Abstract
Severe acute respiratory syndrome virus 2 (SARS-CoV-2) invades host cells by interacting with receptors/coreceptors, as well as with other cofactors, via its spike (S) protein that further mediates fusion between viral and cellular membranes. The host membrane protein, angiotensin-converting enzyme 2 (ACE2), is the major receptor for SARS-CoV-2 and is a crucial determinant for cross-species transmission. In addition, some auxiliary receptors and cofactors are also involved that expand the host/tissue tropism of SARS-CoV-2. After receptor engagement, specific proteases are required that cleave the S protein and trigger its fusogenic activity. Here we discuss the recent advances in understanding the molecular events during SARS-CoV-2 entry which will contribute to developing vaccines and therapeutics.
Keywords: COVID-19, SARS-CoV-2, virus entry, spike protein, receptor recognition, coreceptor, membrane fusion
Coronaviruses and COVID-19 pandemic
In late 2019 a novel coronavirus named SARS-CoV-2 emerged in humans, that causes coronavirus disease 2019 (COVID-19) [1., 2., 3., 4.]. This outbreak has rapidly developed into a worldwide pandemic and has resulted in more than 0.1 billion confirmed cases as of 23 May 2021, including ~3.5 million deaths (www.who.int/emergencies/diseases/novel-coronavirus-2019). SARS-CoV-2 is the seventh human-infecting coronavirus (HCoV) identified so far (Box 1 ), and it is most similar to SARS-CoV which emerged in 2002 [4,5]. However, SARS-CoV-2 exhibits a higher transmission efficiency (see Glossary) compared to SARS-CoV and other HCoVs [6], although it has a relatively lower mortality rate than SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) [6., 7., 8.]. Although several candidate vaccines are being distributed in different countries, the global pandemic situation is far from under control. It is very urgent to promote vaccination among human populations and to develop effective therapeutics.
Box 1. Coronaviruses and related epidemics/pandemics.
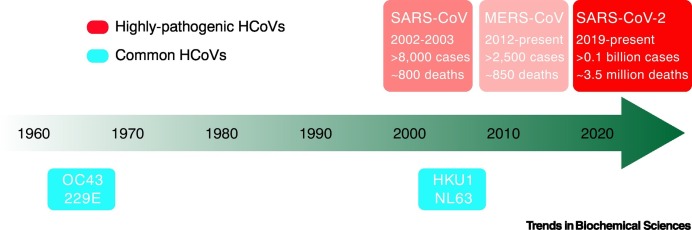
Coronaviruses are a group of enveloped viruses whose surface is decorated with spike (S) proteins, resulting in a crown-shaped morphology. The genome of coronaviruses is a single-stranded positive-sense RNA that can directly serve as an mRNA for translation of viral proteins [11]. Coronaviruses belong to the order of Nidovirales, the family Coronaviridae, and are further classified into Orthocoronavirinae and Letovirinae subfamilies. All human-infecting coronaviruses (HCoVs) are included in the subfamily Orthocoronavirinae, which are further divided into four genera, Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus [9]. So far, a total of seven HCoVs have been identified. Among them, the 229E, NL63, OC43, and HKU1 coronaviruses are commonly found in human populations around the world, resulting in mild symptoms such as common cold and fever. The other three, SARS-CoV, MERS-CoV, and SARS-CoV-2, are categorized as highly pathogenic coronaviruses which have caused epidemics/pandemics in different countries.
The first identified HCoVs are 229E and OC43, reported in the 1960s (Figure I) [107,108]. They are often detected at the same time as other respiratory infections and usually do not lead to severe symptoms. In 2004 the NL63 coronavirus was discovered in a baby with bronchiolitis in The Netherlands [109]. A year later the HKU1 coronavirus was identified in Hong Kong, China from an elderly patient with pneumonia [110]. Since then, this virus has been found in human populations around the world. The first case of SARS-CoV infection was found in Guangdong, China in late 2002 [5]. The epidemic spread to over 30 countries and ended in 2003, resulting in more than 8000 reported cases of infection, including almost 800 deaths. MERS-CoV was first identified in Saudi Arabia in 2012 and has been found in many Middle East countries as well as in some Asian countries [111]. So far, more than 2500 confirmed infection cases have been reported, of which ~850 died from MERS-related disease, and thus has the highest case fatality rate (~35%) of all HCoVs. SARS-CoV-2 infection cases were first reported in Wuhan, China, in late 2019 [3]. This virus has led to the unprecedented ongoing global pandemic that affects almost all countries around the world. As of 23 May 2021, more than 0.1 billion human infection cases have been confirmed, including almost 3.5 million death cases. The case fatality rate of SARS-CoV-2 is much lower than those of SARS-CoV and MERS-CoV, but it seems to be more efficient in transmission among human populations. All three highly pathogenic HCoVs are thought to originate from wild animals, potentially with a common natural host, bats [2,112].
Figure I.
Timeline for the identification of human-infecting coronaviruses (HCoVs).
Alt-text: Box 1
SARS-CoV-2 is a positive-sense RNA virus with a large single-stranded RNA genome of ~30 000 nt [9]. The genome encodes three classes of proteins: two large polyproteins, pp1a and pp1ab, which are cleaved into 16 non-structural proteins (NSPs) that are required for viral RNA synthesis (and probably other functions); four structural proteins (the spike, envelope, membrane, and nucleocapsid proteins) that are essential for viral entry and assembly; and nine accessory proteins that are thought to counteract the host immunity during infection [10,11]. Viral entry is the first step of infection and one of the most important processes in the virus life cycle, which is also the key target for vaccines and therapeutics. This process is executed by the S protein on the envelope of SARS-CoV-2, which recognizes the host cell receptor and mediates membrane fusion to allow the viral genome to be released into the cytoplasm [12].
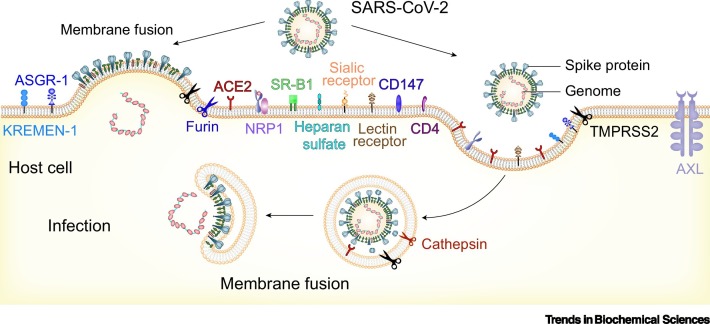
In this review, we summarize the recent functional and structural studies on SARS-CoV-2 entry, with an emphasis on the S protein-mediated receptor binding and membrane fusion processes, as well as on other cellular factors and coreceptors that are potentially involved in the viral entry process (Figure 1 ).
