Abstract
TPPP proteins exhibiting microtubule stabilizing function constitute a eukaryotic protein superfamily, characterized by the presence of the p25alpha domain of various lengths. Vertebrate species possess three TPPP paralogs; all of them possess a full-length p25alpha domain of 160–170 amino acids and are encoded by three exons. Species of Endopterygota (Holometabola) have, besides a full-size TPPP ortholog, a protein with a truncated p25alpha domain as well, where the last coding exon, responsible for microtubule binding, is missing. It is not the result of an alternative splicing but is coded by another gene. In Drosophila melanogaster, they are named as CG45057 (long-type) and CG6709 (truncated). The truncated protein has been found in the Endopterygota orders Diptera, Coleoptera, Hymenoptera, Lepidoptera and Raphidioptera. In Lepidoptera, in several superfamilies (Gelechioidea, Bombycoidea, Noctuoidea, Pyraloidea) two paralogs of the truncated TPPP occur. Truncated orthologs (CG6709) were not found in other insects or in arthropods and are absent in any other organism, as well, while the long-type TPPPs (CG45057 orthologs) occur commonly in all animals. Thus it seems that CG6709 orthologs occur only in insects undergoing on metamorphosis.
Keywords: Endopterygota, Lepidoptera, Phylogenetic tree, p25alpha domain
Endopterygota, Lepidoptera, Phylogenetic tree, p25alpha domain.
1. Introduction
TPPP-like proteins constitute a eukaryotic protein superfamily, characterized by the presence of the p25alpha domain (PF05517, IPR008907) [1]. TPPP-like proteins are named after the first identified member, Tubulin Polymerization Promoting Protein, TPPP/p25 or TPPP1, exhibiting microtubule stabilizing function [2, 3]. This function seems to be conserved in animals from sponge to vertebrates [4]; it is also true for TPPP of Drosophila melanogaster, CG45057 [5]. TPPPs, in the strict sense, contain no other domains but a p25alpha domain of various lengths. Vertebrate species possess generally three TPPP paralogs [6], more precisely, outparalogs [7], i.e., genes/proteins, in the same species, diverged from a common ancestral TPPP as a consequence of genetic duplication in a common ancestor of vertebrates; all of them possess a full-length p25alpha domain of 160–170 amino acids (aa) and are encoded by three exons [8]. Other animals have generally only one TPPP ortholog (orthologs are genes in different species evolved from a common ancestral gene), also with a full-length p25 domain. The number of the coding exons is also three in most cases; however, in some insects, as in various Drosophila species and in Tribolium castaneum, the first two exons are merged. Preliminary BLAST search indicated that some species of the phylum Arthropoda contain an additional TPPP-like protein. Here I show that they are present only in insects which undergo complete metamorphosis (Endopterygota or Holometabola).
2. Methods
BLASTP and TBLASTN analyses [9] were performed on protein and nucleotide sequences available at the NCBI website, http://www.ncbi.nlm.nih.gov/BLAST/ using D. melanogaster NP_648370 protein as a query. If a hit was found in a species of a given phylogenetic branch then its sequence was used as a query within the whole branch. All the protein and nucleotide databases available at this webpage were searched. Nucleotide sequences identified in BLASTN searches were translated in the reading frames denoted in the BLASTN hit, taking frame shifts or introns of genomic sequences into account. Further BLAST search was carried at the Ensemble website, http://metazoa.ensembl.org/, where few additional hits were found. Accession numbers of protein and nucleotide sequences refer to the NCBI GenBank database except otherwise stated.
2.1. Phylogenetic analysis
Multiple alignments of sequences were carried out by the Clustal Omega program [10]. Multiple sequence alignments used for constructing phylogenetic trees are shown in Supplementary Figures 1 and 2. The MEGA5 software [11] was used for maximum parsimony (MP) analysis. The MP tree was obtained using the Close-Neighbor-Interchange algorithm [12] in which the initial trees were obtained with the random addition of sequences (10 replicates). A majority-rule consensus tree was generated from the equally most parsimonious trees using the Consensus Tree option of the program. Internal support was assessed by non-parametric bootstrapping [13]; parsimony bootstrap percentages were based on 1000 replicates. Gaps were treated as missing data. Bayesian analysis was performed using MrBayes v3.1.2 [14]. Default priors and the WAG model [15] were used assuming equal rates across sites. Two independent analyses were run with three heated and one cold chains (temperature parameter 0.2) for 3 × 106 generations, with a sampling frequency of 0.01 and the first 25 % of the generations were discarded as burn-in. The two runs were convergent.
