Abstract
Escherichia coli FadR is a transcription factor regulated by acyl-CoA thioester binding that optimizes fatty acid (FA) metabolism in response to environmental FAs. FadR represses the fad genes of FA degradation (β-oxidation) and activates the fab genes of FA synthesis thereby allowing E. coli to have its cake (acyl chains for phospholipid synthesis) and eat it (degrade acyl chains to acetyl-CoA). Acyl-CoA binding of FadR derepresses the transcription of the fad genes and cancels fab gene transcriptional activation. Activation of fab genes was thought restricted to the fabA and fabB genes of unsaturated FA synthesis, but FadR overproduction markedly increases yields of all FA acyl chains. Subsequently, almost all of the remaining fab genes were shown to be transcriptionally activated by FadR binding, but binding was very weak. Why are the low-affinity sites retained? What effects on cell physiology would result from their conversion to high-affinity sites (thereby mimicking FadR overproduction)? Investigations of E. coli cell size determinants showed that FA synthesis primarily determines E. coli cell size. Upon modest induction of FadR, cell size increases, but at the cost of growth rate and accumulation of intracellular membranes. Greater induction resulted in further growth rate decreases and abnormal cells. Hence, too much FadR is bad. FadR is extraordinarily conserved in γ-proteobacteria but has migrated. Mycobacterium tuberculosis encodes FadR orthologs one of which is functional in E. coli. Strikingly, the FadR theme of acyl-CoA-dependent transcriptional regulation is found in a different transcription factor family where two Bacillus species plus bacterial and archaeal thermophiles contain related proteins of similar function.
Keywords: acyl-CoA, binding sites, FadR, Fatty acid synthesis, thioesterase
1 |. REVIEW
E. coli FadR is a dimeric winged helix-turn-helix transcription factor that regulates two directly opposing pathways (Henry and Cronan, 1991; Henry and Cronan, 1992). FadR represses the transcription of the genes that encode the enzymes of fatty acid (FA) degradation (β-oxidation) and activates the transcription of the genes encoding FA synthesis proteins. In the case of the β-oxidation promoters, FadR generally binds within the promoter sequence motifs whereas FadR binds in the −35 regions of the FA synthesis promoters (Henry and Cronan, 1992). The FadR regulatory ligands are long-chain acyl-CoA thioesters (Henry and Cronan, 1992). Acyl-CoAs bind to the C-terminal region of FadR and distort the N-terminal DNA binding domains by separating the two DNA binding domains by 7Ǻ such that they can no longer bind the operator sequences (van Aalten et al., 2001) (Figure 1). The tetracycline repressor (TetR) uses a similar mechanism where the DNA binding regions also become splayed out upon binding the antibiotic and can no longer bind the operator (Orth et al., 2000). The source of the acyl-CoA ligands is long-chain environmental FAs which are converted to acyl-CoA thioesters by the FadD acyl-CoA synthetase (Figure 1) following fatty acid entry via the FadL outer membrane transporter (Cronan and Subrahmanyam, 1998).
FIGURE 1.