Figure 1.
Schematic diagram of SARS-CoV-2 entry pathways.
Multiple molecules at the cell surface are involved in the entry of SARS-CoV-2, including the major receptor ACE2 [2,19], the membrane protease TMPRSS2, and other potential alternative/auxiliary receptors or cofactors [25,52,53,62,70,71,75,80]. Membrane fusion can take place either at the cell surface (left) or in the endosome (right). Both entry pathways are utilized by SARS-CoV-2 [45,46]. Abbreviations: ACE2, angiotensin-converting enzyme 2; SARS-CoV-2, severe acute respiratory syndrome virus 2; TMPRSS2, transmembrane serine protease 2.
Structure of SARS-CoV-2 S protein
An average of 30–60 S protein trimers protrude from the envelope of SARS-CoV-2 virion, with an average distance of 15 nm from each other [13., 14., 15.]. Each trimeric spike is ~10 nm in length with a long helix stalk hinge that allows the spike to adopt different orientations on the viral envelope [14,15]. The coronavirus S protein is a typical class I viral fusion protein and is the largest viral fusion machine identified so far, containing more than 1200 amino acid residues.
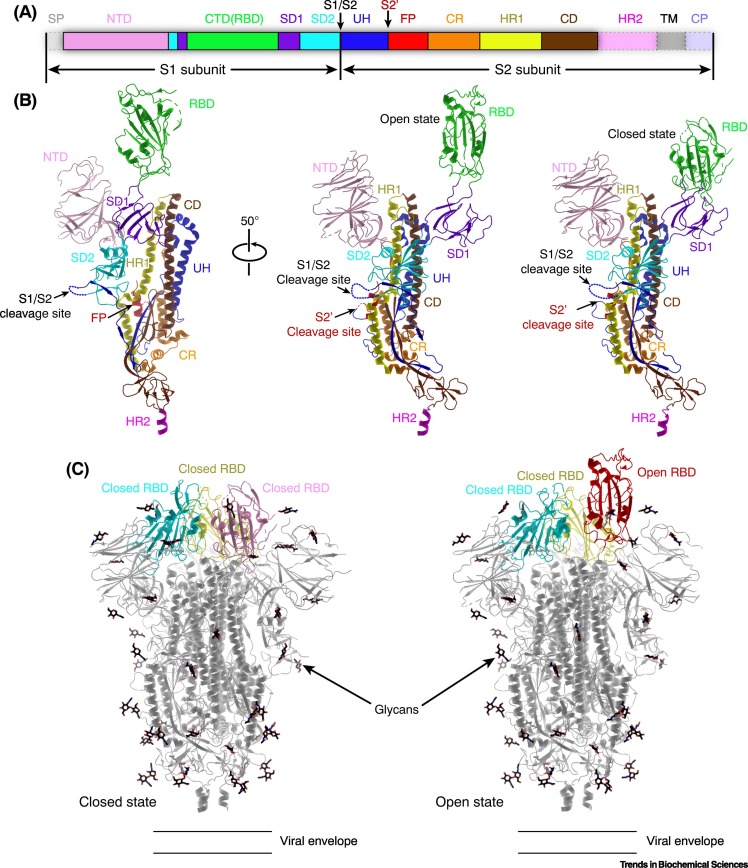
During the cell entry process, the SARS-CoV-2 S protein undergoes proteolytic cleavage by cellular proteases into the S1 and S2 subunits which remain associated and further assemble into trimers of the S1/S2 heterodimer (Figure 2 ) [16,17]. The S1 subunit can be divided into the N-terminal domain (NTD) and the C-terminal domain (CTD), of which the latter is responsible for binding the host receptor angiotensin-converting enzyme 2 (ACE2) and is thus also termed the receptor-binding domain (RBD) [17., 18., 19., 20., 21.]. The S2 subunit is the fusogenic portion of the spike and consists of the upstream helix (UH) region, the fusion peptide (FP), the heptad repeat 1 (HR1), the central domain (CD), the heptad repeat 2 (HR2), the transmembrane domain (TM), and the cytoplasmic tail (CP) (Figure 2A,B). In contrast to most typical class I viral fusion proteins, the FP of coronavirus S protein is not located at the immediate N terminus of the S2 subunit. Instead, it is shielded by the UH domain which therefore requires a second cleavage event to expose the FP [22,23]. Proteolysis at the S2′ cleavage site to remove the UH domain is crucial for activating the fusogenic capacity of S protein, and triggers irreversible conformational changes of the S2 fusion machine to initiate membrane fusion [24., 25., 26.].
Figure 2.
Structure of SARS-CoV-2 spike (S) protein.
(A) Schematic diagram of the domain organization of S protein. Each domain is represented by a unique color. The signal peptide (SP) and the unresolved region at the C terminus are transparent with dashed outlines. (B) Architecture of an S protomer (PDB: 6VXX and 6VYB). The structure is shown in cartoons and colored by domains as in (A). The unresolved regions are represented by broken lines. (C) Structures of the SARS-CoV-2 S trimer in different conformations. The RBD can adopt different conformations (closed or open) in the S trimer, and only the open conformation is competent for binding to the receptor ACE2 [17,18,26]. Abbreviations: ACE2, angiotensin-converting enzyme 2; CD, central domain; CP, cytoplasmic region; CR, connecting region; CTD, C-terminal domain; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; NTD, N-terminal domain; RBD, receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome virus 2; S1/S2 and S2′, cleavage sites within S protein; SD1, subdomain 1; SD2, subdomain 2; TM, transmembrane region; UH, upstream helix.
