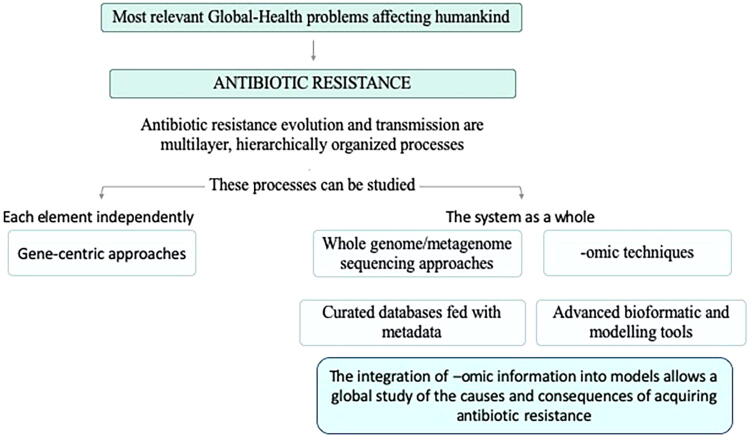
Graphical abstract
Keywords: Antibiotic resistance, Metagenomics, Genomics, One health, Whole genome sequencing, Natural computing, Genome-scale metabolic models
Abstract
Antibiotic resistance has been highlighted by international organizations, including World Health Organization, World Bank and United Nations, as one of the most relevant global health problems. Classical approaches to study this problem have focused in infected humans, mainly at hospitals. Nevertheless, antibiotic resistance can expand through different ecosystems and geographical allocations, hence constituting a One-Health, Global-Health problem, requiring specific integrative analytic tools. Antibiotic resistance evolution and transmission are multilayer, hierarchically organized processes with several elements (from genes to the whole microbiome) involved. However, their study has been traditionally gene-centric, each element independently studied. The development of robust-economically affordable whole genome sequencing approaches, as well as other -omic techniques as transcriptomics and proteomics, is changing this panorama. These technologies allow the description of a system, either a cell or a microbiome as a whole, overcoming the problems associated with gene-centric approaches. We are currently at the time of combining the information derived from -omic studies to have a more holistic view of the evolution and spread of antibiotic resistance. This synthesis process requires the accurate integration of -omic information into computational models that serve to analyse the causes and the consequences of acquiring AR, fed by curated databases capable of identifying the elements involved in the acquisition of resistance. In this review, we analyse the capacities and drawbacks of the tools that are currently in use for the global analysis of AR, aiming to identify the more useful targets for effective corrective interventions.
1. Introduction
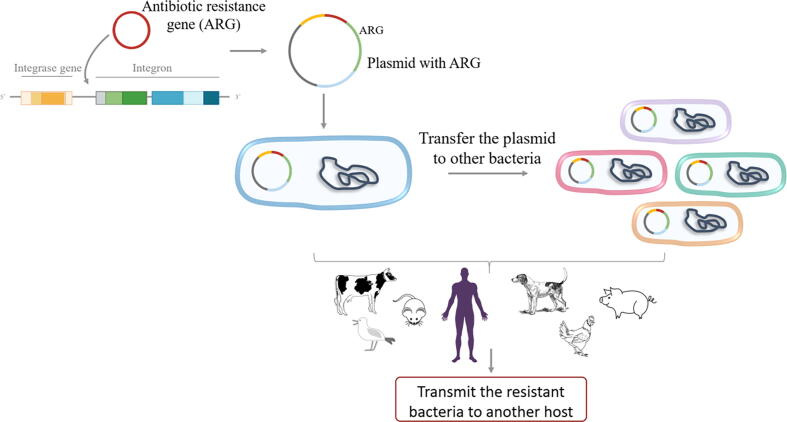
Infections had been historically the most important cause of human death. In this regard, the discovery and further use of antibiotics, and later on antifungal, antiviral and antiparasitic compounds, has been one of the major pharmacotherapeutic contributions to human welfare. The increasing prevalence of antibiotic resistant organisms is hence a relevant problem that has been recognized by several international agencies including, among others, the World Health Organization [1], [2], [3], [4], United Nations (https://www.un.org/pga/71/wp-content/uploads/sites/40/2016/09/DGACM_GAEAD_ESCAB-AMR-Draft-Political-Declaration-1616108E.pdf) and the World Bank (https://www.worldbank.org/en/topic/health/publication/drug-resistant-infections-a-threat-to-our-economic-future). Studies on antibiotic resistance (AR) had traditionally focused on hospitals and health care centres. Further, they had been mainly based on phenotypic tests directed to identify the most appropriate antibiotics for treating the infection suffered by a given patient. Actually, AR is defined at clinical setting using breakpoints of minimal inhibitory concentrations (MICs), which are established based on the likelihood of the success of the treatment [5], no matter the resistance mechanisms involved. Despite its fundamental utility for treating individual patients, it has been revealed that this type of phenotypic tests is not potent enough for advanced epidemiological studies, including the analysis of outbreaks at hospitals. Molecular-based approaches as Multi Locus Sequence Typing (MLST) [6] or typing based on Pulse Field Gel Electrophoresis [7] demonstrated to be useful in the molecular epidemiology analysis or organism causing infections, while the implementation of PCR techniques was a breakthrough for tracking the dissemination of resistance genes and the mobile elements involved in their dispersal [8], [9], [10], [11]. However, each of these techniques allowed to study just a limited aspect of the AR problem. In this regard, it is worth mentioning that AR is an evolutionary process in which different hierarchically interconnected elements participate [12], [13]. An AR gene (ARG) can be acquired by an integron that is further included into a plasmid present in a given bacterium, that can transfer the plasmid to other organisms, each one capable of infecting a different host, which can also transmit the resistant organism to other hosts (Fig. 1) [14], [15]. Analysing each of these levels of complexity and integrating the information to get a clear picture of the roads for the emergence and transmission of AR is not an easy task [16], [17]. Nevertheless, holistic tools, capable of analysing all complexity levels ideally without the need of isolating the resistant organisms, are making this difficult task more feasible.
Fig. 1.
AR Transmission as a multi-layered and hierarchical evolutionary process. ARGs are recruited by gene-capture elements such as integrons that are included in mobile elements as plasmids, which may be in turn acquired by a given bacterium. These clones can transfer the plasmid to other bacteria through HGT; these resistant bacteria may infect different hosts and then can spread among different hosts and environments. Thus, the process of transmission facilitates the evolution of AR at multiple, interconnected levels.
Once the first genome was sequenced, Haemophilus influenzae in 1995, the advances in new technologies, both experimental and computational, for studying biological systems have been enormous [18], ending up in the development of systems biology approaches [19]. Instead of dividing a biological process into pieces as classical molecular biology approaches do, the aim of systems biology is to understand the functioning of a biological system as a whole. In the specific case of AR, the “selfish gene” concept of Dawkins [20] is generally applied; gene-centric (or mutation-centric) approaches are the current rule in the field. However, it is important to recall that the introduction of a new ARG or the selection of AR mutations can produce deep effects in bacterial physiology, altering sometimes the expression of hundreds of genes [21], [22], [23] and hence modifying the bacterial proteome and its metabolism. Besides, it is also relevant to state that the effect of one ARG (or AR mutation) on AR and on bacterial physiology is context-dependent; different in different organisms [24], [25]. Therefore, in order to have a complete understanding of AR mechanisms and the consequences of acquiring resistance for bacterial physiology, bioinformatics tools and techniques able to analyse the increasing data generated in molecular biology through multi-omics technologies - including transcriptomics, proteomics and metabolomics - have become essential. The main objective of the information acquired through “-omics” is to create knowledge from data, providing simultaneous information on the presence and identification of alleles of thousands of genes, analysis of their genetic linkage with other genes, and measurements of their levels of expression as well as of the proteins they encode and cell metabolites [26]. To get this goal, system-scale models able to collect the huge amount of experimental information into mathematical models have been developed to further understand the bacterial cell as a whole [27], [28].
Besides their contribution for analysing the causes and consequences of the acquisition of AR [29], the emergence of easy and non-expensive whole-genome sequencing (WGS) tools (also including transcriptomics studies) had been a hallmark for the study of the phylogenomics of bacterial pathogens [30], [31] and AR [32], [33], [34]. These approaches are currently fundamental in advanced molecular epidemiology studies (including the analysis of human-linked and environmental microbiomes) and had been also proposed as useful diagnostic tools. In the present review, we discuss current information on the advances and limitations of the use of these techniques for studying AR, under the umbrella of One-Health, Global-Health approaches [14], [35], [36].
2. Emergence and spread of antibiotic resistance in a multilevel selection space
By definition, many human bacterial pathogens were susceptible to the antibiotics used in therapy when these antibiotics started to be used (otherwise, the antibiotics would had not been developed by pharma companies). However, resistant variants began to emerge soon after antibiotics were introduced in clinical practice, either due to mutations [37] or as a consequence of the acquisition of ARGs [38]. With few exceptions [39], [40], [41], AR mutations are transferred only vertically; they are relevant just for the clones harbouring the mutation. In this case, clonal selection, and the associated clonal expansion, is the main element involved in AR spread. One aspect to address here is knowing if the acquisition of resistance favours specifically the enrichment of AR clones over the susceptible ones, hence altering the bacterial population structure, or if the most abundant clones are those more prone to acquire AR just by chance, in which case resistance will not necessarily alter the structure of the population. The situation in the case of ARGs acquired by Horizontal Gene Transfer (HGT) is different than in the case of mutational events since, as above stated, their selection implies multiple layers of organization (Fig. 1). Each layer is a unit of selection and, as a matter of fact, the properties of reproduction, inheritance, variation and interaction actually define an individual [42]. Each one of these individuals are the subject of Darwinian evolution. However, since they are hierarchically linked, selection-driven changes [43] in one of them will influence top-down (from microbiomes to genes) or bottom-up (from ARGs to clonal complexes) any one of the others, forming what has been dubbed as a nested living system [44]. Eventually, abundant clones that can acquire ARGs more easily might contribute to the homeostasis of the clonal structure of the species by giving access to minority clones to this “common ARG pool” by means of HGT.
Mutation driven resistance is a problem for the patient infected by bacteria presenting such mutations. Hence, it can become a global public health problem as a consequence of the clonal expansion of such mutants. While this is an obviously dangerous situation, the acquisition, through HGT, of AR determinants can be even more problematic. In addition to clonal expansion, the presence of ARGs in transferable elements allows their spread among different clones and even among different (frequently phylogenetically related) bacterial species [45], [46], [47]. One important aspect of the field of AR applied to human health is risk analysis. The risk of finding a given ARG in non-pathogenic bacteria is not as high as if the same gene is found in a virulent organism [5], [48]. While the detection of gene-host association is easy in the case of isolated bacteria, the situation is not the same when microbiomes are analysed (see below).
Nevertheless, even when isolated bacteria are studied, the impact on human health of AR might be different according to the clone harbouring the ARG and the location of this gene (non-mobile or within a mobile element). Different bacterial isolates from the same species may have different niches and different properties. One example of this situation is Escherichia coli that, despite being a regular human commensal, presents some virulent clones, due to the incorporation in their genome of virulence determinants acquired by HGT. Further, the host range of pathogenic variants can vary; there are clonal complexes able to colonize/infect humans and other animals and there are clones that are animal-specific [49], [50]. When the problem for human health of selection of AR at farms is studied, this feature and the identification of shuttle clones or mobile elements capable of moving from animals to humans has to be taken into consideration [14]. It is important to highlight that the number of genes shared by all members of a given species (the core genome) is much lower than the total number of genes (the pangenome) that the species presents as a whole [51], [52]. Knowing that different clones may behave differently concerning human health, different methodologies have been applied to identify those that most frequently cause infections and/or are involved in the dissemination of ARGs [53], [54]. Currently, the most popular method for phylogenetic analysis is MLST [55], which is based on the identification of specific alleles with phylogenetic value in a relatively low number (typically 7–8) of housekeeping, highly conserved, genes [6]. MLST allows to define sequence types (ST), which are currently used to compare the phylogeny of isolates from different bacterial species all around the word [56], [57]. Comparability of the results among different laboratories is based on the standardization of the methods and loci used for defining STs and in the existence of broad databases as PubMLST (https://pubmlst.org), which contains hundreds of thousands of genomes and millions of alleles. Despite its utility for inferring core-genome based phylogenies, the contribution of the accessory genome, which is critical for the bacterial adaptation to different habitats, cannot be assessed with this technology. In fact, while genome trees based on core genome are consistent with the species ontology, the phylogenetic relationships derived from the analysis of all genes (both core and accessory ones) are better represented as networks [58], [59]. This does not mean that classical MLST analysis is lacking usefulness in a whole WGS era. When linked to PCR-based detection of ARGs, as well as to plasmid typing [8], [60], these studies allow the multilevel analysis of the epidemiology of AR. Note that AR tends to spread and evolve in closely related bacteria lineages, eventually composed by assemblies of particular clones within [61]. In pathogenic bacteria, these clonal complexes frequently constitute the “high-risk” populations for human and animal health, and can be identified by MLST procedures. These types of analyses, mainly based on PCR approaches and Sanger-sequencing of the amplicons, still remain as the golden standard for analysing outbreaks and in regular molecular epidemiology studies dealing with the dissemination of AR. However, as will be discussed later on, WGS price reduction and the development of user-friendly bioinformatic tools for analysing sequencing results in a short time-lapse is changing this panorama.