3. Results
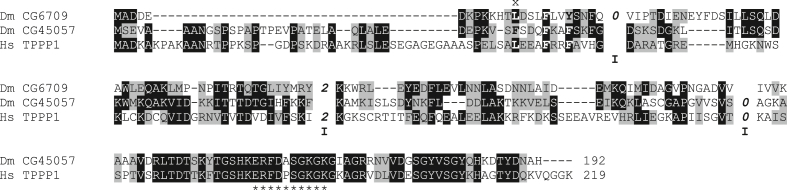
Drosophila melanogaster has, besides the full-size TPPP ortholog, a protein with a truncated p25alpha domain as well, where the last coding exon is missing. It is not the result of an alternative splicing but is coded by another gene. Interestingly, this shorter protein of 117 aa (CG6709) is encoded by a dicistronic gene (The other protein coded by this open reading frame is CG14164). All the Drosophila species, whose genomes have been sequenced, possess an ortholog with the same length. However, in the other species, the CG6709 orthologs are coded by a “normal” gene. Figure 1 shows the sequence alignment of the two D. melanogaster TPPP proteins with the human TPPP1. The p25alpha domain starts with a very conserved LxxxFxxF(Y) motif (See it also in Figure 2). It can be seen that the first two exons of the human protein are merged in the long-type CG45057 fruit fly protein but not in the truncated one (CG6709), while the first exon of the human protein is split in the truncated CG6709 protein. Moreover, at the border of the first and second exon (of the human protein), a conserved lysine is encoded by two nucleotides of the first and one nucleotide of the second exon in both the human and the truncated Drosophila protein (phase 2 introns). The last exon starts in the same position in the long-type CG45057 protein and in the human one but is missing in the truncated CG6709 protein. The 117 aa long CG6709 orthologs are more similar to each other than to the corresponding long-type TPPPs in the same species and vice versa. Based on the comparison of both orthologs in twelve Drosphila species, the identity of CG6709 proteins and CG45057 proteins varies between 65.81% and 99.15% and between 79.37% and 100%, respectively (Supplementary Figure 3). However, the pairwise identity of the long-type (186-194 aa) and “truncated” (117 aa) proteins in the same species is between 21.62%–27.27% (average 24.81%).
Figure 1.
Alignment used for sequence similarity comparisons of human TPPP1 and Drosophila melanogaster TPPP proteins. Intron positions were added manually in front of the affected amino acid position. The intron boundaries are indicated, also giving their phase. Asterisks show the tubulin binding sequence; x denotes the beginning of the p25alpha domain. DmCG6709 - Drosophila melanogaster NP_648370; DmCG45057 - Drosophila melanogaster NP_648881; Hs – Homo sapiens NP_008961.
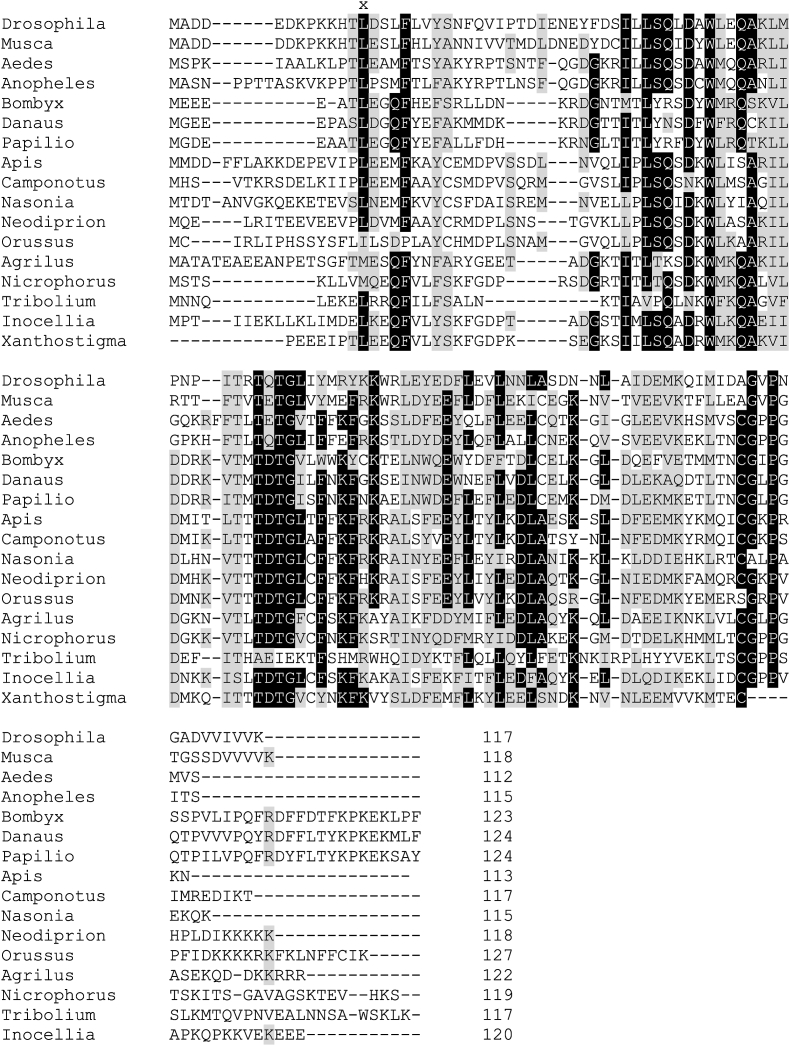
Figure 2.
