ABSTRACT
On and within most sites across an animal's body live complex communities of microorganisms. These microorganisms perform a variety of important functions for their hosts, including communicating with the brain, immune system and endocrine axes to mediate physiological processes and affect individual behaviour. Microbiome research has primarily focused on the functions of the microbiome within the gastrointestinal tract (gut microbiome) using biomedically relevant laboratory species (i.e. model organisms). These studies have identified important connections between the gut microbiome and host immune, neuroendocrine and nervous systems, as well as how these connections, in turn, influence host behaviour and health. Recently, the field has expanded beyond traditional model systems as it has become apparent that the microbiome can drive differences in behaviour and diet, play a fundamental role in host fitness and influence community-scale dynamics in wild populations. In this Review, we highlight the value of conducting hypothesis-driven research in non-model organisms and the benefits of a comparative approach that assesses patterns across different species or taxa. Using social behaviour as an intellectual framework, we review the bidirectional relationship between the gut microbiome and host behaviour, and identify understudied mechanisms by which these effects may be mediated.
KEY WORDS: Gut microbiome, Social behaviour, Physiological mechanisms, Neuroendocrine, Host–microbiome, Free-living animals
Summary: We highlight current and emerging questions about the role of the microbiome in mediating social behaviour and discuss novel mechanisms that may mediate these effects. Non-model organisms are well suited to address these questions.
Introduction: a primer on the microbiome
Within an organism exists a community of living microorganisms that includes commensal, symbiotic and pathogenic bacteria, fungi and archaea, and is termed the microbiota. Their genes and the molecules produced by the microorganisms (e.g. structural elements, metabolites, phages, viruses) are collectively called the microbiome. For most animals, the microbiome plays a critical role in survival (Møller et al., 2009; Williams et al., 2020). Aiding in nutrient acquisition and digestion (Hooper and Gordon, 2001), influencing development (Diaz Heijtz et al., 2011; Erny et al., 2015), affecting host immune system function (Chung et al., 2012; Sylvia and Demas, 2018) and modulating behaviours (Dinan et al., 2015; Sharon et al., 2010; Brucker and Bordenstein, 2013), the microbiome connects many physiological systems (e.g. neuroendocrine, immune and central nervous systems; Garcia-Reyero, 2018; Sylvia and Demas, 2018), and directly affects host fitness (Suzuki, 2017). Because most of the microbiome is within the gastrointestinal tract (i.e. gut; Wallace et al., 2011; Williams et al., 2020), the current Review will focus primarily on the gut microbiome. Investigations into the function of the microbiome associated with other body sites (e.g. skin, vagina, oral cavity) are needed, as these may also affect host behaviour (Rojas et al., 2020; Box 1).
Glossary.
Altruism
Behaviour that benefits another individual that is not closely related to the individual performing the behaviour and is detrimental to the individual performing the behaviour (Trivers, 1971).
Catecholamines
Monoamine neurotransmitters such as dopamine, adrenaline and noradrenaline.
Cooperation/cooperative behaviour
Behaviour that occurs when individuals act in ways that increase the fitness of others at some cost to themselves (Nowak, 2006).
Cooperative breeding
Breeding system in which subordinates (i.e. ‘helpers’) capable of reproducing forgo their own reproduction to assist raising offspring produced by breeders (Cockburn, 1998).
Cytokines
Hormone-like molecules (e.g. interleukins, interferon) produced by the immune system, either in the periphery or in the central nervous system.
Enterochromaffin cells
Endocrine cells lining the gastrointestinal tract.
Germ-free (GF) mice
Mice that are microbiota deficient from birth and are maintained in a microbiota-free environment.
Gnotobiotic mice
Mice born and maintained under aseptic conditions and in which only specific known strains of microbes are present, often introduced by the researcher.
Gut–brain axis
The bidirectional communication between the brain and the gastrointestinal tract.
Kin selection
The process by which traits are favoured because of their beneficial effects on relatives' survival; relatives may be either offspring (direct fitness) or non-descendant offspring (indirect fitness) (Griffin and West, 2002).
Lipopolysaccharide (LPS)
A carbohydrate moiety present on the surface of Gram-negative bacteria, administration of which may be used to induce a sickness response in the absence of infection.
Non-model organisms
Wild or non-captive species typically not used for biomedical research.
Peyer's patches
Lymphatic tissue found in the small intestine.
Probiotics
Live microbial organisms supplemented to an individual that may increase the relative abundance of particular microbial strains.
Pyrogen
A fever-causing agent, typically produced by a bacterium.
Reciprocity
A theory used to explain the evolution of cooperation that highlights the importance of repeating interactions and the benefits incurred by the cooperating individual (Trivers, 1971).
Recolonization
Re-establishing either parts of or the complete gut microbiome via methods such as faecal transfer or probiotic administration; this manipulation typically occurs in GF mice or individuals whose gut microbiome has been depleted.
Specific pathogen-free (SPF) mice
Mice raised with a normal functional microbiota, but without a specific list of pathogens (e.g. Mycoplasma pulmonis) known to affect the health of mice.
Traditional model systems
Defined here as the laboratory rats and mice most commonly used for biomedically relevant research.
Box 1. Beyond the gut.
Animal bodies contain microbial communities that can affect host physiology, behaviour and fitness (Archie and Theis, 2011; Archie and Tung, 2015; Vuong et al., 2017). Within individuals, the microbiota is partitioned across different body sites, and the composition of microbiota differs at different sites (Hyde et al., 2016; Xavier et al., 2019). Like the gut microbiome, these other microbiomes are influenced by host physiology and external factors, and play a critical role in mediating social behaviour.
The skin is the primary physical barrier between vertebrates and their environment, and the skin microbiome can influence development, social behaviour and host health. Amphibian and reptile skin microbiomes are an important component of the host's defences against pathogens (Varga et al., 2018; Hyde et al., 2016), and manipulations of the skin microbiome of these species may help prevent population declines (Harris et al., 2009; Kueneman et al., 2016). The skin microbiome is often affected by changes in the environment (Douglas et al., 2021; Hyde et al., 2016), which could have important consequences for host health and physiological development. In contrast, the skin microbiome of some avian species is unrelated to their shared environment and diet, indicating individual-specific skin microbiomes in these species (Engel et al., 2018). Host- and individual-specific microbial communities may communicate critical information about the host's phenotype and genotype, and even promote individual recognition (Rojas et al., 2020). The skin microbiome can change as a result of changes in host behaviour. For example, Pratte et al. (2018) showed that the mucus of clownfish (Amphiprion clarkii), which display a mutualistic relationship with anemones (Entacmaea quadricolor), changes as a result of associations with the anemone. This change may be ecologically important to both the fish–microbiome and the fish–anemone interactions, specifically helping to sustain the mutualistic relationship.
Another microbiome site that has been investigated is the cloaca. In birds, reptiles, amphibians and some mammals, the cloaca is the end of both the digestive and reproductive tracts, meaning cloacal microbial communities are impacted by both diet and reproductive behaviour (Kulkarni and Heeb, 2007; Maul et al., 2005). The relative influence of these two factors on cloacal microbiome diversity requires further study, as does the effect of the cloacal microbiome on host health, physiology and reproductive behaviour, as well as the offspring microbiome and development. The eggshell microbiome may be affected by the maternal cloacal microbiome or the nesting environment, and this could contribute to the effects on offspring. For example, in woodlarks (Lullula arborea) and skylarks (Alauda arvensis), eggshell microbiomes are influenced by body feathers and female brood patch skin, but not by the maternal gut microbiome (van Veelen et al., 2018). Whether the cloacal microbiome influences the egg microbiome and how this affects offspring colonization requires further study.
In this Review, we highlight how research on traditional model systems (see Glossary; e.g. mice, rats) has laid the foundation for our knowledge of how the gut microbiome interacts with physiological systems to affect behaviour. We argue that the study of the influence of the microbiome on behaviour would benefit from a broader representation of host species that include non-model organisms (see Glossary). Using social behaviours as a framework, we describe how the study of the effects of the microbiome on behaviour can benefit from a comparative approach evaluating host–microbiome relationships across different species to identify patterns or unique features. We discuss how the gut microbiome can affect social behaviour in the context of development, reproduction, parental care and cooperation. Lastly, we identify novel mechanisms and environmental factors that could influence the relationship between the microbiome and social behaviour.
Model systems: laying the foundation
Model systems have provided us with a fundamental understanding of how the gut–brain axis (see Glossary) interacts with the body's physiological systems (e.g. neural, endocrine and immune systems) and affects behaviour (Collins and Bercik, 2014; Cryan and O'Mahony, 2011). Much of our knowledge of the connections between the gut microbiome and behaviour comes from studies using germ-free (GF) mice (see Glossary). Supplementing GF mice with commensal or pathogenic bacteria reveals how the microbes affect host behaviour (Sylvia and Demas, 2018). Using other model systems [e.g. specific pathogen-free (SPF) mice; see Glossary] has shown how manipulations of the host microbiome can affect behaviour. For example, antibiotics, which are used to manipulate the gut microbiome, can have sex-specific effects on the caecal microbiome and exploratory behaviour (Bercik et al., 2011). Additional studies using model systems have revealed sex-specific effects of the parental microbiome on offspring behaviour. For example, antibiotic-induced alterations of the maternal gut microbiome produce long-lasting changes in locomotor activity and exploratory behaviour in male offspring (Tochitani et al., 2016). Studies manipulating parental diet also provide evidence that the parental microbiome may play a role in offspring immune function and behaviour (Sylvia and Demas, 2018). For example, parental diets high in omega-3 fatty acids alter the microbiome of offspring and can affect offspring development (Myles et al., 2014), which could result in differences in offspring behaviour. Furthermore, females transplanted with high-fat diet microbiota produce pups that vocalize less when separated from their mother and male offspring that differ in exploratory, cognitive and compulsive behaviour (Bruce-Keller et al., 2017).
Studying the microbiome
Why consider non-model organisms?
Research using model organisms has demonstrated ways in which the microbiome can modulate behaviour. However, this work provides only a partial view of the role of the microbiome in natural functions because traditional model organisms are often not exposed to natural environmental conditions or lack variation in traits typically observed in wild populations. For example, a complete absence of microbiota, as in GF mice, is not found in wild populations, and GF mice display a variety of abnormalities associated with the lack of a microbiome (e.g. in the development of the gut, immune system and brain; Martin et al., 2016). Additionally, diet, social interactions (or the lack thereof) and other environmental conditions (Hird, 2017) associated with captivity alter the microbiome. For example, captive animals tend to have gut microbiomes that are depleted or imbalanced compared with those of their wild counterparts (Clayton et al., 2016; Kohl et al., 2017; McKenzie et al., 2017) as a result of standard diets (Clayton et al., 2018; Reichelt et al., 2020). Likewise, differences in temperature, light, season (e.g. Ren et al., 2020) or habitat (e.g. Watson et al., 2019; Loo et al., 2019; Davidson et al., 2020; San Juan et al., 2020), which are often minimized in captive studies (e.g. Arentsen et al., 2015; Neufeld et al., 2011), can have important effects on the microbiome (Fig. 1; e.g. Bailey et al., 2010) and behaviour (e.g. foraging innovation; Davidson et al., 2020). Further, the social organization of organisms in the lab often differs from that observed in wild populations, which limits the ability to study the relationship between social interactions and the host microbiome (Archie and Tung, 2015; Soares et al., 2019). The relationship between the gut microbiome and behaviour is bidirectional – behaviour can be influenced by the composition and diversity of the host's microbiome, and in turn, an organism's behaviour can alter their own and other individuals' bacterial communities (Archie and Tung, 2015; Münger et al., 2018) – and is shaped by early development (e.g. parental care, interactions among siblings) and continues into adulthood. Therefore, to further investigate (1) this bidirectional relationship, (2) the role of the gut microbiome in host evolution (Sylvia and Demas, 2018), and (3) population and community dynamics (Corl et al., 2020), the field must broaden to include non-model populations.
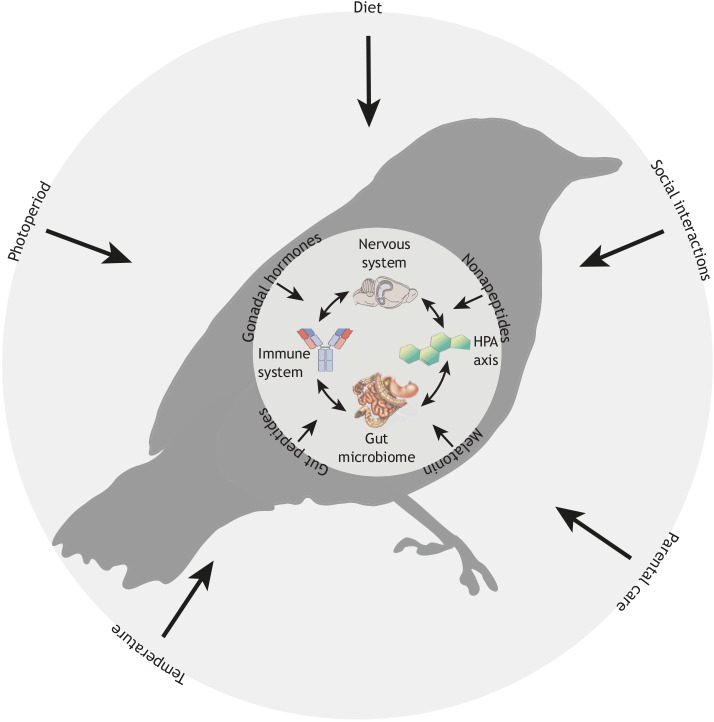
Fig. 1.
