Abstract
Background
Long non-coding RNA (lncRNA) plasmacytoma variant translocation 1 (PVT1) plays a crucial role in non-small cell lung cancer (NSCLC). Nonetheless, regulatory effects of PVT1 on functions of NSCLC cells remain blurry.
Methods
Relative expression levels of PVT1, miR-551b and FGFR1 mRNA in tumor tissues and cells were examined employing quantitative real-time polymerase chain reaction (qRT-PCR); CCK-8 and BrdU assays were utilized for measuring cell viability and proliferation of H1299 and A549 cells; cell migration and invasion were detected deploying Transwell assay; dual-luciferase assay was used for the validation of binding sequence between PVT1 and miR-551b. FGFR1 expression in protein level was quantified employing Western blot.
Results
PVT1 was highly expressed in NSCLC tissues and cell lines, whereas miR-551b expression was down-regulated. Overexpression of PVT1 potentiated viability, proliferation, migration and invasion of NSCLC cells while miR-551b inhibited the biological behaviors mentioned above. MiR-551b was predicted and then confirmed as a direct downstream target of PVT1. Meanwhile, a negative correlation was observed between PVT1 expression and miR-551b expression in NSCLC tissues. Besides, PVT1 could increase FGFR1 expression by repressing miR-551b expression.
Conclusion
PVT1 promotes the proliferation, migration and invasion of NSCLC cells by indirectly mediating FGFR1 via targeting miR-551b.
Keywords: lncRNA PVT, miR-551b, FGFR1, NSCLC
Introduction
Lung cancer tops the list among the leading causes contributing to cancer-related deaths, and non-small cell lung cancer (NSCLC) accounts for more than 80% of all cases.1 About 60% of lung cancer patients are at the advanced stage by the time of diagnosis, which cannot be cured surgically. Due to the resistance of NSCLC cells to chemotherapy, radiotherapy and targeted therapy, the treatment of advanced NSCLC remains particularly challenging.1,2
Long non-coding RNA (lncRNA) plasmacytoma variant translocation 1 (PVT1) can facilitate the progression of some cancers.3 Expression level of PVT1 is significantly increased in NSCLC tissues and cell lines; via targeting miR-361-3p and up-regulating SOX9 expression, PVT1 potentiates the proliferation, migration and invasion of NSCLC cells.4 In another study, knockdown of PVT1 remarkably suppresses the growth of xenograft tumors in nude mice.5 These studies substantiate that PVT1 is a promising therapeutic target of NSCLC, whereas the detailed mechanism of the promoting effects of PVT1 on NSCLC procession warrants further investigation.
MicroRNA (miRNA) also figures prominently in cancer biology. MiR-551b works as a tumor suppressor. In gastric cancer, miR-551b expression is underexpressed and overexpression of miR-551b suppresses the proliferation, migration and invasion of gastric cancer cell lines SGC-7901 and MKN-45.6 Elevated expression of miR-551b indicates a more favorable prognosis of lung adenocarcinoma,7 suggesting the tumor-suppressive function of miR-551b in NSCLC. Nonetheless, the explicit biological function and mechanism of miR-551b in NSCLC are still obscure.
Herein, the biological function of PVT1 and miR-551b in NSCLC and their regulatory mechanism were investigated. We demonstrated that PVT1, which was highly expressed in NSCLC tissues, promoted NSCLC progression via repressing miR-551b expression and up-regulating fibroblast growth factor receptor-1 (FGFR1) expression.
Materials and Methods
Clinical Sample Collection
This study was approved by the Ethics Committees of the First Affiliated Hospital of Zhengzhou University (Zhengzhou, Henan Province, China). Written informed consent was acquired from each patient. All protocols were conducted in line with the principles of the Declaration of Helsinki. Fifty-two NSCLC patients (38 males and 14 females, aged 52.4 ± 6.2 years old) who did not receive radiotherapy, chemotherapy or targeted therapy before surgery were recruited from the First Affiliated Hospital of Zhengzhou University from April 2017 to April 2019, and 40 patients were diagnosed as lung adenocarcinoma and the remaining 12 patients were diagnosed as lung squamous cell carcinoma. Cancer tissues and adjacent tissues were collected during the surgery, and all tissue samples were stored in the refrigerator at −80°C immediately after the resection.
Cell Culture and Transfection
BEAS-2B, H1299, H1975, A549 and H2228 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). They were maintained in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS, Beyotime Biotechnology, Shanghai, China), 100 U/mL penicillin and 100 μg/mL streptomycin (Beyotime Biotechnology, Shanghai, China), and cultured in a thermostatic incubator perfused with 5% CO2 at 37°C. Constructions of overexpression plasmid of PVT1, shRNA targeting PVT1, miR-551b mimic, miR-551b inhibitor and their corresponding controls were performed by GenePharma (Shanghai, China). Before the transfection, H1299 and A549 cells were cultured with RPMI-1640 medium without serum for 24 h. The Lipofectamine® 3000 (Invitrogen, Carlsbad, CA, USA) was mixed with plasmids and oligonucleotide mentioned above, and then the mixture was added to the medium. After 24 h of culture, the medium was changed to complete medium. After another 24 h of culture, the transfection efficiency was detected with quantitative real-time polymerase chain reaction (qRT-PCR).