Multiple alignment of Endopterygota CG6709 orthologs obtained by Clustal Omega program [12]. The alignment was refined manually. “x” denotes the beginning of the p25alpha domain. Residues identical and similar in the majority of the species are indicated by black and grey backgrounds, respectively. Diptera, Brachycera: Drosophila melanogaster NP_648370, Musca domestica XP_005179904; Diptera, Nematocera: Aedes albopictus XP_019531996, Anopheles gambiae XP_556944; Lepidoptera: Bombyx mori XP_004933177, Danaus plexippus XP_032527880, Papilio polytes XP_013136556; Hymenoptera, Apocrita: Apis dorsata XP_006607661, Camponotus floridanus XP_011254991, Nasonia vitripennis XP_008211062; Hymenoptera, Tenthredinoidea: Neodiprion lecontei XP_015522184, Hymenoptera, Orussoidea: Orussus abietinus XP_012277050; Coleoptera: Agrilus planipennis XP_025834993, Nicrophorus vespilloides XP_017778298, Tribolium castaneum XP_008190364; Raphidioptera: Inocellia crassicornis GAZH02002684, Xanthostigma xanthostigma GAUI02021553 (TSA, partial).
Blast [8] searches revealed that the orthologs of CG6709 protein can be found not only in all the Drosophila species but in both suborders of Diptera. The length of the proteins is 116–119 amino acid (aa) and 111-115 aa in Brachycera and Nematocera, respectively (Figure 2). The first exon is split only in the Drosophila species and the very closely related Drosophilini, Zaprionus indianus; thus the other orthologs contain only two exons. Orthologs can also be found in other orders of Endopterygota, namely, in Lepidoptera (butterflies and moths; 123-127 aa), Coleoptera (beetles; 116-122 aa) and Hymenoptera (sawflies, wasps, bees, and ants; 106-128 aa) (Figure 2)(It should be noted that databases name these proteins often erroneously as “CG45057-like”. The correct name ought to be CG6709-like). In three Raphidioptera (snakeflies) species (Fibla maclachlani, Inocellia crassicornis, Xanthostigma xanthostigma) hits were identified as nucleotides in the TSA (Transcriptome Shotgun Assembly) database (A detailed list of the CG6709 orthologs are shown in Supplementary Figure 4). Search in the other Endopterygota orders has not resulted in hits yet; however, it should be noted that far less species have been sequenced from these orders than from the above mentioned ones. E.g., search in the WGS (Whole Genome Shotgun) database suggests that Trrichoptera species also contain this gene and after the annotation of these genomes we can receive an unambiguous answer. I was wondering how strong the correlation is between metamorphosis and the presence of CG6709-like gene/protein. For this purpose, the Endopterygota genome assemblies of the https://www.ncbi.nlm.nih.gov/genome webpage, which lists the completed projects, were analyzed (Table 1). I found that 85% of the fully sequenced Endopterygota genomes contain CG6709 gene/protein. Orthologs were not found in other insects or in arthropods, although the long-type TPPPs (CG45057 orthologs) are common in these taxonomic units. Thus it seems that this protein occurs only in insects that undergo metamorphosis. However, they are absent in any other organisms.