Graphic model identifying potential interactions among environmental factors and physiological systems that can alter the gut microbiome and thus affect behaviour. The inner circle represents the ‘core’ physiological systems regulating microbiome–behaviour interactions, including immune, neural and endocrine factors (HPA axis, hypothalamic–pituitary–adrenal axis), as well as the direct influence of the gut microbiota itself (i.e. metabolic by-products). Our hypothetical bird is nested within a larger environmental context (represented by the outer circle). Around the outer circle, we show examples of environmental variables – ranging from behavioural interactions with conspecifics to the physical environment – that might affect the microbiome–behaviour interaction. Around the perimeter of the inner circle are additional physiological variables that are less often considered but that might play an important role in modulating the effects of environmental variables on the core physiological systems. In conjunction with environmental factors, these molecules serve as ‘environmental input signals’ to the organism. These signals include (but are not limited to) seasonal/photoperiodic information (via melatonin and thyroid hormones), ambient temperature (via thyroid hormones and gonadal steroids), diet and food intake (via gut peptides such as leptin, insulin and ghrelin) and social interactions (via nonapeptides such as oxytocin and vasopressin, and other hormones).
Lessons from non-model organisms
Experiments on non-model populations with naturally occurring microbiomes provide the natural variation needed to assess the complexity of host–microbiome relationships (Brooks et al., 2016; Kohl et al., 2016) under the conditions in which they evolved. In addition, using non-model systems allows for comparative studies across different species or taxa, which provide important opportunities to assess the evolution of the host–microbiome relationship, determine large-scale patterns to explain development and behaviour, and identify unique aspects of the hosts that result in unique functions of the microbiome. Convergence in host phenotypes can provide natural experiments, allowing comparisons between species that share similar traits, but do not share a phylogenetic history. This may reveal important roles of the host microbiome (Box 2).
Box 2. Comparative studies reveal microbiome function.
Convergence in host phenotypes provides natural experiments by which comparisons between species may reveal important roles of the host microbiome. Specialized diets (Song et al., 2019) and the ability to fly (Song et al., 2020) have evolved in multiple lineages across vertebrates, and the microbiome composition reflects these unique adaptations. Comparative studies assessing birds and bats reveal evidence for microbiome convergence (Song et al., 2019, 2020) as a result of specialized diets and flight that make them more similar to each other compared with more closely related species. For example, birds and bats with a haematophagic diet have microbiome compositions that are more similar to each other than to those of non-haematophagic avian and mammalian species (Song et al., 2019). Haematophagy presents many unique nutritional challenges: blood is high in proteins, lipids and salt, but depleted of essential nutrients such as B vitamins. Digestion of blood can release toxic levels of iron and urea. These differences in diet correlate with differences in the microbial taxa found in haematophagic species. For example, Enterobacteriaceae, Helicobacteraceae and Petostreptococaceae are present in haematophagic bat species, but absent in samples from non-haematophagic bat species (Song et al., 2019). Additional evidence of convergent phylogenetic properties among vampire bats and birds is that high-level properties of the overall gut community, especially properties related to function, are similar, as both have a high abundance of the taxon Peptostreptococcaceae (Song et al., 2019).
Similarly, Song et al. (2020) revealed an association between flight and loss of microbiome specificity, even among distant species. Bat microbiomes are more closely related to those of birds than they are to the microbiomes of other mammals (Song et al., 2020). Bats have a lower proportion of Bacteroidetes and a higher proportion of Proteobacteria, which is highly associated with birds and flight. Interestingly, despite similarities between the microbiomes of birds and bats, the strongest association between these two groups is their lack of association with a specific microbiome. Characteristics associated with flight, including shorter intestinal lengths and content retention times, and differences in metabolic demands (Song et al., 2020), may explain the lack of microbiome specificity in these organisms.
To date, approximately 90% of published studies on the host–microbiome relationship are in mammals (Colston and Jackson, 2016), and many are conducted on lab populations. Although these studies have provided many important insights, mammals differ from other taxa in ways that likely influence the host–microbiome relationship (e.g. Hird, 2017; Greene et al., 2019; Song et al., 2020; Sullam et al., 2012). The composition and function of the gut microbiome in other taxa (e.g. birds, fish, amphibians and reptiles) remains understudied (Hird, 2017; Williams et al., 2020). Below, we highlight the qualities of non-mammalian species and the benefits of studying non-model taxa for investigating the interaction between the microbiome and social behaviour.
Birds
Birds represent an excellent system in which to test hypotheses about microbiome effects on behaviour. Avian reproductive biology enables the investigation of the importance of the gut microbiome in reproduction (e.g. does the offspring microbiome differ in altricial nestlings versus precocial nestlings), offspring care [e.g. brood parasitism: Robert and Sorci, 2000; cooperative breeding (see Glossary): Cockburn, 1998] and sociality (e.g. solitary to communal living; Downing et al., 2020). Songbirds also represent an ideal system in which to test how the microbiome interacts with neural systems to affect behaviour. The neural system regulating song learning in birds is a model for studying the neural mechanisms of learning and cognitive plasticity (Brenowitz and Beecher, 2005). The gut microbiome can affect cognitive performance and social learning (e.g. Davidson et al., 2018), but whether this plays a role in avian species is unknown. Variation in song learning and production affect important aspects of social behaviour including mate choice (Nowicki and Searcy, 2005), species recognition (e.g. Soha and Marler, 2000) and the ability to recognize heterospecific alarm calls (Magrath et al., 2015). To date, we know very little about how the microbiome influences these behaviours in avian hosts (Hird, 2017).
Fishes
Teleost fishes provide unique opportunities to advance our understanding of the links between sociality and the microbiome. Fish display complex social behaviours [e.g. deception: Bshary et al., 2002; cooperation (see Glossary): Balshine-Earn et al., 1998] and can live in complex social networks (e.g. Taborsky, 1994). Teleost fish are particularly promising for the study of mating behaviour because they display a variety of mating patterns, as well as parental care (Swenson, 1997). There are also well-established model fish systems that can be studied in the wild and – when brought into the lab – display behaviours comparable to those of wild populations. For example, the zebrafish is a model system in developmental biology for which many behavioural, physiological and molecular tools have been specifically developed. Studies in zebrafish have identified a role for sociality in horizontal bacterial transmission, suggesting that social interactions affect microbial composition (Burns et al., 2017). Furthermore, zebrafish reared in the laboratory may have a gut microbiome comparable to that of animals captured in the wild (Roeselers et al., 2011), which may make this a preferred captive system in which to test the importance of natural environmental factors.
Fish also provide a unique opportunity to investigate sex differences in the composition and diversity of the microbiome and how the microbiome can influence behaviour in sex-specific ways. This is because many fish species are sequential hermaphrodites, beginning life as one sex and changing to the other sex (Gemmell et al., 2019). Sequential hermaphrodites include species capable of transitioning from male to female, female to male, or bidirectionally. These sex changes involve coordinated transformations of many biological systems and are usually cued by changes in social structure, or attainment of age or size (Gemmell et al., 2019) – factors that are also associated with differences in host microbiome composition and diversity (e.g. Rojas et al., 2020; Whittaker et al., 2016). Males and females of the same species often differ in the composition or diversity of their microbiome, which is often attributed to differences in sex hormones (Zha et al., 2018; Chen et al., 2018). Whether the microbiome changes as individuals transition has yet to be explored. Therefore, assessing whether the gut microbiome changes in conjunction with sex changes, whether the microbiome facilitates or triggers those changes, or whether shared underlying physiological mechanisms play a role are important areas for future study.
Amphibians
Amphibians provide an excellent opportunity to examine the importance of critical developmental windows on the gut microbiome–behaviour relationship. Perturbations to the microbiome during ‘sensitive’ phases of development can have long-term consequences on behaviour, and the microbiome may mediate such effects (Sachser et al., 2020). Changes in the microbiota during critical developmental windows can also alter the trajectory of the gut microbiome and have long-term consequences on fitness-related traits (Warne et al., 2019). In amphibians, hatching represents a critical developmental window for gut microbiome colonization (e.g. Warne et al., 2017); metamorphosis, another critical developmental stage, is associated with complex changes in an individual's morphology, physiology, behaviour and composition of the gut microbiome (Chai et al., 2018). For example, during pre-metamorphosis, changes occur in the stress neuropeptide corticotropin releasing factor and the hormone leptin (Denver, 2019), both of which are important in bidirectional gut–brain communication (Holzer, 2016; Rajala et al., 2014). How these factors interact with the gut microbiome during metamorphosis to affect adult behaviour is relatively unknown.
Host microbiome and animal social behaviour
The gut microbiome can both influence and be altered by an individual's behaviour (Archie and Tung, 2015; Münger et al., 2018). Investigating the bidirectional relationship between the gut microbiome and social behaviour is of particular interest given the range and complexity of social interactions, which can include physically distant interactions, repeated contact between social group members and reproductive affiliations (Soares et al., 2019; Tung et al., 2015; Grieneisen et al., 2017; Antwis et al., 2018). Here, we focus on the bidirectional relationship between the gut microbiome and social behaviour in the context of reproduction, development, parental care and cooperation.
Reproduction
Sexual interactions can be directly and indirectly influenced by the composition of the gut microbiome. Recent work in Drosophila melanogaster fruit flies has identified an important role of gut microbes in determining assortative mating. Individuals prefer to mate with partners raised on a diet similar to their own, a preference driven by direct effects of the gut microbiome on fly pheromones, which in turn influence mating behaviour (Leftwich et al., 2018). Additionally, the gut microbiome may have sex-specific effects on the duration of mating behaviour and offspring production (Morimoto et al., 2017). The gut microbiome may also affect mating behaviour indirectly via its effects on social avoidance; the gut microbiome is known to affect social avoidance as recolonization studies (see Glossary) have ameliorated these behaviours in mice (Sampson and Mazmanian, 2015). The gut microbiome may also indirectly influence reproductive behaviour via individual recognition and communication. For example, volatile compounds in preen oil in birds serve as a chemical cue, and signal information about species, sex and breeding condition. Potential mates are attracted to preen oil odours. In the dark-eyed junco (Junco hyemalis), symbiotic bacteria may be responsible for producing behaviourally relevant volatile compounds (Whittaker et al., 2019), indicating that the microbiome can influence behaviours important for mate attraction and reproduction.
Sexual interactions also represent direct means by which an organism's microbiome can be transferred or influence another individual's gut microbiome and other microbial communities (Box 1). For example, in the Mormon cricket (Anabrus simplex), mating behaviour affects the structural communities of the gut. Specifically, the abundance of lactic acid bacteria changes in response to mating (Smith et al., 2017). In lizards (Zootoca vivipara), polyandrous females have significantly more diverse cloacal bacterial communities compared with monandrous females, suggesting the number of sexual partners can influence females' cloacal microbiome (White et al., 2011). In birds, sexual contact between partners also appears to influence the cloacal microbiome (Kreisinger et al., 2015; Hernandez et al., 2020). For example, in barn swallows (Hirundo rustica), repeated contact between sexual partners homogenizes the cloacal microbiome (Kreisinger et al., 2015); pair members have more similar cloacal microbiomes (Ambrosini et al., 2019), and females with numerous partners have more microbial diversity (Levin et al., 2016). Comparative studies evaluating how different mating strategies (e.g. social monogamy, genetic monogamy, polygyny, polyandry, lekking, polygynandry) are influenced by the gut microbiome, how these strategies affect the transmission of bacterial communities, and whether they affect microbial communities across the body differently would advance our knowledge of how these mating strategies evolved and how the gut microbiome influences reproduction.
Development of social behaviour: maternal versus paternal effects
The maternal influence on offspring phenotype is an important source of organismal variation (e.g. maternal stress; Seckl and Meaney, 2004; Duckworth et al., 2015), and it plays a key role in the foundation of the offspring's microbiome. In most mammals, offspring are birthed through the vaginal canal, bacteria from which directly shape the microbiome of the offspring (Dominguez-Bello et al., 2010), affecting offspring behaviour. For example, pregnant mice exposed to antibiotics produce offspring that exhibit less locomotor activity and exploration of the perimeter of an unfamiliar field compared with control offspring (Tochitani et al., 2016). Further, manipulations of the maternal gut microbiome during pregnancy influence offspring neurodevelopment, which alters offspring tactile sensitivity in some somatosensory behavioural tasks, but not in sensorimotor behaviours (Vuong et al., 2020). The maternal gut microbiome can also mediate the effects of prenatal stress (Jašarević et al., 2018) and in sex-specific ways (J.A.C., C.L.W. and G.E.D., unpublished data). For example, in Siberian hamsters, pregnant females exposed to stress produced female offspring that were highly aggressive, whereas pregnant females exposed to stress and antibiotics produced female offspring that displayed aggression comparable to that of controls (J.A.C., C.L.W. and G.E.D., unpublished data).