qRT-PCR
Extraction of total RNA from H1299 and A549 cells was performed utilizing TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Detection of the concentration and purity of the total RNA was conducted employing Thermo NanoDrop 2000, and the integrity of total RNA was detected with gel electrophoresis system Agilent-2100. Reverse transcription of RNA was completed utilizing PrimeScript™ RT Reagent kit (Invitrogen, Carlsbad, CA, USA). qRT-PCR was then performed utilizing SYBR Green PCR kit (Takara, Dalian, China) on ABI 7500 FAST Real-Time PCR system (Applied Biosystems, Waltham, MA, UK). GAPDH was employed as an internal reference for the quantification of PVT1 and FGFR1 mRNA, and U6 was employed as an internal reference for the quantification of miR-551b expression. Calculation of the relative expression level was performed according to 2−ΔΔCt formula, and synthesis of primers was supported by BGI (Shenzhen, China). Detailed primer sequences utilized in this study were as follows: miR-551b forward 5ʹ-GCGACCCATACTTGGTTTCAG-3ʹ, miR-551b reverse 5ʹ-TCGTGAGATGAAGCACTGTAG-3ʹ; U6 forward 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, U6 reverse 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ; PVT1 forward 5ʹ-TTGGCACATACAGCCATCAT-3ʹ, PVT1 reverse 5ʹ-GCAGTAAAAGGGGAACACCA-3ʹ; GAPDH forward 5ʹ-GAAGGTGAAGGTCGGAGTC-3ʹ, GAPDH reverse 5ʹ-GAAGATGGTGATGGGATTTC-3ʹ; FGFR1 forward 5ʹ-CACATCGAGGTGAACGGGAGTAAG-3ʹ, FGFR1 reverse 5ʹ-CGCATCCTCAAAGGAGACATTCC-3ʹ.
Cell Counting Kit-8 (CCK-8) Assay
H1299 or A549 cells in logarithmic growth phase were transferred into 96-well plates (100 μL of cell suspension, 2×103 cells in each well). Ten microliters of CCK-8 solution (Beyotime Biotechnology, Shanghai, China) was added into each well at 24, 48, 72 and 96 h, respectively, and the cells were incubated for 1 h. Following that, the absorbance at 450 nm of each well was detected with a microplate reader.
BrdU Assay
The single-cell suspension was prepared with A549 cells and H1299 cells in the logarithmic growth phase. Subsequently, the cells were inoculated into a 24-well plate at the density of 1×105 cells/well. BrdU reagent (Beyotime Biotechnology, Shanghai, China) was supplemented into each well, and after 12 h of culture, the cells were incubated with anti-BrdU antibody (Beyotime Biotechnology, Shanghai, China) at room temperature for 2 h. After that, the cells were counterstained with DAPI staining solution (Beyotime Biotechnology, Shanghai, China). Then, the number of BrdU positive cells and the total number of DAPI positive cells were counted under a fluorescence microscope (Olympus, Tokyo, Japan). Cell proliferation rate=BrdU positive cells/DAPI positive cells × 100%.
Transwell Assay
In the migration assay, H1299 and A549 cell suspension was prepared with serum-free medium, and the cell density was modulated to 5×105/mL. One hundred microliter of cell suspension was added into the upper compartment of Transwell chamber (Corning, Beijing, China), and 600 μL of RPMI-1640 medium containing 10% FBS was added to the lower compartment. After 24 h, cells remaining in the upper compartment were wiped off with cotton swabs, and the migrated cells were fixed with 4% paraformaldehyde and stained with crystal violet solution. In the invasion assay, the Transwell chambers were pre-coated with Martigel® (30 μg/well; BD, San Jose, CA, USA) and the other procedures were as the same as in the migration assay. Five visual fields of a membrane were randomly selected under the microscope for counting the cells passing through the membranes.
Dual-Luciferase Reporter Assay
The predicted binding sequences between miR-551b and PVT1, or between miR-551b and FGFR1 3ʹ-UTR were amplified and cloned into pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA) for the construction of wild type (WT) reporter vectors (lncPVT1 WT and FGFR1 WT). Then the sequences were mutated with GeneArt™ Site-Directed Mutagenesis PLUS System (Thermo Fisher Scientific, Waltham, MA, USA). The mutated sequences were amplified and cloned into pmirGLO vector (Promega, Madison, WI, USA) for the construction of mutant type (MUT) reporter vectors (PVT1 MUT and FGFR1 MUT). Then the above reporter vectors were co-transfected into H1299 cells or A549 cells with miR-551b mimic, miR-551b inhibitor or their corresponding negative controls with Lipofectamine® 3000 (Invitrogen, Carlsbad, CA, USA). After 48 h, determination of luciferase activity was performed with Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). The luciferase activity of firefly was normalized to that of renilla.