3. Genomic approaches for the study of antibiotic resistance
The appearance of genomics, proteomics and transcriptomics has revolutionized the study of microorganisms. One of the fields that has benefited the most is the study of bacterial pathogens and their resistance to antimicrobials. Nowadays, the sequencing of a bacterial genome (or a metagenome) is affordable and the main constrains for applying this approach to the clinical field are the speed of sequencing (most laboratories outsource WGS) and the time and the skills required for the analysis of the obtained sequence. Although several of the analytical tools require some expertise in informatics, user-friendly bioinformatic approaches are increasingly implemented, which will favour these studies. However, while they are already useful for fundamental studies on AR and for epidemiological purposes [62], WGS-based approaches still have some limitations (see below) regarding their application in fast diagnostic procedures.
The main benefit of WGS-based approaches in the study of AR is that they can address simultaneously all the elements involved in the acquisition of the phenotype and can establish the linkage of each ARG with the element harbouring it, either mobile or non-mobile. Further, WGS allows to develop genome-wide studies based on the analysis of several genes/mutations [63], [64], [65], [66], [67], [68], [69], [70]. Furthermore, WGS is fundamental for performing phylogenomic studies including the accessory genome that go beyond classical MLST analyses, which as stated above, take into consideration just the core genome. Since bacterial evolution, and specifically the evolution of virulence and of AR, largely relies on the acquisition of virulence determinants/ARGs [71], [72], the availability of WGS tools is particularly relevant for understanding the elements driving the evolution of virulence and AR. Besides the aspects dealing with epidemiology, which will be discussed later on, WGS approaches are useful tools not only to describe novel mechanisms of resistance but also to predict how resistance (even to drugs not present yet in the market yet) can emerge. To note here, epidemiological studies must necessarily rely on what is known. The identification of novel resistance mechanisms is frequently cumbersome and WGS approaches are helping to solve this problem. Indeed, these technologies are increasingly being used for the identification of genes associated with AR [73]; allowing the development of genome collections and the annotation of genes from clinically relevant strains [74]. Bioinformatics approaches to study antimicrobial resistance include computing programs able to predict, detect, infer and analyse ARGs from isolated culture data. These programs are used jointly with the systematic compilations of previous knowledge taken from databases (Table 1). Unlike other prediction methods, new detection methods employ rule-based inference, which takes advantage of already known AR phenotypes features. There are other methods that combine knowledge-based detection with machine-learning and mathematical inference approaches (hybrid methods) [75]. Specifically, Resfams addresses this remote homology detection problem employing hidden Markov models for AR protein identification [76]. Despite the potency of these methods, it is important to point out that predictions require validations.
Table 1.
Tools for analysing antibiotic resistance from genomes and metagenomes.
| Tool | Tool type | Database-link | Access | Approach | Status | Input description | Requirements | References |
|---|---|---|---|---|---|---|---|---|
| AMRFinderPlus | Detection-database-based | Reference Gene Catalog | Web and standalone | NA; EA; Assembly-based and read based tool | Active | Protein search, protein.fa and nucleotide | HMMER, BLAST+, Linux, and Perl | [78] |
| ResFinder | Detection-database-based | resfinder_db | Web and standalone | NA; EA; Assembly-based and read based tool | Active | Whole genome sequencing, isolate or annotated genome, preassembled partial, complete genomes, reads | [79] | |
| PointFinder | Detection-database-based | Pointfinder_db | Web and standalone | NA; EA; Assembly-based and read based tool | Active | Sequence file in FASTA | BioEdit platform (http://www.mbio.ncsu.edu/bioedit/ | [80] |
| Antibiotic Resistance Gene-ANNOTation (ARG-ANNOT) | Detection-database-based | ARG-ANNOT | Standalone | NA; EA; Assembly-based and read based tool | Active | Analysing genomes, genomes assemblies, metagenomic contigs or proteomes | Prodigal, DIAMON | [81] |
| The Resistance Gene Identifier (RGI) | Detection-database-based | CARD | Web and standalone | NA; EA; Assembly-based and read based tool | Active | Metagenomic | R | [82] |
| Online Analysis Pipeline for Anti-biotic Resistance Genes Detection (ARGs-OAP v2) | Detection-database-based | Web and standalone | NA; EA; Assembly-based and read based tool | Active | Sequence reads, allele db, gene db | python (v2.7.5) | [83] | |
| Short Read Sequence Typing for Bacterial Pathogens (SRST2) | Detection-database-based | Standalone | NA; EA Read-based tool | Active | Metagenomic and db | Unix server with ~2 GB of disk space for reference data 2X the input FASTQ file size in both RAM and disk space for temporary file storage :Perl , r, usearch, bwa, tophat incl., bam2fastx, samtools incl. bcftools and vcfutils.pl, ncbi blast, seqtk | [84] | |
| Search Engine for Antimicrobial Resistance (SEAR) | Detection-database-based | Web and standalone | NA; EA Read-based tool | Last update 2017 | Sequence reads | Linux | [85] | |
| Antimicrobial Resistance Identification By Assembly (ARIBA) | Detection-database-based | ARG-ANNOT, CARD, MEGAres and ResFinder | Standalone | NA; EA; Assembly-based and read based tool | Active | Sequence reads | Linux | [86] |
| SSTAR | Detection-database-based | SEED (annotator), AMR-related proteins that have been curated at ARDB and CARD | Standalone | NA; EA assembly-based and read based tool | Active | Two sequence files in FASTA format, one containing the bacterial genome assembly and the other the AR gene collection | NS | [73] |
| AdaBoost (PATRIC) | Detection and classification | Web | ML; EA Read-based tool | Active | Whole genome sequencing reads | NA | [87] | |
| PARGT | Detection-database-based | Standalone | NA | Active | Protein sequences | R and Python | [88] | |
| Resfams | Databased and AMR protein predictor | Resfams | Web and standalone | NA | Last update 2018 | NA | None | [89] |
| Antibiotic Resistance Genes Database (ARDB) | Database | ARDB | Web and standalone | NA | Available | NA | None | [90] |
| NCBI Bacterial Antimicrobial Resistance Reference Gene Database (BARRGD) | Database | PRJNA313047 | Web | NA | Active | NA | None | |
| NCBI Pathogen Detection | Database | Pathogens | Web | NA | Active | NA | None | |
| National Database of Antibiotic Resistant Organisms (NDARO) | Database | NDARO | Web | NA | Active | NA | None | |
| Isolates Browser | Database | Insolates Browser | Web | NA | Active | NA | None | |
| RefSeq | Database | refseq | Web | NA | Active | NA | None | [91] |
| How to Request New Alleles for Beta-Lactamase, MCR, and Qnr Gene | Database | Home page | Web | NA | Active | NA | None | |
| Microbial Browser for Identification of Genetic and Genomic Elements (MicroBIG-E) | Database | microbigge | Web | NA | Active | NA | None | |
| HMDARG | Database | HMDARG | Web | Assembly-based tool and hierarchical classification | Active | Raw sequence encoding | None | [120] |
NA: Not applicable EA: Exploratory approaches.
When current methods for predicting AR are compared [76], they differ in aspects as their link to previous knowledge sources (database-based methods), tool access (standalone or web service) and the range of data used for the analysis (from gene or protein sequences to whole genome or proteome data). Several database-based prediction methods are assembly-based; however, some applications admit raw sequences or protein sequences as input data. While for analysing the structure of the genetic elements harbouring ARGs contig generation is needed, the use of raw sequences can be more appropriate for quantification purposes. Standardization of methodologies and databases is still ongoing, although regulatory agencies as FDA have raised initiatives in this direction [77].
As stated in [33] and credited to Gerry Wright from McMaster University, ARGs can be classified in known knowns (those already analysed), known unknowns (those that have not been analysed yet, but are homologous to the knowns) and unknown unknowns. Although AR can be context-dependent [24], we have the tools needed to link genotype with phenotype for the first category, at least in most cases. Reads-based methods use different strategies for detecting ARGs from raw reads. For example, SRST2 [84] and GeneFinder [92] use efficient read alignment; and Mykrobe [93] creates Bruijn graphs of contigs from raw data and matches them against known ARGs; however, it omits the generation of a consensus sequence. Regarding the second category of known unknowns, machine-learning approaches enable strong predictions since these algorithms work by finding the relevant features in complex datasets. This approach uses a “rules-based” classification based on the presence of one or more known ARGs or mutations [76].
The PATRIC annotation system links bacterial genomes with AR metadata, which favours genotype/phenotype linkage. This project has also implemented a prediction method called AdaBoost, a machine learning classifier that identifies specific ARGs using whole-genome sequences. A balanced dataset between susceptible and resistant genomes influences the algorithm accuracy. Unfortunately, there are insufficient numbers of genomes from clearly susceptible bacteria to build these classifiers because several sequenced organisms are resistant to antibiotics, due to their epidemiological relevance [94].
PARGT is another machine learning prediction method. It uses protein characteristics to identify ARGs in Gram-negative and Gram-positive bacteria. Results obtained from a simulation carried out by the authors showed that PARGT could predict AMR sequences in Gram-positive bacteria with 87 to 90% accuracy. Since not all AR determinants contribute equally to the final AR phenotype, noise in phenotype predictions can often be reduced and accuracy increased by weighting each locus using a machine-learning model. These models can also be trained to consider potential interactions between loci. It is worth noting that there is compensation between the accuracy and the computational time when large-scale reference databases are used to generate the models [88]. Despite these advances in predictive methods, we must be aware that a single mutation can change the activity profile of an ARG, as it happens in the case of extended spectrum beta-lactamases [95]. Besides, synonymous mutations might have cryptic effects on the evolution of a global phenotype (resistance, fitness cost, and compensatory evolution), hence forming part of the “whisper mutations” set [96], [97]. Altogether, this implies that a phenotype cannot be always directly inferred from a consensus sequence. However, once the novel mutational variant is detected, analysing the associated phenotype, and incorporate this information in the analysis of AR, is an easier task. The main problem lies in the unknown unknowns category and, despite advances in the field, novel ARGs that may emerge always require experimental validation of their activity. Examples of this situation are qnr [98] or mcr-1 [99] conferring resistance to quinolones and polymyxin respectively and presenting sequences that did not resemble, at the time of their discovery, any previously known ARG.
Before WGS approaches were available, the identification of AR mutations relied on mechanisms-knowledge-driven approaches. Mutations in antibiotic transporters, targets or elements involved in the regulation of the expression of resistance determinants (as antibiotic inactivating enzymes or efflux pumps) had been searched under the idea that they should be the main cause of resistance. Based on this knowledge, specific applications such as the web tool PointFinder [80] able to identify known point mutations after mapping reads against reference genomes, as well as dedicated databases focusing on SNPs known to be involved in the acquisition of resistance in specific organisms have been developed [100]. However, efficient blind analysis of novel elements involved in the acquisition of resistance has been feasible just when WGS has become available. Massively parallel sequencing combined with traditional transposon mutagenesis [101], as well as high-throughput screening of transposon insertion libraries, have been used to track genes whose inactivation modifies the susceptibility to antibiotics [102], [103]. This method has been used to analyse the resistome of different pathogens, including Pseudomonas aeruginosa [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114], [115], [116], [117], [118], [119], E. coli [110], Staphylococcus aureus [111], [112] or Klebsiella pneumoniae [113], among other bacteria [114], [115].
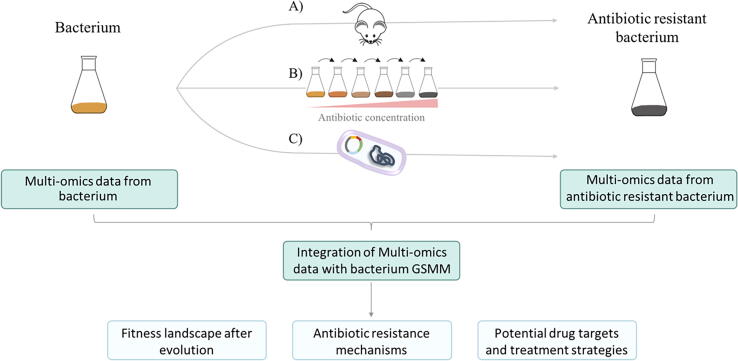
It is worth mentioning that the aforementioned studies only allow the identification of inactivating mutations that alter the susceptibility to antibiotics. However, AR is frequently achieved by mutations that do not inactivate the protein but just modify its activity. The use of adaptive laboratory evolution (ALE) experiments (Fig. 2) followed by WGS has allowed to describe AR mutations even before they are found in clinical settings. A major concern of these approaches is the differences between in vivo and in vitro growing conditions; it might be possible that in vitro-selected antibiotic resistant mutants are not selected in vivo, because fitness costs associated to the acquisition of resistance might be habitat-dependent. However, on several occasions, the same genetic events involved in the acquisition of AR in vitro have been also encountered in clinical isolates [116], [117], [118], [119], validating the efficacy of these approaches.
Fig. 2.
Integrated research of AR by multi-omics data and pathogen-specific genome-scale metabolic models. Multi-omics data are collected from a parental wild-type strain and a derived and antibiotic-resistant bacterium. After that, pairwise comparison between both strains is performed. Resistance can be selected during infection upon in vivo evolution (a), from adaptive laboratory evolution linages, which can be explored at different time points along evolution (b), or through the introduction of an ARG in the wild-type strain (c). The comparison of isogenic bacteria, with different levels of resistance, and isolated from the same patient provides the most accurate information on the evolution of AR during treatment [120], [121]. High throughput -omics data obtained from the analysis can be computationally integrated onto genome-scale metabolic models. Analyses of metabolic changes and mechanisms of AR allow the discovery of potential drug targets and treatment strategies against antibiotic-resistant pathogens. Fitness landscapes show the fitness cost of the AR, that influences the rate of development, stability and transmission of resistance, and the rate of resistance loss in the absence of the selecting antibiotic.