The major receptor ACE2
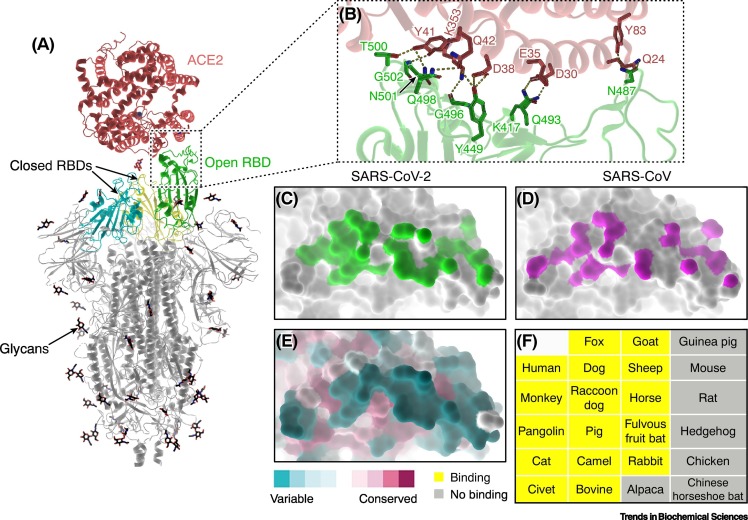
Soon after the outbreak, several groups promptly identified ACE2 as the major receptor for SARS-CoV-2, similar to SARS-CoV that emerged in 2002–2003 [2,17,25] (Figure 3 ). ACE2, a carboxypeptidase that cleaves polypeptides from the renin/angiotensin system, is essential for cardiac function and is widely expressed in various tissues and organs, suggesting the potential capacity of SARS-CoV-2 to develop systematic infections in patients [27,28]. The RBDs of SARS-CoV-2 and SARS-CoV share a high degree of sequence identity (74%) and exhibit highly similar interaction profiles with ACE2 [19., 20., 21.]. However, some substitutions in the key interacting residues in the RBD lead to more atomic contacts between SARS-CoV-2 S protein and ACE2, potentially resulting in the higher binding affinity compared to SARS-CoV (~fourfold difference) (Figure 3C,D) [19]. This property may contribute to the highly efficient human-to-human transmission of SARS-CoV-2 [6,7]. In addition, ACE2 orthologs are widely distributed in various domestic and wild mammals such as cats, dogs, pigs, camels, horses, pangolins, and bats, indicating that SARS-CoV2 is likely to have a broad host spectrum [29]. Some closely related coronaviruses to SARS-CoV-2 have been isolated in pangolins and bats [2,30]. Two recent studies have shown that the ACE2 orthologs of a wide range of animals can bind SARS-CoV-2 RBD and mediate the cell entry of S pseudotyped viruses, although the S-binding interface displays significant diversity (Figure 3E,F) [31,32]. These findings strongly imply that SARS-CoV-2 may have experienced multiple spillover events in adaptation to the diverse molecular determinants in different animals, which enabled its host-jump across different intermediate hosts and finally allowed it to infect humans.
Figure 3.
Interactions between SARS-CoV-2 spike (S) protein and ACE2.
(A) Overall structure of a trimeric S bound to ACE2 (PDB: 7KNB). ACE2 binds to the RBD of S in the open conformation, while the other two closed RBDs are inaccessible by the receptor. (B) Close-up view of the S–ACE2 contacting interface (PDB: 6LZG). The key interacting residues are shown as sticks. Hydrogen bonds and salt bridges are represented by dashed lines. (C,D) The S-binding footprint on ACE2 for SARS-CoV-2 (C; PDB: 6LZG) and SARS-CoV (D; PDB: 2AJF). The interaction between SARS-CoV-2 (green) and ACE2 involves more atomic contacts than for SARS-CoV (magenta) [19., 20., 21.,28]. (E) Conservation of the S-binding interface on ACE2. The interface for SARS-CoV-2 S binding displays a high degree of variation among ACE2 orthologs from different animal species. (F) A list of animals tested whose ACE2 can (yellow) or cannot (grey) bind to the RBD of SARS-CoV-2 S protein [29,31,32]. Abbreviations: ACE2, angiotensin-converting enzyme 2; RBD, receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome virus 2.
Conformational dynamics of SARS-CoV-2 S during virus entry
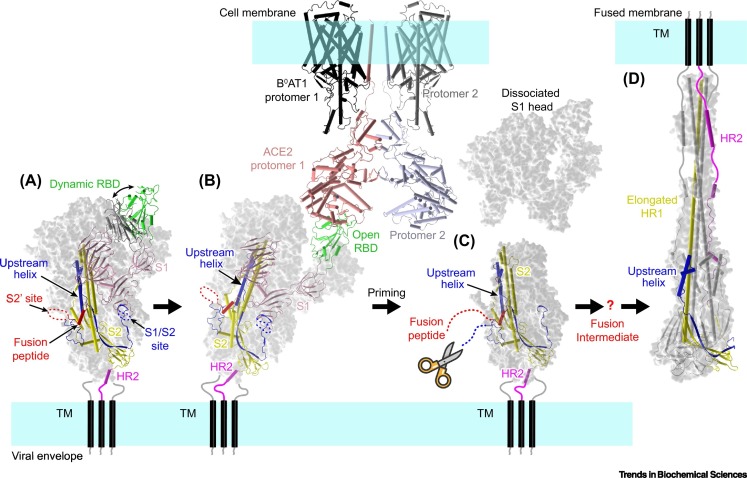
Cryogenic electron microscopy (cryo-EM) studies have determined the structures of SARS-CoV-2 S protein in various conformations, both before and after membrane fusion [17,18,26], as well as its complex with the receptor ACE2 [33., 34., 35.]. These structural snapshots enable the deduction of a complete scenario for the conformational changes of S protein during SARS-CoV-2 entry (Figure 4 ). The RBD of S protein can adopt different conformations at the prefusion state in which the receptor binding interface is buried by the adjacent protomer (closed conformation) or exposed for the access by ACE2 (open conformation) (Figures 2C and 4A), similar to other known coronavirus S proteins [17,18,36]. The three RBDs within a trimeric spike are not synchronized, implying asymmetric interactions with the receptor. A recent study revealed that binding of ACE2 to an open RBD can promote the conformational transition of the other closed RBDs to make them accessible by the receptor (Figure 4B) [33]. Therefore, a trimeric spike can bind to 1–3 copies of ACE2, depending on the conformation of each individual RBD [33,35]. The binding of ACE2 modulates the local conformation of S1 subunit to disrupt its interactions with the S2 fusion core, which involves a key salt bridge contributed by residue D614 [33]. Progressive interactions with ACE2 molecules will lead to dissociation of the S1 head from the fusogenic S2 stalk, which facilitates fusion activation by further proteolysis at the S2′ site (Figure 4C) [33]. Of note, a SARS-CoV-2 variant harboring a D614G substitution in S protein was identified in Europe in mid-2020. This mutation makes the RBD much more flexible, and increases the probability of adopting the open conformation so as to be accessible by the receptor, and reduces the stability of the prefusion structure of S trimer, thus facilitating fusion activation [33,37,38]. This may explain the higher transmission efficiency of the variant strain compared to the earlier isolates [39,40]. In addition, the S protein is extensively decorated by glycans on both the S1 and S2 subunits (Figure 2C). The glycan shield not only changes the antigenicity of S protein but may also alter the conformation of specific domains. It has been shown that the N-linked glycans at residues N165 and N234 may modulate the conformational dynamics of the RBD, thus affecting the interactions with the receptor [41,42]. In addition, removal of the glycans on SARS-CoV-2 S renders it more sensitive to proteolysis during biogenesis in cells and thus compromises its stability, which would reduce the number of functional spikes presented on the viral envelope and thus inhibit viral infectivity [43].