Less is known about how manipulating the maternal microbiome affects offspring behaviour in non-mammalian species, highlighting the need for hypothesis-driven research across taxa. In birds, eggs pass through the cloaca; whether cloacal microbes can penetrate the semipermeable barrier of the eggshell to shape the offspring microbiome or mediate the effects of prenatal conditions is not fully understood. Thus, in birds, the foundation of the offspring's microbiome may be more heavily influenced by maternal behaviour (e.g. nest building, incubation behaviour and temperature maintenance; Evans et al., 2017). For example, limited evidence suggests incubation may promote beneficial bacteria while eliminating pathogenic bacteria (Shawkey et al., 2009). Chicks may also acquire bacteria through indirect maternal transmission (e.g. passive uptake from the nesting environment) and parental care (Chen et al., 2020). Similarly, in fish and reptiles, parental skin microbiome diversity (Soares et al., 2019), specifically through skin-to-skin contact with the female, is critical in establishing the offspring microbiome (Banning et al., 2008). Questions relating to how the microbiome is established from birth in non-mammalian species and the cascading effects of the gut–brain axis on host behaviour would benefit from a comparative approach.
Recent studies have focused on the role of the paternal microbiome on offspring development and behaviour. Previous work has shown that paternal presence and condition can have sex-specific effects on the health and behaviour of their offspring (e.g. Tabbaa et al., 2017; Hellmann et al., 2020). Fathers may directly transmit information to their partners or offspring through their microbiome (Rando and Simmons, 2015). For example, Javurek et al. (2016) revealed that, in some strains, male mice have unique seminal fluid microbiomes that differ in composition from their gut microbiomes. Therefore, it is possible that males' seminal fluid microbiome could influence offspring development during embryogenesis, a critical stage during development linked to postnatal differences in social learning, cognitive development and memory recall that later can impact social behaviours (Desbonnet et al., 2014; Davidson et al., 2018). This might occur via many different mechanisms, described in detail below, and could include changes in neuronal hyperactivity, serotonergic systems in the hippocampus and gene expression in the amygdala (Davidson et al., 2018).
Paternal experience could also affect offspring behaviour via changes in the gut microbiome of offspring. Callaghan et al. (2016) observed that, in rats, emotional learning – including the retention of aversive associations – emerges earlier in offspring produced by stress-exposed fathers. This effect is reversed by the administration of probiotics (see Glossary) to offspring, suggesting a link between paternal experience, offspring microbiome and offspring behaviour. The effect of paternal stress on offspring behaviour has been observed in non-model systems as well. For example, in three-spined sticklebacks (Gasterosteus aculeatus), predator-exposed fathers produce sons that differ in how risk prone they are (Hellmann et al., 2020). Considering whether the microbiome plays a role in mediating these effects in non-model systems is an important next step.
Parental care
Parental care behaviours may be affected by the gut microbiome. In honey bees, differences in the gut microbiome are associated with different behavioural tasks, including nursing tasks inside the colony, suggesting maternal care may be mediated partly by the gut microbiome (Jones et al., 2018). Paternal care may also be influenced by the gut microbiome. A recent theoretical study suggested that microbes associated with increased paternal care could be favoured by natural selection (Gurevich et al., 2020), and may help to explain the occurrence of paternal care when commonly proposed explanations are insufficient (e.g. Hamilton, 1964). Whether the gut microbiome influences parental care and the mechanisms by which this occurs has yet to be experimentally evaluated. Comparative studies using closely related species that display different forms of parental care (e.g. Phodopus sungorus and Phodopus campbelli, which show single-parent and biparental care, respectively) or studies involving species where parental care sex roles are reversed (e.g. jacanas: Jenni and Betts, 1978; Emlen and Wrege, 2004; tidewater goby, Eucyclogobius newberryi: Swenson, 1997) provide natural experiments for testing the role of the gut microbiome in parental care behaviour.
Differences in parental care behaviour have been associated with differences in hormones (e.g. da Silva Mota et al., 2006; Riedman, 1982) and other behavioural traits (e.g. aggression; Mutzel et al., 2013), and these relationships may be influenced by the host microbiome. For example, changes in corticosterone concentrations are associated with differences in parental care (e.g. Dantzer et al., 2017). The gut–brain axis is tightly linked with the hypothalamic–pituitary–adrenal (HPA) axis (Davidson et al., 2018), and investigating whether the gut microbiome mediates this link between glucocorticoids and parental care is an important next step. Differences in parental care are also associated with differences in other behavioural traits. For example, more aggressive individuals often contribute less to parental care behaviours (e.g. Kopachena and Falls, 1993; Mutzel et al., 2013), and aggressive behaviour can be influenced by the gut microbiome. In dogs, specific groups of bacteria (e.g. Lactobacillus, Dorea, Blautia, Turicibacter, Bacteroides) have been used to stratify aggressive and non-aggressive individuals (Kirchoff et al., 2019). In Siberian hamsters, perturbations of the gut microbiome result in sex-specific changes in aggressive behaviour (e.g. Sylvia et al., 2017). These data suggest that the gut microbiome may affect parental care via its effect on aggression.
Parental care can also alter the foundation of the microbiome in offspring (e.g. nursing; Hassiotou and Geddes, 2015). In the maritime earwig (Anisolabis maritima), egg attendance (i.e. maternal care) significantly affects the bacterial microbiome of the eggs (Greer et al., 2020), which could result in cascading effects on offspring behaviour. Variation in maternal care can alter the expression of genes that influence behaviour, stress responses and cognition in offspring (Meaney, 2001). Microbes are heritable through parental care behaviour (e.g. Kulkarni and Heeb, 2007; Banning et al., 2008), supporting the idea that parental care can influence offspring microbiome, development and behaviour.
Cooperation
Cooperative behaviour (see Glossary) is observed across taxa – from bacteria to mammals (West et al., 2007) – and across a variety of contexts, including foraging and hunting (Boesch, 2002), displaying to and acquiring mates (Krakauer, 2005; DuVal, 2007), and offspring care (e.g. Koenig and Mumme, 1987; Cusick et al., 2018). But the evolution of cooperation via natural selection remains a puzzle (Pennisi, 2005; Barta, 2016; Kingma, 2017). Explanatory theories include kin selection (see Glossary; Hamilton, 1964) and reciprocity (see Glossary; Trivers, 1971), which emphasize the importance of interactions among kin and repeated interactions, respectively. However, cooperation and altruism (see Glossary) also occur between unrelated individuals, which cannot be fully explained by these theories (e.g. Kingma, 2017). Recent theoretical work suggests that microbes can facilitate the evolution of host altruism and that cooperation induced by microbes can evolve in a wide variety of host populations (Lewin-Epstein et al., 2017; Lewin-Epstein and Hadany, 2020). In addition, empirical work on how the gut microbiome influences other social behaviours related to cooperation provides additional evidence for its role in mediating cooperation.
Kin selection theory suggests that individuals assisting related individuals derive indirect fitness benefits. This is a potential explanation for the evolution of cooperative breeding (Cockburn, 1998). How individuals recognize and differentiate related and non-related individuals could be mediated partly by the gut microbiome. For example, in gopher tortoises (Gopherus polyphemus), the gut microbiome is partially determined by kinship (Yuan et al., 2015). In the Japanese termite (Reticulitermes speratus), intestinal bacteria are colony specific and play an important role in nestmate recognition; in this species, the administration of antibiotics changes the composition of the intestinal bacteria and alters nestmate recognition (Matsuura, 2001). Testing whether the gut microbiome plays a role in kin recognition could provide an explanation for how individuals recognize relatives to form cooperative breeding groups.
A major challenge in the study of cooperation is understanding why individuals vary in cooperative behaviour (Komdeur, 2006), especially when helping occurs among unrelated individuals. Individual differences in behavioural (Barta, 2016) and physiological mechanisms (Cusick, 2019) provide alternative explanations for individual variation in cooperative behaviour. For example, variation in glucocorticoids has been linked to individual differences in cooperative contributions (Dantzer et al., 2019, 2017; Raynaud et al., 2015), cooperative tendency (Cusick, 2019) and other social behaviours that may affect cooperation (e.g. De Fraipoint et al., 2000; Raulo and Dantzer, 2018; Anacker et al., 2016). Glucocorticoids can affect the composition of the gut microbiome (e.g. Noguera et al., 2018), and the microbiome influences the development of the HPA axis. For example, the maternal microbiome is related to the offspring HPA axis and there is evidence for bidirectional communication between the HPA axis and the gut microbiome (Williams et al., 2020). Hypothesis-driven studies assessing the role of the microbiome in mediating the relationship between glucocorticoids and cooperation are especially important for determining whether glucocorticoids directly or indirectly affect cooperative behaviour. The gut microbiome may also influence variation in cooperative behaviour or related behaviours. For example, Lactobacillus decreases stress-induced and anxiety-like behaviour (e.g. in mice; Bravo et al., 2011; Sgritta et al., 2019), which could increase the host's interactions with other individuals, a necessary component of cooperation. The composition of the gut microbiome is also associated with group aggregation (e.g. German cockroach, Blattella germanica; Wada-Katsumata et al., 2015) and social isolation (Ntranos and Casaccia, 2018), which may affect whether individuals interact with conspecifics to form cooperative groups. Additionally, individual differences in aggressive behaviour may affect whether individuals form cooperative groups, and differences in aggressive behaviour are correlated with differences in gut composition (e.g. Canis familiaris; Kirchoff et al., 2019).
The microbiome may also be an important determinant of the benefits and costs of cooperation. For example, in cooperatively breeding species, helpers benefit breeders by caring for offspring (e.g. Koenig and Walters, 2011; Cusick et al., 2018) and assisting in nest construction (Leighton and Vander Meiden, 2016) and nest or offspring defence (e.g. Hellmann and Hamilton, 2014; Cusick et al., 2021). In some cooperatively breeding species, breeders assisted by helpers reduce their investment in production and/or care of their current offspring, potentially conserving resources for future reproductive efforts (e.g. Russell et al., 2007). Helpers may behave in ways that affect the microbiome of both breeders and offspring because they are often in direct contact with both. For example, helpers can affect offspring growth, development (e.g. Cusick et al., 2018) and stress physiology (Cusick, 2019). As a result, offspring raised by groups with helpers may differ in their gut microbiome, which could have important consequences for their development, health and behaviour as adults. In some cooperatively breeding species, the beneficial effects of helpers are difficult to detect, or ‘concealed’ (Russell et al., 2007), making it hard to assess how selection could favour cooperation. Assessing how the presence of helpers affects the microbiome of both breeders and offspring could reveal unexplored ‘concealed’ benefits to cooperation and provide explanations for why cooperation occurs in some species.
Taxa that share similarities in behavioural traits can also be used to elucidate the bidirectional relationship between the host microbiome and cooperative behaviour. For example, eusociality and cooperative breeding behaviour are observed in many mammalian (e.g. Clutton-Brock et al., 2001; Hodge, 2007), avian (Koenig and Walters, 2011; Woolfenden, 1975) and piscine (e.g. Kasper et al., 2017) species, making a comparative approach an ideal way to identify the role of the host microbiome in this behaviour.
Gut–brain–behaviour communication: proximate mechanisms
A wide variety of physiological mechanisms have been suggested to explain how the gut microbiome might communicate with the brain to mediate behaviour (Cryan et al., 2019). Such mechanisms include input from the vagus nerve, neuroendocrine regulation by the HPA axis, neurotransmitter or neuropeptide regulation, microbial byproducts of the gut microbiota and direct influences of the immune system. Most studies investigating these mechanisms have been conducted on model systems and focus on behavioural abnormalities and their relationship with human health. Similar studies in non-model systems that display natural variations in behaviour will provide a more comprehensive understanding of the proximate links between physiological mechanisms and the brain that affect behaviour. Although space constraints prevent an exhaustive overview of all the possible mechanisms (for a comprehensive review, see Cryan et al., 2019), here we highlight some of the likely critical players. We also provide a brief introduction to some less well-studied mechanisms that may regulate gut microbiome–behaviour interactions via influences from environmental factors (see Fig. 1). Non-model organisms may be particularly well suited to study the latter given their relatively robust responsiveness to environmental variables.
The immune system
The ways in which microbes communicate with the brain to influence behaviour are not completely understood, although the immune system may play an important role (Lee and Mazmanian, 2010). The immune system interacts extensively with the nervous system; microglia and astrocytes, the resident immune cells of the nervous system, directly modulate central nervous system (CNS) activity (Vezzani and Viviani, 2015). Furthermore, cytokines (see Glossary) can indirectly modulate CNS activity via interactions with the autonomic nervous system and the HPA axis (Bilbo and Schwarz, 2012). In addition, some cytokines and chemokines can pass through the blood–brain barrier (BBB) and directly interact with neurons in the CNS, whereas others, such as interleukin-(IL)-1, a key pro-inflammatory cytokine and endogenous pyrogen (see Glossary), can signal through the microvasculature of the BBB, inducing release of prostaglandins in the CNS, which can alter neuronal activity (Rivest, 2010). Thus, microbiome-induced alterations in immune function could mediate effects on brain and behaviour. Indeed, the microbiome can influence the immune system locally by affecting lymph nodes within the digestive tract, while also influencing immune system development in both the brain and periphery. For example, the development of the immune system is altered in GF rats: they have fewer, smaller and often inactive lymph nodes and Peyer's patches (see Glossary), which hinders the immune response (Hoshi et al., 1992). Within the gut, various microbe-associated molecular patterns (MAMPs), including lipopolysaccharide (LPS; see Glossary), bacterial lipoprotein and flagellin may also influence immune system activity. These MAMPs activate different parts of the immune system, which affects the production of pro-inflammatory cytokines and can cause changes in physiology and behaviour (reviewed in Sampson and Mazmanian, 2015). For example, increases in pro-inflammatory cytokines such as IL1-β and tumour-necrosis factor (TNF)α mediate sickness behaviour induced by LPS administration (e.g. Breder et al., 1994). Connections between the gut microbiome and immune system function have been identified in non-model systems as well. For example, in the Chinese mitten crab (Eriocheir sinensis), changes in the composition and diversity of the gut microbiome are closely associated with the severity of infection with white spot syndrome virus, an emerging pathogen in the aquaculture industry (Ding et al., 2017). In honey bees (A. mellifera), antimicrobial peptides – key components of insect innate immunity – are strongly upregulated by inoculation with microbiota, suggesting the microbiota exert a universal immune effect (Kwong et al., 2017), and may protect against future infections and survival. Investigating the role of the immune system in mediating the cross-talk between the brain and the gut microbiome in non-model organisms and considering potential contributions of other microbial communities (e.g. the skin microbiome; Box 1) are exciting areas of further research.