Besides genomic studies, transcriptomic, proteomic and, eventually, metabolomic analyses have been implemented with the aim of deciphering novel mechanisms of resistance [122], [123], [124], [125], or to detect the substrates, besides antibiotics, of known resistance determinants as multidrug efflux pumps [126]. The main drawback of these approaches is that, although the acquisition of AR usually implies changes in the level of expression of several genes/proteins, most of these changes are not associated with the mechanism of resistance, but with the fitness costs due associated with such resistance (see below). In this respect, unless the expression of an already known resistance determinant changes, expression-based approaches alone do not always provide accurate information on the mechanisms of resistance; experimental validation is needed to distinguish between correlation and causality [127], [128]. Further, it has been shown that antibiotics may induce an uncoordinated response in which transcriptionally and phenotypically important genes are not linked [129]. Nevertheless, this decoupling does not mean that transcriptomic/proteomic data are irrelevant for analysing AR, and there are different examples of their application [130], [131], [132], [133], [134]. Indeed, these methods have been used to describe new mechanisms of resistance, which have been experimentally validated [135], and it has been proposed that transcriptome profiling can be a useful tool in the analysis of AR in organisms such as P. aeruginosa [136].
In addition to the study of antibiotic resistant mutants, holistic expression-based studies (proteomics and transcriptomics), based on the analysis of the differential expression of some genes in presence of antibiotics, have been performed with the aim of describing novel mechanisms of resistance [137]. It is important to note that while it is true that transient resistance can be achieved via changes in gene expression, not all genes whose expression changes upon antibiotic stress are involved in the response to this injury [138], [139], [140], [141]. Careful analyses, taking into consideration available information and experimental validation of predictions based on changes in gene expression, are needed in order to establish the relevance of the findings derived from these studies. Besides its potential application for discovering novel mechanisms of resistance, transcriptomic-based assays in the presence of antibiotics have been proposed as a useful approach for the fast diagnosis of AR. In a first step, conserved differences of gene expression between susceptible and resistant strains are searched. Afterward, a small set of genes (around 10) are chosen and their expression in presence of antibiotics measured using fast hybridization techniques that allow to determine resistance in just a few hours [142].
4. Whole-genome based diagnosis and epidemiology of antibiotic resistance
The use of -omic techniques has been certainly a milestone in any area of biology, AR included. Besides their contribution in basic laboratory-based analysis, they are revolutionizing clinical microbiology studies [143] in two different areas: molecular epidemiology and diagnostic procedures.
Concerning the first, the full understanding of all the elements driving the emergence of resistance requires global approaches so that WGS is increasingly replacing current PCR-based techniques (see above). Nowadays, WGS studies that include hundreds or even thousands of strains are not uncommon. Whole-genome MLST (wgMLST) or core genome MLST (cgMLST) approaches have allowed, without the need of a reference genome, the taxonomical classification of isolates based on more than 1000 genes instead of the few ones analysed in classical MLST studies [63], [64], [65], [66], [67], [68], [69], [70]. Although the fundamentals of MLST and wgMLST are the same, wgMLST (and cgMLST) have a much higher discrimination power, allowing a much detailed, fine grained, description of the phylogenetic structures of bacterial populations 'from domain to strain' [69]. The linkage of this information with specific databases containing controlled reference genomes for diagnostic use and regulation as FDA-ARGOS [77], can help for studying in more detail the population structures of microorganisms with relevance for human health Concerning AR determinants, several approaches for finding ARGs and resistance mutations have been implemented (see above) and are in use for epidemiological studies. However, besides finding the resistance genes, it is also relevant to describe the mobile elements carrying them. It is important to note that, among all genomes deposited in databases, only a minor part of them is fully assembled; most are found as a variable number of different contigs, making in occasions difficult to distinguish if an ARG is present in a plasmid or in a chromosome. However, methods that allow getting long reads are helping to solve this problem; together with assemblers as SPAdes that include pipelines (plasmidSPAdes) for assembling plasmids [144] and informatic tools as PLACNET that allow the reconstruction of plasmids sequences obtained de novo or already present at WGS databases [145]. These tools can be used in order to analyse the genome/mobilome integration as well as to reconstruct the history of plasmid evolution and spread among different bacterial lineages [145]. More recently, a Perl application (Accessory genome Constellation Network-AcCNET) has been developed to compare the accessory genome (eventually containing ARGs) of several organisms, using the proteomes deduced from the analysed genomes [146]. Novel developments along this line as Pangenome Analysis Toolkit (PATO) allow to create, using conventional computers, clusters of thousands of genomes (pan-genome) using a list of pre-defined clusters, eventually from external sources as MLST; in addition, pan-genomes can be filtered by genome frequency (i.e. the number of genomes that belong to the pangenome), and the frequency of ARGs [147].
While the methodologies for genome-based molecular epidemiology studies are already robust, a problem exists in databases, particularly in their reutilization for meta-analysis purposes. This problem consists of the general weakness (eventually absence) of metadata associated with the deposited sequences. Information regarding inclusion criteria, point of isolation, treatment, disease or age in the case of patients; chemical composition or potential pollution of the habitat in the case of isolates from natural ecosystems; and any other information that might influence emergence, evolution and spread of AR is nearly absent and should be included in databases. In addition, databases are biased by the overrepresentation of strains derived from epidemic events. Although there are some databases that incorporate these metadata [148], [149], their development is still in its infancy.
WGS has also been proposed as a good tool for simultaneously determine the phenotype of resistance and the mechanisms involved in this resistance. For this purpose, WGS databases that include in their metadata phenotypic information are valuable tools [150], [151]. The use of machine-learning approaches able to predict AR based on sequence and expression data has been as well explored [152]. However, phenotypic tests are still superior to genome-based approaches, despite the utility of the latter in the case of slow-growing, non-cultivable or difficult-to-sample bacteria. One of these microorganisms is Mycobacterium tuberculosis, WGS has been proposed as a diagnostic tool that may be useful to analyse AR [153], [154] and the in vivo evolution of resistance even prior to tuberculosis diagnosis [155], and dedicated databases comprising hundreds of AR mutations have been implemented. Although they are becoming affordable, time and resources required for genome analysis are still a problem for their use at clinical setting. An important drawback to the general application of these approaches as diagnostic tools is that they just provide predictions. Although genotypic-phenotypic linkage seems to be strong in the case of mutation driven resistance [156] and stand-alone informatic tools have been developed to link genotypic-phenotypic information [157], in the case of resistance genes the phenotype of resistance depends not only on the ARG, but also on its genomic context [24]. Further, sequence-based approaches cannot identify as yet unknown mechanisms of resistance that may contribute to the phenotype. Another drawback of WGS-based approaches is that they usually require the isolation of the organism. To avoid this problem, metagenomic analysis using techniques as Nanopore, which is portable, fast and renders long sequence reads, can be implemented at least in low-complexity microbiomes, as those involved in infections at orthopaedic implants [158]. Whole genome enriching methods targeting specific organisms might be also applied in the case that the most probable causal agent of the disease is known. Indeed, these methods have been used for improving the recovery of M. tuberculosis DNA from sputum [159] in tuberculosis patients.
5. Antibiotic resistance beyond boundaries
The problem of AR goes beyond a single resistant bacterium or an infected patient. It is a global problem that involves the transfer of ARGs among bacteria forming part of gene-exchange communities [160], which are present in different ecosystems, including not only human-associated microbiomes, but also animal and environmental microbiomes [14], [35], [36], [118], [161], [162], [163], [164], [165], [166]. Culture-based approaches just allow studying a minority of the populations present in a microbiome. Consequently, the implementation of metagenomic techniques, which do not require culture, as well as the development of tools for the analysis of metagenomes such as MEGAN [167] have been fundamental for a deep understanding of the taxonomic composition and the overall genetic capacities (including AR) of any microbiome.
AR is a global problem, in which any ecosystem (from natural environments to human hosts) may participate [168], [169], [170]. The socioeconomic issues that modulate this transmission [171], and the mathematical models addressing this aspect of the problem [172], [173], [174], [175] will not be reviewed here. However, tools for tracking the transmission of ARGs among different ecosystems and geographical allocations [176], as well as detailed analyses of the effect of the selection of AR in a given microbiome are needed. In this regard, comparison of the resistomes of interconnected microbiomes [177] can provide clues of the transmission routes of resistance genes [35], and of the presence of these genes in bacteria colonizing ecological distinct niches as soil or human host [178]. Extremely randomized tree algorithm, optimized with Bayesian approaches, have been used to determine the variability of ARGs in different ecosystems and to track the ones that are discriminatory [179]. Besides, the analysis of the transmission of ARGs among ecosystems under strong selective pressure will certainly provide information on AR transmission. The microbiomes that are under stronger antibiotic selection are those from hospitalized humans and farms' animals. Accordingly, it has been stated that the use of antibiotics in farming may have important consequences for the spread of resistance in human populations [180], [181], [182]. ARGs selected in farms might be transferred to human pathogens or, eventually, the same pathogen can infect humans and other animals [183]. While this is generally true, there are some aspects of the problem that still require fine-tuning studies. Although, the species that infect humans and other animals, particularly in the case of mammals, can be the same, for some of them, the clonal lineages infecting each of the hosts are different [184]. Linking ARGs with the host is hence of relevance for estimating the risk to human health of such genes found in non-human-linked microbiomes. Besides, specific studies focusing on the mobilome shared by humans and other animals, may also help to analyse animal/human ARGs transmission [46], [185], [186].
Despite tools such as SqueezeMeta [187], a pipeline that integrates a set of programs required for metagenomic analysis, as well as the implementation of different AR databases (Table 1), are certainly helping in metagenomic studies on AR, some basic issues must be afforded when these studies are performed. The bioinformatic categorization of ARGs is one of them. This definition is usually carried out using DNA-based Blast searches oriented to already known resistance genes present in databases. In this regard, it is worth mentioning that existing ARG databases are skewed and biased towards beta-lactams resistance and aerobic/facultative aerobic bacteria, mainly those causing human infections. This bias has to be taken into consideration when studies of the global resistome of a given habitat -particularly environmental ones- is performed [36]. In addition to in-house Blast search approaches, tools linked to specific databases as ARIBA [86], ARG-ANNOT [81], AMRFinder [188], MEGARes [189], CARD [82] or Resfinder [190] have been fruitfully used for finding putative resistance genes, and AR mutations, in a variety of genomes and metagenomes. Besides classical pairwise comparison approaches, probabilistic annotation algorithms, such as hidden Markov models, have demonstrated to be useful for detecting ARGs [79], [191]. Although the predictive potential of these tools has largely improved, there are some issues that need to be taken into consideration when interpreting the results of the analyses of resistomes; in terms of the risk that detecting these putative resistance genes may have for human health [5], [48]. The strength of the constraints used for in silico searches is one of them. In order to get a comprehensive view of the resistome, some authors use relaxed parameters for ARGs search. While this can be valid for creating a picture of the whole potential resistome, the results can be confusing to specifically track the resistance genes acquired by human pathogens after the discovery of antibiotics, which are the ones currently being a risk for human health [5].
The use of the above-mentioned approaches leads to the description of several ARGs in any studied metagenome [192], [193], [194], [195], [196], [197], [198], [199]. However, most of them are unlikely to be a problem for human health because they are not present in human pathogens. Further, as discussed in [200], ARGs abundance does not necessarily correlates with risks to human health. Even more; the genomic context, which may drive the expression levels of a given ARG is fundamental for its contribution to the AR phenotype in its host [24]. It is true that all genes acquired by human pathogens were previously present in the genome of another microorganism [201], but the fact that the number of resistance genes present in the so far studied microbiomes is orders of magnitude above those currently moving among pathogenic bacteria, indicates that the probability of each of the detected genes to jump inside a human pathogen is low, unless it is already present in a mobile element [5]. In this regard, it has been shown that the resistome is linked to both ecology and phylogeny [202], [203]. This is an indication that most detected ARGs belong to the intrinsic resistome, formed by the ensemble of genes present in all (or most) members of a given bacterial species, which contribute to its characteristic phenotype of antibiotic susceptibility and had not been recently acquired by HGT [103], [104], [204], [205], [206], [207]. As discussed in [5], these genes have to be considered as phylogenetic markers, more than a risk of for human health. Specific databases on mobile elements as ACLAME [208], INTEGRALL [207], or mMGE (specifically focusing on human metagenomic mobile genetic elements) [209] are certainly helping in distinguishing between intrinsic and mobile ARGs.
While DNA-based approaches are useful for detecting already known ARGs, functional metagenomics [178], [199], [203], [210], [211] or structural based approaches can be used for predicting fully novel resistance genes [212]. Using the later, it has been established that most ARGs present in the human gut microbiome belong to the intrinsic resistome. Hence, more than being a risk for the acquisition of resistance by bacterial pathogens, they are resilience genes, able to reduce the perturbations due to antibiotic use and help to keep the homeostasis of human gut microbiome [213].
For the sake of comprehensibility, most AR databases contain intrinsic and acquired ARGs and, on several occasions, AR mutations are also included. When automatic analysis of these databases is performed, manual curation of the results is needed [204]. For instance, the finding of intrinsic resistance genes as acrAB, which encode an enterobacterial efflux pump [214], in a metagenome just means that Enterobacteriaceae are present in the microbiome. Similarly, the finding of genes as marA (encoding a global regulator) [215] or gyrA (coding for the target of quinolones) [216], [217] is not a sign of AR unless the mutant allele (that confers resistance) is found. Information on whether the cause of resistance is a mutation in the gene included in the database and not just to the presence of the gene, if the gene can confer by itself resistance or is a regulator of the expression of the actual ARG, as well as if the gene has been found in mobile elements or is just an intrinsic ARG, must be included in annotations to allow an accurate analysis of resistomes. Hierarchically organized databases, based on well-curated data, including information on resistance antibiotic class, resistance mechanism and gene mobility [218] can help solve this problem.