Table 1.
Occurrence of CG6709-like gene/protein in Endopterygota.
| Order | Family | Genus | Species | |
|---|---|---|---|---|
| Diptera | 1/1 | 12/15 | 25/30 | 85/90 |
| Coleoptera | 1/1 | 6/10 | 9/19 | 9/20 |
| Hymenoptera | 1/1 | 15/16 | 39/56 | 54/75 |
| Lepidoptera | 1/1 | 19/20 | 64/65 | 85/86 |
| Siphonaptera | 0/1 | 0/1 | 0/1 | 0/1 |
| Endopterygota | 4/5 | 52/62 | 127/171 | 233/272 |
| 80.0% | 83.9% | 74.3% | 85.7% |
The 554 Endopterygota genome assemblies of the https://www.ncbi.nlm.nih.gov/genome webpage, which lists the completed projects (date: 28-02-2021), were analyzed; only the genomes where the protein count was given (272) were used for analysis. Numbers of genomes containing CG6709 versus all genomes are given.
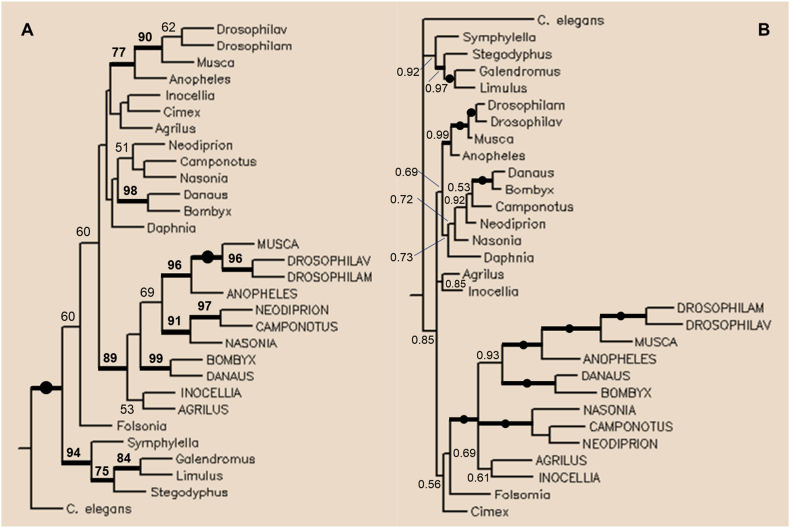
The phylogenetic analysis included long-type arthropod TPPPs as well as truncated TPPPs found in Endopterygota. Maximum parsimony analysis shows that truncated TPPPs are separated from the full-length (long-type) TPPPs and they are sister groups of each other's (Figure 3A). The N- and C-terminal parts of the long-type proteins were omitted from the alignment used for the construction of the tree since they are absent from the “truncated” ones; i.e., their p25alpha domain was used without the last exon (cf. Figure 1). Thus the tree is based on the common part of the proteins. Bayesian phylogenetic analysis using the program MrBayes v3.1.2 [15] also shows that truncated and long-type TPPPs form a separate clade (Figure 3B). It indicates that the presence of two kinds of TPPPs in Endopterygota species is not the result of in-species/family/order gene duplications but the consequence of an event occurring earlier, in their common Endopterygota ancestor. This is the reason for what can be seen in Supplementary Figure 3: there is a greater evolutionary distance between long-type – truncated TPPP pairs in the same Drosophila species than between truncated TPPPs in different Drosophila species.
Figure 3.