Neurotransmitters and neuropeptides
Many neurotransmitters, such as serotonin (5-hydroxytryptamine, 5-HT), dopamine, noradrenaline (norepinephrine), adrenaline (epinephrine), gamma-aminobutyric acid (GABA) and acetylcholine, are produced in the gut, by enterochromaffin cells (see Glossary), enteric neurons or the gut microbiota (Sampson and Mazmanian, 2015), and may alter physiology and behaviour (Gareau, 2014). For example, compared with SPF mice, GF mice exhibit decreased expression of 5-HT receptor 1A (5-HT1A) and increased expression of brain-derived neurotrophic factor (BDNF) in the hippocampus, which is associated with greater anxiety-like behaviour (Neufeld et al., 2011). The decrease in 5-HT1A receptors can affect the activity of serotonergic neurons. Given that 5-HT1A receptors play a role in regulating anxiety-like behaviours (Li et al., 2004; Savitz et al., 2009), the decrease in 5-HT1A receptors might be responsible for the increase in anxiety-like behaviour in GF mice.
Catecholamines (see Glossary) may also play a role in the cross-talk. For example, inoculating either SPF or gnotobiotic mice (see Glossary) with a mixture of Clostridium species increases levels of biologically active dopamine and noradrenaline in the gut. Conversely, catecholamines are present at lower levels and in biologically inactive forms in GF mice (Asano et al., 2012). Given that catecholamines are chemical mediators of a wide range of critical behaviours, ranging from ingestion, thermoregulation and reproduction to reward, motivation and memory (Bloom et al., 1989), alteration of catecholamines via dysregulation of the microbiome can play an important role in a number of behavioural disorders.
The gut microbiome is important in maintaining the integrity of the BBB during development. In GF mice, the permeability of the BBB is greater during both development and adulthood, suggesting that particular microbes may facilitate development of the BBB. This increased permeability of the BBB in GF mice may be caused by fewer tight junction proteins (Braniste et al., 2014), and could allow immune cells to enter the CNS and stimulate an inflammatory response (Moretti et al., 2015). Given the important role of immune cells – such as microglia – and cytokines in sculpting the developing nervous system (Thion and Garel, 2017), this could dramatically alter neural development, producing neurobehavioral alterations in adulthood.
Additionally, changes in serotonin and its precursors might also mediate the interaction between the gut microbiome and the brain. Increased permeability of the BBB during development could allow 5-HT to cross through. Alternatively, given that the amino acid tryptophan (the precursor of 5-HT) can cross the BBB more readily – especially if the permeability of the BBB is increased by alterations in the gut microbiome – increased tryptophan may increase 5-HT synthesis in the brain (O'Mahony et al., 2015). As 5-HT is involved in modulating neural development, increased 5-HT production could greatly influence physiology and behaviour.
Neuropeptides – including neuropeptide Y (NPY), substance P (SP) and vasoactive intestinal polypeptide (VIP) – may also be involved in the cross-talk between the brain, microbiome and immune system (reviewed in Holzer and Farzi, 2014). These compounds often function in similar ways, and it is therefore challenging to clarify their role in this cross-talk. In rats, intracerebroventricular injections of NPY increase anti-depressive behaviours, an effect that is partially prevented by NPY receptor blockade (Ishida et al., 2007). We also know that antibiotic treatment increases SP expression in the colon, and administering the probiotic Lactobacillus paracasei normalizes SP expression in antibiotic-treated mice (Verdu et al., 2006). VIP can protect against colitis-induced epithelial damage by maintaining the integrity and distribution of tight junction proteins in the intestinal lining. This indicates that VIP may be particularly important in the prevention of diseases related to gut permeability (Conlin et al., 2009). Whether neuropeptides affect the gut–brain axis in a dose-dependent manner remains unknown.
The vagus nerve
The vagus nerve serves as the primary neural pathway connecting the gastrointestinal tract to the brainstem, thus representing the primary bidirectional neural connection between the brain and the gut. The vagus nerve provides both sensory and motor innervation that regulates several key functions, especially homeostatic functions. Although the vagus nerve does not directly interact with the microbiome, it can detect changes in the microbiota via signals in the form of bacterial metabolites or by-products, or via enteroendocrine cells in the gut epithelium. A series of studies employing surgical vagotomy in mice have provided strong evidence that the vagus nerve mediates at least some of the effects of microbiome alterations on behaviour. For example, the anxiolytic effects of either Lactobacillus rhamnosus (Bravo et al., 2011) or Bifidobacterium longum (Bercik et al., 2011) treatment are reduced in vagotomized mice. Furthermore, the probiotic Lactobacillus reuteri increases the central release of oxytocin (Poutahidis et al., 2013), a nonapeptide implicated in the formation of social bonds in mammals; vagotomy eliminates this effect (Bercik et al., 2011).
HPA endocrine axis
The HPA axis is one of the primary neuroendocrine systems involved in the regulation of homeostatic functions (e.g. energy, reproduction, feeding, immune function, stress responses) in vertebrates, and thus is a key mediator of neuroendocrine–microbiome communication. In response to an external stimulus – for instance, a perceived threat – corticotropin releasing hormone (CRH) is released by the paraventricular nucleus of the hypothalamus. CRH acts on the pituitary gland to regulate the release of adrenocorticotropic hormone (ACTH). ACTH stimulates the adrenal glands in the periphery to release glucocorticoid hormones (e.g. corticosterone, cortisol). Regulation of the HPA axis by the microbiome has been explored in a variety of contexts. For example, the maternal microbiome, and the timing and progression of initial colonization, are linked to offspring development and HPA axis functioning (de Weerth, 2017). How an individual responds to stress may also be affected by the gut microbiome. In GF mice, the HPA response is more sensitive to a restraint stressor than in SPF mice with normal functioning microbiota (Sudo et al., 2004). In wild red squirrels (Tamiasciurus hudsonicus), bacterial diversity is lower at higher levels of faecal glucocorticoid metabolites (FGMs) and, over time, changes in bacterial relative abundance are related to increases in FGMs, highlighting the link between the HPA axis and microbial communities (Stothart et al., 2016).
Mounting evidence from laboratory animals suggests there is bidirectional communication between the gut microbiome and the HPA axis. However, there is little relevant research in non-model systems (Williams et al., 2020). How the HPA and the gut microbiome communicate probably differs across species, partly because the primary stress hormones vary across taxa. For example, cortisol is the primary glucocorticoid hormone in most mammals (including humans) and fish, whereas corticosterone is the primary glucocorticoid hormone in rodents, birds, amphibians and reptiles (Baker et al., 2013). The interactions among different hormone axes and the gut microbiome are also likely to be complex. For instance, the type of stressor or hormonal manipulation can alter the composition of the gut microbiota in different ways (Williams et al., 2020). Additionally, the gut microbial community may help metabolize steroid hormones and convert steroid precursors (e.g. dietary cholesterol) into active glucocorticoids (Williams et al., 2020). Therefore, there are many areas of research to pursue to discern the relationship between the HPA axis and the gut microbiome in non-model organisms.
Microbial by-products
As mentioned above, microbial (particularly bacterial) by-products and metabolites can affect the brain and behaviour, although there is very little research on this topic. However, we know that short-chain fatty acids (SFAs; e.g. acetate, butyrate, propionate) can affect immune responses and the function of the CNS (Borre et al., 2014a). SFAs also facilitate the proper development of neurons and glia in the brain (Borre et al., 2014b); this is particularly true of microglia, as SFAs also contribute to the maintenance of this cell type. For example, microglia of GF mice appear immature, but this can be corrected with SFA treatment (Erny et al., 2015). Thus, microbial by-products present in the gut may interact with the immune system and the CNS. Outstanding questions include identifying the source of these by-products, where they act and how they function to influence species-typical behaviours.
Considering new physiological and environmental factors
In addition to the more commonly studied mechanisms discussed above, several physiological variables and environmental factors are less often considered but are key areas for future research (Fig. 1). Although these physiological mechanisms do not necessarily directly modulate interactions between the gut microbiome and behaviour, they serve as important transducers of environmental information (e.g. season, food availability, social context), allowing hosts to modify their physiology and behaviour according to their individual ecological context.
The pineal gland hormone melatonin and hypothalamic thyroid converting enzymes (e.g. diododinases) transduce photoperiodic information that is responsible for modulating behaviours including aggression (e.g. Munley et al., 2020) and photoperiod-dependent seasonal responses (e.g. reproduction; Yoshimura et al., 2003). Photoperiod modulates the gut microbiome and behaviour (Ren et al., 2020), but the role that melatonin and thyroid hormones play in this process remains unclear, especially in non-model organisms. There is also increasing evidence for an effect of environmental temperature on gut microbial communities (e.g. Fontaine et al., 2018; Kohl and Yahn, 2016), but our understanding of this relationship remains limited and may benefit from a comparative approach. For example, in endotherms, the effect of temperature on the microbiome may be due to physiological changes caused by temperature changes (e.g. Chevalier et al., 2015). However, the temperature of ectotherms can vary widely, and changes in temperature may have a more direct effect on their microbiomes (Moeller et al., 2020). Ambient temperature also interacts with physiological systems, such as gonadal hormones, to affect behaviour (e.g. Silverin et al., 2008). In addition, temperature is an important environmental factor affecting sex determination in many species, and the role of the microbiome in this process requires further study. Diet and food intake (Box 2) via gut peptides (Fig. 1) and social interactions via nonapeptides and other hormones (Fig. 1) are likely to modulate the effects of the microbiome on social behaviour and have rarely been studied in non-model organisms.
Conclusions
The microbiome performs a variety of functions for its host, including coordinating interactions with the brain, immune system and endocrine axes to mediate physiological processes and affect individual behaviour. Techniques for sampling and quantifying the microbiome have become more widely available, more sophisticated and less costly. Additionally, methodologies for collecting and preserving samples from wild populations are more readily available (Hird, 2017). Collectively, these advances make it possible to investigate the role of the host microbiome in non-model populations.
Current and emerging questions about the role of the microbiome in mediating social behaviour would benefit greatly from hypothesis-driven research using a wider range of non-model systems. We have identified important areas for future research in terms of how social behaviour can alter the gut microbiome and how the microbiome can affect social behaviour, both of which could play important roles in host fitness and evolution. Further investigation into how the gut microbiome can influence variation in social behaviour, especially reproduction, parental care, offspring development and cooperation, will be important. In addition, we have identified newer, less-appreciated mechanisms by which the effects of the microbiome on social behaviour may be mediated, including mechanisms that are driven in part by environmental factors. Non-model organisms are particularly well suited for investigating these mechanisms. Gut microbiome communities within non-model populations are likely to represent natural variation of a species or population. In addition, non-model organisms are exposed to and have evolved under many of the external factors that may indirectly modulate interactions between the gut microbiome and behaviour. Finally, future work should investigate the role of traditionally studied physiological mechanisms in non-model organisms, which will be critical if we are to determine whether findings are generalizable across species. Incorporating microbiome assessments into the study of animal behaviour adds critical components to the complex network of biological interactions, generating new ultimate and proximate questions and potentially providing answers to questions that remain a biological puzzle in the fields of ecology, behavioural ecology and evolutionary biology.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by National Science Foundation grant IOS-1656414 (G.E.D., C.L.W.), National Institutes of Health training grant T32HD049336 (J.A.C.) and generous funds provided by Indiana University. Deposited in PMC for release after 12 months.