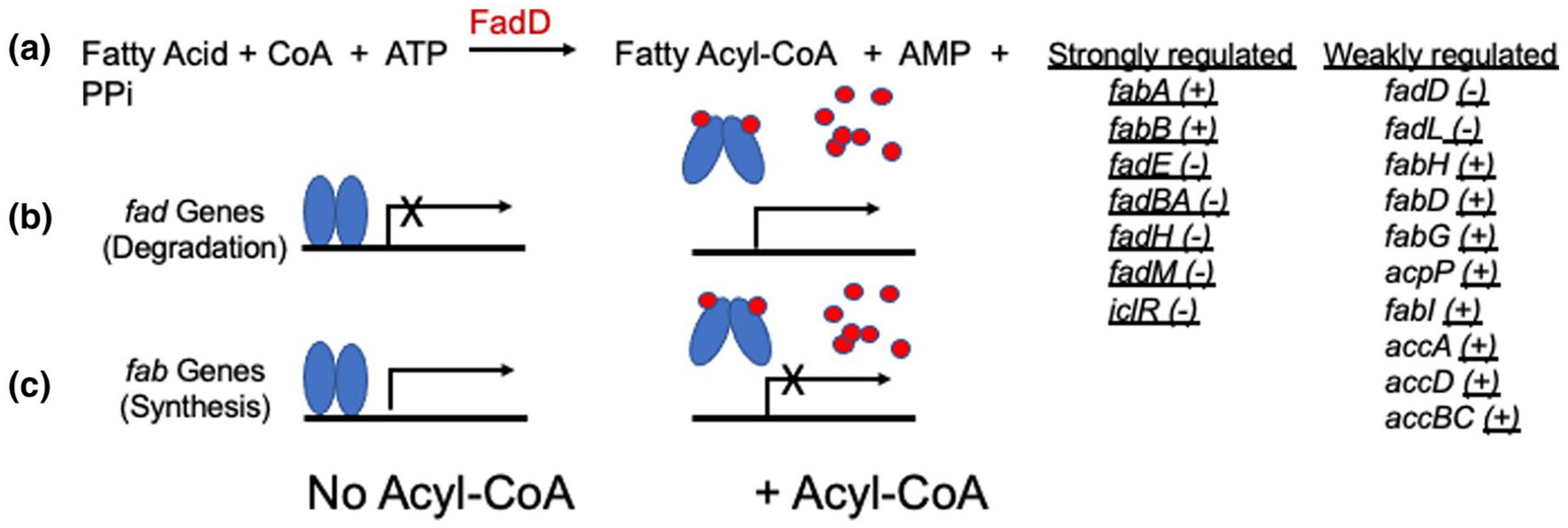
Fad R Regulates Two Opposing Pathways in E. coli. Fad R is a repressor of the fad genes and an activator of the fab genes. When present, environmental long-chain fatty acids are converted to their acyl-CoA thioesters after entering the cytosol. The acyl-CoAs (red dots) bind to FadR (blue ovals) resulting in the formation of a helix piston that spreads the N-terminal FadR DNA binding domains thereby eliminating the ability to cooperatively bind to the operator sites of the genes. In the absence of exogenous fatty acids (hence no acyl-CoAs) FadR is bound to the operators of both then fad and fab genes. The strongly regulated genes (+ and − denote positive and negative regulation, respectively) are those regulated by wild-type levels of FadR whereas the weakly related genes fall into two classes. The fatty acid uptake genes (fadD and fadL) are weakly negatively regulated by wild-type FadR whereas the fab, acc, and acpP genes show positive regulation only when FadR is overexpressed. Note that the genes of the anaerobic fatty acid degradation pathway are not under FadR regulation (Campbell et al., 2003)
The mode of derepression of the β-oxidation enzymes was first uncovered by Overath and coworkers (Overath et al., 1969) who discovered FadR mutants by their ability to grow on short-chain FAs (e.g., decanoate) that cannot induce the β-oxidation enzymes. Activation of unsaturated FA (UFA) synthesis by FadR was discovered later through the synthetic lethality of FadR mutants with temperature-sensitive mutations in fabA, the gene encoding the enzyme that inserts the cis-double bond of the UFAs (Nunn et al., 1983). Subsequent work has shown that fabB, the second gene required for UFA synthesis is also activated by FadR binding (Campbell and Cronan, 2001). The picture that emerged was one of noticeably sophisticated expediency (Cronan and Subrahmanyam, 1998). In the presence of UFA supplementation, the cell could both use the fatty acid as a carbon source via β-oxidation and replace endogenous UFA synthesis by the incorporation of oleic acid into phospholipids.