Phylogenetic tree of long and truncated TPPPs of Arthropoda obtained by Maximum Parsimony (A) and Bayesian (B) analysis. (A) Numbers above internal branches indicate bootstrap values shown as percentages (A) and Bayesian posterior probabilities (BPP) (B). Branches that received maximum support are indicated by full circles. Branches with bootstrap support higher than 70% or 0.95 BPP are indicated by thickened lines. Values lower than 50% are not indicated. For easier comparison, truncated TPPPs are labeled by capital letters. Proteins (TSAs∗) used for the construction of the tree: Hexapoda, Insecta, Endopterygota: Drosophila melanogaster NP_001246792, NP_648370; Drosophila virilis XP_015031007, XP_002047114; Musca domestica XP_005178632, XP_005179904; Anopheles gambiae XP_308808, XP_556944; Danaus plexippus XP_032520671, XP_032527880; Bombyx mori XP_004931507, XP_004933177; Nasonia vitripennis XP_001604263, XP_008211062; Camponotus floridanus XP_011250777. XP_011254991; Neodiprion lecontei XP_015521829, XP_015522184; Agrilus planipennis XP_018319221; XP_025834993; Inocellia crassicornis GAZH02007654∗; GAZH02002684∗; Hexapoda, Insecta, Paraneoptera: Cimex lectularius XP_014248365; Hexapoda; Collembola: Folsomia candida OXA61843; Crustacea: Daphnia pulex EFX79744; Myriopoda: Symphylella vulgaris GAKX01025293∗; Chelicerata: Galendromus (Metaseiulus) occidentalis XP_003743482; Limulus polyphemus XP_013794809; Stegodyphus mimosarum KFM57015; Nematoda: Caenorhabditis elegans NP_491219 (outgroup).
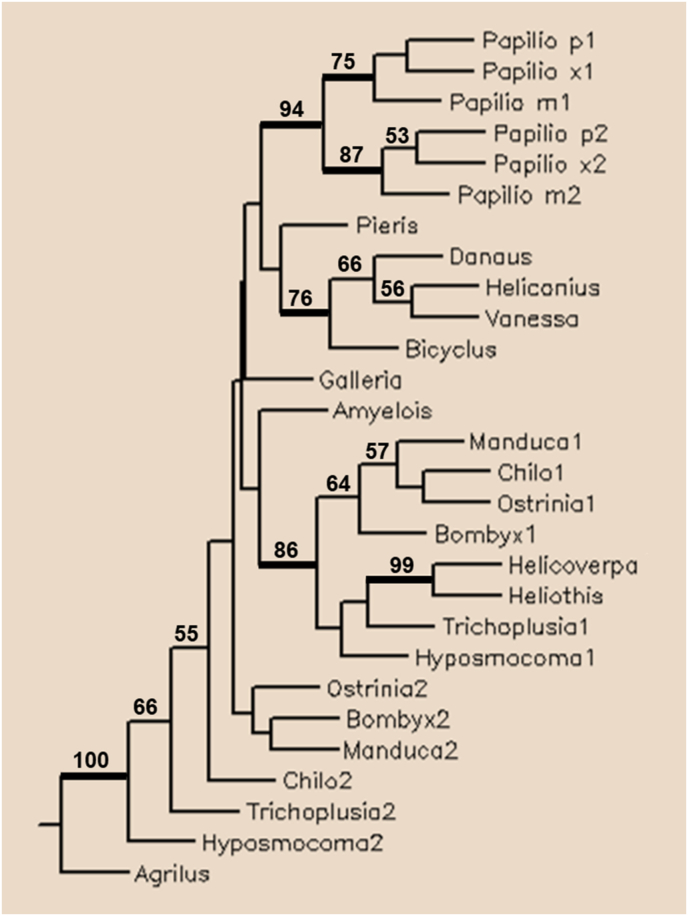
In Lepidoptera, in several superfamilies (Gelechioidea, Bombycoidea, Noctuoidea, Pyraloidea) two paralogs of the truncated TPPP occur. According to the phylogenetic tree, it seems to be the consequence of an early gene duplication within the order since the two paralogs form separate clades (Figure 4). They can be considered as outparalogs [7] since the duplication event happened earlier than the species speciation. Interestingly, in butterflies (Papilionoidea superfamily), only one truncated TPPP can be found except in Papilio species. However, the duplicated presence of the gene/protein in this genus seems to be consequence of a genus (family)-specific gene duplication (Figure 4); i.e., they are inparalogs [7].
Figure 4.