Western Blot
Extraction of total protein of A549 and H1299 cells was performed with RIPA lysis buffer (Beyotime, Shanghai, China), protein concentration was determined by BCA method, and then the protein samples were mixed with loading buffer and denatured in boiling water. Subsequently, 15 μL of protein sample in each group was loaded into each well and the protein was dissolved by SDS-PAGE. After the protein was transferred to PVDF membrane (Millipore, Bedford, MA, USA), the PVDF membrane was blocked with 5% skimmed milk at room temperature for 1 h. Then the primary antibody (rabbit anti-FGFR1 monoclonal antibody, ab76464, Abcam, Shanghai, China, 1:1000) was used to incubate the PVDF membrane at 4°C overnight and then rinsed with TBST 3 times with 5 min each time. The corresponding secondary antibody (goat anti-rabbit IgG, ab205718, Abcam, Shanghai, China. 1:2000) was then applied to incubate the membrane at room temperature for 1.5 h before the membrane. After that, the membrane was rinsed with TBST 3 times with 5 min each time. Next, the protein band was developed employing ECL kit (Beyotime Biotechnology, Shanghai, China), and the gray values of protein bands were quantitatively analyzed utilizing ImageJ (National Institutions of Health, Bethesda, MD, USA). GAPDH was used as the internal reference.
In vivo Experiments
The in vivo experiments were reviewed and approved by the Animal Experimentation Ethics Committee of the First Affiliated Hospital of Zhengzhou University (Approval Number: 2021-06). Twenty male BALB/c nude mice (6–8 weeks old, average weight: 22.5 ± 2.0 g) were purchased from Zhejiang Province Experimental Animal Center (Hangzhou, China). Mice were maintained in standard housing conditions (23 °C, 40% humidity, 12 h light cycles, and free access to food and water). The animals were randomly divided into four groups (Blank vector group, PVT1 overexpression group, PVT1 + NC mimics group, PVT1 + miR-511b, n = 5 in per group). H1299 cells (2×107 cells/per mouse) were injected into each mouse via tail vein. Three weeks later, the mice were euthanized, and lung tissues were obtained. The lung tissues were fixed in 10% neutral formalin, and embedded in paraffin. Then hematoxylin/eosin staining was performed for pathological examination of lung metastatic nodules of the mice.
Statistical Analysis
SPSS 23.0 statistical analysis software (SPSS Inc., Chicago, IL, USA) was applied for statistical analysis. The measurement data were expressed by mean ± standard deviation (x±s). For data with normal distribution, Student’s t-test was used for the comparison between the two groups; for data with skewed distribution, the rank test was performed. One-way ANOVA was used for the comparison among multiple groups. P < 0.05 indicated statistical significance.
Results
Expression of PVT1 in NSCLC Tissues and Cell Lines Was Aberrantly Up-Regulated
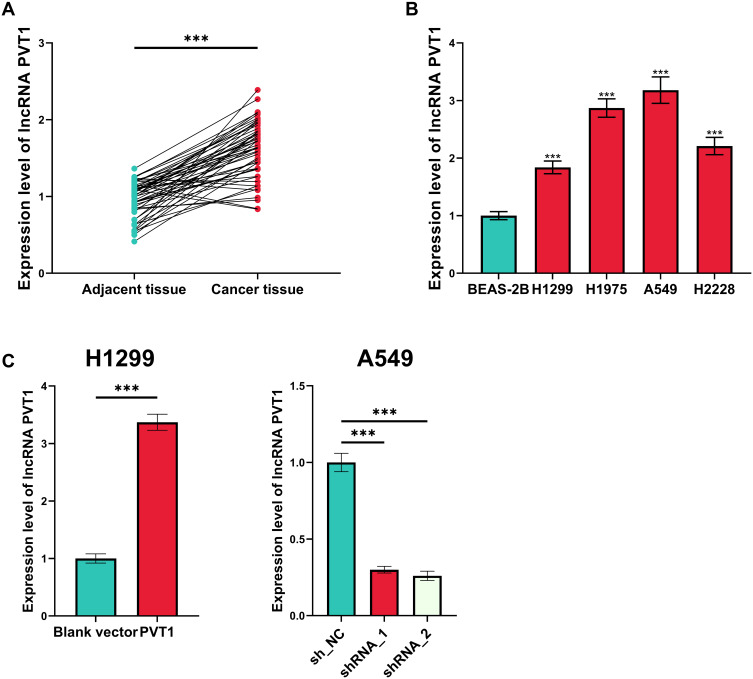
First of all, the expression level of PVT1 in NSCLC tissues and cell lines was evaluated, and as shown, in NSCLC tissues (and NSCLC cell lines), it was significantly higher than that in adjacent tissues and normal pulmonary epithelial cell line BEAS-2B (Figure 1A and B). Among the NSCLC cells lines, the expression level of PVT1 was the highest in A549 cells and the lowest in H1299 cells, so PVT1 knockdown and overexpression cell models were established with A549 and H1299 cells, respectively, which was validated by qRT-PCR (Figure 1C).
Figure 1.
Expression of PVT1 was up-regulated in NSCLC. (A) qRT-PCR was performed for quantifying the expression of PVT1 in NSCLC tissues and adjacent tissues. (B) qRT-PCR was conducted for detecting the expression of PVT1 in normal lung epithelial cells and NSCLC cell lines. (C) Transfection efficiency of PVT1 overexpression plasmid and shRNAs was detected utilizing qRT-PCR. ***Denotes P < 0.001.