This picture was believed complete until 2012 when a team of biofuel investigators reported that overproduction of FadR resulted in a significant increase in FAs released to the medium via thioesterase cleavage of the FA synthetic intermediates, acyl-acyl carrier proteins (ACPs) (Zhang et al., 2012). This paper was followed by the analyses of the effects of FadR overproduction on the transcription of each of the fab genes (My et al., 2015; My et al., 2013). The first gene examined was fabH, which encodes 3-ketoacyl-ACP III, the enzyme that catalyzes the first step of acyl chain growth and is cotranscribed with the fadD and fabG genes (My et al., 2013). Expression of fabH was strongly dependent on FadR in that expression in a ΔfadR strain was seven-fold lower than wild-type levels (My et al., 2013). Overproduction of FadR in a wild-type strain resulted in an about 10-fold increase in fabH expression relative to the empty plasmid control (My et al., 2013). Hence, both in vivo and in vitro transcription analyses clearly demonstrate that the fabH promoter is activated by FadR. Mutagenesis showed that the site of FadR binding was in the expected position for transcriptional activation and DNase footprinting analyses demonstrated that FadR bound this fabH promoter region (My et al., 2013). However, 1 μM FadR was required to give a strong fabH footprint (My et al., 2013) whereas only 4 nM FadR was required to give an equivalent result with the fabA promoter (DiRusso et al., 1993). Hence, the fabH promoter binds FadR with very low affinity relative to the paradigm fabA promoter which explains the requirement for FadR overproduction.
In a subsequent paper, transcription of the other genes of FA synthesis was examined (My et al., 2015). In comparisons of wild-type and ΔfadR strains, none of these genes showed the strong FadR dependence of fabH. Most genes showed no dependence on FadR (the error bars overlap) and the effects were modest (≤2-fold) in those that were stimulated. However, upon FadR overproduction the genes encoding the four subunits of acetyl-CoA carboxylase were markedly stimulated. This enzyme is responsible for the synthesis of malonyl-CoA, the building block of FA synthesis (Cronan and Waldrop, 2002). Activation of these genes, accA, accD, and accB-accC (a two-gene operon), is noteworthy since acetyl-CoA carboxylase is a limiting step in FA synthesis (Davis et al., 2000; Lennen et al., 2011; Leonard et al., 2008). My and coworkers went on to document the FadR binding site of each gene by mutagenesis and mapped the transcripts of the more complex promoter regions (My et al., 2015). However, as was seen with fabH expression, these promoters bind FadR very poorly. In EMSA experiments 1 μM FadR gave only barely detectable shifts for probes containing the promoter regions of accB, accD and fabI (My et al., 2015) whereas 5–10 nM FadR gave complete gel shifts of the fabA and fabB promoter regions (DiRusso et al., 1993; Raman et al., 1997).
How can the presence of validated but very weak FadR binding sites be rationalized? My and coworkers argue that this is a means to optimize the allocation of enzyme levels in response to the environment (My et al., 2015; My et al., 2013). This argues that there must be conditions that require greatly increased synthesis of FA synthetic proteins or their products. The effects of FadR overproduction indicate that this increased synthesis would almost certainly require increased FadR levels. One possibility is that attacks on the outer membrane that activate the PldA outer membrane phospholipase could require increased FA synthesis to “heal” the sites of attack. However, FadD would convert the free FA released by PldA to CoA esters (May and Silhavy, 2018) which would bind FadR and prevent the activation of the FA synthesis genes. However, if FadD activity was limiting, increased synthesis of fatty acyl-ACPs could be of advantage because those acyl chains would be directly incorporated into phospholipids. Moreover, when the lesion has been repaired, FadD could convert the free FA to their CoA esters which would shut down FadR activation of the FA synthesis genes.
The mode (if any) of fadR regulation is unknown. Although the fadR promoter lacks a convincing −35 motif (DiRusso, 1988), FadR is far more abundant than dedicated transcription factors such as LacY. When grown in minimal media there are 300–400 molecules per cell, a level that increases 5 to 6-fold in cells grown in rich medium (Ishihama et al., 2008; Li et al., 2014). Although a transcription database has reported that FadR regulates its own transcription, the only published data on this point failed to detect autoregulation (Henry and Cronan, 1992). FadR transcription has been reported to be modestly inhibited (≤2-fold) by ppGpp in vivo and in vitro (My et al., 2013). A similarly modest inhibition of FadR expression by a small RNA called co293 was reported (Madikonda et al., 2020). The small co293 RNA, which base pairs within the fadR coding sequence 23 bases downstream of the initiation codon, is encoded within the e14 defective lambdoid prophage found in a minority of E. coli K-12 strains (e14 is readily cured). Many of the effects of small RNAs are seen only upon high-level overproduction and this is the case with small RNA co293 which was expressed from a strong promoter (Madikonda et al., 2020). Determination of FadR levels in isogenic strains with and without e14 should be performed to understand the role of co293.