References
- Ambrosini, R., Corti, M., Franzetti, A., Caprioli, M., Rubolini, D., Motta, V. M., Costanzo, A., Saino, N. and Gandolfi, I. (2019). Cloacal microbiomes and ecology of individual barn swallows. FEMS Microbiol. Ecol. 95, fiz061. 10.1093/femsec/fiz061 [DOI] [PubMed] [Google Scholar]
- Anacker, A. M. J., Reitz, K. M., Goodwin, N. L. and Beery, A. K. (2016). Stress impairs new but not established relationships in seasonally social voles. Horm. Behav. 79, 52-57. 10.1016/j.yhbeh.2016.01.004 [DOI] [PubMed] [Google Scholar]
- Antwis, R. E., Lea, J. M. D., Unwin, B. and Shultz, S. (2018). Gut microbiome composition is associated with spatial structuring and social interactions in semi-feral Welsh Mountain ponies. Microbiome 6, 207. 10.1186/s40168-018-0593-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archie, E. A. and Theis, K. R. (2011). Animal behaviour meets microbial ecology. Anim. Behav. 82, 425-436. 10.1016/j.anbehav.2011.05.029 [DOI] [Google Scholar]
- Archie, E. A. and Tung, J. (2015). Social behavior and the microbiome. Curr. Opin. Behav. Sci. 6, 28-34. 10.1016/j.cobeha.2015.07.008 [DOI] [Google Scholar]
- Arentsen, T., Raith, H., Qian, Y., Forssberg, H. and Diaz Heijtz, R. (2015). Host microbiota modulates development of social preference in mice. Microb. Ecol. Health Dis. 26, 29719. 10.3402/mehd.v26.29719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, Y., Hiramoto, T., Nishino, R., Aiba, Y., Kimura, T., Yoshihara, K., Koga, Y. and Sudo, N. (2012). Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G1288-G1295. 10.1152/ajpgi.00341.2012 [DOI] [PubMed] [Google Scholar]
- Bailey, M. T., Walton, J. C., Dowd, S. E., Weil, Z. M. and Nelson, R. J. (2010). Photoperiod modulates gut bacteria composition in male Siberian hamsters (Phodopus sungorus). Brain Behav. Immun. 24, 577-584. 10.1016/j.bbi.2009.12.010 [DOI] [PubMed] [Google Scholar]
- Baker, M. E., Funder, J. W. and Kattoula, S. R. (2013). Evolution of hormone selectivity in glucocorticoid and mineralocorticoid receptors. J. Steroid Biochem. Mol. Biol. 137, 57-70. 10.1016/j.jsbmb.2013.07.009 [DOI] [PubMed] [Google Scholar]
- Balshine-Earn, S., Neat, F. C., Reid, H. and Taborsky, M. (1998). Paying to stay or paying to breed? Field evidence for direct benefits of helping behavior in a cooperatively breeding fish. Behav. Ecol. 9, 432-438. 10.1093/beheco/9.5.432 [DOI] [Google Scholar]
- Banning, J. L., Weddle, A. L., Wahl, G. W., III, Simon, M. A., Lauer, A., Walters, R. L. and Harris, R. N. (2008). Antifungal skin bacteria, embryonic survival, and communal nesting in four-toed salamanders, Hemidactylium scutatum. Oecologia 156, 423-429. 10.1007/s00442-008-1002-5 [DOI] [PubMed] [Google Scholar]
- Barta, Z. (2016). Individual variation behind the evolution of cooperation. Phil. Trans. R. Soc. B 371, 20150087. 10.1098/rstb.2015.0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik, P., Denou, E., Collins, J., Jackson, W., Lu, J., Jury, J., Deng, Y., Blennerhassett, P., Macri, J., McCoy, K. D.et al. (2011). The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology 141, 599-609, e1–3. 10.1053/j.gastro.2011.04.052 [DOI] [PubMed] [Google Scholar]
- Bilbo, S. D. and Schwarz, J. M. (2012). The immune system and developmental programming of brain and behavior. Front. Neuroendocrinol. 33, 267-286. 10.1016/j.yfrne.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, F. E., Schulman, J. A. and Koob, G. F. (1989). Catecholamines and Behavior. In Catecholamines II. Handbook of Experimental Pharmacology, Vol. 90/2 (eds Trendelenburg U. and Weiner N.), pp. 27-88. Berlin: Springer. [Google Scholar]
- Boesch, C. (2002). Cooperative hunting roles among Taï chimpanzees. Human Nature 13, 27-46. 10.1007/s12110-002-1013-6 [DOI] [PubMed] [Google Scholar]
- Borre, Y. E., Moloney, R. D., Clarke, G., Dinan, T. G. and Cryan, J. F. (2014a). The impact of microbiota on brain and behavior: mechanisms & therapeutic potential. In Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease (ed. Lyte M. and Cryan J. F.), p. 403. New York, NY: Springer. [DOI] [PubMed] [Google Scholar]
- Borre, Y. E., O'Keeffe, G. W., Clarke, G., Stanton, C., Dinan, T. G. and Cryan, J. F. (2014b). Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol. Med. 20, 509-518. 10.1016/j.molmed.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Braniste, V., Al-Asmakh, M., Kowal, C., Anuar, F., Abbaspour, A., Tóth, M., Korecka, A., Bakocevic, N., Ng, L. G., Kundu, P.et al. (2014). The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 6, 263ra158. 10.1126/scitranslmed.3009759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo, J. A., Forsythe, P., Chew, M. V., Escaravage, E., Savignac, H. M., Dinan, T. G., Bienenstock, J. and Cryan, J. F. (2011). Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl Acad. Sci. USA 108, 16050-16055. 10.1073/pnas.1102999108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breder, C. D., Hazuka, C., Ghayur, T., Klug, C., Huginin, M., Yasuda, K., Teng, M. and Saper, C. B. (1994). Regional induction of tumor necrosis factor alpha expression in the mouse brain after systemic lipopolysaccharide administratio. Proc. Natl. Acad. Sci. USA 91, 11393-11397. 10.1073/pnas.91.24.11393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz, E. A. and Beecher, M. D. (2005). Song learning in birds: diversity and plasticity, opportunities and challenges. Trends Neurosci. 28, 127-132. 10.1016/j.tins.2005.01.004 [DOI] [PubMed] [Google Scholar]
- Brooks, A. W., Kohl, K. D., Brucker, R. M., van Opstal, E. J. and Bordenstein, S. R. (2016). Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol., 14, e2000225. 10.1371/journal.pbio.2000225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce-Keller, A. J., Fernandez-Kim, S.-O., Townsend, R. L., Kruger, C., Carmouche, R., Newman, S., Salbaum, J. M. and Berthoud, H.-R. (2017). Maternal obese-type gut microbiota differentially impact cognition, anxiety and compulsive behavior in male and female offspring in mice. PLoS ONE 12, e0175577, 1–20. 10.1371/journal.pone.0175577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucker, R. M. and Bordenstein, S. R. (2013). The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science 341, 667-669. 10.1126/science.1240659 [DOI] [PubMed] [Google Scholar]
- Bshary, R., Wickler, W. and Fricke, H. (2002). Fish cognition: a primate's eye view. Anim. Cogn. 5, 1-13. 10.1007/s10071-001-0116-5 [DOI] [PubMed] [Google Scholar]
- Burns, A. R., Miller, E., Agarwal, M., Rolig, A. S., Milligan-Myhre, K., Seredick, S., Guillemin, K. and Bohannan, B. J. M. (2017). Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model. Proc. Natl Acad. Sci. USA 114, 11181-11186. 10.1073/pnas.1702511114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan, B. L., Cowan, C. S. M. and Richardson, R. (2016). Treating generational stress: effect of paternal stress on development of memory and extinction in offspring is reversed by probiotic treatment. Psychol. Sci. 27, 1171-1180. 10.1177/0956797616653103 [DOI] [PubMed] [Google Scholar]
- Chai, L., Dong, Z., Chen, A. and Wang, H. (2018). Changes in intestinal microbiota of Bufo gargarizans and its association with body weight during metamorphosis. Arch. Microbiol. 200, 1087-1099. 10.1007/s00203-018-1523-1 [DOI] [PubMed] [Google Scholar]
- Chen, L., Guo, Y., Hu, C., Lam, P. K. S., Lam, J. C. W. and Zhou, B. (2018). Dysbiosis of gut microbiota by chronic coexposure to titanium dioxide nanoparticles and bisphenol A: Implications for host health in zebrafish. Environ. Pollut. 234, 307-317. 10.1016/j.envpol.2017.11.074 [DOI] [PubMed] [Google Scholar]
- Chen, C.-Y., Chen, C.-K., Chen, Y.-Y., Fang, A., Shaw, G. T.-W., Hung, C.-M. and Wang, D. (2020). Maternal gut microbes shape the early-life assembly of gut microbiota in passerine chicks via nests. Microbiome 8, 129. 10.1186/s40168-020-00896-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier, C., Stojanović, O., Colin, D. J., Suarez-Zamorano, N., Tarallo, V., Veyrat-Durebex, C., Rigo, D., Fabbiano, S., Stevanović, A., Hagemann, S.et al. (2015). Gut microbiota orchestrates energy homeostasis during cold. Cell 163, 1360-1374. 10.1016/j.cell.2015.11.004 [DOI] [PubMed] [Google Scholar]
- Chung, H., Pamp, S. J., Hill, J. A., Surana, N. K., Edelman, S. M., Troy, E. B., Reading, N. C., Villablanca, E. J., Wang, S., Mora, J. R.et al. , (2012). Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149, 1578-1593. 10.1016/j.cell.2012.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, J. B., Vangay, P., Huang, H., Ward, T., Hillmann, B. M., All-Ghalith, G. A., Travis, D. A., Long, H. T., Tuan, B. V., Minh, V. V.et al. (2016). Captivity humanizes the primate microbiome. Proc. Natl Acad. Sci. USA 113, 10376-10381. 10.1073/pnas.1521835113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, J. B., Al-Ghalith, G. A., Long, H. T., Tuan, B. V., Cabana, F., Huang, H., Vangay, P., Ward, T., Minh, V. V., Tam, N. A.et al. (2018). Associations between nutrition, gut microbiome, and health in a novel nonhuman primate model. Sci. Rep. 8, 11159. 10.1038/s41598-018-29277-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock, T. H., Russell, A. F., Sharpe, L. L., Brotherton, P. N. M., McIlrath, G. M., White, S. and Cameron, E. Z. (2001). Effects of helpers on juvenile development and survival in meerkats. Science 293, 2446-2449. 10.1126/science.1061274 [DOI] [PubMed] [Google Scholar]
- Cockburn, A. P. (1998). Evolution of helping in cooperatively breeding birds. Annu. Rev. Ecol. Syst. 29, 141-177. 10.1146/annurev.ecolsys.29.1.141 [DOI] [Google Scholar]
- Collins, S. M. and Bercik, P. (2014). The relationship between intestinal microbiota and the central nervous system in normal gastrointestinal function and disease. Gastroenterology 136, 2003-2014. 10.1053/j.gastro.2009.01.075 [DOI] [PubMed] [Google Scholar]
- Colston, T. J. and Jackson, C. R. (2016). Microbiome evolution along divergent branches of the vertebrate tree of life: what is known and unknown. Mol. Ecol. 25, 3776-3800. 10.1111/mec.13730 [DOI] [PubMed] [Google Scholar]
- Conlin, V. S., Wu, X., Nguyen, C., Dai, C., Vallance, B. A., Buchan, A. M. J., Boyer, L. and Jacobson, K. (2009). Vasoactive intestinal peptide ameliorates intestinal barrier disruption associated with Citrobacter rodentium-induced colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G735-G750. 10.1152/ajpgi.90551.2008 [DOI] [PubMed] [Google Scholar]
- Corl, A., Charter, M., Rozman, G., Toledo, S., Turjeman, S., Kamath, P. L., Getz, W. M., Nathan, R. and Bowie, R. C. K. (2020). Movement ecology and sex are linked to barn owl microbial community composition. Mol. Ecol. 29, 1358-1371. 10.1111/mec.15398 [DOI] [PubMed] [Google Scholar]
- Cryan, J. F. and O'Mahony, S. M. (2011). The microbiome-gut-brain axis: from bowel to behavior. Neurogastroenterol. Motil. 23, 187-192. 10.1111/j.1365-2982.2010.01664.x [DOI] [PubMed] [Google Scholar]
- Cryan, J. F., O'Riordan, K. J., Cowan, C. S. M., Sandhu, K. V., Bastiaanssen, T. F. S., Boehme, M., Codagnone, M. G., Cussotto, S., Fulling, C., Golubeva, A. V.et al. (2019). The microbiota-gut-brain axis. Physiol. Rev. 99, 1877-2013. 10.1152/physrev.00018.2018 [DOI] [PubMed] [Google Scholar]
- Cusick, J. A. (2019). Proximate mechanisms influencing individual variation in cooperative behavior. PhD thesis, ProQuest: Florida State University. [Google Scholar]
- Cusick, J. A., de Villa, M., DuVal, E. H. and Cox, J. A. (2018). How do helpers help? Helper contributions throughout the nesting cycle in the cooperatively breeding brown-headed nuthatch. Behav. Ecol. Sociobiol. 72, 1-13. 10.1007/s00265-018-2470-1 [DOI] [Google Scholar]
- Cusick, J. A., DuVal, E. H. and Cox, J. A. (2021). Breeder aggression does not predict current or future cooperative group formation in a cooperatively breeding bird. Ethology 127,1-12. 10.1111/eth.13141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Mota, M. T., Franci, C. R. and de Sousa, M. B. (2006). Hormonal changes related to paternal and alloparental care in common marmosets (Callithrix jacchus). Hormones Behav. 49, 293-302. 10.1016/j.yhbeh.2005.07.012 [DOI] [PubMed] [Google Scholar]