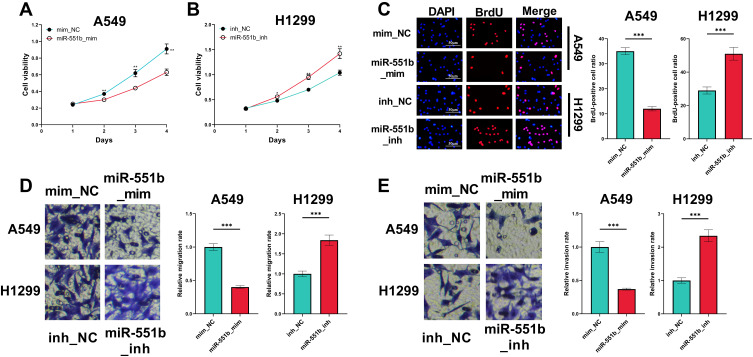
PVT1 Promoted Proliferation, Migration and Invasion of NSCLC Cells
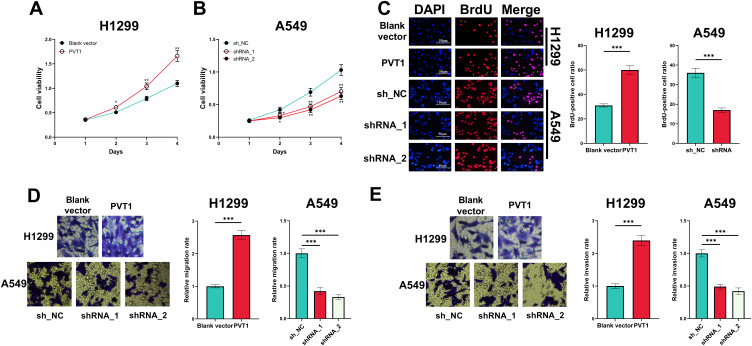
In CCK-8 experiment, overexpression of PVT1 significantly potentiated the viability of H1299 cells whereas PVT1 silencing suppressed the growth of A549 cells (Figure 2A and B). Moreover, BrdU assay indicated that PVT1 overexpression significantly promoted the proliferation of H1299 cells, whereas after the knockdown of PVT1, the proliferation of A549 cells was suppressed (Figure 2C). Additionally, overexpression of PVT1 promoted the migration and invasion of H1299 cells while PVT1 knockdown worked oppositely (Figure 2D and E).
Figure 2.
Effect of PVT1 on the proliferation, migration and invasion of NSCLC cells. (A and B) CCK-8 assay was performed for detecting effects of overexpression and knockdown of PVT1 on the viability of H1299 and A549 cells. (C) BrdU assay was used for detecting effects of overexpression and knockdown of PVT1 on the proliferation of H1299 and A549 cells. (D and E) The effects of overexpression and knockdown of PVT1 on migration and invasion of H1299 and A549 cells were analyzed utilizing Transwell assay. *, ** and ***Represent P < 0.05, P < 0.01 and P < 0.001, respectively.
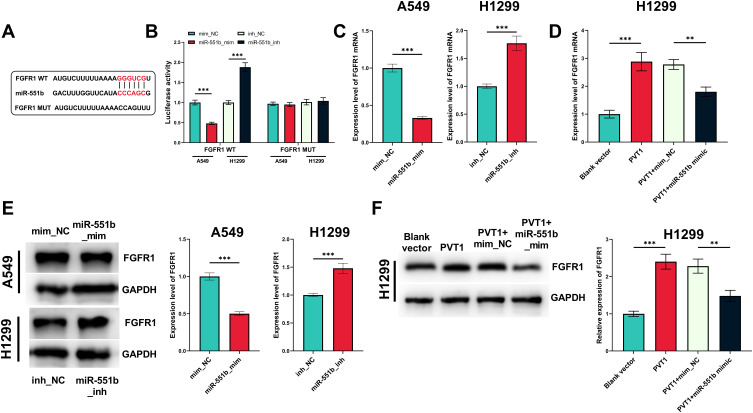
MiR-551b Was a Downstream Target of PVT1
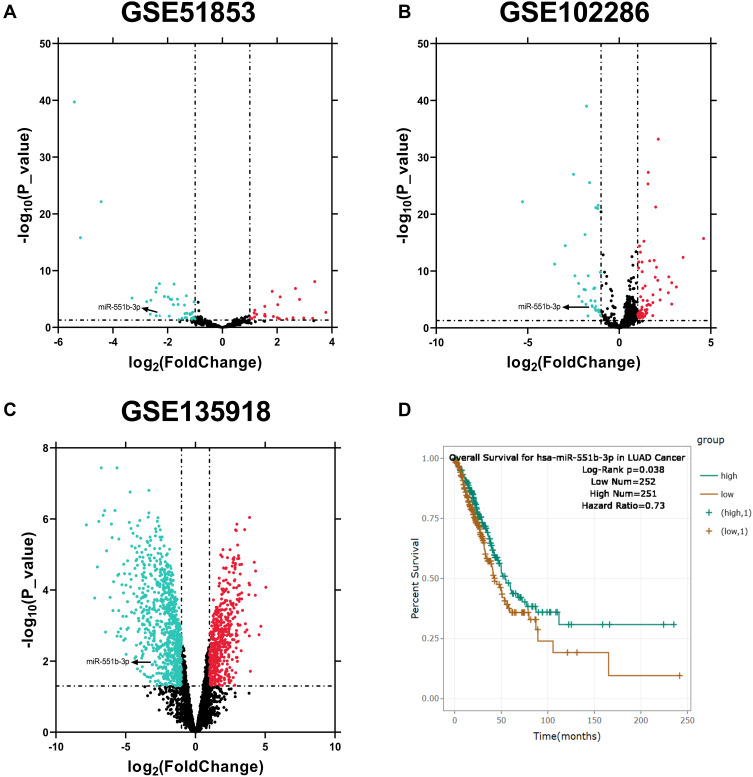
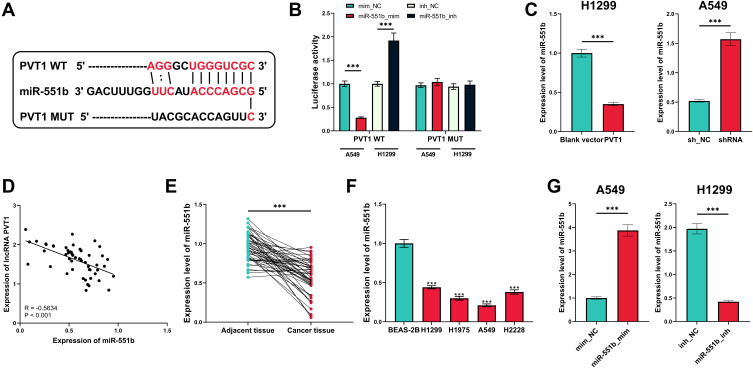
The alterations of miRNA expression profile in NSCLC tumorigenesis were detected through reanalyzing Gene Expression Omnibus (GEO) datasets, including GSE51853, GSE102286 and GSE135918. It was indicated that, in all of the three datasets, the expression of miR-551b in NSCLC tissues was markedly down-regulated by the comparison with that in normal lung tissues (Figure 3A–C). Meanwhile, through searching ENCORI database, it was suggested that miR-551b high expression was a favorable indicator of longer survival time in patients with lung adenocarcinoma (Figure 3D). These data suggested that miR-551b might be a tumor suppressor in NSCLC. Intriguingly, miR-551b was predicted as one of direct downstream targets of PVT1 by ENCORI database (Figure 4A). Through dual-luciferase assay, the predicted binding sequence between miR-551b and PVT1 was confirmed (Figure 4B). Furthermore, qRT-PCR suggested that overexpression of PVT1 induced a significant reduction of miR-551b expression in H1299 cells while knockdown of PVT1 up-regulated the expression of miR-551b in A549 