Hartline and coworkers performed mathematical modeling analyses which argued that the negative autoregulation of FadR would have metabolic benefits over the effects of constitutive expression or positive autoregulation (Hartline et al., 2020). These authors buttressed their metabolic modeling by the construction of a positively regulated fadR gene. This was performed by the insertion of the fabA FadR and RNA polymerase binding sites into the fadR promoter. They also constructed a negatively regulated fadR gene in which fadR transcription was driven by the fabD promoter. Unfortunately, despite having two FadR binding sites, repression of fabD transcription is weak, only 2- to 2.5-fold, rather than the >10-fold repression of the other fad promoters (e.g., the fadBA promoter) (Feng and Cronan, 2012; Overath et al., 1969). Choice of this promoter limited the observable regulatory amplitude, but more importantly, introduced a known CRP binding site. Since CRP would activate the promoter under the culture conditions utilized (Feng and Cronan, 2012), the designed negative regulation may have been offset by positive regulation by CRP-cyclic-AMP.
Given that the overproduction of FadR in E. coli results in significantly increased rates of FA synthesis, why is this synthetic capacity not normally utilized? In a remarkably well-documented paper Vadia, Levin and coworkers reported that cells overproducing FadR are ~75% larger than wild-type cells and that the cytosols become filled with phospholipids (a 25-fold overproduction) that appear to be organized into intracellular membranes (Vadia et al., 2017). Increased cell size was dependent on increases in the phospholipid content because coexpression of a cytosolic thioesterase that intercepted the fatty acyl ACPs before the acyl groups could be incorporated into phospholipids restored cell size to almost normal (Vadia et al., 2017). Moreover, FadR mutant strains defective in DNA binding had almost normal cell sizes. Interestingly, the FadR overproduction strains grow more slowly than the wild-type strain (2.09 doublings/h versus 2.88 doublings/h) even under conditions of very modest FadR induction (1, 5 or 10 μM IPTG versus the 500–1000 μM IPTG that gives full induction (Vadia et al., 2017). At 10 μM IPTG the differing doubling times of the wild type and IPTG-induced FadR overproduction strains argues that in cultures that began with equal numbers of each strain, the wild type cells would become dominant (87% of the total cells) in 8 hr of exponential growth. Hence, further growth would essentially eliminate the FadR overproduction strain from the culture. IPTG concentrations greater than 10 μM blocked growth more strongly and resulted in dramatic morphological effects including widespread bulging and perhaps lysis (S. Vadia and P. Levin, personal communication). Therefore, the accumulation of excess phospholipid strongly compromises cell growth and fitness. A plausible scenario is that increased phospholipid synthesis results in larger cells up to the point where another component (e.g., peptidoglycan) becomes limiting.
FA overproduction significantly stresses the overproducing cells even when thioesterase action allows FA to be released to the medium (Lennen et al., 2011; Zhang et al., 2012). Stress is indicated by the induction of the pspA-G genes that cope with membrane damage from phage infection, osmotic shock, temperature increase, osmotic shock, and exposure to organic solvents plus the periplasmic stress genes spy and cpxP (Zhang et al., 2012. Since upon coexpression with a thioesterase, FadR overproducing strains largely return to normal size and growth rates (Vadia et al., 2017), cells lacking thioesterase seem likely to undergo much greater stress. Why should intracellular membrane accumulation be stressful? A likely explanation is that the E. coli cytosol is an extremely crowded environment that contains high concentrations of biopolymers which occupy 20%–30% of the available volume (Golding and Cox, 2006). A subset of these proteins must diffuse through the cytosol to allow cell division, DNA replication, transcription, regulation, etc. (Golding and Cox, 2006) and it would be surprising if intracellular membranes did not impede diffusion.