Phylogenetic tree of truncated TPPPs of Lepidoptera obtained by Maximum Parsimony analysis. Numbers above internal branches indicate bootstrap values. Only values higher than 50% are indicated. Branches with bootstrap support higher than 70% are indicated by thickened lines. Proteins (TSAs∗) used for the construction of the tree: Gelechioidea, Cosmopterigidae: Hyposmocoma kahamanoa XP_026315514, XP_026315526; Bombycoidea, Bombycidae: Bombyx mori XP_004933177, FS874530∗; Bombycoidea, Sphingidae: Manduca sexta XP_030040282, XP_030040281; Noctuoidea, Noctuidae: Trichoplusia ni XP_026742116, XP_026742114; Heliothis virescens PCG65904; Helicoverpa armigera XP_021192998; Papilionoidea, Nymphalidae: Bicyclus anynana XP_023933890; Danaus plexippus XP_032527880; Heliconius melpomene HMEL012067 Ensemble.; Vanessa tameamea XP_026490343; Papilionoidea, Papilionidae: Papilio machaon KPJ10293, KPJ10294; Papilio polytes XP_013136554, XP_013136556; Papilio xuthus XP_013162360, XP_013162358; Papilionoidea, Pieridae: Pieris rapae XP_022125790; Pyraloidea, Pyralidae: Amyelois transitella XP_013189465; Chilo suppressalis RVE51050, RVE42175; Galleria mellonella XP_026762893; Ostrinia furnacalis XP_028172140, XP_028169773; Coleoptera: Agrilus planipennis XP_025834993 (outgroup).
4. Discussion
The function of this truncated TPPP protein is not known. In the case of other members of the TPPP-like proteins, their binding to the microtubules and their role in stabilizing/organizing cytoskeletal structures were shown. Long-type TPPPs bind tubulin and promote its polymerization into microtubules; and bundle microtubules [2, 6]. This function is conserved in animals [4], including TPPP of D. melanogaster, CG45057. The fruit fly protein regulates microtubule stabilization and axonal extension during embryonic development [5], as well as synaptic microtubule organization via the acetylation level of the microtubule network [16]; and acts probably as a hub for microtubule regulators [17].
The amino acid sequences needed for tubulin/microtubule binding are located in the C-terminus of long-type TPPPs [4, 18, 19, 20]. In human TPPP1, there is an additional binding site at the N-terminus; the central part lacks any microtubule binding properties [18, 19, 20]. The insect-specific shorter protein contains practically only the “core” part but not the N- and C termini of the long-type TPPP (cf. Figure 1). The amino acid sequences needed for tubulin/microtubule binding located in the C-terminus of long-type TPPPs can be found in another TPPP-like protein, apicortin, occurring mostly in apicomplexan parasites [21]. Indeed, the necessity of this protein for the formation of the structure of the conoid, the nontubular polymeric form of tubulin, was proven [22]. Since the “truncated” TPPP lacks these amino acids thus it is a logical conclusion that it very probably is not able to bind microtubules.
One can speculate, on the basis of its specific phylogenetic occurrence, i.e., that it is present only in Endopterygota, and, otherwise, practically all Endopterygota orders seem to contain this gene, that its function may be related somehow to metamorphosis. According to the gene expression data of the Bgee database (https://bgee.org/; [23]), CG6709 is most abundantly expressed in testis, in pupa and in imaginal discs. During the pupal stage, many larval structures are broken down, and adult structures, including the discs, undergo rapid development [24]. Its abundance in the pupa in general and in the imaginal discs corroborates its potential role during metamorphosis. However, since about 15% of Endopterygota species seem to lack the CG6709 gene/protein, it cannot be excluded that it may have another role. It would mean that Endopterygota species might find a new, yet unknown, function for an old sequence. Obviously, experimental verification of this hypothesis is necessary.
Declarations
Author contribution statement
Ferenc Orosz: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The author thanks Dr. Judit Oláh for the careful reading of the manuscript.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
File 1: Multiple sequence alignments of TPPP proteins by Clustal Omega [10] used for constructing the phylogenetic trees in Figure 3.
File 2: Multiple sequence alignments of TPPP proteins by Clustal Omega [10] used for constructing the phylogenetic tree in Figure 4.
File 3: The percent identity matrix of CG6709 proteins and CG45057 proteins from twelve Drosophila species obtained by Clustal Omega program [10].
File 4: List of CG6709 orthologs.