cells (Figure 4C). Additionally, with Pearson’s correlation analysis, a negative correlation between expression levels of PVT1 and miR-551b was observed in NSCLC tissues (Figure 4D). We also evaluated the expression level of miR-551b in NSCLC, and as shown, qRT-PCR suggested that miR-511b expression was markedly down-regulated in NSCLC tissues and cell lines (Figure 4E and F). Given that miR-551b was the lowest in A549 cells and the highest in H1299 cells among NSCLC cell lines, miR-551b mimics were transfected into A549 cells, and miR-551b inhibitors were transfected into H1299 cells for the subsequent experiments (Figure 4G).
Figure 3.
MiRNAs with aberrant expression in NSCLC were screened through multiple datasets. (A–C) MiRNAs with aberrant expression in NSCLC tissues compared with normal lung tissues were analyzed, respectively, in GSE51853, GSE102286 and GSE135918. (D) The correlation between miR-551b expression and the overall survival rate of patients with lung adenocarcinoma was analyzed with ENCORI database.
Figure 4.
MiR-551b was confirmed to be one of the downstream targets of PVT1. (A) The binding sequence between PVT1 and miR-551b was predicted by StarBase database. (B) The binding site between miR-551b and PVT1 was verified by dual-luciferase reporter gene assay. (C) qRT-PCR was conducted for detecting effects of overexpression and knockdown of PVT1 on miR-551b expression in NSCLC cells. (D) Correlation between PVT1 expression and miR-551b expression in NSCLC samples. (E) Expression of miR-551b in NSCLC tissues and normal tissues adjacent to cancer was detected by qRT-PCR. (F) Expression of miR-551b in human normal lung epithelial cells and NSCLC cell lines was quantified by qRT-PCR. (G) qRT-PCR was utilized for verifying the transfection efficiency of miR-551b mimic and miR-551b inhibitor. ***Denotes P < 0.001.
MiR-551b Impeded the Proliferation, Migration and Invasion of NSCLC Cells
CCK-8 and BrdU assays indicated that the overexpression of miR-551b in A549 cells repressed the viability and proliferation, while the inhibition of miR-551b expression facilitated the viability and proliferation of H1299 cells (Figure 5A–C). In Transwell experiment, migration and invasion of A549 cells transfected with miR-551b mimics were remarkably impeded, whereas in H1299 cells transfected with miR-551b inhibitors, the migration and invasion were promoted (Figure 5D and E). These results suggested that miR-551b was a tumor suppressor in NSCLC.
Figure 5.