FadR has long been considered as purely a γ-proteobacterial protein and indeed FadR seems totally conserved in the myriad of Escherichia, Shigella, Salmonella, Citrobacter genome sequences, and this extends to Haemophilus, Vibrio, Pasteurella, Erwinia, Yersinia, and Shewanella, all of which show strong (>60%) sequence conservation with E. coli FadR, although there are variations on the theme.
Notably, Vibrios have developed an atypical FadR protein that contains two acyl-CoA binding sites rather than a single site found in E. coli FabR (Feng and Cronan, 2011; Gao et al., 2017; Shi et al., 2015) that gives a 5- to 8-fold increased affinity for long-chain acyl-CoAs (Iram and Cronan, 2005). The “hair-trigger” imparted by the extra binding site seems likely to play a role in vibrio pathogenesis by sensing low concentrations of host FAs as their CoA esters. In Vibrio vulnificus deletion of the fadR gene resulted in strains that grew poorly and were highly attenuated in a mouse infection model (Brown and Gulig, 2008). V. vulnificus FadR was shown to regulate the cognate fabA, fabB, and fadB genes but not fadD. Addition of the unsaturated FA, oleic acid, to the medium restored normal growth to the ΔfadR strain and also partially restored pathogenesis when added to the infection inoculum (Brown and Gulig, 2008). In V. cholerae, a ΔfadR mutation in the El Tor biotype (but not in the classical biotype) prevents the expression of the virulence cascade by influencing both the transcription and the post-translational regulation of the master virulence regulator ToxT (Kovacikova et al., 2017). This was shown to be due to the lack of FadR activation of fabA gene transcription. Indeed, in a mutant strain unable to carry out FadR-mediated activation of fabA due to an operator mutation, ectopic expression of fabA restored the levels of ToxT and virulence gene expression (Kovacikova et al., 2017). A more diverse γ-proteobacterium, Shewanella oneidensis, which is an unusually metabolically proficient γ-proteobacterium capable of reducing metals, has a FadR that is the functional equivalent of E. coli FadR. It binds both the E. coli fabA and fabB operators, regulates its cognate fabA (but not its fabB) gene and long chain acyl-CoAs abolish DNA binding (Luo et al., 2014; Zhang et al., 2015).
However, the parochial γ-proteobacterial view of FadR distribution has been overturned by the Mce2R/Rv0586 protein of Mycobacterium tuberculosis. Expression of this protein in E. coli blocks growth of a ΔfadR strain on decanoate (Yousuf et al., 2018). This result argues that the M. tuberculosis protein recognizes the E. coli FadR operator sequence and that its native operator must have a similar sequence. This is the case, the Mce2R/Rv0586 protein binds to an extended version of the E. coli FadR binding site and several DNA binding residues of E. coli FadR are both conserved in the mycobacterial protein and were shown to be required for DNA binding (Yousuf et al., 2018). Finally, long-chain acyl-CoAs at physiologically reasonable concentrations (25–50 μM) prevented DNA binding by Mce2R/Rv0586 demonstrating it to be a valid functional homolog of E. coli FadR (Yousuf et al., 2018). M. tuberculosis encodes a second FadR homolog, Rv0494 which has a greater sequence similarity to E. coli FadR than does Rv0586 (24% vs 20%) and is also a DNA-binding protein whose binding is disrupted by long-chain acyl-CoAs. However, Rv0494 recognizes an operator unrelated to the E. coli operators and thus was unable to complement the E. coli ΔfadR strain (Yousuf et al., 2018).