In the case of metagenomic samples, a particular problem concerns the identification of allelic forms of a resistance gene that differ just in a few nucleotides, as TEM-1-derived extended spectrum beta-lactamases [96]. Most metagenomic works are based in Illumina, short-reads, technologies. The assemble of these short reads may produce chimeric genes, not really present in an organism within the microbiome. The application of sequencing technologies allowing long reads seems to be a better approach to solve this problem [219]. While PacBio [220] is accurate enough to distinguish among alleles, Nanopore [221], and associated analytic tools as NanoGalaxy, which can be extremely useful to analyse arranges of ARGs, mobile elements [222], [223], or the presence of acquired ARGs in a given microorganism (see below), are not accurate enough for the analysis of alleles presenting few differences in their sequences. Nevertheless, the portability and the fast generation of results of Nanopore devices make this technology an excellent choice for in situ metagenomic studies [224].
The risk for human health of an ARG depends on the organism in which this gene is present, ranging from environmental (lower) to commensals or pathogenic (higher). Binning approaches, based either on abundance or on nucleotide composition, are then needed for linking gene/bacterial host. [225], [226], even rescuing potential novel species from metagenomic data (metagenomic species, see [227]). Applications such as GroopM [228], CONCOCT [229], SCIMM [230], Metawatt [231], MetaBAT [232], MaxBin [233], and others have been used for this purpose. However, although whole genome reconstruction from metagenomic data is feasible in low-complexity microbiomes, when a large number of sequence reads is available [234] or when the purpose is to analyse the pangenome of a given microorganism [235], these approaches present some limitations, particularly when commensal and pathogenic sublineages of the same species (as it happens in the case of E. coli) may co-exist in the same microbiome. The situation is even more difficult for analysing genomic islands and plasmids, presenting different copy numbers, with a nucleotide composition different to the one of the chromosome of the bacterial host, and eventually harboured by different hosts. In these cases, it has been reported that the percentage of mobile elements that are reconstructed and rightly binned to the chromosome of their bacterial host from metagenomic data is low [236]. Despite these problems, it has been recently stated that advanced sequencing techniques able to detect bacterium-specific methylation patterns allow a more accurate binning of chromosomes and plasmid sequences Further, non-bioinformatic, laboratory-based techniques, as EPIC-PCR [237]or Hi-C [238], may help to solve the problem of plasmid-chromosome binning.
A final concern refers to the abundance of ARGs reads in metagenomic data, since these reads can fall below the detection limits using regular metagenomic sequence depth. This is particularly relevant in the case of complex and/or environmental microbiomes, in which the resistant bacteria carrying the mobile ARGs constituting a risk for human health represent a very low fraction. Gene-capture systems, which allow targeted metagenomic studies, have been developed to enrich ARGs sequences in metagenomic analysis of complex microbiomes [239].
6. Natural computing applied to study hierarchically organized systems
As stated before, AR develops in the whole biosphere as a hierarchically organized complex system with several layers of organization, each one affecting the others. Its performance is based on homeostatic rules able to digest and integrate entropy (randomness) to keep identity, while allowing variation and evolution [240]. In Platonic terms, a cell is model of Nature; a biological microcosm reflecting the natural macrocosm. This has inspired the development of natural computing, nature-inspiring computational methods [241]. Among them, cellular computing includes approaches as artificial neuronal networks, or cellular automata, where ensembles of cells produce emergent qualities using communication strategies (“bacterial computing”), and bioinspired processors [242]. The latter includes network evolutionary processors transducers to simulate chemical transformations of substances or network genetic processors.
If the progress of genomic techniques has provided a powerful analytic tool to understand the biological complexity of the microbiosphere, new methods of synthesis are required to understand evolutionary functioning, including AR. Synthesis has benefited from the concept “cells as teachers”, giving rise to new computational engineering methods that can be applied at higher levels [205], [206].
One of the branches of cellular computing is membrane computing, taking advantage of the knowledge of the cellular organization to approach modelling of hierarchically structured biological systems. A cell is a space delimited by a membrane, which allows interaction with the environment while providing cellular individualization and identity; in fact, death essentially consists in the loss of individuality by membrane disruption [243]. Inside this space, there is a set of independent “objects”, surrounded by membranes in the case of eukaryotic cells -as nucleus, mitochondria, or chloroplasts- or discrete self-assembled (in a sense, compartmentalized by a virtual membrane) entities -as ribosomes, or efflux pumps-; and in bacteria, independent self-replicative genetic elements as plasmids. This nested (tree-like) ontology of individuals inside individuals, membranes inside membranes, can be expanded downwards to other independent discrete units, as genes themselves, or associated ensembles of genes (operons). However, it can be also expanded upwards, as cells are individuals forming part (“inside”) of individual supra-structures -as tissues, organs, species, microbiotas or holobionts- and finally ecosystems, which can be also treated as surrounded by functional membranes. Many of these intracellular or supracellular objects are independent units of selection [42], [244], [245].
The problem to address is how the evolution of a lower rank object influences the evolution of a higher rank object in the hierarchy, and vice versa. Indeed, the analytic (reductionistic) approach to study the complexity of natural systems and to ascertain the risks of their variation for human health has evident experimental boundaries. Empirical science is limited by the number of testable variables and by the practical impossibility of combining different ranges of them at various levels of the natural world. Mathematical modelling of AR has been limited until recently to “between-hosts” or “within-treated host” studies, but unfrequently using both dimensions [175]. Nevertheless, cellular computing allows the combination of both dimensions, at the same time that adds the “between-cells” and “within cells” dynamics of ARGs [246].
The series of “objects” defined by physical or virtual membranes in membrane computing models have interactive rules, integrating their action to sustain life. This view constitutes the basis of P-systems, one of the computational models used in membrane computing, developed by Gheorghe Păun [247]. Membranes (including whole cells, or higher structures as microbiota) have rules by which they can replicate, propagate, dissolve, exchange information according to flexible rules, mutate, be selected by external agents, or be transferred into other membranes. For instance, an ARGs-containing plasmid moves from one cell to another, or a resistant clone moves from one microbiota to another, based on probabilistic events. Alterations in an object, as those derived from different mutation rates, are quantitatively reflected by differences in membrane fitness. Another kind of “objects” can be introduced (or excluded) from the system to influence the rules of the membrane-contained objects; for instance, certain concentrations of antibiotic might reduce the rate of replication or the rate of plasmid transfer between cells.
7. Membrane computing and the dynamics of plasmid-mediated lateral transfer of antibiotic resistance genes
The full analysis of the emergence, spread and evolution of AR requires to consider a highly complex multi-parametric and multihierarchical landscape [248]. Membrane computing allows not only to study the effect of changing each one of these parameters in the model and observe the effects this change causes, but also to combine, in a single computational run, the associate effects of changes in multiple parameters [249], [250]. Tools as ARES [250] have been developed to analyse the trans-hierarchical evolution dynamics of AR in nested ecological compartments. Despite the intention of such tools is being user-friendly, their utilization still requires some bioinformatic knowledge, particularly on basic principles of membrane computing. The main advantages of these studies are: 1) the possibility of performing modelling à la carte, using the real data from a hospital, communities, or regions; 2) to use “range of possible parametric values” when some data are unavailable; 3) to evaluate the influence of a wide range of probabilities in the events influencing AR; 4) to assess the possible efficiency of interventions directed to reduce the burden of AR. In fact, membrane computing offers not only a way of representing processes but also a way to virtually reproduce (as a microworld) the process in a system.
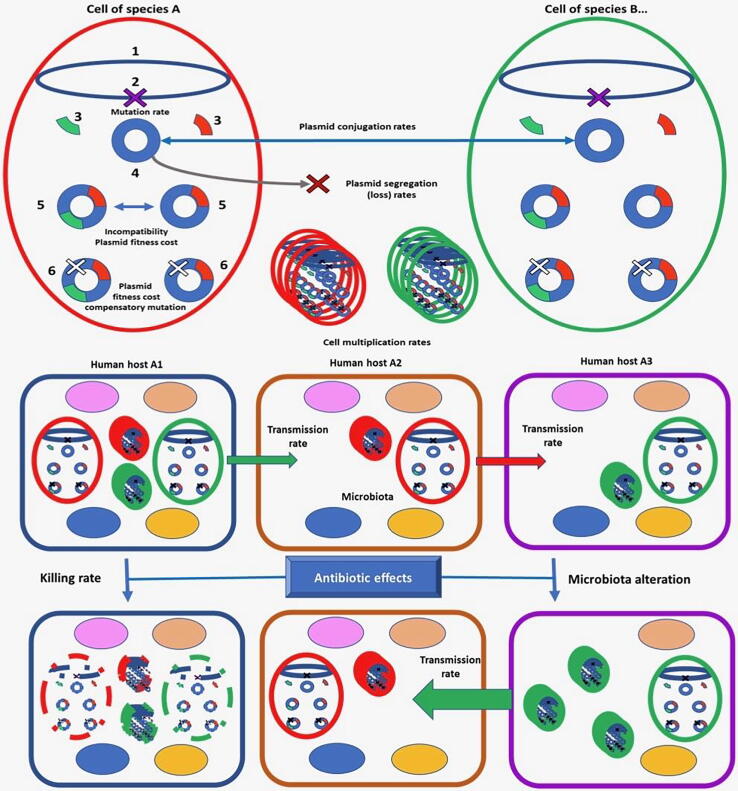
Several factors influence the dynamics of AR plasmids transfer among bacterial populations. These factors have been analysed under in vitro conditions, but determining their relative weight in the dissemination and maintenance of AR in the real world is an impossible task using classical tools. Membrane computing is an appropriate method to cast light in this question, because of the nested nature of the biological units involved: genes into plasmids, plasmids into cells, bacterial cells among bacterial populations and microbiotas in a hospital or community (Fig. 3). The problem of plasmid-driven dissemination of AR has been addressed investigating the independent effect of these factors in the evolution of resistance, using the landscape described in the former paragraph [251].
Fig. 3.
Thought scheme of a membrane computing model to simulate the evolution of AR. The main factors influencing evolution of resistance are shown in the figure. Numbered parts inside the cell: 1) bacterial chromosome; 2) mutational event leading to AR; 3) ARGs; 4) mobile genetic element (plasmid); 5) compensatory mutation decreasing plasmid fitness cost for the cell. In the lower part, rounded squares represented different human hosts containing species (ovals) in the microbiota; bacteria are transmitted between hosts (green and red arrows, transmission of red or green strains). A given antibiotic can eliminate (lower, left) some types of bacterial cells (green, red) and their contents (broken lines) in host 1, and another one might also eliminate members of the microbiota in host 3 (lower, right), favouring the multiplication of resistant cells to this antibiotic (green), increasing the possibilities of being transmitted to untreated host 2. AR depends on “within and between cells” and “within and between hosts” evolution, in a complex nested system. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
This study has shown that plasmid conjugation frequencies significantly influence the spread of AR strains whose resistance determinants are encoded by the element only at transfer rates of 10-3. Coexistence or exclusion (incompatibility) between plasmids with similar replication mechanisms was tested, and it has been described that coexistence might occur if the host strain contains two copies of these plasmids. Spontaneous plasmid loss has been found to have a small influence, being only of certain relevance at frequencies of loss of 10-4-10-5. Harbouring a plasmid might produce a reduction in fitness for thebacterial host; however, the model indicates that only reductions in growth rate above 6% influences the prevalence of plasmid carrying strains in a complex reality-like scenario. Plasmid fitness cost can be alleviated by “compensatory mutations”, but the effect of such compensation is extremely low under the analysed conditions, except in strong hypermutable strains (10–5 of mutation frequency). All in all, this study [251] indicates that changes in evolutionary units located at sub-cellular scale might have some consequences in the “big-real” world constituted by higher hierarchical level units.
8. Membrane computing and the epidemiology of antibiotic resistance
Membrane computing models have been applied to understand the multilevel dynamics of antimicrobial resistance within a hospital [252]. The evolution of several different bacterial species and clones introduced at the beginning of the process at different abundances and presenting different AR phenotypes was studied. Given the time-lapse (see below) and the involvement of patients, this type of study cannot be experimentally addressed.
To get this goal, different species and strains were included in the model. E. coli was included with four initial phenotypes: fully susceptible, plasmid-mediated aminopenicillin-resistance, ciprofloxacin-resistance, and both aminopenicillin and ciprofloxacin-resistance; K. pneumoniae, which is intrinsically resistant to aminopenicillins, was plasmid-mediated cefotaxime-resistance and ciprofloxacin-resistance; Enterococcus faecium (intrinsically resistant to cefotaxime) was included with two phenotypes: aminopenicillin-susceptible and aminopenicillin-resistance (encoded in a conjugative, transmissible element); and P. aeruginosa (intrinsically resistant). The model allows to treat modelled individual patients with different antibiotics at different dosages and was implemented for mimicking 20,000 steps (one hour each), representing a total of around 2.3 years. New resistance phenotypes emerged during the modelization process, due to the transmission of ampicillin and cefotaxime from resistant to susceptible populations, and/or mutational events leading to ciprofloxacin-resistance. Notably, these resistance phenotypes are more frequent in hospital patients than in community ones, and the evolution of resistance depends on the basal R situation.