Figure 4.
Model of the SARS-CoV-2 entry process.
(A) The trimeric spike on viral envelope can adopt different conformations (PDB: 6VXX and 6VYB). The RBD undergoes dynamic transition between the closed and open conformations. The SARS-CoV-2 S protein is prone to be cleaved into the S1 and S2 subunits during biosynthesis or cell entry process, and the two subunits remain noncovalently associated to form trimeric spikes [16,17,45]. (B) Only the RBD in the open conformation can bind to the receptor ACE2 (PDB: 7KNB and 6M17). Binding of ACE2 can promote the transition of adjacent RBDs to the open conformation, and thus may facilitate the binding of more ACE2 molecules [33., 34., 35.]. (C) ACE2 binding induces conformational changes that destabilize the interactions between S1 and S2 subunits, thus probably triggering the dissociation of the S1 head [33]. The second proteolysis event at the S2′ site, by TMPRSS2 and other proteases [25,52], produces a free N terminus for the FP and triggers conformational changes to expose the FP for cellular membrane targeting. The question mark indicates the fusion intermediate structures that are not well understood. It is thought that the central domain will refold to form an elongated helix stalk with the HR1 helix, which facilitates FP reaching the target membrane [26]. (D) The HR2 helices then fold upwards to contact the elongated HR1 stalk helices, transforming into the six-helix bundle postfusion conformation [24,26]. This rearrangement draws the viral and cellular membranes into close proximity and induces fusion. Abbreviations: ACE2, angiotensin-converting enzyme 2; B0AT1, sodium-dependent amino acid transporter 1, also known as SLC6A19; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; RBD, receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome virus 2; TM, transmembrane region.
Binding of ACE2 to S protein induces endocytosis of the virion, after which the viral envelope fuses with the endosomal membrane to enable the release of the viral genome into the cytoplasm [44., 45., 46.]. Alternatively, membrane fusion can also occur at the plasma membrane after receptor engagement. Both entry mechanisms have been reported for SARS-CoV-2, probably with a preference for the endosomal pathway (Figure 1) [45]. This evidence also suggests that the low-pH environment is not a crucial determinant for the entry of SARS-CoV-2, similar to the previous observations on SARS-CoV, MERS-CoV, and mouse hepatitis virus (MHV) [12]. After receptor engagement, the cleavage at the S2′ site will induce the exposure of the FP and initiate conformational changes in the S2 subunit to form a long HR1 for membrane targeting [26,33]. Once activated, the HR2 helices will fold back to contact the HR1 helix stalk, generating a six-helix bundle hairpin which, as a result, draws the viral envelope and the host membrane into close proximity and induces fusion (Figure 4D) [26].
The cofactors TMPRSS2 and furin proteases
A remarkable feature of SARS-CoV-2 S protein is the insertion of a polybasic residue motif at the boundary between the S1 and S2 subunits, which renders it prone to be cleaved by furin/furin-like proteases during biogenesis and cell entry [17,47]. The trimeric spikes on the viral envelope are thus mostly cleaved S1/S2 complexes, in contrast to the uncleaved S0 form observed in SARS-CoV and other SARS-CoV-related viruses [17,25]. This property suggests that SARS-CoV-2 S protein may be more readily primed for membrane fusion than SARS-CoV S because the latter requires two proteolytic events after receptor binding. Because furin-like proteases are almost ubiquitously distributed in different tissues and organs, the presence of furin-type cleavage sites in viral fusion proteins may be related to the broad cell tropism and even the high pathogenicity of related viruses [48,49]. This may partially explain the highly efficient transmissibility of SARS-CoV-2 among human populations that has resulted in the unprecedented global pandemic of coronavirus. In line with this hypothesis, a recent study reported that mutations in this furin cleavage site in SARS-CoV-2 S led to reduced viral replication in a human respiratory cell line and attenuated its pathogenesis in both hamster and mouse models [50].
Following receptor engagement, the second step of proteolysis of SARS-CoV-2 S is thought to be executed mainly by the transmembrane serine protease 2 (TMPRSS2), which cleaves the S2 fusion machine at the S2′ site to expose the FP [51]. The TMPRSS2 is a primary serine protease expressed in many epithelial cells, and has also been shown to be an ideal antiviral target for inhibiting SARS-CoV-2 entry [25]. In addition to furin/furin-like proteases and TMPRSS2, other proteases may also be involved in the SARS-CoV-2 entry process, such as serine endoprotease proprotein convertase 1 (PC1), trypsin, matriptase (trypsin-like integral membrane serine peptidase), and cathepsins [52., 53., 54.] (Figure 1). The broad spectrum of protease usage by SARS-CoV-2 may significantly facilitate its infectivity and transmissibility among humans and other hosts, and may lead to systematic multi-organ infections in infected patients [55].