Other bacterial regulatory proteins often confusingly called FadR have been reported in the Bacilli (Matsuoka et al., 2007; Yeo et al., 2017) and in the thermophile, Thermus thermophilus HB8, a bacterium that belongs to the Deinococcus-Thermus Phylum (Agari et al., 2011). Although these proteins are TetR family proteins and thus are very different in sequence and structure from the canonical FadRs of the GntR family, they function in a remarkably similar manner. To date, all TetR FadR-like proteins repress the transcription of FA degradation genes (in Bacillus subtilis, a different transcription factor regulates the FA synthesis genes). As is the case with E. coli FadR, long-chain acyl-CoAs eliminate DNA binding although high (μM) acyl-CoA concentrations are required for release. This is consistent with the role of these proteins in the regulation of β-oxidation since there is little point in derepressing the pathway unless there is a substantial FA supply. Finally, TetR FadR-like proteins have migrated to the Archaea. The hot spring thermophile, Sulfolobus acidocaldarius, regulates β-oxidation and other pathways of lipid degradation by a TetR FadR-like protein Saci_1107, which has structural and functional similarity to the Bacillus and T. thermophilus proteins (Wang et al., 2019). In the Archaea, such proteins are found only in certain clades of thermophilic Archaea isolated from hot springs where bacteria such as T. thermus are also present. Hence, it seems plausible that the archaeal system was acquired from co-resident hot spring bacteria by horizontal gene transfer.
A scenario in which positive regulation of the acyltransferases, PlsB and PlsC by FadR would seem physiologically advantageous because this would expedite incorporation of acyl chains into phospholipids. However, neither the plsB nor plsC gene promoter regions contain a recognizable FadR binding site (although the plsC promoter remains unmapped). Vibrio cholerae transcription of the plsB gene is under FadR control and control is reversed by exogenous oleic acid (Feng and Cronan, 2011). However, the regulation is negative, FadR decreases transcription. This seems perverse if the V. cholerae fab genes are positively controlled as is the case in E. coli. However, data on the expression of the V. cholerae fab genes are lacking.
Finally, it should be noted that E. coli cells containing intracellular membranes have been reported upon the overexpression of several integral inner membrane proteins. A recent synopsis is found in Carranza and coworkers and references therein (Carranza et al., 2017). Most germane to this review is the overproduction of PlsB (Wilkison et al., 1986). However, this is a single report and it is difficult to ascribe the membrane formation to PlsB enzyme activity since overproduction of other proteins, the AtpF subunit of F-ATPase, fumarate reductase, mannitol permease, the Tsr chemotaxis receptor, and mycoplasma glucosyltransferases also elicit the intracellular membrane formation. Hence, the formation of intracellular membranes can be triggered either by phospholipid overproduction or by the overexpression of membrane proteins. Hence, like FadR overexpression, inner membrane protein overexpression must somehow increase the rate of phospholipid synthesis. Only in the cases of fumarate reductase (Elmes et al., 1986), AtpF (Carranza et al., 2017) and glucosyltransferases (Eriksson et al., 2009) have the membranes been isolated and characterized in some detail. In the case of fumarate reductase case the intracellular membranes were reported to branch from the inner membrane rather than arising by de novo nucleation in the cytosol. One possible means to decide between these alternatives would be to assay the isolated intracellular membranes for quinones. “Branched” membranes may contain quinones which are thought to diffuse freely within the inner membrane whereas membranes resulting from nucleation in the cytosol may not. In this regard, the protein and quinone composition of intracellular membranes resulting from FadR overexpression should be very interesting.
