1169–74. [PubMed] [Google Scholar]
- 7. He T, Kaplan S, Kamboj M, Tang YW. Laboratory diagnosis of central nervous system infection. Curr Infect Dis Rep 2016; 18:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramachandran PS, Wilson MR. Metagenomics for neurological infections — expanding our imagination. Nat Rev Neurol 2020; 16:547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilson MR, Sample HA, Zorn KC, et al. . Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med 2019; 380:2327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schlaberg R, Chiu CY, Miller S, et al. ; Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology; Microbiology Resource Committee of the College of American Pathologists . Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med 2017; 141:776–86. [DOI] [PubMed] [Google Scholar]
- 11. Thomson J, Sucharew H, Cruz AT, et al. . Cerebrospinal fluid reference values for young infants undergoing lumbar puncture. Pediatrics 2018; 141:e20173405. [DOI] [PubMed] [Google Scholar]
- 12. Thompson BT, Schoenfeld D. Usual care as the control group in clinical trials of nonpharmacologic interventions. Proc Am Thorac Soc 2007; 4:577–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schlaberg R, Queen K, Simmon K, et al. . Viral pathogen detection by metagenomics and pan-viral group polymerase chain reaction in children with pneumonia lacking identifiable etiology. J Infect Dis 2017; 215:1407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graf EH, Simmon KE, Tardif KD, et al. . Unbiased detection of respiratory viruses by use of RNA sequencing-based metagenomics: a systematic comparison to a commercial PCR panel. J Clin Microbiol 2016; 54:1000–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hennig C. fpc: Flexible Procedures for Clustering. Available at: https://CRAN.R-project.org/package=fpc. Accessed 18 August 2020.
- 17. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 1995; 57:289–300. [Google Scholar]
- 18. Alexa A, Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. Bioconductor version: Release (3.11). Available at: https://bioconductor.org/packages/topGO/. Accessed 18 August 2020.
- 19. Liberzon A, Subramanian A, Pinchback R, et al. . Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011; 27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fabregat A, Jupe S, Matthews L, et al. . The Reactome Pathway Knowledgebase. Nucleic Acids Res 2018; 46:D649–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heinz S, Benner C, Spann N, et al. . Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005; 4:Article17. [DOI] [PubMed] [Google Scholar]
- 23. Brohée S, van Helden J. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinformatics 2006; 7:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Green DA, Pereira M, Miko B, et al. . Clinical significance of human herpesvirus 6 positivity on the FilmArray meningitis/encephalitis panel. Clin Infect Dis 2018; 67:1125–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hong NTT, Anh NT, Mai NTH, et al. . Performance of metagenomic next-generation sequencing for the diagnosis of viral meningoencephalitis in a resource-limited setting. Open Forum Infect Dis 2020; 7:ofaa046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haston JC, Rostad CA, Jerris RC, et al. . Prospective cohort study of next-generation sequencing as a diagnostic modality for unexplained encephalitis in children. J Pediatric Infect Dis Soc 2020; 9:326–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lepennetier G, Hracsko Z, Unger M, et al. . Cytokine and immune cell profiling in the cerebrospinal fluid of patients with neuro-inflammatory diseases. J Neuroinflammation 2019; 16:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coimbra RS, Voisin V, de Saizieu AB, et al. . Gene expression in cortex and hippocampus during acute pneumococcal meningitis. BMC Biol 2006; 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coutinho LG, Grandgirard D, Leib SL, Agnez-Lima LF. Cerebrospinal-fluid cytokine and chemokine profile in patients with pneumococcal and meningococcal meningitis. BMC Infect Dis 2013; 13:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kastenbauer S, Angele B, Sporer B, et al. . Patterns of protein expression in infectious meningitis: a cerebrospinal fluid protein array analysis. J Neuroimmunol 2005; 164:134–9. [DOI] [PubMed] [Google Scholar]
- 31. Bastos MS, Coelho-dos-Reis JG, Zauli DAG, et al. . Divergent cerebrospinal fluid cytokine network induced by non-viral and different viral infections on the central nervous system. BMC Infect Dis 2015; 15:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Read RC, Camp NJ, di Giovine FS, et al. . An interleukin-1 genotype is associated with fatal outcome of meningococcal disease. J Infect Dis 2000; 182:1557–60. [DOI] [PubMed] [Google Scholar]
- 33. Wittwer M, Grandgirard D, Rohrbach J, Leib SL. Tracking the transcriptional host response from the acute to the regenerative phase of experimental pneumococcal meningitis. BMC Infect Dis 2010; 10:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee KT, Hong J, Lee DG, et al. . Fungal kinases and transcription factors regulating brain infection in Cryptococcus neoformans. Nat Commun 2020; 11:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Casselli T, Qureshi H, Peterson E, et al. . MicroRNA and mRNA transcriptome profiling in primary human astrocytes infected with Borrelia burgdorferi. PLoS One 2017; 12:e0170961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leong WF, Chow VT. Transcriptomic and proteomic analyses of rhabdomyosarcoma cells reveal differential cellular gene expression in response to enterovirus 71 infection. Cell Microbiol 2006; 8:565–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Belogurov AA Jr, Ivanova OM, Lomakin YA, et al. . Mediators and biomarkers of inflammation in meningitis: cytokine and peptidome profiling of cerebrospinal fluid. Biochemistry (Mosc) 2016; 81:1293–302. [DOI] [PubMed] [Google Scholar]
- 38. Granerod J, Ambrose HE, Davies NW, et al. ; UK Health Protection Agency (HPA) Aetiology of Encephalitis Study Group. Causes of encephalitis and differences in their clinical presentations in England: a multicentre, population-based prospective study. Lancet Infect Dis 2010; 10:835–44. [DOI] [PubMed] [Google Scholar]
- 39. Pillai SC, Hacohen Y, Tantsis E, et al. . Infectious and autoantibody-associated encephalitis: clinical features and long-term outcome. Pediatrics 2015; 135:e974–84. [DOI] [PubMed] [Google Scholar]
- 40. Fleischer E, Neuman MI, Wang ME, et al. ; Febrile Young Infant Research Collaborative. Cerebrospinal fluid profiles of infants ≤60 days of age with bacterial meningitis. Hosp Pediatr 2019; 9:979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.