- Dantzer, B., Goncalves, I. B., Spence-Jones, H. C., Bennett, N. C., Heistermann, M., Ganswindt, A., Dubuc, C., Gaynor, D., Manser, M. B. and Clutton-Brock, T. H. (2017). The influence of stress hormones and aggression on cooperative behaviour in subordinate meerkats. Proc. R. Soc. B 284, 20171248. 10.1098/rspb.2017.1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer, B., Dubuc, C., Goncalves, I. B., Cram, D. L., Bennett, N. C., Ganswindt, A., Heistermann, M., Duncan, C., Gaynor, D. and Clutton-Brock, T. H. (2019). The development of individual differences in cooperative behaviour: maternal glucocorticoid hormones alter helping behaviour of offspring in wild meerkatsPhil. Trans. R. Soc. B 3742 018011720180117. 10.1098/rstb.2018.0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, G. L., Cooke, A. C., Johnson, C. N. and Quinn, J. L. (2018). The gut microbiome as a driver of individual variation in cognition and functional behaviour. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20170286. 10.1098/rstb.2017.0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, G., Wiley, N., Cooke, A. C., Johnson, C. N., Fouhy, F., Reichert, M. S., de la Hera, I., et al. (2020). Diet induces parallel changes to the gut microbiota and problem solving performance in a wild bird. Sci. Rep. 10, 20783. 10.1038/s41598-020-77256-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fraipoint, M., Colbert, J., John, H., Adler, and Meylan, S. (2000). Increased pre-natal maternal corticosterone promotes philopatry of offspring in common lizards Lacerta vivipara. J. Anim. Ecol. 69, 404-413. 10.1046/j.1365-2656.2000.00405.x [DOI] [Google Scholar]
- de Weerth, C. (2017). Do bacteria shape our development? Crosstalk between intestinal microbiota and HPA axis. Neurosci. Biobehav. Rev. 83, 458-471. 10.1016/j.neubiorev.2017.09.016 [DOI] [PubMed] [Google Scholar]
- Denver, R. J. (2019). Tadpole behavior and metamorphosis. In Encyclopedia of Animal Behavior (ed. Choe J. C.), pp. 375-378. Elsevier. [Google Scholar]
- Desbonnet, L., Clarke, G., Shanahan, F., Dinan, T. G. and Cryan, J. F. (2014). Microbiota is essential for social development in the mouse. Mol. Psychiatry 19, 146-148. 10.1038/mp.2013.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz, R., Wang, S., Anuar, F., Qian, Y., Bjorkholm, M., Samuelsson, A., Hibberd, M. L., Forssberg, H. and Pettersson, S. (2011). Normal gut microbiota modulates brain development and behavior. Proc. Natl Acad. Sci. USA 108, 3047-3052. 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan, T. G., Stilling, R. M., Stanton, C. and Cryan, J. F. (2015). Collective unconscious: how gut microbes shape human behavior. J. Psychiatr. Res. 63, 1-9. 10.1016/j.jpsychires.2015.02.021 [DOI] [PubMed] [Google Scholar]
- Ding, Z. F., Cao, M. J., Zhu, X. S., Xu, G. H. and Wang, R. L. (2017). Changes in the gut microbiome of the Chinese mitten crab (Eriocheir sinensis) in response to White spot syndrome virus (WSSV) infection. J. Fish Dis. 40, 1561-1571. 10.1111/jfd.12624 [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello, M. G., Costello, E. K., Contreras, M., Magris, M., Hidalgo, G., Fierer, N. and Knight, R. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971-11975. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, A. J., Hug, L. A. and Katzenback, B. A. (2021). Composition of the North American Wood Frog (Rana sylvatica) Bacterial Skin Microbiome and Seasonal Variation in Community Structure. Microb. Ecol. 81, 78-92. 10.1007/s00248-020-01550-5 [DOI] [PubMed] [Google Scholar]
- Downing, P. A., Griffin, A. S. and Cornwallis, C. K. (2020). Group formation and the evolutionary pathway to complex sociality in birds. Nat. Ecol. Evol. 4, 479-486. 10.1038/s41559-020-1113-x [DOI] [PubMed] [Google Scholar]
- Duckworth, R. A., Belloni, V. and Anderson, S. R. (2015). Cycles of species replacement emerge from locally induced maternal effects on offspring behavior in a passerine bird. Science 347, 875-877. 10.1126/science.1260154 [DOI] [PubMed] [Google Scholar]
- DuVal, E. H. (2007). Cooperative display and lekking behavior of the lance-tailed Manakin (Chiroxiphia Lanceolata). The Auk 124, 1168-1185. 10.1093/auk/124.4.1168 [DOI] [Google Scholar]
- Emlen, S. T. and Wrege, P. H. (2004). Division of labour in parental care behaviour of a sex-role-reversed shorebird, the wattled jacana. Anim. Behav. 68, 847-855. 10.1016/j.anbehav.2003.08.034 [DOI] [Google Scholar]
- Engel, K., Sauer, J., Jünemann, S., Winkler, A., Wibberg, D., Kalinowski, J., Tauch, A. and Caspers, B. A. (2018). Individual- and species-specific skin microbiomes in three different estrildid finch species revealed by 16S amplicon sequencing. Microb. Ecol. 76, 518-529. 10.1007/s00248-017-1130-8 [DOI] [PubMed] [Google Scholar]
- Erny, D., Hrabě de Angelis, A. L., Jaitin, D., Wieghofer, P., Staszewski, O., David, E., Keren-Shaul, H., Mahlakoiv, T., Jakobshagen, K., Buch, T.et al. (2015). Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18, 965-977. 10.1038/nn.4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J. K., Buchanan, K. L., Griffith, S. C., Klasing, K. C. and Addison, B. A. (2017). Ecoimmunology and microbial ecology: contributions to avian behavior, physiology, and life history. Horm. Behav. 88, 112-121. 10.1016/j.yhbeh.2016.12.003 [DOI] [PubMed] [Google Scholar]
- Fontaine, S. S., Novarro, A. J. and Kohl, K. D. (2018). Environmental temperature alters the digestive performance and gut microbiota of a terrestrial amphibian. J. Exp. Biol. 221, jeb187559. 10.1242/jeb.187559 [DOI] [PubMed] [Google Scholar]
- Garcia-Reyero, N. (2018). The clandestine organs of the endocrine system. Gen. Comp. Endocrinol. 257, 264-271. 10.1016/j.ygcen.2017.08.017 [DOI] [PubMed] [Google Scholar]
- Gareau, M. G. (2014). Microbiota-gut-brain axis and cognitive function. Adv. Exp. Med. Biol 817, 357-371. 10.1007/978-1-4939-0897-4_16 [DOI] [PubMed] [Google Scholar]
- Gemmell, N. J., Todd, E. V., Goikoetxea, A., Ortega-Recalde, O. and Hore, T. A. (2019). Natural sex change in fish. Curr. Top. Dev. Biol. 134, 71-117. 10.1016/bs.ctdb.2018.12.014 [DOI] [PubMed] [Google Scholar]
- Greene, L. K., Bornbusch, S. L., McKenney, E. A., Harris, R. L., Gorvetzian, S. R., Yoder, A. D. and Drea, C. M. (2019). The importance of scale in comparative microbiome research: new insights from the gut and glands of captive and wild lemurs. Am. J. Primatol., 81, e22974. 10.1002/ajp.22974 [DOI] [PubMed] [Google Scholar]
- Greer, J. A., Swei, A., Vredenburg, V. T. and Zink, A. G. (2020). Parental care alters the egg microbiome of maritime earwigs. Microb. Ecol. 80, 920-934. [DOI] [PubMed] [Google Scholar]
- Grieneisen, L. E., Livermore, J., Alberts, S., Tung, J. and Archie, E. A. (2017). Group living and male dispersal predict the core gut microbiome in wild baboons. Integr. Comp. Biol. 57, 770-785. 10.1093/icb/icx046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, A. S. and West, S. A. (2002). Kin selection: fact and fiction. Trends Ecol. Evol. 17, 15-21. 10.1016/S0169-5347(01)02355-2 [DOI] [Google Scholar]
- Gurevich, Y., Lewin-Epstein, O. and Hadany, L. (2020). The evolution of paternal care: a role for microbes? Phil. Trans. R. Soc. B 375, 20190599. 10.1098/rstb.2019.0599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, W. D. (1964). The genetical evolution of social behaviour. I. J. Theoret. Biol. 7, 1-16. 10.1016/0022-5193(64)90038-4 [DOI] [PubMed] [Google Scholar]
- Harris, R. N., Lauer, A., Simon, M. A., Banning, J. L. and Alford, R. A. (2009). Addition of antifungal skin bacteria to salamanders ameliorates the effects of chytridiomycosis. Dis. Aquat. Organ. 83, 11-16. 10.3354/dao02004 [DOI] [PubMed] [Google Scholar]
- Hassiotou, F. and Geddes, D. T. (2015). Immune cell-mediated protection of the mammary gland and the infant during breastfeeding. Adv. Nutr. 6, 267-275. 10.3945/an.114.007377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, J. K. and Hamilton, I. M. (2014). The presence of neighbors influences defense against predators in a cooperatively breeding cichlid. Behav. Ecol. 25, 386-391. 10.1093/beheco/aru001 [DOI] [Google Scholar]
- Hellmann, J. K., Bukhari, S. A., Deno, J. and Bell, A. M. (2020). Sex-specific plasticity across generations I: Maternal and paternal effects on sons and daughters. J. Anim. Ecol. 89, 2788–2799. 10.1111/1365-2656.13364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, J., Escallón, C., Medina, D., Vernasco, B. J., Walke, J. B., Belden, L. K. and Moore, I. T. (2020). Cloacal bacterial communities of tree swallows (Tachycineta bicolor): Similarity within a population, but not between pair-bonded social partners. PLoS ONE 15, e0228982. 10.1371/journal.pone.0228982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird, S. M. (2017). Evolutionary biology needs wild microbiomes. Front. Microbiol. 8, 725. 10.3389/fmicb.2017.00725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, S. J. (2007). Counting the costs: the evolution of male-biased care in the cooperatively breeding banded mongoose. Anim. Behav. 74, 911-919. 10.1016/j.anbehav.2006.09.024 [DOI] [Google Scholar]
- Holzer, P. (2016). Neuropeptides, microbiota, and behavior. Int. Rev. Neurobiol. 131, 67-89. 10.1016/bs.irn.2016.08.005 [DOI] [PubMed] [Google Scholar]
- Holzer, P. and Farzi, A. (2014). Neuropeptides and the microbiota-gut-brain axis. In Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease (ed. Lyte M. and Cryan J. F.), pp. 195-219. New York, NY: Springer. [Google Scholar]
- Hooper, L. V. and Gordon, J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292, 1115-1118. 10.1126/science.1058709 [DOI] [PubMed] [Google Scholar]
- Hoshi, H., Aijima, H., Horie, K., Nagata, H., Kaneko, T. and Ikeda, T. (1992). Lymph follicles and germinal center in popliteal lymph nodes and other lymphoid tissues of germ-free and conventional rats. Tohoku J. Exp. Med 166, 297-307. 10.1620/tjem.166.297 [DOI] [PubMed] [Google Scholar]
- Hyde, E. R., Navas-Molina, J. A., Song, S. J., Kueneman, J. G., Ackermann, G., Cardona, C., Humphrey, G., Boyer, D., Weaver, T., Mendelson, J. R.III. et al. (2016). The oral and skin microbiomes of captive komodo dragons are significantly shared with their habitat. mSystems 1, e00046-16. 10.1128/mSystems.00046-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida, H., Shirayama, Y., Iwata, M., Katayama, S., Yamamoto, A., Kawahara, R. and Nakagome, K. (2007). Infusion of neuropeptide Y into CA3 region of hippocampus produces antidepressant-like effect via Y1 receptor. Hippocampus 17, 271-280. 10.1002/hipo.20264 [DOI] [PubMed] [Google Scholar]
- Jašarević, E., Howard, C. D., Morrison, K., Misic, A., Weinkopff, T., Scott, P., Hunter, C., Beiting, D. and Bale, T. L. (2018). The maternal vaginal microbiome partially mediates the effects of prenatal stress on offspring gut and hypothalamus. Nat. Neurosci. 21, 1061-1071. 10.1038/s41593-018-0182-5 [DOI] [PubMed] [Google Scholar]
- Javurek, A. B., Spollen, W. G., Ali, A. M. M., Johnson, S. A., Lubahn, D. B., Bivens, N. J., Bromert, K. H., Ellersieck, M. R., Givan, S. A. and Rosenfeld, C. S. (2016). Discovery of a novel seminal fluid microbiome and influence of estrogen receptor alpha genetic status. Sci. Rep. 6, 23027. 10.1038/srep23027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenni, D. A. and Betts, B. J. (1978). Sex differences in nest construction, incubation, and parental behaviour in the polyandrous American jaçana (Jacana spinosa). Anim. Behav. 26, 207-218. 10.1016/0003-3472(78)90020-9 [DOI] [Google Scholar]
- Jones, J. C., Fruciano, C., Marchant, J., Hildebrand, F., Forslund, S., Bork, P., Engel, P. and Hughes, W. O. H. (2018). The gut microbiome is associated with behavioural task in honey bees. Insectes Soc. 65, 419-429. 10.1007/s00040-018-0624-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper, C., Kolliker, M., Postma, E. and Taborsky, B. (2017). Consistent cooperation in a cichlid fish is caused by maternal and developmental effects rather than heritable genetic variation. Proc. Biol. Sci. 284, 20170369. 10.1098/rspb.2017.0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma, S. A. (2017). Direct benefits explain interspecific variation in helping behaviour among cooperatively breeding birds. Nat. Commun. 8, 1094. 10.1038/s41467-017-01299-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchoff, N. S., Udell, M. A. R. and Sharpton, T. J. (2019). The gut microbiome correlates with conspecific aggression in a small population of rescued dogs (Canis familiaris). PeerJ 7, e6103. 10.7717/peerj.6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig, W. D. and Mumme, R. L. (1987). Population Ecology of the Cooperatively Breeding Acorn Woodpecker. Princeton, NJ: Princeton University Press. [Google Scholar]
- Koenig, W. D. and Walters, E. L. (2011). Brooding, provisioning, and compensatory care in the cooperatively breeding acorn woodpecker. Behav. Ecol. 23, 181-190. 10.1093/beheco/arr172 [DOI] [Google Scholar]