Alternative receptors, coreceptors, and cofactors
The alternative receptors AXL, KREMEN1, and ASGR1
Although both SARS-CoV and SARS-CoV-2 utilize ACE2 as the main receptor for cell entry, cells in the main target organs (e.g., the lungs and bronchi) display low levels of ACE2 expression as revealed by single-cell sequencing analyses [56,57]. Moreover, SARS-CoV-2 can also efficiently infect the upper respiratory tract and other tissues/organs, including the pharynx, heart, liver, brain, kidneys, and the gastrointestinal tract [55,58]. This evidence suggests that other molecules are involved in the SARS-CoV-2 entry process (Table 1 ). As an explanation for the paradox, Wang et al. identified AXL (tyrosine-protein kinase receptor UFO) as a potential receptor for SARS-CoV-2 entry in lung and bronchial tissues [59], and Gu et al. reported that KREMEN1 (Kringle-containing protein marking the eye and the nose protein 1) and ASGR1 (asialoglycoprotein receptor 1) may contribute to multiple organ invasion in COVID-19 patients [60] (Figure 1). Each of the three molecules may potentially mediate SARS-CoV-2 entry independently of ACE2 [59,60], providing alternative pathways for SARS-CoV-2 infection in different tissues. Even though knocking out AXL significantly inhibited SARS-CoV-2 entry in human pulmonary cell lines, it only moderately reduced viral replication in lung organoids [59]. Besides, overexpression of KREMEN1 or ASGR1 in ACE2-knockout cells only partially restored the infectivity of SARS-CoV-2, with a lower efficiency compared to ACE2-expressing cells [60]. Therefore, the exact contributions of these three candidate receptors to SARS-CoV-2 entry into human cells remain to be established. Biochemical data reveal that AXL binds to the NTD of S protein, whereas KREMEN1 and ASGR1 interact with both the NTD and RBD [59,60]. These observations provide a compelling explanation for the underlying mechanisms of some protective neutralizing antibodies (nAbs) that target the NTD but do not interfere with ACE2–S interactions, and imply that the NTD of SARS-CoV-2 S is a potent antigen for developing vaccines and therapeutic antibodies [61].
Table 1.
Receptors, coreceptors, and cofactors involved in SARS-CoV-2 entry
| Molecule | Category | Functional annotation | Refs |
|---|---|---|---|
| ACE2 | Receptor | The major entry receptor | [2,19] |
| AXL | Receptor | A potential alternative receptor independent of ACE2 | [59] |
| KREMEN1 | Receptor | A potential alternative receptor independent of ACE2 | [60] |
| ASGL1 | Receptor | A potential alternative receptor independent of ACE2 | [60] |
| CD147 | Receptor | A potential alternative receptor independent of ACE2 | [80] |
| Heparan sulfate | Coreceptor | Auxiliary attachment receptor, dependent on ACE2 | [62] |
| Sialic acid | Coreceptor | Auxiliary attachment receptor, dependent on ACE2 | [65,66] |
| Lectin receptors | Coreceptor | Auxiliary attachment receptor, dependent on ACE2 | [64] |
| Neuropilin 1 | Coreceptor | Auxiliary attachment receptor, dependent on ACE2 | [70,71] |
| CD4 | Coreceptor | Potential auxiliary attachment receptor, dependent on ACE2 | [81] |
| SR-B1/ cholesterol |
Cofactor | The S protein binds to cholesterol, and SR-B1 increases virion endocytosis by promoting cholesterol uptake | [75] |
| Furin | Cofactor | Proteolysis of S protein at the S1/S2 site | [17,25,45,53] |
| PC-1 | Cofactor | Proteolysis of S protein at the S1/S2 site | [52,53] |
| Trypsin | Cofactor | Proteolysis of S protein at the S1/S2 site | [52., 53., 54.] |
| Matriptase | Cofactor | Proteolysis of S protein at the S1/S2 site | [52., 53., 54.] |
| Cathepsins | Cofactor | Proteolysis of S protein at the S1/S2 and S2′ sites | [45,54] |
| TMPRSS2 | Cofactor | Proteolysis of S protein at the S2′ site | [25] |
The coreceptors heparan sulfate (HS), sialic acid, and lectin receptors
A recent study reported that HS is a necessary attachment receptor that promotes SARS-CoV-2 infection in various target cells [62]. Biochemical assays revealed that HS interacts with the RBD of SARS-CoV-2 S protein at a non-overlapping site with the ACE2 contacting interface. The binding of HS significantly promotes the structural transition of SARS-CoV-2 S from the closed conformation to the open conformation, which increases the accessibility of the RBD for ACE2 engagement [62]. This interaction is essential for SARS-CoV-2 entry into many target cells. The therapeutic unfractionated heparin (UFH), non-anticoagulant heparin, and heparan sulfate proteoglycans (HSPGs) derived from human lung and other tissues can effectively block the cell entry of S-pseudotyped virus and authentic SARS-CoV-2 [62,63]. Further, because the S protein is extensively decorated by glycans, some lectin receptors have been shown to promote SARS-CoV-2 infection by interacting with the glycan shield, serving as non-specific attachment receptors for viral infections [64]. In addition, the NTD of SARS-CoV-2 S protein contains a sialic acid-binding pocket that can mediate viral attachment by interactions with various sialoproteins, glycoproteins, and gangliosides on the cell membrane [65,66]. This feature has been observed in many other coronaviruses and might indicate a universal mechanism for coronavirus entry using sialic acid receptors [67., 68., 69.]. This evidence underscores the importance of these ubiquitous non-specific molecules which facilitate the attachment of SARS-CoV-2 to the cell surface to promote interactions with the major entry receptor ACE2, thus increasing the efficiency of virus entry (Figure 1).
The coreceptor neuropilin 1 (NRP1)
Two additional studies reported that the membrane protein NRP1 promotes SARS-CoV-2 entry [70,71] (Figure 1). Neuropilins are a family of membrane-anchored coreceptors for a panel of molecules such as vascular endothelial growth factors (VEGFs) and semaphorins [72]. Both NRP1 and NRP2 bind to furin-cleaved C-terminal peptides of VEGFs, which are accommodated within a pocket in the b1 domains of the NRPs [73]. This C-terminal motif generally follows the rule Arg/Lys-X-X-Arg/Lys (R/K-XX-R/K, where X is any amino acid) [74]. Interestingly, proteolysis of SARS-CoV-2 S by furin/furin-like proteases generates a polybasic Arg-Arg-Ala-Arg motif at the C terminus of the S1 subunit, which conforms to the rule for interactions with NRPs [17,53]. Structural and biochemical evidence has demonstrated that the S1 C-terminal motif of SARS-CoV-2 directly binds to NRP1 [70,71]. Downregulating the expression of NRP1 by RNA interference or blockade of NRP1–S interactions by inhibitors can effectively reduce the cell entry and infectivity of SARS-CoV-2 [70,71]. Therefore, NRP1 serves as a coreceptor for SARS-CoV-2 infection that may complement the low expression levels of ACE2 in some target cells.