The effects of miR-551b on the malignant phenotypes of NSCLC cells. (A and B) The effects of overexpression or inhibition of miR-551b on the viability of A549 and H1299 cells were detected by CCK-8 method. (C) The effects of overexpression or inhibition of miR-551b on the proliferation of A549 and H1299 cells were assessed utilizing BrdU assay. (D and E) The effects of overexpression or inhibition of miR-551b on migration and invasion of A549 and H1299 cells were detected employing Transwell assay. *, ** and ***Represent P < 0.05, P < 0.01 and P < 0.001, respectively.
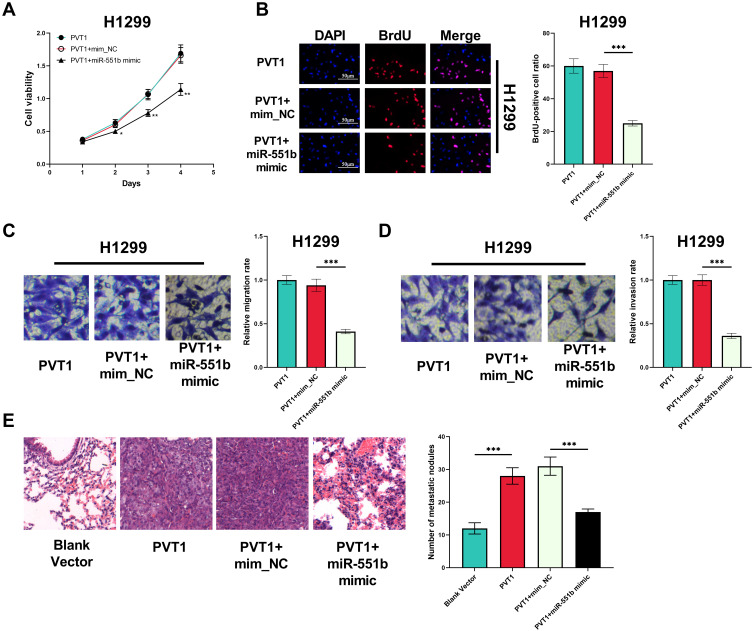
MiR-551b Counteracted the Effects of PVT1 in NSCLC
Next, we transfected miR-551b mimics into H1299 cells with PVT1 overexpression. CCK-8 and BrdU experiments indicated that the enhancement of viability and proliferation of H1299 cells induced by PVT1 overexpression was abrogated by miR-551b mimics (Figure 6A and B). Meanwhile, the promoting effect of PVT1 on the migration and invasion of H1299 was reversed by miR-551b mimics (Figure 6C and D). Additionally, the metastatic potential of H1299 cells was investigated in vivo with mice model with lung metastasis. As shown, PVT1 overexpression markedly promoted the lung metastasis of H1299 cells (Figure 6E). Based upon these results, it was concluded that PVT1 could exhibit its cancer-promoting role by specifically attenuating the expression of miR-551b.
Figure 6.
MiR-551b counteracted the biological function of PVT1 in NSCLC cells. (A) CCK-8 assay was conducted for detecting the effect of co-transfection of PVT1 overexpression plasmid and miR-551b mimic on the viability of H1299 cells. (B) The effects of co-transfection of PVT1 overexpression plasmid and miR-551b mimic on the proliferation of H1299 cells were assessed utilizing BrdU assay. (C and D) The effects of co-transfection of PVT1 overexpression plasmid and miR-551b mimic on the migration and invasion of H1299 cells were detected by Transwell assay. (E) The metastatic potential of H1299 cells in each group was examined with mice model, and HE staining was used to detect the metastatic nodules in the lung tissues of the mice. *, ** and ***Represent P < 0.05, P < 0.01 and P < 0.001, respectively.
PVT1 Indirectly Regulated FGFR1 via miR-551b
Next, the downstream mechanism of miR-551b was explored. The prediction of downstream target genes pertaining to miR-551b was performed utilizing miRanda and miRmap databases. Thirty-four genes were co-predicted to be potential targets of miR-551b (Supplementary Figure 1A). FGFR1 is among them (Supplementary Figure 1B and C) and is confirmed to promote NSCLC progression.8 In dual-luciferase reporter assay, miR-551b mimics remarkably repressed the luciferase activity of FGFR1 WT reporter vector while miR-551b inhibitors worked oppositely; no significant alteration was observed in the luciferase activity of FGFR1 MUT reporter vector (Figure 7A and B). Furthermore, qRT-PCR indicated that miR-551b negatively regulated the expression of FGFR1 at both mRNA and protein expression levels; PVT1 positively regulated FGFR1 expression in NSCLC cells, which was partly abolished by the co-transfection of miR-551b (Figure 7C–F). Collectively, it was confirmed that FGFR1 was a target gene of miR-551b and miR-551b indirectly increased the expression of FGFR1 through repressing miR-551b expression.
Figure 7.
The regulatory functions of PVT1 and miR-551b on FGFR1. (A) The binding sequence between miR-551b and FGFR1 was predicted utilizing miRanda and miRmap. (B) The binding site between miR-551b and FGFR1 was verified using a dual-luciferase reporter gene experiment. (C and D) The effects of miR-551b and PVT1 on FGFR1 mRNA were detected by qRT-PCR. (E and F) The effects of miR-551b and PVT1 on FGFR1 protein expression level were detected employing Western blot. ** and ***Represent P < 0.01 and P < 0.001, respectively.