The study shows the importance of hospital admission-discharge rates in hospital; if this parameter decreases, resistance increases in the hospital. The reduction in hospital bacterial transmission (measured as the proportion of individuals that acquired any kind of bacteria from another individual per hour), also reduces the evolution of AR even though less than expected, probably because of the transmission of susceptible bacteria. The transmitted bacterial load was shown to facilitate colonization. More important than hospital transmission was the proportion of hospital patients treated with antibiotics, enhancing the proportion and complexity of AR. The effect of rapid or slow bactericidal effect of antibiotics was tested and it was shown that slow killing facilitates resistance. The importance of the intensity of the “ecological effect” of different antibiotics in decreasing susceptible populations of the gut, and therefore the colonization-resistance of the microbiota (favouring colonization, abundance, and transmission of resistant organisms), was also supported by the study. Finally, the strength of antibiotic selection on resistance traits was investigated; for instance, the possibility that ampicillin selects for clones harbouring ESBLs, even though this antibiotic is not the most clinically-relevant substrate of these antibiotic inactivating enzymes, was approached. This summary of results is just an example of the possibilities of membrane computing in predicting the evolution of AR at different levels and under disparate situations.
9. Data integration for tackling antibiotic resistance
It has been generally assumed that the acquisition of AR produces a fitness cost that is mainly associated with an impaired bacterial growth. However, the situation is by far more complex; it is known that AR may alter the expression of several genes, not always associated with the acquisition of the resistance phenotype [23], [253], [254], [255], [256]. Besides altering the expression of virulence factors or signalling networks, the acquisition of AR may alter bacterial metabolism. These changes could be the Achilles' heel to specifically fight AR. Examples of metabolic rewiring associated with the acquisition of resistance are, among others, the anaerobic respiratory chain increased expression of P. aeruginosa multidrug resistant mutants [22], [23], or the increasing expression of an rRNA methylase in capreomycin resistant mycobacteria [257]. Knowing the effects of the acquisition of AR can hence provide evolution-based information for tackling this problem [258]. Conversely, the understanding of the bacterial metabolism can provide clues about the mechanisms and the effects of AR acquisition.
All the above-mentioned multi-omics technologies are increasingly used for the global analysis of AR. They simultaneously provide measurements of the expression of genes, proteins and metabolites that can be used to compare the physiology of susceptible and resistant pathogens at different time points [26]. However, information from these types of analysis is frequently disconnected, and the integration of this information may benefit from the establishment of cell models. The availability of an increasing amount of sequenced genomes and transcriptomes, together with experimental work on the associated phenotypes, metabolic fluxes and biochemical studies, has allowed the reconstruction of genome-scale metabolic models (GSMMs) to elucidate the organisms' metabolisms and thus deduce metabolic capabilities for different species, several of them causing infections [26] (Fig. 2).
GSMMs interconnect metabolic reactions, their metabolites, genes and enzymes into mathematical models to study cellular mechanisms. Thereby, they predict metabolic responses linking gaps between transcriptional and phenotypic responses, a feature particularly relevant to understand the bacterial response to antibiotics [259], [260], [261], [262], [263], [264], [265], [266]. In this regard, GSMMs together with multi-omics data are the first step to a quantitative analysis of pathogens physiology, including their metabolism, resistance and virulence mechanisms, hence constituting novel approaches for metabolism-based drug target studies [26].
An example of how -omic data can be used to determine metabolic changes in response to antibiotics is provided by the analysis of P. aeruginosa biochemical pathways perturbed by polymyxin treatment [260]. To get this information, the P. aeruginosa iPAO1 metabolic model together with RNA-Seq data was used. Apart from the known lipid A modifications associated with the response to polymyxin injury, numerous biochemical pathways involved in central, amino acid and fatty acid metabolism were perturbed, which enhances oxygen uptake and decreases growth rate [260], [267]. Another example is the in-silico analysis of S. aureus small colony variants (SCVs). A GSMM of this organism showed that an in silico-generated hemB SCV mutant should have an essentially fermentative behaviour, should secrete lactate and its growth would be, at least partially, recovered in presence of glutamate, glutamine or arginine [28]. It remains to be established if this growth recovery means a reduction in AR of SCVs, in which case, these amino acids might be used as antibiotic adjuvants. Within the same study, the effect of in silico deletions of different genes on bacterial metabolism was analysed. Notably, all lethal gene deletions that had been identified experimentally were correctly predicted by the model, supporting its reliability, and allowing to propose enzymes from glycan biosynthesis and lipid metabolism as promising targets for the search of S. aureus inhibitors.
The construction of GSMMs is a tedious labour, in which four main stages – draft reconstruction, manual curation, conversion to mathematical model and network analysis – are needed to create a metabolic model [268], [269]. There are different tools to create metabolic reconstructions [270], [271], [272], [273], [274] and among them, new automated tools, such as CarveMe universal model, allow the reconstruction of both species and community metabolisms, creating amazing metabolic model collections [275]. This automated tool overcomes the problems of previous methods, such as gap-filling, correction of elemental balance or directionality of reactions, detection of futile cycles and removal of blocked reactions or dead-end metabolites [275]. In addition, a flexible tool capable of combining information of GSMMs to identify promising targets as well as multi-target treatments is the web application FindTargetsWEB, which has been used to evaluate metabolic networks of both Gram-negative and Gram-positive multidrug-resistant pathogens such as P. aeruginosa or S. aureus [276].
Together with the use of -omic approaches, GSMMs across multiple strains and species or communities provide knowledge of pathogenic shifts due to AR at system-level [26] (Fig. 2). As an example, the use of GSMMs together with metabolomics data has been used to study streptomycin and chloramphenicol resistance in Chromobacterium violaceum. This study revealed that, while chloramphenicol resistance enhanced acetate production, streptomycin resistance increased acetate and formate secretion [259]. In addition to describe common metabolic pathways, GSMMs are essential to describe specific metabolic pathways for each microorganism, including AR ones, as well as virulence factor synthesis pathways [26]. This knowledge, together with transcriptomic, proteomic and metabolomic data can be used to understand the physiology of the bacterial pathogen under different conditions, including those with relevance for infection. For instance, this type of models has allowed to elucidate key elements associated to biofilm type of growth [277], [278], to identify virulence factors [279] or to simulate the metabolic dynamic of P. aeruginosa in cystic fibrosis patients [280]. Further, recent works have identified common metabolic signatures associated with AR, virulence and clinical outcome in P. aeruginosa causing cystic fibrosis chronic infections [281] or metabolic profiles of multidrug resistant M. tuberculosis [282]. All this information is valuable for diagnosis/prognosis purposes, as well as for finding pathways whose metabolic reprogramming may tackle resistance [283], [284], [285], [286], [287] or impair virulence aside from identifying metabolites that might be used for improving antibiotics' activity [284], [288].
Even though transcriptomic/proteomic data have been used for target identification, the absence of correlation between transcriptional and phenotypical responses triggered by antibiotics calls into question if these studies could be generalized [129]. Transcriptional response related to antibiotics is unrelated to stress and confers no fitness advantage, thus a more useful approach to predict drug targets and eradicate pathogens would include network topological analyses to put into realistic models high-throughput experiments [129]. It is for these reasons that the development of GSMMs linking the metabolic phenotype to environmental and genetic perturbations is central to drug discovery [275], [289].
There are different computational methods used to test and validate GSMMs in the field of drug target discovery. Firstly, flux balance analysis (FBA) is used to represent the possible behaviour of microbial metabolism and to predict gene essentiality [290]. One of the objectives of FBA is the use of biomass production studies to detect new elements able to compromise pathogen’s growth. This approach has provided insights into Porphyromonas gingivalis essential growth and has postulated LPS production as a potential drug target [291]. Moreover, FBA is used to determine if the deletion of a given gene can arrest the selected function in the bacterial metabolism, being the gene knockout stimulation using FBA the most popular method employed for drug discovery [289], [276]. Secondly, flux variability analysis (FVA) determines the range of alternate routes able to get the same objective, identifying other potential drug targets [292]. Thirdly, flux sampling calculates all solutions in a statistical meaningful way when the objective is not clear [293].
Traditionally, the most common method to identify drug targets has been the prediction of essential genes whose inactivation leads to flux redistribution, being the enzymes they encode potential drug targets. GSMMs are highly effective in gene deletion analysis studies, since this type of computational analyses, performed in a few seconds, save time and effort in comparison to experimental work [26]. Consequently, GSMMs have allowed to identify essential enzymes that can be exploited as targets of broad-spectrum antibiotics against different pathogens. For instance, these studies have revealed that Yersinia pestis essential genes are mainly genes encoding proteins involved in amino acid transport and metabolism and that some of these metabolic genes are also identified as essential in E. coli and Salmonella typhimurium [294]. The use of GSMMs for the identification of the organisms' catabolic cores might then be a useful tool for identifying pathways predicted to be critical for biomass generation, being even essential. The experimental validation of such prediction shows that this approach is indeed useful [295].
Besides the identification of essential pathways, another approach is the prediction of essential metabolites [296]; when a metabolite is removed within the model, all reactions in which it is involved disappear and the consequences are studied. The enzymes found to be involved in the consumption and production of these metabolites can be studied as new drug targets [263], [289], and the information on the metabolites themselves can be used to search for analogues of them that might inhibit bacterial growth [289]. These approaches can also be used to explore the chemical space that may be eventually useful to develop combination antibiotic therapies [297] or multi-target antibiotics. For the later, sets of reactions that act together may be targeted as an unique pathway [298] and consequently, altering the flux of one of them by targeting one enzyme results in an altered flux of all the reactions. For instance, 147 reactions were found to be coupled in M. tuberculosis models and 25 of these reactions account with previously known drug targets; thus, lethal combinations of these reactions could be assessed as potential targets [299].
GSMMs may also allow to simulate different infection scenarios, predict cellular phenotypes by changing nutrient sources and deduce infection features in different infected locations. This information on the metabolic situation of bacteria at different stages of infection might serve for identifying specific biomarkers and putative drug targets not so easily identified through in vitro approaches. For instance, pathogens may require specific nutrients and resources from the host cells. One example of this situation is Leishmania major, which obtains certain amino acids from the host macrophages since it is unable to synthesize them. The inhibition of this kind of pathways associated to infective situations and host characteristics, which may lead to environment perturbations, may help in defining new targets and therapeutic options based in metabolism remodelling [300], [301]. Among others, fatty acid and lipid metabolic reactions have been recently proposed as promising antimicrobial targets [26].
All in all, conventional methods to detect possible targets are expensive and slow. Accordingly, computational studies are a more efficient alternative to detect essential genes and their products as possible drug targets, facilitating, upon validation of the generated predictions, the development of new drugs targeting at different stages of infection [26]. Despite the fact that metabolic modelling is a new approach, it has allowed to detect targets that are consistent with previous data [28] as well as to provide new hypotheses [296] to explore.
Once drug targets are identified, drugs capable of inhibiting these targets should be screened. The most evident category for developing new drugs interfering with bacterial metabolism would be the structural analogues of the essential metabolites found by metabolic reconstruction and reprogramming through GSMM. These analogues would be able to replace and to compete with the essential metabolite, disrupting metabolism and causing cell death [289]. For instance, lysine analogues that target lysine riboswitches inhibit Bacillus subtilis growth by repressing lysine biosynthesis pathway. Some of these riboswitches regulate essential genes whose proteins produce metabolites that cannot be obtained from the external medium, being promising targets for the development of new RNA-binding antibiotics [302]. In addition to the search of new antibacterial compounds, these approaches could be used to identify metabolite analogues able to re-sensitize resistant pathogens to existing antibiotics. As mentioned before, C. violaceum resistance is induced by metabolic shifts, more specifically by a switch to fermentative metabolism thanks to an increased NADH/NAD ratio. GSMM studies have allowed to identify 2-oxoadipate as a compound able to re-sensitize these resistant populations, inducing susceptibility to existing antibiotics and death in C. violaceum [259]. Nevertheless, and despite these advances in the field, no novel antibiotic based on these approximations is in the market yet.
10. Summary and outlook
Before the worldwide COVID pandemic emerged, AR was considered as the most relevant One Heath, Global Health problem that might compromise human health and welfare in the next years [303], [304]. Traditionally, AR studies have focused on infected patients. However, it is now clear that the problem goes outside hospitals and that AR evolution and spread involves a variety of ecosystems that, besides humans, include other animals (farm animals, pets and also wild-animals) and natural, non-clinical, ecosystems [36], [200]. Most common methods to analyse the genetic and molecular bases of AR are gene-centric: the presence of a specific gene or mutation is sought and, eventually, the association of this gene with mobile genetic elements is determined. While relevant for detecting specific mechanisms of resistance, these approaches are not sufficient to provide a global picture of the elements involved in the emergence and spread of AR. In this regard, it is relevant to state that AR is a multihierachical process with different interconnected elements (from genes to global population) involved and the simultaneous analysis of these processes requires the implementation of holistic approaches. WGS and other -omic techniques (transcriptomics, proteomics and metabolomics) have been implemented in the last years as the most appropriate tools for the integrative analysis of AR. These tools have already demonstrated to be valuable in basic studies on AR and in molecular epidemiology studies. However, they still present some limitations in their use as diagnostic tools (see below). Further, several of the tools require some bioinformatic training by users. This is not necessarily a drawback if the teams include bioinformaticians and experts in AR, but the fast translation of the results into clinics requires the development of user-friendly tools, not always available.