Funding information
National Institute of Allergy and Infectious Diseases, Grant/Award Number: AI15650
REFERENCES
- Agari Y, Agari K, Sakamoto K, Kuramitsu S and Shinkai A (2011) TetR-family transcriptional repressor Thermus thermophilus FadR controls FA degradation. Microbiology, 157,1589–1601. [DOI] [PubMed] [Google Scholar]
- Brown RN and Gulig PA (2008) Regulation of fatty acid metabolism by FadR is essential for Vibrio vulnificus to cause infection of mice. Journal of Bacteriology, 190, 7633–7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JW and Cronan JE (2001) Escherichia coli FadR positively regulates transcription of the fabB fatty acid biosynthetic gene. Journal of Bacteriology, 183, 5982–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JW, Morgan-Kiss RM and Cronan JE (2003) A new Escherichia coli metabolic competency: growth on fatty acids by a novel anaerobic β-oxidation pathway. Molecular Microbiology, 47, 793–805. [DOI] [PubMed] [Google Scholar]
- Carranza G, Angius F, Ilioaia O, Solgadi A, Miroux B and Arechaga I (2017) Cardiolipin plays an essential role in the formation of intracellular membranes in Escherichia coli. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1859,1124–1132. [DOI] [PubMed] [Google Scholar]
- Cronan JE and Subrahmanyam S (1998) FadR, transcriptional co-ordination of metabolic expediency. Molecular Microbiology, 29,937–943. [DOI] [PubMed] [Google Scholar]
- Cronan JE and Waldrop GL (2002) Multi-subunit acetyl-CoA carboxylases. Progress in Lipid Research, 41,407–435. [DOI] [PubMed] [Google Scholar]
- Davis MS, Solbiati J and Cronan JE (2000) Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. Journal of Biological Chemistry, 275, 28593–28598. [DOI] [PubMed] [Google Scholar]
- DiRusso CC (1988) Nucleotide sequence of the fadR gene, a multifunctional regulator of fatty acid metabolism in Escherichia coli. Nucleic Acids Research, 16, 7995–8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRusso CC, Metzger AK and Heimert TL (1993) Regulation of transcription of genes required for fatty acid transport and unsaturated fatty acid biosynthesis in Escherichia coli by FadR. Molecular Microbiology, 7,311–322. [DOI] [PubMed] [Google Scholar]
- Elmes ML, Scraba DG & Weiner JH (1986) Isolation and characterization of the tubular organelles induced by fumarate reductase overproduction in Escherichia coli. Journal of General Microbiology, 132, 1429–1439. [DOI] [PubMed] [Google Scholar]
- Eriksson HM, Wessman P, Ge C, Edwards K and Wieslander A (2009) Massive formation of intracellular membrane vesicles in Escherichia coli by a monotopic membrane-bound lipid glycosyltransferase. Journal of Biological Chemistry, 284,33904–33914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y and Cronan JE (2011) The Vibrio cholerae fatty acid regulatory protein, FadR, represses transcription of plsB, the gene encoding the first enzyme of membrane phospholipid biosynthesis. Molecular Microbiology, 81,1020–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y and Cronan JE (2012) Crosstalk of Escherichia coli FadR with global regulators in expression of fatty acid transport genes. PLoS One, 7, e46275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Li D, Lin Y, Lin J, Xia X, Wang H et al. (2017) Structural and functional characterization of the FadR regulatory protein from Vibrio alginolyticus. Frontiers in Cellular and Infection Microbiology, 7, 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding I and Cox EC (2006) Physical nature of bacterial cytoplasm. Physical Review Letters, 96, 098102. [DOI] [PubMed] [Google Scholar]
- Hartline CJ, Mannan AA, Liu D, Zhang F & Oyarzún DA (2020) Metabolite sequestration enables rapid recovery from fatty acid depletion in Escherichia coli. mBio, 11, e03112–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry MF and Cronan JE (1991) Escherichia coli transcription factor that both activates fatty acid synthesis and represses fatty acid degradation. Journal of Molecular Biology, 222, 843–849. [DOI] [PubMed] [Google Scholar]
- Henry MF and Cronan JE (1992) A new mechanism of transcriptional regulation: release of an activator triggered by small molecule binding. Cell, 70, 671–679. [DOI] [PubMed] [Google Scholar]