The cofactor scavenger receptor B type 1 (SR-B1)
In the sequence of SARS-CoV-2 S protein, some putative cholesterol recognition consensus motifs were identified in both the NTD and CTD of S1 subunit. Biochemical studies confirmed that SARS-CoV-2 S protein and RBD can directly bind cholesterol, and potentially high-density lipoprotein (HDL) components [75]. These interactions may promote the endocytosis of SARS-CoV-2 virion via SR-B1-mediated cholesterol uptake (Figure 1). Thus, the expression of SR-B1 can enhance the internalization of SARS-CoV-2, which further increases the efficiency of viral entry via an ACE2-dependent mechanism [75]. Because the binding of HDL to SARS-CoV-2 S involves both the NTD and RBD, NTD-targeting monoclonal antibodies that block the cholesterol-binding site on SARS-CoV-2 S can significantly inhibit HDL-enhanced SARS-CoV-2 infection. A similar inhibitory effect can also be observed by treatment with SR-B1 antagonists which impair cholesterol uptake [75]. Because SR-B1 is coexpressed with ACE2 in human pulmonary tissue and several extrapulmonary tissues, SR-B1 may play an important role in the multi-organ damage resulting from SARS-CoV-2 infection [76,77].
The complementary receptor CD147 and auxiliary receptor CD4
In addition to visceral organs, SARS-CoV-2 has also been shown to invade the immune cells and neurons [78,79]. Some of these cells do not express ACE2, and thus might be infected via other alternative receptors. Two recent studies reported that the SARS-CoV-2 S protein may also bind CD147 [80] and CD4 [81] molecules (Figure 1). CD147 is a type I transmembrane protein that interacts with several extracellular and intracellular molecules, and plays crucial roles in the infection of Plasmodium falciparum and SARS-CoV [82,83]. It is expressed in many cells including epithelial, neuronal, lymphoid, and myeloid cells, which may facilitate SARS-CoV-2 invasion of the immune and nervous systems. Indeed, Wang et al. showed that CD147 directly binds to the RBD of SARS-CoV-2 S protein with a high affinity [80]. The expression of human CD147 allows SARS-CoV-2 to infect the non-permissive BHK-21 cells, and loss of CD147 or treatment with anti-CD147 antibody or soluble CD147 extracellular domain inhibits SARS-CoV-2 replication [80]. These observations suggest that CD147 may be a potential alternative receptor for SARS-CoV-2 entry, which may further expand the tissue tropism of the virus [84,85]. Of note, a recent study reported contradictory findings that CD147 does not interact with S protein and plays no roles in SARS-CoV-2 entry [86]. Further studies will be necessary to verify the functions of CD147 in SARS-CoV-2 infection.
In contrast to other fully functional receptors, the CD4 molecule may be insufficient to mediate SARS-CoV-2 infection on its own. It potentially interacts with the RBD of S protein to assist viral attachment, facilitating the virus to infect CD4+ T helper cells through an ACE2/TMPRSS2-dependent mechanism [81]. The vulnerability of CD4+ T cells for SARS-CoV-2 infection may explain the poor adaptive immune responses of many COVID-19 patients [87,88]. In addition, KREMEN1 and ASGR1 are also well expressed in some immune cell populations, which may also contribute to immune invasion during SARS-CoV-2 infections [60].
Antibodies, vaccines, and inhibitors
Given the crucial role of S protein for virus entry, it is a very important target for antiviral intervention and is also the major antigen for developing vaccines. Many nAbs targeting different epitopes of SARS-CoV-2 S protein have been reported, and some of them have shown promising efficacies in clinical trials [61,89., 90., 91., 92.]. Most of potent nAbs target the RBD of S protein, making it a hotspot for eliciting antibodies and an ideal molecular entity for developing subunit vaccines [91,93,94]. Some antibodies targeting the NTD can also achieve effective neutralization by different mechanisms [61,75], such as causing steric hindrance for receptor engagement by the RBD or by impairing interactions with potential (co)receptors/cofactors that bind to the NTD. In addition, some antibodies may target the S2 subunit or the S1/S2 interface to prevent the conformational changes of S protein for membrane fusion [93]. More alternative or auxiliary mechanisms for SARS-CoV-2 entry are being discovered, and the number of candidate nAbs intercepting different steps of SARS-CoV-2 entry will continue to increase, which thus increases the likelihood of identifying highly potent therapeutic antibodies for clinical application.
Apart from antibodies, some small-molecule inhibitors are also promising drug candidates for blocking SARS-CoV-2 entry. For example, the anti-influenza drug Arbidol was able to neutralize SARS-CoV-2 by interfering with viral attachment to host cells, and has been used for clinical treatment of COVID-19 in China [95]. Molecular dynamics simulation analyses revealed that Arbidol may bind to both RBD and ACE2 at the contacting interface, with a higher affinity for RBD. The presence of Arbidol may impair the interactions between S and ACE2 to prevent viral entry [96]. Moreover, several fusion-inhibitory peptides have been developed with potent antiviral efficacies in vitro which bind to the HR1 domain of S2 subunit and prevent the conformational changes required for membrane fusion [97., 98., 99., 100., 101., 102.]. In addition, developing inhibitors for the host proteases that proteolytically activate the S protein, such as furin and TMPRSS2 inhibitors, is also a promising strategy to block SARS-CoV-2 infection of various susceptible cells [25,103., 104., 105.].
Concluding remarks
The entry of SARS-CoV-2 into host cells is a complex process and involves a panel of different molecules that may constitute multiple pathways for viral infection. The diversity of receptors, coreceptors, and regulatory cofactors is likely to greatly expand the host/tissue tropism of SARS-CoV-2, which may explain its high efficiency of transmission among humans and potentially other intermediate hosts. In addition to the aforementioned (co-)receptors/cofactors, a panel of other molecules have also been reported to interact with SARS-CoV-2 S protein, including CD207, leukocyte immunoglobulin-like receptor B2 (LILRB2), epidermal growth factor receptor (EGFR), low-density lipoprotein receptor (LDLR), and multiple innate immune receptors [59,60,106], but lack virology-related evidence to demonstrate their roles in SARS-CoV-2 infection. This evidence suggests the SARS-CoV-2 is well adapted to different cell types and that this may maximize its propagation and transmission among the various host populations. Therefore, continuous and extensive surveillance for SARS-CoV-2 and related coronaviruses is of particular importance for the prevention and control of the potential outbreaks. In spite of remarkable advances, several unresolved issues remain (see Outstanding questions) for achieving a comprehensive understanding in the mechanisms of SARS-CoV-2 entry. These insights would provide valuable clues for developing potent therapeutics and vaccines to counteract the raging global pandemic.