There two areas that still require novel tools and information; one is the accurate prediction of AR based in WGS data, which is needed for implementing sequence-based diagnostic approaches. Mutation-driven resistance is likely an affordable task, although it still requires a more comprehensive information of all mutations leading to AR resistance in the different organisms under study. Indeed, recent studies on experimental evolution in presence of antibiotics have shown that there are still hidden AR mutations that should be incorporated into the models [116], [305], [306], [307]. Comprehensive, organism-specific databases as MUBII-TB-DB, developed for predicting WGS-based M. tuberculosis AR have shown to be effective for this type of analysis [100]. The situation concerning acquired ARGs is more complex. Although there exist specific databases on mobile elements as ACLAME [208], INTEGRALL [207], or mMGE [209], most databases contain information on mutations, intrinsic and acquired ARGs. This information is fundamental for manual annotation of the results, but is less amenable for the automatic analysis of ARGs. Hierarchically organized databases as HMD-ARG [218] may help in solving this problem. However, even in these cases further refinement in the annotation is needed to distinguish between phenotypically validated ARGs and predicted ones. Rankings as those stablished in some genomic databases, as the one of P. aeruginosa, distinguishing among genes, which function has been validated in the organism under study, genes that are orthologs to genes with a validated function in another organism and genes with a function inferred just by homology [308] can help to distinguish between functionally validated and predicted AR, a feature with relevance for risks analysis [5], [48].
The other area requiring further development is the binning between the identified ARGs and the organism carrying them (commensal or pathogen) when metagenomes are studied. The use of powerful, automatic analytical tools as PhyloPhlAn 3.0 [309], which uses hundreds of thousands of genomes and tens of thousands of metagenomes for genome reconstruction may help in this endeavour. However, even in this case, the incorporation of HGT-acquired genes into the genomes is still cumbersome, unless the genomes containing ARGs are highly represented in the metagenome, because they present a different nucleotide composition and, if present in plasmid a different abundance. Sequence methods that provide long reads can help to solve this problem, and the combination of long- and short-reads information for assembling seems to be superior than using independently either short-reads or long-reads based methodologies, however costs will be higher too [310]. Hybrid methods making use of information on abundance and composition are the best option for binning the generated contigs [311]. However, even in this case, plasmid-chromosome binning can be cumbersome. The above-mentioned analysis of DNA methylation patterns that are specific of restriction/modification systems may help in these studies [312].
Once omic-based global information on AR is becoming accessible, time has come to implement models for its synthesis. The future perspective in the advancement of knowledge on complex biological systems will be highly dependent on the progresses in the integration of throughput analytic methods (-omics) and powerful synthetic tools (as natural multihierarchical computing, GSMMs, Big Data, machine-learning and metadata management, or artificial intelligence for systems engineering). In the case of AR, both detailed and global genomic information should become increasingly available, feeding integrative models to extract the predominant trends and to associate them with anthropogenic interventions. The information will constitute an important step forward for stablishing rational, ecological, and evolutionary approaches to tackle this relevant health problem.
CRediT authorship contribution statement
Teresa Gil-Gil: Writing - review & editing. Luz Edith Ochoa-Sánchez: Writing - review & editing. Fernando Baquero: Writing - review & editing. José Luis Martínez: Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Work in JLM laboratory is supported by grants from the Instituto de Salud Carlos III (Spanish Network for Research on Infectious Diseases [RD16/0016/0011]), from the Spanish Ministry of Economy, Industry and Competitivity (BIO2017-83128-R) and from the Autonomous Community of Madrid B2017/BMD-3691. Work in FB laboratory is supported by grants AC16/00043 ST131S, InGEMICS-CM, funded by Comunidad de Madrid (Spain) and European Structural and Investment Funds, CIBER in Epidemiology and Public Health, CIBERESP; CB06/02/0053, 2013-2016 and co-funded by Instituto de Salud Carlos III and the European Regional Development Fund (ERDF, ’A way to achieve Europe’). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- 1.WHO . World Health Organization; Geneva: 2014. Antimicrobial resistance: global report on surveillance. [Google Scholar]
- 2.WHO . World Health Organization Report in Infectious Disease. 2010. Antimicrobial resistance. [Google Scholar]
- 3.WHO . World Health Organization Report in Infectious Disease. 2000. Overcoming antibiotics resistance. [Google Scholar]
- 4.WHO . World Health Organization; Geneva: 1999. Removing obstacles for healthy development. [Google Scholar]
- 5.Martinez J.L., Coque T.M., Baquero F. What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol. 2015;13(2):116–123. doi: 10.1038/nrmicro3399. [DOI] [PubMed] [Google Scholar]
- 6.Maiden M.C., Bygraves J.A., Feil E., Morelli G., Russell J.E., Urwin R. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. PNAS. 1998;95(6):3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sader H.S., Hollis R.J., Pfaller M.A. The use of molecular techniques in the epidemiology and control of infectious diseases. Clin Lab Med. 1995;15(2):407–431. [PubMed] [Google Scholar]
- 8.Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K.L., Threlfall E.J. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Carattoli A., Zankari E., Garcia-Fernandez A., Voldby Larsen M., Lund O., Villa L. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibata N., Doi Y., Yamane K., Yagi T., Kurokawa H., Shibayama K. PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J Clin Microbiol. 2003;41(12):5407-13. doi: 10.1128/JCM.41.12.5407-5413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., Yang L., Fu J., Yan M., Chen D., Zhang L. Genotyping and high flux sequencing of the bacterial pathogenic elements - integrons. Microb Pathog. 2018;116:22–25. doi: 10.1016/j.micpath.2017.12.073. [DOI] [PubMed] [Google Scholar]
- 12.Baquero F. From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat Rev Microbiol. 2004;2(6):510–518. doi: 10.1038/nrmicro909. [DOI] [PubMed] [Google Scholar]
- 13.Baquero F., Coque T.M., Canton R. Allodemics. Lancet Infect Dis. 2002;2(10):591–592. doi: 10.1016/s1473-3099(02)00393-6. [DOI] [PubMed] [Google Scholar]
- 14.Hernando-Amado S., Coque T.M., Baquero F., Martinez J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat Microbiol. 2019;4(9):1432–1442. doi: 10.1038/s41564-019-0503-9. [DOI] [PubMed] [Google Scholar]
- 15.Martinez J.L. General principles of antibiotic resistance in bacteria. Drug Discov Today Technol. 2014;11:33–39. doi: 10.1016/j.ddtec.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Martinez J.L., Baquero F., Andersson D.I. Predicting antibiotic resistance. Nat Rev Microbiol. 2007;5(12):958–965. doi: 10.1038/nrmicro1796. [DOI] [PubMed] [Google Scholar]
- 17.Martinez J.L., Baquero F., Andersson D.I. Beyond serial passages: new methods for predicting the emergence of resistance to novel antibiotics. Curr Opin Pharmacol. 2011;11(5):439–445. doi: 10.1016/j.coph.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Fleischmann R.D., Adams M.D., White O., Clayton R.A., Kirkness E.F., Kerlavage A.R. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269(5223):496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 19.Sandoval-Motta S., Aldana M. Adaptive resistance to antibiotics in bacteria: a systems biology perspective. Wiley Interdiscip Rev Syst Biol Med. 2016;8(3):253–267. doi: 10.1002/wsbm.1335. [DOI] [PubMed] [Google Scholar]
- 20.The D.R., Gene S. Anniversary edition. Oxford University Press; New York: 2006. 30th. [Google Scholar]
- 21.Alcalde-Rico M., Olivares-Pacheco J., Halliday N., Cámara M., Martínez J.L. The impaired quorum sensing response of Pseudomonas aeruginosa MexAB-OprM efflux pump overexpressing mutants is not due to non-physiological efflux of 3-oxo-C12-HSL. Environ Microbiol. 2020 doi: 10.1111/1462-2920.15177. [DOI] [PubMed] [Google Scholar]
- 22.Olivares J., Alvarez-Ortega C., Martinez J.L. Metabolic compensation of fitness costs associated with overexpression of the multidrug efflux pump MexEF-OprN in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58(7):3904–3913. doi: 10.1128/AAC.00121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivares Pacheco J., Alvarez-Ortega C., Alcalde Rico M., Martínez J.L. Metabolic Compensation of Fitness Costs Is a General Outcome for Antibiotic-Resistant Pseudomonas aeruginosa Mutants Overexpressing Efflux Pumps. mBio. 2017;8(4) doi: 10.1128/mBio.00500-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dantas G., Sommer M.O. Context matters - the complex interplay between resistome genotypes and resistance phenotypes. Curr Opin Microbiol. 2012;15(5):577–582. doi: 10.1016/j.mib.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Luo N., Pereira S., Sahin O., Lin J., Huang S., Michel L. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Nat Acad Sci USA. 2005;102(3):541–546. doi: 10.1073/pnas.0408966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sertbas M., Ulgen K.O. Genome-Scale Metabolic Modeling for Unraveling Molecular Mechanisms of High Threat Pathogens. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.566702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendriksen R.S., Bortolaia V., Tate H., Tyson G.H., Aarestrup F.M., McDermott P.F. Using Genomics to Track Global Antimicrobial Resistance. Front Public Health. 2019;7:242. doi: 10.3389/fpubh.2019.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinemann M., Kummel A., Ruinatscha R., Panke S. In silico genome-scale reconstruction and validation of the Staphylococcus aureus metabolic network. Biotechnol Bioeng. 2005;92(7):850–864. doi: 10.1002/bit.20663. [DOI] [PubMed] [Google Scholar]
- 29.Sukhum K.V., Diorio-Toth L., Dantas G. Genomic and Metagenomic Approaches for Predictive Surveillance of Emerging Pathogens and Antibiotic Resistance. Clin Pharmacol Ther. 2019;106(3):512–524. doi: 10.1002/cpt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez-Losada M., Arenas M., Castro-Nallar E. Microbial sequence typing in the genomic era. Infect Genet Evol : J Mol Epidemiol Evol Genet Infect Dis. 2018;63:346–359. doi: 10.1016/j.meegid.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klemm E., Dougan G. Advances in Understanding Bacterial Pathogenesis Gained from Whole-Genome Sequencing and Phylogenetics. Cell Host Microbe. 2016;19(5):599–610. doi: 10.1016/j.chom.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Crofts T.S., Gasparrini A.J., Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol. 2017;15(7):422–434. doi: 10.1038/nrmicro.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McArthur A.G., Tsang K.K. Antimicrobial resistance surveillance in the genomic age. Ann N Y Acad Sci. 2017;1388(1):78–91. doi: 10.1111/nyas.13289. [DOI] [PubMed] [Google Scholar]
- 34.Schürch A.C., van Schaik W. Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance. Ann N Y Acad Sci. 2017;1388(1):108–120. doi: 10.1111/nyas.13310. [DOI] [PubMed] [Google Scholar]
- 35.Baquero F., Coque T.M., Martínez J.L., Aracil-Gisbert S., Lanza V.F. Gene Transmission in the One Health Microbiosphere and the Channels of Antimicrobial Resistance. Front Microbiol. 2019;10:2892. doi: 10.3389/fmicb.2019.02892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berendonk T.U., Manaia C.M., Merlin C., Fatta-Kassinos D., Cytryn E., Walsh F. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol. 2015;13(5):310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 37.Martinez J.L., Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother. 2000;44(7):1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boto L., Martinez J.L. Ecological and Temporal Constraints in the Evolution of Bacterial Genomes. Genes. 2011;2:804–828. doi: 10.3390/genes2040804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balsalobre L., Ferrandiz M.J., Linares J., Tubau F., de la Campa A.G. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob Agents Chemother. 2003;47(7):2072–2081. doi: 10.1128/AAC.47.7.2072-2081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrandiz M.J., Fenoll A., Linares J., De La Campa A.G. Horizontal transfer of parC and gyrA in fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 2000;44(4):840–847. doi: 10.1128/aac.44.4.840-847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coffey T.J., Dowson C.G., Daniels M., Zhou J., Martin C., Spratt B.G. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5(9):2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 42.Gould S.J., Lloyd E.A. Individuality and adaptation across levels of selection: how shall we name and generalize the unit of Darwinism? Proc Nat Acad Sci USA. 1999;96(21):11904–11909. doi: 10.1073/pnas.96.21.11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez JL. Effect of antibiotics on bacterial populations: a multi-hierachical selection process. F1000Research. 2017;6:51. [DOI] [PMC free article] [PubMed]
- 44.Günther F., Folke C. Characteristics of nested living systems. J Biol Syst. 1993;1:257–274. [Google Scholar]
- 45.Jorgensen T.S., Kiil A.S., Hansen M.A., Sorensen S.J., Hansen L.H. Current strategies for mobilome research. Front Microbiol. 2014;5:750. doi: 10.3389/fmicb.2014.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez J.L., Coque T.M., Lanza V.F., de la Cruz F., Baquero F. Genomic and metagenomic technologies to explore the antibiotic resistance mobilome. Ann N Y Acad Sci. 2017;1388(1):26–41. doi: 10.1111/nyas.13282. [DOI] [PubMed] [Google Scholar]