- Iram SH and Cronan JE (2005) Unexpected functional diversity among FadR fatty acid transcriptional regulatory proteins. Journal of Biological Chemistry, 280, 32148–32156. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ et al. (2008) Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics, 9, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova G, Lin W, Taylor RK and Skorupski K (2017) The fatty acid regulator FadR influences the expression of the virulence cascade in the El Tor Biotype of Vibrio cholerae by modulating the levels of ToxT via two different mechanisms. Journal of Bacteriology, 199, e00762–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennen RM, Kruziki MA, Kumar K, Zinkel RA, Burnum KE, Lipton MS et al. (2011) Membrane stresses induced by overproduction of free fatty acids in Escherichia coli. Applied and Environment Microbiology, 77, 8114–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard E, Yan Y, Fowler ZL, Li Z, Lim CG, Lim KH & et al. (2008) Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Molecular Pharmaceutics, 5,257–265. [DOI] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross C and Weissman JS (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell, 157, 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Shi M, Ren Y and Gao H (2014) Transcription factors FabR and FadR regulate both aerobic and anaerobic pathways for unsaturated fatty acid biosynthesis in Shewanella oneidensis. Frontiers in Microbiology, 5, 736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madikonda AK, Shaikh A, Khanra S, Yakkala H, Yellaboina S, Lin-Chao S and Siddavattam D (2020) Metabolic remodeling in Escherichia coli MG1655. A prophage el4-encoded small RNA, co293, post-transcriptionally regulates transcription factors HcaR and FadR. FEBS J. 10.1111/febs.15247 Feb, 2020. [DOI] [PubMed] [Google Scholar]
- Matsuoka H, Hirooka K and Fujita Y (2007) Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. Journal of Biological Chemistry, 282, 5180–5194. [DOI] [PubMed] [Google Scholar]
- May KL and Silhavy TJ (2018) The Escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. mBio, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- My L, Ghandour Achkar N, Viala JP & Bouveret E (2015) Reassessment of the genetic regulation of fatty acid synthesis in Escherichia coli: Global positive control by the dual functional regulator FadR. Journal of Bacteriology, 197, 1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- My L, Rekoske B, Lemke JJ, Viala JP, Gourse RL and Bouveret E (2013) Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR and inhibited by ppGpp. Journal of Bacteriology, 195,3784–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn WD, Giffin K, Clark D and Cronan JE (1983) Role for fadR in unsaturated fatty acid biosynthesis in Escherichia coli. Journal of Bacteriology, 154, 554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth P, Schnappinger D, Hillen W, Saenger W and Hinrichs W (2000) Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Natural Structural Biology, 7,215–219. [DOI] [PubMed] [Google Scholar]
- Overath P, Pauli G and Schairer HU (1969) Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old-mutations, and the isolation of regulatory mutants. European Journal of Biochemistry, 7, 559–574. [PubMed] [Google Scholar]
- Raman N, Black PN and DiRusso CC (1997) Characterization of the fatty acid-responsive transcription factor FadR. Biochemical and genetic analyses of the native conformation and functional domains. Journal of Biological Chemistry, 272,30645–30650. [DOI] [PubMed] [Google Scholar]
- Shi W, Kovacikova G, Lin W, Taylor RK, Skorupski K and Kull FJ (2015) The 40-residue insertion in Vibrio cholerae FadR facilitates binding of an additional fatty acyl-CoA ligand. Nature Communications, 6,6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadia S, Tse JL, Lucena R, Yang Z, Kellogg DR, Wang JD et al. (2017) Fatty acid availability sets cell envelope capacity and dictates microbial cell size. Current Biology, 27,1757–1767.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aalten DM, DiRusso CC and Knudsen J (2001) The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO Journal, 20, 2041–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Sybers D, Maklad HR, Lemmens L, Lewyllie C, Zhou X et al. (2019) A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius. Nature Communications, 10, 1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkison WO, Walsh JP, Corless JM and Bell RM (1986) Crystalline arrays of the Escherichia coli sn-glycerol-3-phosphate acyltransferase, an integral membrane protein. Journal of Biological Chemistry, 261, 9951–9958. [PubMed] [Google Scholar]
- Yeo HK, Park YW and Lee JY (2017) Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR. Nucleic Acids Research, 45,4244–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousuf S, Angara RK, Roy A, Gupta SK, Misra R and Ranjan A (2018) Mce2R/Rv0586 of Mycobacterium tuberculosis is the functional homologue of FadR E. coli. Microbiology, 164, 1133–1145. [DOI] [PubMed] [Google Scholar]
- Zhang F, Ouellet M, Batth TS, Adams PD, Petzold CJ, Mukhopadhyay A et al. (2012) Enhancing fatty acid production by the expression of the regulatory transcription factor FadR. Metabolic Engineering, 14, 653–660. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zheng B, Gao R and Feng Y (2015) Binding of Shewanella FadR to the fabA fatty acid biosynthetic gene: implications for contraction of the fad regulon. Protein Cell, 6, 667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]