Outstanding questions.
How many alternative receptors or coreceptors, beyond the major receptor ACE2, exist for SARS-CoV-2 entry?
Are ACE2/TMPSS2 alone sufficient to mediate SARS-CoV-2 infection? Are other coreceptors/cofactors necessary to facilitate ACE2-dependent viral entry?
What is the role of CD4 in mediating SARS-CoV-2 entry into T helper cells in cooperation with ACE2? How does CD4 interact with S protein?
How is CD147 involved in SARS-CoV-2 entry, and what is its specific function?
How many copies of ACE2 actually bind to a S trimer during the authentic viral entry process? Are the different ACE2 molecules in sufficient close proximity to allow simultaneous interactions with a trimeric spike?
How many trimeric spikes are required for producing the fusion pore, further triggering a productive fusion event?
What is the trigger signal for S1 head dissociation – the binding of multiple ACE2 molecules, or proteolysis at the S2′ site, or both?
How do the NTD-interacting receptors (e.g., AXL) modulate the conformation of S protein and induce membrane fusion?
What is the host spectrum of SARS-CoV-2 in various domestic and wild animals?
How many SARS-CoV-2 related coronaviruses are there in animal reservoir, and what is their potential to infect humans?
What are the key determinants for the host-jump of SARS-CoV-2, and potentially related viruses, that enable human infection?
How can we design antibodies and vaccines to counteract the emerging SARS-CoV-2 variants?
Can we develop broad-spectrum coronavirus inhibitors or vaccines based on the S proteins?
Alt-text: Outstanding questions
Acknowledgments
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2016YFD0500305), and the Multiscale Studies on Structure, Function, and Regulation of Biological Supra-macromolecular Complexes (XDB37030204). R.P. is supported by the Young Elite Scientist Sponsorship Program (YESS) of the China Association for Science and Technology (CAST) (2018QNRC001).
Declaration of interests
We declare no competing interests.
Glossary
- Antigen
a molecule presented on the outside of a pathogen that can induce host immunity. Antigens can be any form of pathogen-derived molecules, and can be recognized by host immune cells to induce the production of antibodies or specific T cells.
- Efficacy
the capacity of therapeutics to treat a specific disease – the effectiveness of therapeutics or drugs.
- Epitope
a portion of the antigen that can be recognize by the immune system, either antibodies or T cells. For antibodies, the epitope is the region within the antigen that directly interacts with the antibody.
- Mortality rate
a measure of the ratio of deaths within an infected population that is an indicative parameter of the virulence of a specific pathogen.
- Neutralizing antibody (nAbs)
a group of antibodies that can protect host cells from invasion by pathogens. nAbs bind to the pathogen to inactivate its infectivity.
- Pathogenicity
the capacity of a pathogen to cause a disease in its host. The term is similar to the virulence of viral pathogens, and both are indicators of the potential harm to the infected host.
- Positive-sense RNA virus
a group of viruses with a single-stranded RNA genome whose sequence is consistent with that of the mRNA that encodes the viral proteins.
- Small-molecule inhibitor
a group of inhibitory molecules of low molecular weight, in contrast large-molecule inhibitors such as antibodies. They bind to specific targets to block their activities or functions.
- Transmission efficiency
different viruses may transmit with different degrees of efficiency among their hosts. Generally, a reproductive number (R0) is used to evaluate the efficiency of transmission, and indicates the average number of cases that become infected from a single infected case.
- Tropism
a biological term describing the permissive host cell types of viruses. Different viruses only infect specific types of host cells within their spectrum of tropism.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., et al. A novel coronavirus from patients with pneumonia in China, 2019. New Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong N.S., et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madewell Z.J., et al. Household transmission of SARS-CoV-2: a systematic review and meta-analysis. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.31756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z., et al. The assessment of transmission efficiency and latent infection period in asymptomatic carriers of SARS-CoV-2 infection. Int. J. Infect. Dis. 2020;99:325–327. doi: 10.1016/j.ijid.2020.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zumla A., et al. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siddell S.G., et al. Coronaviridae. Intervirology. 1983;20:181–189. doi: 10.1159/000149390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.V'Kovski P., et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belouzard S., et al. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao H., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke Z., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turonova B., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan L., et al. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q.H., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 21.Shang J., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuyama S. Protease-dependent cell entry mechanism of coronaviruses. Uirusu. 2011;61:109–116. doi: 10.2222/jsv.61.109. (article in Japanese) [DOI] [PubMed] [Google Scholar]
- 23.Walls A.C., et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan X., et al. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat. Commun. 2020;11:3618. doi: 10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai Y., et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuba K., et al. Multiple functions of angiotensin-converting enzyme 2 and its relevance in cardiovascular diseases. Circ. J. 2013;77:301–308. doi: 10.1253/circj.cj-12-1544. [DOI] [PubMed] [Google Scholar]
- 28.Yan R., et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigues J., et al. Insights on cross-species transmission of SARS-CoV-2 from structural modeling. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1008449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao K.P., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 31.Liu K., et al. Cross-species recognition of SARS-CoV-2 to bat ACE2. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2020216118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L., et al. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020;6:68. doi: 10.1038/s41421-020-00210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benton D.J., et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu C., et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 2020;7 doi: 10.1126/sciadv.abe5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou T.Q., et al. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe. 2020;28:867–879. doi: 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mansbach R.A., et al. The SARS-CoV-2 spike variant D614G favors an open conformational state. Sci. Adv. 2020;7 doi: 10.1126/sciadv.abf3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yurkovetskiy L., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korber B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou Y.J., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casalino L., et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henderson R., et al. Glycans on the SARS-CoV-2 spike control the receptor binding domain conformation. BioRxiv. 2020 doi: 10.1101/2020.06.26.173765. Published online June 30, 2020. [DOI] [Google Scholar]