References
- 1.Orosz F. A new protein superfamily: TPPP-like proteins. PloS One. 2012;7 doi: 10.1371/journal.pone.0049276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hlavanda E., Kovács J., Oláh J. Brain-specific p25 protein binds to tubulin and microtubules and induces aberrant microtubule assemblies at substoichiometric concentrations. Biochemistry. 2002;41:8657–8664. doi: 10.1021/bi020140g. [DOI] [PubMed] [Google Scholar]
- 3.Tirián L., Hlavanda E., Oláh J. TPPP/p25 promotes tubulin assemblies and blocks mitotic spindle formation. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13976–13981. doi: 10.1073/pnas.2436331100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oláh J., Szénási T., Szabó A. Tubulin binding and polymerization promoting properties of TPPP proteins are evolutionarily conserved. Biochemistry. 2017;56:1017–1024. doi: 10.1021/acs.biochem.6b00902. [DOI] [PubMed] [Google Scholar]
- 5.Mino R.E., Rogers S.L., Risinger A.L. Drosophila Ringmaker regulates microtubule stabilization and axonal extension during embryonic development. J. Cell Sci. 2016;129:282–294. doi: 10.1242/jcs.187294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vincze O., Tőkési N., Oláh J. Tubulin polymerization promoting proteins (TPPPs): members of a new family with distinct structures and functions. Biochemistry. 2006;45:13818–13826. doi: 10.1021/bi061305e. [DOI] [PubMed] [Google Scholar]
- 7.Sonnhammer E.L., V Koonin E. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 8.Štifanić M., Batel R., Müller W.E.G. Tubulin polymerization promoting protein TPPP ortholog from Suberites domuncula and comparative analysis of TPPP/p25 gene family. Biologia. 2011;66:111–120. [Google Scholar]
- 9.Altschul S.F., Madden T.L., Schäffer A.A. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sievers F., Wilm A., Dineen D. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura K., Peterson D., Peterson N. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nei M., Kumar S. Oxford University Press; New York: 2000. Molecular Evolution and Phylogenetics. [Google Scholar]
- 13.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixture models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 15.Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 16.Shi Q., Lin Y.Q., Saliba A. Tubulin Polymerization Promoting Protein, Ringmaker, and MAP1B homolog Futsch coordinate microtubule organization and synaptic growth. Front. Cell. Neurosci. 2019;13:192. doi: 10.3389/fncel.2019.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vargas E.J.M., Matamoros A.J., Qiu J. The microtubule regulator ringer functions downstream from the RNA repair/splicing pathway to promote axon regeneration. Genes Dev. 2020;34:194–208. doi: 10.1101/gad.331330.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hlavanda E., Klement E., Kókai E. Phosphorylation blocks the activity of tubulin polymerization-promoting protein TPPP: identification of sites targeted by different kinases. J. Biol. Chem. 2007;282:29531–29539. doi: 10.1074/jbc.M703466200. [DOI] [PubMed] [Google Scholar]
- 19.Tőkési N., Oláh J., Hlavanda E. Identification of motives mediating alternative functions of the neomorphic moonlighting TPPP/p25. Biochim. Biophys. Acta. 2014;1842:547–557. doi: 10.1016/j.bbadis.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 20.DeBonis S., Neumann E., Skoufias D. Self protein-protein interactions are involved in TPPP/p25 mediated microtubule bundling. Sci. Rep. 2015;5:13242. doi: 10.1038/srep13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orosz F. Wider than thought phylogenetic occurrence of apicortin, a characteristic protein of apicomplexan parasites. J. Mol. Evol. 2016;82:303–314. doi: 10.1007/s00239-016-9749-5. [DOI] [PubMed] [Google Scholar]
- 22.Leung J.M., Nagayasu E., Hwang Y.C. A doublecortin-domain protein of Toxoplasma and its orthologs bind to and modify the structure and organization of tubulin polymers. BMC Mol. Cell Biol. 2020;21:8. doi: 10.1186/s12860-020-0249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bastian F.B., Roux J., Niknejad A. The Bgee suite: integrated curated expression atlas and comparative transcriptomics in animals. Nucleic Acids Res. 2021;49(D1):D831–D847. doi: 10.1093/nar/gkaa793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beira J.V., Paro R. The legacy of Drosophila imaginal discs. Chromosoma. 2016;125:573–592. doi: 10.1007/s00412-016-0595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File 1: Multiple sequence alignments of TPPP proteins by Clustal Omega [10] used for constructing the phylogenetic trees in Figure 3.
File 2: Multiple sequence alignments of TPPP proteins by Clustal Omega [10] used for constructing the phylogenetic tree in Figure 4.
File 3: The percent identity matrix of CG6709 proteins and CG45057 proteins from twelve Drosophila species obtained by Clustal Omega program [10].
File 4: List of CG6709 orthologs.
Data Availability Statement
Data included in article/supplementary material/referenced in article.