- Kohl, K. D. and Yahn, J. (2016). Effects of environmental temperature on the gut microbial communities of tadpoles. Environ. Microbiol. 18, 1561-1565. 10.1111/1462-2920.13255 [DOI] [PubMed] [Google Scholar]
- Kohl, K. D., Stengel, A. and Dearing, M. D. (2016). Inoculation of tannin-degrading bacteria into novel hosts increases performance on tannin-rich diets. Environ. Microbiol. 18, 1720-1729. 10.1111/1462-2920.12841 [DOI] [PubMed] [Google Scholar]
- Kohl, K. D., Brun, A., Magallanes, M., Brinkerhoff, J., Laspiur, A., Acosta, J. C., Caviedes-Vidal, E. and Bordenstein, S. R. (2017). Gut microbial ecology of lizards: Insights into diversity in the wild, effects of captivity, variation across gut regions and transmission. Mol. Ecol. 26, 1175-1189. 10.1111/mec.13921 [DOI] [PubMed] [Google Scholar]
- Komdeur, J. (2006). Variation in individual investment strategies among social animals. Ethology 112, 729-747. 10.1111/j.1439-0310.2006.01243.x [DOI] [Google Scholar]
- Kopachena, J. and Falls, J. (1993). Re-evaluation of morph-specific variations in parental behavior of the white-throated sparrow. Wilson Bull. 105, 48-59. [Google Scholar]
- Krakauer, A. H. (2005). Kin selection and cooperative courtship in wild turkeys. Nature 434, 69-72. 10.1038/nature03325 [DOI] [PubMed] [Google Scholar]
- Kreisinger, J., Cížková, D., Kropácková, L. and Albrecht, T. (2015). Cloacal microbiome structure in a long-distance migratory bird assessed using deep 16sRNA pyrosequencing. PLoS ONE 10, e0137401. 10.1371/journal.pone.0137401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueneman, J. G., Woodhams, D. C., Harris, R., Archer, H. M., Knight, R. and McKenzie, V. J. (2016). Probiotic treatment restores protection against lethal fungal infection lost during amphibian captivity. Proc. Biol. Sci. 283, 20161553. 10.1098/rspb.2016.1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, S. and Heeb, P. (2007). Social and sexual behaviours aid transmission of bacteria in birds. Behav. Processes 74, 88-92. 10.1016/j.beproc.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Kwong, W. K., Mancenido, A. L. and Moran, N. A. (2017). Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 4, 170003. 10.1098/rsos.170003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. K. and Mazmanian, S. K. (2010). Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330, 1768-1773. 10.1126/science.1195568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leftwich, P. T., Hutchings, M. I. and Chapman, T. (2018). Diet, Gut Microbes and Host Mate Choice: understanding the significance of microbiome effects on host mate choice requires a case by case evaluation. BioEssays 40, e1800053. 10.1002/bies.201800053 [DOI] [PubMed] [Google Scholar]
- Leighton, G. M. and Vander Meiden, L. N. (2016). Sociable weavers increase cooperative nest construction after suffering aggression. PLoS ONE 11, e0150953. 10.1371/journal.pone.0150953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, I. I., Zonana, D. M., Fosdick, B. K., Song, S. J., Knight, R. and Safran, R. J. (2016). Stress response, gut microbial diversity and sexual signals correlate with social interactions. Biol. Lett. 12, 20160352. 10.1098/rsbl.2016.0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin-Epstein, O. and Hadany, L. (2020). Host-microbiome coevolution can promote cooperation in a rock-paper-scissors dynamics. Proc. Biol. Sci. 287, 20192754. 10.1098/rspb.2019.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin-Epstein, O., Aharonov, R. and Hadany, L. (2017). Microbes can help explain the evolution of host altruism. Nat. Commun. 8, 14040. 10.1038/ncomms14040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Holmes, A., Ma, L., Van de Kar, L. D., Garcia, F. and Murphy, D. L. (2004). Medial hypothalamic 5-hydroxytryptamine (5-HT)1A receptors regulate neuroendocrine responses to stress and exploratory locomotor activity: application of recombinant adenovirus containing 5-HT1A sequences. J. Neurosci. 24, 10868-10877. 10.1523/JNEUROSCI.3223-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo, W. T., García-Loor, J., Dudaniec, R. Y., Kleindorfer, S. and Cavanaugh, C. M. (2019). Host phylogeny, diet, and habitat differentiate the gut microbiomes of Darwin's finches on Santa Cruz Island. Sci. Rep. 9, 18781. 10.1038/s41598-019-54869-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrath, R. D., Haff, T. M., McLachlan, J. R. and Igic, B. (2015). Wild birds learn to eavesdrop on heterospecific alarm calls. Curr. Biol. 25, 2047-2050. 10.1016/j.cub.2015.06.028 [DOI] [PubMed] [Google Scholar]
- Martin, R., Bermudez-Humaran, L. G. and Langella, P. (2016). Gnotobiotic rodents: an in vivo model for the study of microbe–microbe interactions. Front. Microbiol. 7, 1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, K. (2001). Nestmate recognition mediated by intestinal bacteria in a termite, Reticulitermes speratus. Oikos 92, 20-26. 10.1034/j.1600-0706.2001.920103.x [DOI] [Google Scholar]
- Maul, J. D., Gandhi, J. P. and Farris, J. L. (2005). Community-level physiological profiles of cloacal microbes in songbirds (order: Passeriformes): variation due to host species, host diet, and habitat. Microb. Ecol. 50, 19-28. 10.1007/s00248-004-0076-9 [DOI] [PubMed] [Google Scholar]
- McKenzie, V. J., Song, S. J., Delsuc, F., Prest, T. L., Oliveria, A. M., Korpita, T. M., Alexiev, A., Amato, K. R., Metcalf, J. L., Kowalewski, M.et al. (2017). The effects of captivity on the mammalian gut microbiome. Integr. Comp. Biol. 57, 690-704. 10.1093/icb/icx090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney, M. J. (2001). Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu. Rev. Neurosci. 24, 1161-1192. 10.1146/annurev.neuro.24.1.1161 [DOI] [PubMed] [Google Scholar]
- Moeller, A. H., Ivey, K., Cornwall, M. B., Herr, K., Rede, J., Taylor, E. N. and Gunderson, A. R. (2020). The lizard gut microbiome changes with temperature and is associated with heat tolerance. Appl. Environ. Microbiol. 86, e01181-20. 10.1128/AEM.01181-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller, A. P., Czirjak, G. A. and Heeb, P. (2009). Feather micro-organisms and uropygial antimicrobial defences in a colonial passerine bird. Funct. Ecol. 23, 1097-1102. 10.1111/j.1365-2435.2009.01594.x [DOI] [Google Scholar]
- Moretti, R., Pansiot, J., Bettati, D., Strazielle, N., Ghersi-Egea, J.-F., Damante, G., Fleiss, B., Titomanlio, L. and Gressens, P. (2015). Blood-brain barrier dysfunction in disorders of the developing brain. Front. Neurosci. 9, 40. 10.3389/fnins.2015.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, J., Simpson, S. J. and Ponton, F. (2017). Direct and trans-generational effects of male and female gut microbiota in Drosophila melanogaster. Biol. Lett. 13, 20160966. 10.1098/rsbl.2016.0966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münger, E., Montiel-Castro, A. J., Langhans, W. and Pacheco-López, G. (2018). Reciprocal interactions between gut microbiota and host social behavior. Front. Integr. Neurosci. 12, 21. 10.3389/fnint.2018.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munley, K. M., Deyoe, J. E., Ren, C. C. and Demas, G. E. (2020). Melatonin mediates seasonal transitions in aggressive behavior and circulating androgen profiles in male Siberian hamsters. Horm. Behav. 117, 104608. 10.1016/j.yhbeh.2019.104608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutzel, A., Dingemanse, N. J., Araya-Ajoy, Y. G. and Kempenaers, B. (2013). Parental provisioning behaviour plays a key role in linking personality with reproductive success. Proc. R. Soc. B 280, 20131019. 10.1098/rspb.2013.1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles, I. A., Pincus, N. B., Fontecilla, N. M. and Datta, S. K. (2014). Effects of parental omega-3 fatty acid intake on offspring microbiome and immunity. PLoS ONE 9, e87181. 10.1371/journal.pone.0087181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld, K. M., Kang, N., Bienenstock, J. and Foster, J. A. (2011). Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 23, 255-264.e119. 10.1111/j.1365-2982.2010.01620.x [DOI] [PubMed] [Google Scholar]
- Noguera, J. C., Aira, M., Pérez-Losada, M., Domínguez, J. and Velando, A. (2018). Glucocorticoids modulate gastrointestinal microbiome in a wild bird. R. Soc. Open Sci. 5, 171743. 10.1098/rsos.171743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak, M. A. (2006). Five rules for the evolution of cooperation. Science 314, 1560-1563. 10.1126/science.1133755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki, S. and Searcy, W. A. (2005). Song and mate choice in birds: how the development of behavior helps us understand function. The Auk 122, 1-14. [Google Scholar]
- Ntranos, A. and Casaccia, P. (2018). The microbiome-gut-behavior axis: crosstalk between the gut microbiome and oligodendrocytes modulates behavioral responses. Neurotherapeutics 15, 31-35. 10.1007/s13311-017-0597-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony, S. M., Clarke, G., Borre, Y. E., Dinan, T. G. and Cryan, J. F. (2015). Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav. Brain Res. 277, 32-48. 10.1016/j.bbr.2014.07.027 [DOI] [PubMed] [Google Scholar]
- Pennisi, E. (2005). How did cooperative behavior evolve? Science 309, 93. 10.1126/science.309.5731.93 [DOI] [PubMed] [Google Scholar]
- Poutahidis, T., Kearney, S. M., Levkovich, T., Qi, P., Varian, B. J., Lakritz, J. R., Ibrahim, Y. M., Chatzigiagkos, A., Alm, E. J. and Erdman, S. E. (2013). Microbial symbionts accelerate wound healing via the neuropeptide hormone oxytocin. PLoS One 8, e78898. 10.1371/journal.pone.0078898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratte, Z. A., Patin, N. V., McWhirt, M. E., Caughman, A. M., Parris, D. J. and Stewart, F. J. (2018). Association with a sea anemone alters the skin microbiome of clownfish. Coral Reefs 37, 1119-1125. 10.1007/s00338-018-01750-z [DOI] [Google Scholar]
- Rajala, M. W., Patterson, C. M., Opp, J. S., Foltin, S. K., Young, V. B. and Myers, M. G.Jr. (2014). Leptin acts independently of food intake to modulate gut microbial composition in male mice. Endocrinology 155, 748-757. 10.1210/en.2013-1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando, O. J. and Simmons, R. A. (2015). I'm eating for two: parental dietary effects on offspring metabolism. Cell 161, 93-105. 10.1016/j.cell.2015.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulo, A. and Dantzer, B. (2018). Associations between glucocorticoids and sociality across a continuum of vertebrate social behavior. Ecol. Evol. 8, 7697-7716. 10.1002/ece3.4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud, J., Schradin, C. and Fusani, L. (2015). Corticosterone levels correlate with alloparental care in a sex-dependent manner in African striped mice, Rhabdomys pumilio. Ethology 121, 57-67. 10.1111/eth.12317 [DOI] [Google Scholar]
- Reichelt, A. C., Loughman, A., Bernard, A., Raipuria, M., Abbott, K. N., Dachtler, J., Van, T. T. H. and Moore, R. J. (2020). An intermittent hypercaloric diet alters gut microbiota, prefrontal cortical gene expression and social behaviours in rats. Nutr. Neurosci. 23, 613-627. 10.1080/1028415X.2018.1537169 [DOI] [PubMed] [Google Scholar]
- Ren, C. C., Sylvia, K. E., Munley, K. M., Deyoe, J. E., Henderson, S. G., Vu, M. P. and Demas, G. E. (2020). Photoperiod modulates the gut microbiome and aggressive behavior in Siberian hamsters. J. Exp. Biol. 223, jeb212548. 10.1242/jeb.212548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedman, M. L. (1982). The evolution of alloparental care and adoption in mammals and birds. Q Rev. Biol. 57, 405-435. 10.1086/412936 [DOI] [Google Scholar]
- Rivest, S. (2010). Interactions between the immune and neuroendocrine systems. In Neuroendocrinology: The Normal Neuroendocrine System (ed. Martini L.), pp. 43-53. Elsevier. [DOI] [PubMed] [Google Scholar]
- Robert, M. and Sorci, G. (2000). The evolution of obligate interspecific brood parasitism in birds. Behav. Ecol. 12, 128-133. 10.1093/beheco/12.2.128 [DOI] [Google Scholar]
- Roeselers, G., Mittge, E. K., Stephens, W. Z., Parichy, D. M., Cavanaugh, C. M., Guillemin, K. and Rawls, J. F. (2011). Evidence for a core gut microbiota in the zebrafish. ISME J. 5, 1595-1608. 10.1038/ismej.2011.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, C. A., Holekamp, K. E., Winters, A. D. and Theis, K. R. (2020). Body site-specific microbiota reflect sex and age-class among wild spotted hyenas. FEMS Microbiol. Ecol. 96, fiaa007. 10.1093/femsec/fiaa007 [DOI] [PubMed] [Google Scholar]