- 47.Perry J.A., Wright G.D. The antibiotic resistance “mobilome”: searching for the link between environment and clinic. Front Microbiol. 2013;4:138. doi: 10.3389/fmicb.2013.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez J.L., Coque T.M., Baquero F. Prioritizing risks of antibiotic resistance genes in all metagenomes. Nat Rev Microbiol. 2015;13(6):396. doi: 10.1038/nrmicro3399-c2. [DOI] [PubMed] [Google Scholar]
- 49.Price L.B., Hungate B.A., Koch B.J., Davis G.S., Liu C.M. Colonizing opportunistic pathogens (COPs): The beasts in all of us. PLoS Pathog. 2017;13(8) doi: 10.1371/journal.ppat.1006369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheppard S.K., Guttman D.S., Fitzgerald J.R. Population genomics of bacterial host adaptation. Nat Rev Genet. 2018;19(9):549–565. doi: 10.1038/s41576-018-0032-z. [DOI] [PubMed] [Google Scholar]
- 51.Domingo-Sananes M.R., McInerney J.O. Mechanisms That Shape Microbial Pangenomes. Trends Microbiol. 2021 doi: 10.1016/j.tim.2020.12.004. [DOI] [PubMed] [Google Scholar]
- 52.Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15(6):589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 53.Willems R.J., Hanage W.P., Bessen D.E., Feil E.J. Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):872–900. doi: 10.1111/j.1574-6976.2011.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woodford N., Turton J.F., Livermore D.M. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011 doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 55.Tümmler B. Molecular epidemiology in current times. Environ Microbiol. 2020;22(12):4909–4918. doi: 10.1111/1462-2920.15238. [DOI] [PubMed] [Google Scholar]
- 56.Manges A.R., Geum H.M., Guo A., Edens T.J., Fibke C.D., Pitout J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin Microbiol Rev. 2019;32(3) doi: 10.1128/CMR.00135-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bianchi-Jassir F., Paul P., To K.N., Carreras-Abad C., Seale A.C., Jauneikaite E. Systematic review of Group B Streptococcal capsular types, sequence types and surface proteins as potential vaccine candidates. Vaccine. 2020;38(43):6682–6694. doi: 10.1016/j.vaccine.2020.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bapteste E., Lopez P., Bouchard F., Baquero F., McInerney J.O., Burian R.M. Evolutionary analyses of non-genealogical bonds produced by introgressive descent. PNAS. 2012;109(45):18266–18272. doi: 10.1073/pnas.1206541109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toussaint A., Chandler M. Prokaryote genome fluidity: toward a system approach of the mobilome. Methods Mol Biol. 2012;804:57–80. doi: 10.1007/978-1-61779-361-5_4. [DOI] [PubMed] [Google Scholar]
- 60.Alvarado A., Garcillan-Barcia M.P., de la Cruz F. A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS ONE. 2012;7(7) doi: 10.1371/journal.pone.0040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baquero F, Martínez JL, Lanza VF, Rodríguez-Beltrán J, Galán JC, San Millán Á, et al. Evolutionary Pathways and Trajectories in Antibiotic Resistance. Clinical microbiology reviews. In the press. [DOI] [PMC free article] [PubMed]
- 62.Hwang S.M., Cho H.W., Kim T.Y., Park J.S., Jung J., Song K.H. Whole-Genome Sequencing for Investigating a Health Care-Associated Outbreak of Carbapenem-Resistant Acinetobacter baumannii. Diagnostics (Basel. Switzerland) 2021;11(2) doi: 10.3390/diagnostics11020201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmid D., Allerberger F., Huhulescu S., Pietzka A., Amar C., Kleta S. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011–2013. Clin Microbiol Infect : Off Publ the Eur Soc Clin Microbiol Infect Dis. 2014;20(5):431–436. doi: 10.1111/1469-0691.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mellmann A., Harmsen D., Cummings C.A., Zentz E.B., Leopold S.R., Rico A. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leopold S.R., Goering R.V., Witten A., Harmsen D., Mellmann A. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol. 2014;52(7):2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kohl T.A., Diel R., Harmsen D., Rothganger J., Walter K.M., Merker M. Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J Clin Microbiol. 2014;52(7):2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bratcher H.B., Corton C., Jolley K.A., Parkhill J., Maiden M.C. A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics. 2014;15:1138. doi: 10.1186/1471-2164-15-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Been M., Pinholt M., Top J., Bletz S., Mellmann A., van Schaik W. Core Genome Multilocus Sequence Typing Scheme for High- Resolution Typing of Enterococcus faecium. J Clin Microbiol. 2015;53(12):3788–3797. doi: 10.1128/JCM.01946-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maiden M.C., Jansen van Rensburg M.J., Bray J.E., Earle S.G., Ford S.A., Jolley K.A. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol. 2013;11(10):728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Antwerpen M.H., Prior K., Mellmann A., Hoppner S., Splettstoesser W.D., Harmsen D. Rapid high resolution genotyping of Francisella tularensis by whole genome sequence comparison of annotated genes (“MLST+”) PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0123298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martinez J.L. Bacterial pathogens: from natural ecosystems to human hosts. Environ Microbiol. 2013;15(2):325–333. doi: 10.1111/j.1462-2920.2012.02837.x. [DOI] [PubMed] [Google Scholar]
- 72.Martinez J.L. Ecology and Evolution of Chromosomal Gene Transfer between Environmental Microorganisms and Pathogens. Microbiol Spectr. 2018;6(1) doi: 10.1128/microbiolspec.mtbp-0006-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Man TJB, Limbago BM. SSTAR, a Stand-Alone Easy-To-Use Antimicrobial Resistance Gene Predictor. mSphere. 2016;1(1):mSphere.00050-15, e-15. [DOI] [PMC free article] [PubMed]
- 74.Van Camp P.-J., Haslam D.B., Porollo A. Bioinformatics Approaches to the Understanding of Molecular Mechanisms in Antimicrobial Resistance. IJMS. 2020;21(4):1363. doi: 10.3390/ijms21041363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang S., McCormick T.H., Leek J.T. Methods for correcting inference based on outcomes predicted by machine learning. PNAS. 2020;117(48):30266–30275. doi: 10.1073/pnas.2001238117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ransom E.M., Potter R.F., Dantas G., Burnham C.-A.-D. Genomic Prediction of Antimicrobial Resistance: Ready or Not, Here It Comes! Clin Chem. 2020;66(10):1278–1289. doi: 10.1093/clinchem/hvaa172. [DOI] [PubMed] [Google Scholar]
- 77.Sichtig H., Minogue T., Yan Y., Stefan C., Hall A., Tallon L. FDA-ARGOS is a database with public quality-controlled reference genomes for diagnostic use and regulatory science. Nat Commun. 2019;10(1):3313. doi: 10.1038/s41467-019-11306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feldgarden M., Brover V., Haft D.H., Prasad A.B., Slotta D.J., Tolstoy I. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob Agents Chemother. 2019;63(11) doi: 10.1128/AAC.00483-19. e00483-19, /aac/63/11/AAC.-19.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bortolaia V., Kaas R.S., Ruppe E., Roberts M.C., Schwarz S., Cattoir V. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zankari E., Allesøe R., Joensen K.G., Cavaco L.M., Lund O., Aarestrup F.M. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother. 2017;72(10):2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gupta S.K., Padmanabhan B.R., Diene S.M., Lopez-Rojas R., Kempf M., Landraud L. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58(1):212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin X., Jiang X.T., Chai B., Li L., Yang Y., Cole J.R. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics. 2018;34(13):2263–2270. doi: 10.1093/bioinformatics/bty053. [DOI] [PubMed] [Google Scholar]
- 84.Inouye M., Dashnow H., Raven L.-A., Schultz M.B., Pope B.J., Tomita T. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6(11):90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rowe W., Baker K.S., Verner-Jeffreys D., Baker-Austin C., Ryan J.J., Maskell D. Search Engine for Antimicrobial Resistance: A Cloud Compatible Pipeline and Web Interface for Rapidly Detecting Antimicrobial Resistance Genes Directly from Sequence Data. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0133492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hunt M., Mather A.E., Sánchez-Busó L., Page A.J., Parkhill J., Keane J.A. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genomics. 2017;3(10) doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wattam A.R., Abraham D., Dalay O., Disz T.L., Driscoll T., Gabbard J.L. PATRIC, the bacterial bioinformatics database and analysis resource. Nucl Acids Res. 2014;42(Database issue):D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chowdhury A.S., Call D.R., Broschat S.L. PARGT: a software tool for predicting antimicrobial resistance in bacteria. Sci Rep. 2020;10(1):11033. doi: 10.1038/s41598-020-67949-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gibson M.K., Forsberg K.J., Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2015;9(1):207–216. doi: 10.1038/ismej.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu B., Pop M. ARDB–Antibiotic Resistance Genes Database. Nucl Acids Res. 2009;37(Database issue):D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tatusova T., Ciufo S., Fedorov B., O'Neill K. Tolstoy I. RefSeq microbial genomes database: new representation and annotation strategy. Nucl Acids Res. 2014;42(1):D553–D599. doi: 10.1093/nar/gkt1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mason A., Foster D., Bradley P., Golubchik T., Doumith M., Gordon N.C. Accuracy of Different Bioinformatics Methods in Detecting Antibiotic Resistance and Virulence Factors from <i>Staphylococcus aureus</i> Whole-Genome Sequences. J Clin Microbiol. 2018;56(9) doi: 10.1128/JCM.01815-17. e01815-17, /jcm/56/9/e-17.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bradley P., Gordon N.C., Walker T.M., Dunn L., Heys S., Huang B. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun. 2015;6(1):10063. doi: 10.1038/ncomms10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Davis J.J., Boisvert S., Brettin T., Kenyon R.W., Mao C., Olson R. Antimicrobial Resistance Prediction in PATRIC and RAST. Sci Rep. 2016;6(1):27930. doi: 10.1038/srep27930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bradford P.A. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14(4):933–951. doi: 10.1128/CMR.14.4.933-951.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fåhraeus R., Marin M., Olivares-Illana V. Whisper mutations: cryptic messages within the genetic code. Oncogene. 2016;35(29):3753–3759. doi: 10.1038/onc.2015.454. [DOI] [PubMed] [Google Scholar]
- 97.Supek F. The Code of Silence: Widespread Associations Between Synonymous Codon Biases and Gene Function. J Mol Evol. 2016;82(1):65–73. doi: 10.1007/s00239-015-9714-8. [DOI] [PubMed] [Google Scholar]
- 98.Tran J.H., Jacoby G.A. Mechanism of plasmid-mediated quinolone resistance. PNAS. 2002;99(8):5638–5642. doi: 10.1073/pnas.082092899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 100.Flandrois J.P., Lina G., Dumitrescu O. MUBII-TB-DB: a database of mutations associated with antibiotic resistance in Mycobacterium tuberculosis. BMC Bioinf. 2014;15:107. doi: 10.1186/1471-2105-15-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Opijnen T., Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol. 2013;11(7):435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Olivares J., Bernardini A., Garcia-Leon G., Corona F., Maria Blanca S., Martinez J.L. The intrinsic resistome of bacterial pathogens. Front Microbiol. 2013;4:103. doi: 10.3389/fmicb.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fajardo A., Martinez-Martin N., Mercadillo M., Galan J.C., Ghysels B., Matthijs S. The neglected intrinsic resistome of bacterial pathogens. PLoS One. 2008;3(2) doi: 10.1371/journal.pone.0001619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Breidenstein E.B., Khaira B.K., Wiegand I., Overhage J., Hancock R.E. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob Agents Chemother. 2008;52(12):4486–4491. doi: 10.1128/AAC.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schurek K.N., Marr A.K., Taylor P.K., Wiegand I., Semenec L., Khaira B.K. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52(12):4213–4219. doi: 10.1128/AAC.00507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alvarez-Ortega C., Wiegand I., Olivares J., Hancock R.E., Martinez J.L. The intrinsic resistome of Pseudomonas aeruginosa to beta-lactams. Virulence. 2011;2(2):144–146. doi: 10.4161/viru.2.2.15014. [DOI] [PubMed] [Google Scholar]
- 107.Fernandez L., Alvarez-Ortega C., Wiegand I., Olivares J., Kocincova D., Lam J.S. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:110–119. doi: 10.1128/AAC.01583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krahn T., Gilmour C., Tilak J., Fraud S., Kerr N., Lau C.H. Determinants of intrinsic aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2012;56(11):5591–5602. doi: 10.1128/AAC.01446-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dotsch A., Becker T., Pommerenke C., Magnowska Z., Jansch L., Haussler S. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(6):2522–2531. doi: 10.1128/AAC.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu A., Tran L., Becket E., Lee K., Chinn L., Park E. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob Agents Chemother. 2010;54(4):1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Blake K.L., O'Neill A.J. Transposon library screening for identification of genetic loci participating in intrinsic susceptibility and acquired resistance to antistaphylococcal agents. J Antimicrob Chemother. 2013;68(1):12–16. doi: 10.1093/jac/dks373. [DOI] [PubMed] [Google Scholar]
- 112.Vestergaard M., Leng B., Haaber J., Bojer M.S., Vegge C.S., Ingmer H. Genome-Wide Identification of Antimicrobial Intrinsic Resistance Determinants in Staphylococcus aureus. Front Microbiol. 2016;7:2018. doi: 10.3389/fmicb.2016.02018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bernardini A., Cuesta T., Tomas A., Bengoechea J.A., Martinez J.L., Sanchez M.B. The intrinsic resistome of Klebsiella pneumoniae. Int J Antimicrob Agents. 2019;53(1):29–33. doi: 10.1016/j.ijantimicag.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 114.Gomez M.J., Neyfakh A.A. Genes involved in intrinsic antibiotic resistance of Acinetobacter baylyi. Antimicrob Agents Chemother. 2006;50(11):3562–3567. doi: 10.1128/AAC.00579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Krawczyk-Balska A., Markiewicz Z. The intrinsic cephalosporin resistome of Listeria monocytogenes in the context of stress response, gene regulation, pathogenesis and therapeutics. J Appl Microbiol. 2016;120(2):251–265. doi: 10.1111/jam.12989. [DOI] [PubMed] [Google Scholar]