Discussion
The complicated molecular mechanism of NSCLC progression is far away from being fully clarified.9 In recent years, non-coding RNAs draw a lot of attention in the research of cancer biology.10–12 In previous studies, the role of lncRNA PVT1 in NSCLC has been explored preliminarily.4,5,13,14 It is reported that the expression level of PVT1 is enhanced in NSCLC tissues and cell lines, and knockdown of PVT1 impedes proliferation, migration and invasion of NSCLC cells.13 Additionally, knockdown of PVT1 sensitizes A549 cells to cisplatin.14 In our current study, we demonstrated that the expression of lncRNA PVT1 was markedly increased in NSCLC tissues and cell lines; in vitro experiments indicated that PVT1 could facilitate proliferation, migration and invasion of NSCLC cells. Our demonstrations were consistent with previous reports, which suggested that PVT1 was an oncogenic lncRNA in NSCLC and could probably be a promising therapeutic target for this disease.
The role of miR-551b is explored in cancer biology in some previous studies. For example, the expression of miR-551b is significantly down-regulated in colorectal cancer tissues and cell lines, and low expression of miR-551b is confirmed as an independent predictor of detrimental prognosis of colorectal cancer patients; in vitro experiments suggested that miR-551b repressed proliferation, migration and invasion of colorectal cancer cells.15 In ovarian cancer, the expression of miR-551b is positively correlated with the disease progression; miR-551b expedites the proliferation of ovarian cancer cells in vitro and accelerates the growth of tumors in vivo by inhibiting FOXO3 and TRIM31 expression levels.16 These studies suggest that miR-551b exerts distinct biological effects in different cancers. Highly expressed miR-551b is, reportedly, associated with better prognosis of patients with lung adenocarcinoma,7 suggesting miR-551b may be an oncomiR. In this study, we confirmed that miR-551b could suppress the malignant biological behaviors of NSCLC cells and was a downstream target of PVT1, and the tumor-promoting function of PVT1 was partially dependent on its regulatory function on miR-551b. Our data validated the tumor-suppressive properties of miR-551b in NSCLC and partly explained the mechanism of its dysregulation.
FGFR1, a promising therapeutic target for NSCLC, can facilitate the progression of NSCLC. After FGFR1 is silenced with siRNA, the colony formation, migration and invasion of H522 cells and H460 cells are remarkably inhibited while apoptosis is promoted.17 Besides, the expression of FGFR1 in NSCLC is significantly increased and high expression of FGFR1 implies an unfavorable prognosis of NSCLC patients; knockdown of FGFR1 sensitizes PC9 cells to gefitinib.18 FGFR1 is proved to activate ERK signaling pathway, thereupon up-regulating expression of SOX2 and promoting the proliferation and migration of NSCLC cells.19 Unfortunately, although the role of FGFR1 in NSCLC progression is confirmed and its inhibitors are designed to treat NSCLC, most of the studies are still at preclinical stage and the application of FGFR1 inhibitors in the clinical treatment of NSCLC is still facing great challenges.20,21 In the present study, FGFR1 was validated to be a novel target of miR-551b and PVT1 was proved to indirectly increase the expression of FGFR1 through repressing miR-551b expression. Our data suggested that the competitive endogenous RNA (ceRNA) network formed by PVT1, miR-551b and FGFR1 contributed to the dysregulation of FGFR1 in NSCLC.
All in all, our work demonstrates that PVT1 can indirectly increase FGFR1 expression through targeted inhibition of miR-551b expression, thereupon exhibiting its oncogenic role in promoting proliferation, migration and invasion of NSCLC cells. Our current study further explains the underlying mechanism by which PVT1 facilitates NSCLC progression and provides theoretical basis for developing novel treatment strategies for NSCLC. Nonetheless, the ceRNA mechanism presented by this study is only based on in vitro experiments and in vivo studies are required in the following studies to make our conclusion more convincing. Additionally, the regulatory effects of PVT1 on the downstream pathways and genes of FGFR1 should be investigated in the future. What’s more, the values of PVT1 in predicting the prognosis of NSCLC should be explored in the following work, with more patients enrolled.
Acknowledgment
We thank Hubei Yican Health Industry Co., Ltd for its linguistic assistance during the preparation of this manuscript.
Funding Statement
The study was funded by the National Natural Science Foundation of China (NO. U1904142).