- Russell, A. F., Langmore, N. E., Cockbur, A. P., Astheimer, L. B. and Kilner, R. M. (2007). Reduced egg investment can conceal helper effects in cooperatively breeding birds. Science 317, 941-944. 10.1126/science.1146037 [DOI] [PubMed] [Google Scholar]
- Sachser, N., Zimmermann, T. D., Hennessy, M. B. and Kaiser, S. (2020). Sensitive phases in the development of rodent social behavior. Curr. Opin. Behav. Sci. 36, 63-70. 10.1016/j.cobeha.2020.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, T. R. and Mazmanian, S. K. (2015). Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 17, 565-576. 10.1016/j.chom.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Juan, P. A., Hendershot, J. N., Daily, G. C. and Fukami, T. (2020). Land-use change has host-specific influences on avian gut microbiomes. ISME J. 14, 318-321. 10.1038/s41396-019-0535-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitz, J., Lucki, I. and Drevets, W. C. (2009). 5-HT(1A) receptor function in major depressive disorder. Prog. Neurobiol. 88, 17-31. 10.1016/j.pneurobio.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seckl, J. R. and Meaney, M. J. (2004). Glucocorticoid programming. Ann. N. Y. Acad. Sci. 1032, 63-84. 10.1196/annals.1314.006 [DOI] [PubMed] [Google Scholar]
- Sgritta, M., Dooling, S. W., Buffington, S. A., Momin, E. N., Francis, M. B., Britton, R. A. and Costa-Mattioli, M. (2019). Mechanisms underlying microbial-mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron 101, 246-259.e6. 10.1016/j.neuron.2018.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon, G., Sega, D., Ringo, J. M., Hefetz, A., Zilber-Rosenberg, L. and Rosenberg, E. (2010). Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl Acad. Sci. USA 107, 20051-20056. 10.1073/pnas.1009906107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawkey, M. D., Firestone, M. K., Brodie, E. L. and Beissinger, S. R. (2009). Avian incubation inhibits growth and diversification of bacterial assemblages on eggs. PLoS ONE 4, e4522. 10.1371/journal.pone.0004522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverin, B., Wingfield, J., Stokkan, K.-A., Massa, R., Jarvinen, A., Andersson, N. A., Lambrechts, M., Sorace, A. and Blomqvist, D. (2008). Ambient temperature effects on photo induced gonadal cycles and hormonal secretion patterns in Great Tits from three different breeding latitudes. Horm. Behav. 54, 60-68. 10.1016/j.yhbeh.2008.01.015 [DOI] [PubMed] [Google Scholar]
- Smith, C. C., Srygley, R. B., Dietrich, E. I. and Mueller, U. G. (2017). Partitioning the effects of mating and nuptial feeding on the microbiome in gift-giving insects. Environ. Microbiol. Rep. 9, 104-112. 10.1111/1758-2229.12506 [DOI] [PubMed] [Google Scholar]
- Soares, M. C., Cable, J., Lima-Maximino, M. G., Maximino, C. and Xavier, R. (2019). Using fish models to investigate the links between microbiome and social behaviour: The next step for translational microbiome research? Fish. Fisheries 20, 640-652. 10.1111/faf.12366 [DOI] [Google Scholar]
- Soha, J. A. and Marler, P. (2000). A species-specific acoustic cue for selective song learning in the white-crowned sparrow. Anim. Behav. 60, 297-306. 10.1006/anbe.2000.1499 [DOI] [PubMed] [Google Scholar]
- Song, S. J., Sanders, J. G., Baldassarre, D. T., Chaves, J. A., Johnson, N. S., Piaggio, A. J., Stuckey, M. J., Nováková, E., Metcalf, J. L., Chomel, B. B.et al. (2019). Is there convergence of gut microbes in blood-feeding vertebrates? Philos. Trans. R. Soc. B Biol. Sci. 374, 20180249. 10.1098/rstb.2018.0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S. J., Sanders, J. G., Delsuc, F., Metcalf, J., Amato, K., Taylor, M. W., Mazel, F., Lutz, H. L., Winker, K., Graves, G. R.et al. (2020). Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. MBio 11, e02901-19. 10.1128/mBio.02901-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothart, M. R., Bobbie, C. B., Schulte-Hostedde, A. I., Boonstra, R., Palme, R., Mykytczuk, N. C. S. and Newman, A. E. M. (2016). Stress and the microbiome: linking glucocorticoids to bacterial community dynamics in wild red squirrels. Biol. Lett. 12, 20150875. 10.1098/rsbl.2015.0875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo, N., Chida, Y., Aiba, Y., Sonoda, J., Oyama, N., Yu, X.-N., Kubo, C. and Koga, Y. (2004). Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 558, 263-275. 10.1113/jphysiol.2004.063388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullam, K. E., Essinger, S. D., Lozupone, C. A., O'Connor, M. P., Rosen, G. L., Knight, R., Kilham, S. S. and Russell, J. A. (2012). Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol. Ecol. 21, 3363-3378. 10.1111/j.1365-294X.2012.05552.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T. A. (2017). Links between natural variation in the microbiome and host fitness in wild mammals. Integr. Comp. Biol. 57, 756-769. 10.1093/icb/icx104 [DOI] [PubMed] [Google Scholar]
- Swenson, R. O. (1997). Sex-role reversal in the tidewater goby, Eucyclogobius newberryi. Environ. Biol. Fishes 50, 27-40. 10.1023/A:1007352704614 [DOI] [Google Scholar]
- Sylvia, K. E. and Demas, G. E. (2018). A gut feeling: microbiome-brain-immune interactions modulate social and affective behaviors. Horm. Behav. 99, 41-49. 10.1016/j.yhbeh.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvia, K. E., Jewell, C. P., Rendon, N. M., St John, E. A. and Demas, G. E. (2017). Sex-specific modulation of the gut microbiome and behavior in Siberian hamsters. Brain Behav. Immun. 60, 51-62. 10.1016/j.bbi.2016.10.023 [DOI] [PubMed] [Google Scholar]
- Tabbaa, M., Lei, K., Liu, Y. and Wang, Z. (2017). Paternal deprivation affects social behaviors and neurochemical systems in the offspring of socially monogamous prairie voles. Neuroscience 343, 284-297. 10.1016/j.neuroscience.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taborsky, M. (1994). Sneakers, satellites, and helpers: parasitic and cooperative behavior in fish reproduction. Adv. Study Behav. 23, 1-100. 10.1016/S0065-3454(08)60351-4 [DOI] [Google Scholar]
- Thion, M. S. and Garel, S. (2017). On place and time: microglia in embryonic and perinatal brain development. Curr. Opin. Neurobiol. 47, 121-130. 10.1016/j.conb.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Tochitani, S., Ikeno, T., Ito, T., Sakurai, A., Yamauchi, T. and Matsuzaki, H. (2016). Administration of non-absorbable antibiotics to pregnant mice to perturb the maternal gut microbiota is associated with alterations in offspring behavior. PLoS ONE 11, e0138293. 10.1371/journal.pone.0138293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivers, R. L. (1971). The evolution of reciprocal altruism. Q Rev. Biol. 46, 35-57. 10.1086/406755 [DOI] [Google Scholar]
- Tung, J., Barriero, L. B., Burns, M. B., Grenier, J.-C., Lynch, J., Grieneisen, L. E., Altmann, J., Alberts, S. C., Blekhman, R. and Archie, E. A. (2015). Social networks predict gut microbiome composition in wild baboons. eLife 16, e05224. 10.7554/eLife.05224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veelen, H. P. J., Salles, J. F. and Tieleman, B. I. (2018). Microbiome assembly of avian eggshells and their potential as transgenerational carriers of maternal microbiota. ISME J. 12, 1375-1388. 10.1038/s41396-018-0067-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, J. F. A., Bui-Marinos, M. P. and Katzenback, B. A. (2018). Frog skin innate immune defences: sensing and surviving pathogens. Front. Immunol. 9, 3128. 10.3389/fimmu.2018.03128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu, E. F., Bercik, P., Verma-Gandhu, M., Huang, X. X., Blennerhassett, P., Jackson, W., Mao, Y., Wang, L., Rochat, F. and Collins, S. M. (2006). Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut 55, 182-190. 10.1136/gut.2005.066100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzani, A. and Viviani, B. (2015). Neuromodulatory properties of inflammatory cytokines and their impact on neuronal excitability. Neuropharmacology 96, 70-82. 10.1016/j.neuropharm.2014.10.027 [DOI] [PubMed] [Google Scholar]
- Vuong, H. E., Yano, J. M., Fung, T. C. and Hsiao, E. Y. (2017). The microbiome and host behavior. Annu. Rev. Neurosci. 40, 21-49. 10.1146/annurev-neuro-072116-031347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong, H. E., Pronovost, G. N., Williams, D. W., Coley, E. J. L., Siegler, E. L., Qiu, A., Kazantsev, M., Wilson, C. J., Rendon, T. and Hsiao, E. Y. (2020). The maternal microbiome modulates fetal neurodevelopment in mice. Nature 586, 281-286. 10.1038/s41586-020-2745-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada-Katsumata, A., Zurke, L., Nalyanya, G., Roelofs, M. L., Zhang, A. and Schal, C. (2015). Gut bacteria mediate aggregation in the German cockroach. Proc. Natl. Acad. Sci. USA 112, 15678-15683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, T. C., Guarner, F., Madsen, K., Cabana, M. D., Gibson, G., Hentges, E. and Sanders, M. E. (2011). Human gut microbiota and its relationship to health and disease. Nutr. Rev. 69, 392-403. 10.1111/j.1753-4887.2011.00402.x [DOI] [PubMed] [Google Scholar]
- Warne, R. W., Kirschman, L. and Zeglin, L. (2017). Manipulation of gut microbiota reveals shifting community structure shaped by host developmental windows in amphibian larvae. Integr. Comp. Biol. 57, 786-794. 10.1093/icb/icx100 [DOI] [PubMed] [Google Scholar]
- Warne, R. W., Kirschman, L. and Zeglin, L. (2019). Manipulation of gut microbiota during critical developmental windows affects host physiological performance and disease susceptibility across ontogeny. J. Anim. Ecol. 88, 845-856. 10.1111/1365-2656.12973 [DOI] [PubMed] [Google Scholar]
- Watson, S. E., Hauffe, H. C., Bull, M. J., Atwood, T. C., McKinney, M. A., Pindo, M. and Perkins, S. E. (2019). Global change-driven use of onshore habitat impacts polar bear faecal microbiota. ISME J. 13, 2916-2926. 10.1038/s41396-019-0480-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, S. A., Griffin, A. S. and Gardner, A. (2007). Evolutionary explanations for cooperation. Curr. Biol. 17, R661-R672. 10.1016/j.cub.2007.06.004 [DOI] [PubMed] [Google Scholar]
- White, J., Richard, M., Massot, M. and Meylan, S. (2011). Cloacal bacterial diversity increases with multiple mates: evidence of sexual transmission in female common lizards. PLoS ONE 6, e22339. 10.1371/journal.pone.0022339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker, D. J., Gerlach, N. M., Slowinski, S. P., Corcoran, K. P., Winters, A. D., Soini, H. A., Novotny, M. V., Ketterson, E. D. and Theis, K. R. (2016). Social environment has a primary influence on the microbial and odor profiles of a chemically signaling songbird. Front. Ecol. Evol. 4, 90. 10.3389/fevo.2016.00090 [DOI] [Google Scholar]
- Whittaker, D. J., Slowinski, S. P., Greenberg, J. M., Alian, O., Winters, A. D., Ahmad, M. M., Burrell, M. J. E., Soini, H. A., Novotny, M. V., Ketterson, E. D.et al. (2019). Experimental evidence that symbiotic bacteria produce chemical cues in a songbird. J. Exp. Biol. 222, jeb202978. 10.1242/jeb.202978 [DOI] [PubMed] [Google Scholar]
- Williams, C. L., Garcia-Reyero, N., Martyniuk, C. J., Tubbs, C. W. and Bisesi, J. H.Jr. (2020). Regulation of endocrine systems by the microbiome: perspectives from comparative animal models. Gen. Comp. Endocrinol. 292, 113437. 10.1016/j.ygcen.2020.113437 [DOI] [PubMed] [Google Scholar]
- Woolfenden, G. E. (1975). Florida scrub jay helpers at the nest. The Auk 92, 1-15. 10.2307/4084414 [DOI] [Google Scholar]
- Xavier, R., Mazzei, R., Pérez-Losada, M., Rosado, D., Santos, J. L., Veríssimo, A. and Soares, M. C. (2019). A risky business? habitat and social behavior impact skin and gut microbiomes in caribbean cleaning gobies. Front. Microbiol. 10, 716. 10.3389/fmicb.2019.00716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, T., Yasuo, S., Watanabe, M., Iigo, M., Yamamura, T., Hirunagi, K. and Ebihara, S. (2003). Light-induced hormone conversion of T4 to T3 regulates photoperiodic response of gonads in birds. Nature 426, 178-181. 10.1038/nature02117 [DOI] [PubMed] [Google Scholar]
- Yuan, M. L., Dean, S. H., Longo, A. V., Rothermel, B. B., Tuberville, T. D. and Zamudio, K. R. (2015). Kinship, inbreeding and fine-scale spatial structure influence gut microbiota in a hindgut-fermenting tortoise. Mol. Ecol. 24, 2521-2536. 10.1111/mec.13169 [DOI] [PubMed] [Google Scholar]
- Zha, Y., Eiler, A., Johansson, F. and Svanbäck, R. (2018). Effects of predation stress and food ration on perch gut microbiota. Microbiome 6, 28. 10.1186/s40168-018-0400-0 [DOI] [PMC free article] [PubMed] [Google Scholar]