- 43.Yang Q., et al. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. eLife. 2020;9 doi: 10.7554/eLife.61552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y.L., et al. Virus-free and live-cell visualizing SARS-CoV-2 cell entry for studies of neutralizing antibodies and compound inhibitors. Small Methods. 2020;5 doi: 10.1002/smtd.202001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ou X.Y., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bayati A., et al. SARS-CoV-2 infects cells following viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2020;296 doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffmann M., et al. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klenk H.D., Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2:39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 49.Steinhauer D.A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 50.Johnson B.A., et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bestle D., et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3 doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mykytyn A.Z., et al. SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. eLife. 2021;10 doi: 10.7554/eLife.64508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang T., et al. Proteolytic activation of SARS-CoV-2 spike at the S1/S2 boundary: potential role of proteases beyond furin. ACS Infect. Dis. 2021;7:264–272. doi: 10.1021/acsinfecdis.0c00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaimes J.A., et al. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience. 2020;23 doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puelles V.G., et al. Multiorgan and renal tropism of SARS-CoV-2. New Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lukassen S., et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39 doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sungnak W., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrison A.G., et al. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang S., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu Y., et al. Interaction network of SARS-CoV-2 with host receptome through spike protein. BioRxiv. 2020 doi: 10.1101/2020.09.09.287508. Published online September 13, 2020. [DOI] [Google Scholar]
- 61.Chi X., et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clausen T.M., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tandon R., et al. Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J. Virol. 2020;95 doi: 10.1128/JVI.01987-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thépaut M., et al. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. PLoS Pathog. 2020;17 doi: 10.1371/journal.ppat.1009576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seyran M., et al. The structural basis of accelerated host cell entry by SARS-CoV-2 dagger. FEBS J. 2020 doi: 10.1111/febs.15651. Published online December 2, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fantini J., et al. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Ag. 2020;55 doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W.T., et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park Y.J., et al. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat. Struct. Mol. Biol. 2019;26:1151–1157. doi: 10.1038/s41594-019-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szczepanski A., et al. Canine respiratory coronavirus, bovine coronavirus, and human coronavirus OC43: receptors and attachment factors. Viruses. 2019;11:328. doi: 10.3390/v11040328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cantuti-Castelvetri L., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Daly J.L., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zachary I. Neuropilins: role in signalling, angiogenesis and disease. Chem. Immunol. Allergy. 2014;99:37–70. doi: 10.1159/000354169. [DOI] [PubMed] [Google Scholar]
- 73.Parker M.W., et al. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 2012;287:11082–11089. doi: 10.1074/jbc.M111.331140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zanuy D., et al. Sequence dependence of C-end rule peptides in binding and activation of neuropilin-1 receptor. J. Struct. Biol. 2013;182:78–86. doi: 10.1016/j.jsb.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei C., et al. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2020;2:1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- 76.Zhang H., et al. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int. J. Infect. Dis. 2020;96:19–24. doi: 10.1016/j.ijid.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu F.R., et al. ACE2 expression in pancreas may cause pancreatic damage after SARS-CoV-2 infection. Clin. Gastroenterol. Hepatol. 2020;18:2128–2130. doi: 10.1016/j.cgh.2020.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song E., et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 2021;218 doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pontelli M.C., et al. Infection of human lymphomononuclear cells by SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.07.28.225912. Published online August 7, 2020. [DOI] [Google Scholar]
- 80.Wang K., et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Tar. 2020;5:283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davanzo G.G., et al. SARS-CoV-2 uses CD4 to infect T helper lymphocytes. MedRxiv. 2020 doi: 10.1101/2020.09.25.20200329. Published online September 28, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Z., et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2005;191:755–760. doi: 10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang M.Y., et al. Disrupting CD147-RAP2 interaction abrogates erythrocyte invasion by Plasmodium falciparum. Blood. 2018;131:1111–1121. doi: 10.1182/blood-2017-08-802918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qiao J., et al. The expression of SARS-CoV-2 receptor ACE2 and CD147, and protease TMPRSS2 in human and mouse brain cells and mouse brain tissues. Biochem. Biophys. Res. Commun. 2020;533:867–871. doi: 10.1016/j.bbrc.2020.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ulrich H., Pillat M.M. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev. Rep. 2020;16:434–440. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shilts J., et al. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci. Rep. 2021;11:413. doi: 10.1038/s41598-020-80464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robbiani D.F., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Long Q.X., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 89.Wu Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi R., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 91.Ju B., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 92.Lv Z., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cao Y., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Du S., et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183:1013–1023. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang X., et al. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 2020;6:28. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Padhi A.K., et al. Unraveling the mechanism of arbidol binding and inhibition of SARS-CoV-2: insights from atomistic simulations. Eur. J. Pharmacol. 2020;894 doi: 10.1016/j.ejphar.2020.173836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.de Vries R.D., et al. Intranasal fusion inhibitory lipopeptide prevents direct contact SARS-CoV-2 transmission in ferrets. Science. 2020;371:1379–1382. doi: 10.1126/science.abf4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xia S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun H., et al. Structural basis of HCoV-19 fusion core and an effective inhibition peptide against virus entry. Emerg. Microbes Infect. 2020;9:1238–1241. doi: 10.1080/22221751.2020.1770631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Musarrat F., et al. The anti-HIV drug nelfinavir mesylate (Viracept) is a potent inhibitor of cell fusion caused by the SARS-CoV-2 spike (S) glycoprotein warranting further evaluation as an antiviral against COVID-19 infections. J. Med. Virol. 2020;92:2087–2095. doi: 10.1002/jmv.25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu Y., et al. Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J. Virol. 2020;94 doi: 10.1128/JVI.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ling R., et al. In silico design of antiviral peptides targeting the spike protein of SARS-CoV-2. Peptides. 2020;130 doi: 10.1016/j.peptides.2020.170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheng Y.W., et al. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hoffmann M., et al. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maggio R., Corsini G.U. Repurposing the mucolytic cough suppressant and TMPRSS2 protease inhibitor bromhexine for the prevention and management of SARS-CoV-2 infection. Pharmacol. Res. 2020;157 doi: 10.1016/j.phrs.2020.104837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gao C., et al. SARS-CoV-2 spike protein interacts with multiple innate immune receptors. BioRxiv. 2020 doi: 10.1101/2020.07.29.227462. Published online July 30, 2020. [DOI] [Google Scholar]
- 107.Tyrrell D.A., Bynoe M.L. Cultivation of a novel type of common-cold virus in organ cultures. Br. Med. J. 1965;1:1467–1470. doi: 10.1136/bmj.1.5448.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 109.van der Hoek L., et al. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Woo P.C., et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zaki A.M., et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 112.Hu B., et al. Bat origin of human coronaviruses. Virol. J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]