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17:637–658. doi: 10.1038/nrc.2017.84 [DOI] [PubMed] [Google Scholar]
- 2.Osmani L, Askin F, Gabrielson E, Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52:103–109. doi: 10.1016/j.semcancer.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu D, Luo P, Wang Q, Ye Y, Wang B. lncRNA PVT1 in cancer: a review and meta-analysis. Clin Chim Acta. 2017;474:1–7. doi: 10.1016/j.cca.2017.08.038 [DOI] [PubMed] [Google Scholar]
- 4.Qi G, Li L. Long non-coding RNA PVT1 contributes to cell growth and metastasis in non-small-cell lung cancer by regulating miR-361-3p/SOX9 axis and activating Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2020;126:110100. doi: 10.1016/j.biopha.2020.110100 [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Zhang Q, Sun Y, Shao F. Long non-coding RNA PVT1 regulates BAMBI to promote tumor progression in non-small cell lung cancer by sponging miR-17-5p. Onco Targets Ther. 2020;13:131–142. doi: 10.2147/OTT.S217335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan H, Chen Z, Bai S, et al. Molecular mechanisms of lncRNA SMARCC2/miR-551b-3p/TMPRSS4 axis in gastric cancer. Cancer Lett. 2018;418:84–96. doi: 10.1016/j.canlet.2018.01.032 [DOI] [PubMed] [Google Scholar]
- 7.Lin K, Xu T, He BS, et al. MicroRNA expression profiles predict progression and clinical outcome in lung adenocarcinoma. Onco Targets Ther. 2016;9:5679–5692. doi: 10.2147/OTT.S111241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan Q, Wang Z, Wang Q, et al. A novel FGFR1-binding peptide exhibits anti-tumor effect on lung cancer by inhibiting proliferation and angiogenesis. Int J Biol Sci. 2018;14(10):1389–1398. doi: 10.7150/ijbs.24739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu Z, He D, Deng X, et al. An eight-long non-coding RNA signature as a candidate prognostic biomarker for lung cancer. Oncol Rep. 2016;36:215–222. doi: 10.3892/or.2016.4817 [DOI] [PubMed] [Google Scholar]
- 10.Fang C, Wang L, Gong C, Wu W, Yao C, Zhu S. Long non-coding RNAs: how to regulate the metastasis of non-small-cell lung cancer. J Cell Mol Med. 2020;24:3282–3291. doi: 10.1111/jcmm.15054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonetti A, Assaraf YG, Veltsista PD, El Hassouni B, Tiseo M, Giovannetti E. MicroRNAs as a drug resistance mechanism to targeted therapies in EGFR-mutated NSCLC: current implications and future directions. Drug Resist Updat. 2019;42:1–11. doi: 10.1016/j.drup.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 12.Osielska MA, Jagodziński PP. Long non-coding RNA as potential biomarkers in non-small-cell lung cancer: what do we know so far? Biomed Pharmacother. 2018;101:322–333. doi: 10.1016/j.biopha.2018.02.099 [DOI] [PubMed] [Google Scholar]
- 13.Wei CM, Zhao XF, Qiu HB, Ming Z, Liu K, Yan J. The long non-coding RNA PVT1/miR-145-5p/ITGB8 axis regulates cell proliferation, apoptosis, migration and invasion in non-small cell lung cancer cells. Neoplasma. 2020;67(4):802–812. doi: 10.4149/neo_2020_190723N657 [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Han X, Hu Z, Chen L. The PVT1/miR-216b/Beclin-1 regulates cisplatin sensitivity of NSCLC cells via modulating autophagy and apoptosis. Cancer Chemother Pharmacol. 2019;83(5):921–931. doi: 10.1007/s00280-019-03808-3 [DOI] [PubMed] [Google Scholar]
- 15.Sun HY, Qu ZC, Liu DM, Zhang W, Liu G, Zhu L. Decreased expression of miR-551b predicts poor prognosis and promotes tumorigenesis by targeting PTP4A3 in human colorectal cancer. Eur Rev Med Pharmacol Sci. 2019;23:5741–5751. doi: 10.26355/eurrev_201907_18311 [DOI] [PubMed] [Google Scholar]
- 16.Wei Z, Liu Y, Wang Y, et al. Downregulation of Foxo3 and TRIM31 by miR-551b in side population promotes cell proliferation, invasion, and drug resistance of ovarian cancer. Med Oncol. 2016;33(11):126. doi: 10.1007/s12032-016-0842-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Li J, Li S, Zhou C, Qin Y, Li X. miR-802 inhibits the aggressive behaviors of non-small cell lung cancer cells by directly targeting FGFR1. Int J Oncol. 2019;54:2211–2221. doi: 10.3892/ijo.2019.4765 [DOI] [PubMed] [Google Scholar]
- 18.Zhang D, Han LL, Du F, et al. FGFR1 induces acquired resistance against gefitinib by activating AKT/mTOR pathway in NSCLC. Onco Targets Ther. 2019;12:9809–9816. doi: 10.2147/OTT.S220462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Ji W, Yu Y, et al. FGFR1-ERK1/2-SOX2 axis promotes cell proliferation, epithelial-mesenchymal transition, and metastasis in FGFR1-amplified lung cancer. Oncogene. 2018;37:5340–5354. doi: 10.1038/s41388-018-0311-3 [DOI] [PubMed] [Google Scholar]
- 20.Xie FJ, Lu HY, Zheng QQ, et al. The clinical pathological characteristics and prognosis of FGFR1 gene amplification in non-small-cell lung cancer: a meta-analysis. Onco Targets Ther. 2016;9:171–181. doi: 10.2147/OTT.S91848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Z, Cheng D, Luo L, et al. Design, synthesis and biological evaluation of 4-bromo-N-(3,5-dimethoxyphenyl)benzamide derivatives as novel FGFR1 inhibitors for treatment of non-small cell lung cancer. J Enzyme Inhib Med Chem. 2018;33:905–919. doi: 10.1080/14756366.2018.1460824 [DOI] [PMC free article] [PubMed] [Google Scholar]