- 116.Sanz-Garcia F., Hernando-Amado S., Martinez J.L. Mutational Evolution of Pseudomonas aeruginosa Resistance to Ribosome-Targeting Antibiotics. Front Genet. 2018;9:451. doi: 10.3389/fgene.2018.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gil-Gil T., Laborda P., Sanz-García F., Hernando-Amado S., Blanco P., Martínez J.L. Antimicrobial resistance: A multifaceted problem with multipronged solutions. Microbiologyopen. 2019;8(11) doi: 10.1002/mbo3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lopez-Causape C., Rubio R., Cabot G., Oliver A. Evolution of the Pseudomonas aeruginosa aminoglycoside mutational resistome in vitro and in the cystic fibrosis setting. Antimicrob Agents Chemother. 2018 doi: 10.1128/AAC.02583-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wardell S.J.T., Rehman A., Martin L.W., Winstanley C., Patrick W.M., Lamont I.L. A large-scale whole-genome comparison shows that experimental evolution in response to antibiotics predicts changes in naturally evolved clinical Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2019 doi: 10.1128/AAC.01619-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lopez-Causape C., Sommer L.M., Cabot G., Rubio R., Ocampo-Sosa A.A., Johansen H.K. Evolution of the Pseudomonas aeruginosa mutational resistome in an international Cystic Fibrosis clone. Sci Rep. 2017;7(1):5555. doi: 10.1038/s41598-017-05621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Folkesson A., Jelsbak L., Yang L., Johansen H.K., Ciofu O., Hoiby N. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10(12):841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 122.Peng B., Li H., Peng X. Proteomics approach to understand bacterial antibiotic resistance strategies. Exp Rev Proteomics. 2019;16(10):829–839. doi: 10.1080/14789450.2019.1681978. [DOI] [PubMed] [Google Scholar]
- 123.Lin Y., Li W., Sun L., Lin Z., Jiang Y., Ling Y. Comparative metabolomics shows the metabolic profiles fluctuate in multi-drug resistant Escherichia coli strains. J Proteomics. 2019;207 doi: 10.1016/j.jprot.2019.103468. [DOI] [PubMed] [Google Scholar]
- 124.Liu S.R., Peng X.X., Li H. Metabolic mechanism of ceftazidime resistance in Vibrio alginolyticus. Infect Drug Res. 2019;12:417–429. doi: 10.2147/IDR.S179639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zampieri M., Szappanos B., Buchieri M.V., Trauner A., Piazza I., Picotti P. High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds. Sci Transl Med. 2018;10(429) doi: 10.1126/scitranslmed.aal3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang-Kan X., Rodríguez-Blanco G., Southam A.D., Winder C.L., Dunn W.B., Ivens A. Metabolomics Reveal Potential Natural Substrates of AcrB in Escherichia coli and Salmonella enterica Serovar Typhimurium. mBio. 2021;12(2) doi: 10.1128/mBio.00109-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bernardini A., Corona F., Dias R., Sanchez M.B., Martinez J.L. The inactivation of RNase G reduces the Stenotrophomonas maltophilia susceptibility to quinolones by triggering the heat shock response. Front Microbiol. 2015;6:1068. doi: 10.3389/fmicb.2015.01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gil-Gil T., Corona F., Martínez J.L., Bernardini A. The Inactivation of Enzymes Belonging to the Central Carbon Metabolism Is a Novel Mechanism of Developing Antibiotic Resistance. mSystems. 2020;5(3) doi: 10.1128/mSystems.00282-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jensen P.A., Zhu Z., van Opijnen T. Antibiotics Disrupt Coordination between Transcriptional and Phenotypic Stress Responses in Pathogenic Bacteria. Cell Rep. 2017;20(7):1705–1716. doi: 10.1016/j.celrep.2017.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sulaiman J.E., Hao C., Lam H. Specific Enrichment and Proteomics Analysis of <i>Escherichia coli</i> Persisters from Rifampin Pretreatment. J Proteome Res. 2018;17(11):3984–3996. doi: 10.1021/acs.jproteome.8b00625. [DOI] [PubMed] [Google Scholar]
- 131.Sulaiman J.E., Lam H. Proteomic Investigation of Tolerant <i>Escherichia coli</i> Populations from Cyclic Antibiotic Treatment. J Proteome Res. 2020;19(2):900–913. doi: 10.1021/acs.jproteome.9b00687. [DOI] [PubMed] [Google Scholar]
- 132.Chang K.-C., Kuo H.-Y., Tang C., Chang C.-W., Lu C.-W., Liu C.-C. Transcriptome profiling in imipenem-selected Acinetobacter baumannii. BMC Genomics. 2014;15(1):815. doi: 10.1186/1471-2164-15-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wright M.S., Suzuki Y., Jones M.B., Marshall S.H., Rudin S.D., van Duin D. Genomic and Transcriptomic Analyses of Colistin-Resistant Clinical Isolates of Klebsiella pneumoniae Reveal Multiple Pathways of Resistance. Antimicrob Agents Chemother. 2015;59(1):536–543. doi: 10.1128/AAC.04037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tsakou F., Jersie-Christensen R., Jenssen H., Mojsoska B. The Role of Proteomics in Bacterial Response to Antibiotics. Pharmaceuticals. 2020;13(9):214. doi: 10.3390/ph13090214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liang Y., Deng F., Mu P., Wen J., Deng Y. Quantitative proteomics implicates YggT in streptomycin resistance in Salmonella enterica serovar Enteritidis. Biotechnol Lett. 2021 doi: 10.1007/s10529-021-03083-4. [DOI] [PubMed] [Google Scholar]
- 136.Khaledi A., Schniederjans M., Pohl S., Rainer R., Bodenhofer U., Xia B. Transcriptome Profiling of Antimicrobial Resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60(8):4722–4733. doi: 10.1128/AAC.00075-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Molina-Mora J.A., Chinchilla-Montero D., Chavarría-Azofeifa M., Ulloa-Morales A.J., Campos-Sánchez R., Mora-Rodríguez R. Transcriptomic determinants of the response of ST-111 Pseudomonas aeruginosa AG1 to ciprofloxacin identified by a top-down systems biology approach. Sci Rep. 2020;10(1):13717. doi: 10.1038/s41598-020-70581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fajardo A., Martinez J.L. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 2008;11(2):161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 139.Linares J.F., Gustafsson I., Baquero F., Martinez J.L. Antibiotics as intermicrobial signaling agents instead of weapons. PNAS. 2006;103(51):19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Davies J., Spiegelman G.B., Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9(5):445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 141.Davies J. Are antibiotics naturally antibiotics? J Ind Microbiol Biotechnol. 2006;33(7):496–499. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 142.Martinsen M.A., Jaramillo Cartagena A., Bhattacharyya R.P. Core Antibiotic-Induced Transcriptional Signatures Reflect Susceptibility to All Members of an Antibiotic Class. Antimicrob Agents Chemother. 2021 doi: 10.1128/AAC.02296-20. AAC.02296-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bertelli C., Greub G. Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clinical Microbiol Infect : Off Publ Eur Soc Clin Microbiol Infect Dis. 2013;19(9):803–813. doi: 10.1111/1469-0691.12217. [DOI] [PubMed] [Google Scholar]
- 144.Antipov D., Hartwick N., Shen M., Raiko M., Lapidus A., Pevzner P.A. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 145.Lanza V.F., de Toro M., Garcillan-Barcia M.P., Mora A., Blanco J., Coque T.M. Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences. PLoS Genet. 2014;10(12) doi: 10.1371/journal.pgen.1004766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lanza V.F., Baquero F., de la Cruz F., Coque T.M. AcCNET (Accessory Genome Constellation Network): comparative genomics software for accessory genome analysis using bipartite networks. Bioinformatics. 2017;33(2):283–285. doi: 10.1093/bioinformatics/btw601. [DOI] [PubMed] [Google Scholar]
- 147.Fernández-de-Bobadilla MD, Talavera-Rodríguez A, Chacón L, Baquero F, Coque TM, Lanza VF. PATO: Pangenome Analysis Toolkit. bioRxiv. 2021:2021.01.30.428878. [DOI] [PubMed]
- 148.Chekabab S.M., Lawrence J.R., Alvarado A., Predicala B., Korber D.R. A health metadata-based management approach for comparative analysis of high-throughput genetic sequences for quantifying antimicrobial resistance reduction in Canadian hog barns. Comput Struct Biotechnol J. 2020;18:2629–2638. doi: 10.1016/j.csbj.2020.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Singh I., Kuscuoglu M., Harkins D.M., Sutton G., Fouts D.E., Nelson K.E. OMeta: an ontology-based, data-driven metadata tracking system. BMC Bioinf. 2019;20(1):8. doi: 10.1186/s12859-018-2580-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Břinda K., Callendrello A., Ma K.C., MacFadden D.R., Charalampous T., Lee R.S. Rapid inference of antibiotic resistance and susceptibility by genomic neighbour typing. Nat Microbiol. 2020;5(3):455–464. doi: 10.1038/s41564-019-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lees J.A., Mai T.T., Galardini M., Wheeler N.E., Horsfield S.T., Parkhill J. Improved Prediction of Bacterial Genotype-Phenotype Associations Using Interpretable Pangenome-Spanning Regressions. mBio. 2020;11(4) doi: 10.1128/mBio.01344-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Khaledi A., Weimann A., Schniederjans M., Asgari E., Kuo T.H., Oliver A. Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics. EMBO Mol Med. 2020;12(3) doi: 10.15252/emmm.201910264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Moreno-Molina M., Comas I., Furió V. The Future of TB Resistance Diagnosis: The Essentials on Whole Genome Sequencing and Rapid Testing Methods. Arch Bronconeumol. 2019;55(8):421–426. doi: 10.1016/j.arbres.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 154.Satta G., Lipman M., Smith G.P., Arnold C., Kon O.M., McHugh T.D. Mycobacterium tuberculosis and whole-genome sequencing: how close are we to unleashing its full potential? Clin Microbiol Infect : Off Publ the Eur Soc Clin Microbiol Infect Dis. 2018;24(6):604–609. doi: 10.1016/j.cmi.2017.10.030. [DOI] [PubMed] [Google Scholar]
- 155.Zhang D., Gomez J.E., Chien J.Y., Haseley N., Desjardins C.A., Earl A.M. Genomic Analysis of the Evolution of Fluoroquinolone Resistance in Mycobacterium tuberculosis Prior to Tuberculosis Diagnosis. Antimicrob Agents Chemother. 2016;60(11):6600–6608. doi: 10.1128/AAC.00664-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Knopp M., Andersson D.I. Predictable Phenotypes of Antibiotic Resistance Mutations. MBio. 2018;9(3) doi: 10.1128/mBio.00770-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Muzondiwa D., Mutshembele A., Pierneef R.E., Reva O.N. Resistance Sniffer: An online tool for prediction of drug resistance patterns of Mycobacterium tuberculosis isolates using next generation sequencing data. Int J Med Microbiol. 2020;310(2) doi: 10.1016/j.ijmm.2020.151399. [DOI] [PubMed] [Google Scholar]
- 158.Noone J.C., Helmersen K., Leegaard T.M., Skråmm I., Aamot H.V. Rapid Diagnostics of Orthopaedic-Implant-Associated Infections Using Nanopore Shotgun Metagenomic Sequencing on Tissue Biopsies. Microorganisms. 2021;9(1) doi: 10.3390/microorganisms9010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Soundararajan L., Kambli P., Priyadarshini S., Let B., Murugan S., Iravatham C. Whole genome enrichment approach for rapid detection of Mycobacterium tuberculosis and drug resistance-associated mutations from direct sputum sequencing. Tuberculosis (Edinburgh, Scotland) 2020;121 doi: 10.1016/j.tube.2020.101915. [DOI] [PubMed] [Google Scholar]
- 160.Skippington E., Ragan M.A. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol Rev. 2011;35(5):707–735. doi: 10.1111/j.1574-6976.2010.00261.x. [DOI] [PubMed] [Google Scholar]
- 161.Pärnänen K.M.M., Narciso-da-Rocha C., Kneis D., Berendonk T.U., Cacace D., Do T.T. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Science. Advances. 2019;5(3):eaau9124. doi: 10.1126/sciadv.aau9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Durso L.M., Cook K.L. One Health and Antibiotic Resistance in Agroecosystems. EcoHealth. 2018 doi: 10.1007/s10393-018-1324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jean B.P. One Health and the Politics of Antimicrobial Resistance. Emerging Infect Dis J. 2017;23(4):724. [Google Scholar]
- 164.Segawa T., Takeuchi N., Rivera A., Yamada A., Yoshimura Y., Barcaza G. Distribution of antibiotic resistance genes in glacier environments. Environ Microbiol Rep. 2013;5(1):127–134. doi: 10.1111/1758-2229.12011. [DOI] [PubMed] [Google Scholar]
- 165.Martinez J.L., Fajardo A., Garmendia L., Hernandez A., Linares J.F., Martinez-Solano L. A global view of antibiotic resistance. FEMS Microbiol Rev. 2009;33(1):44–65. doi: 10.1111/j.1574-6976.2008.00142.x. [DOI] [PubMed] [Google Scholar]
- 166.Baquero F., Alvarez-Ortega C., Martinez J.L. Ecology and evolution of antibiotic resistance. Environ Microbiol Rep. 2009;1:469–476. doi: 10.1111/j.1758-2229.2009.00053.x. [DOI] [PubMed] [Google Scholar]
- 167.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17(3):377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Martinez J.L. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321(5887):365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 169.Davies J.E. Origins, acquisition and dissemination of antibiotic resistance determinants. Ciba Found Symp. 1997;207:15–27. [PubMed] [Google Scholar]
- 170.Martinez J.L. Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Front Microbiol. 2012;3 doi: 10.3389/fmicb.2011.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hernando-Amado S., Coque T.M., Baquero F., Martínez J.L. Antibiotic Resistance: Moving From Individual Health Norms to Social Norms in One Health and Global Health. Front Microbiol. 2020;11:1914. doi: 10.3389/fmicb.2020.01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Levin B.R., Udekwu K.I. Population dynamics of antibiotic treatment: a mathematical model and hypotheses for time-kill and continuous-culture experiments. Antimicrob Agents Chemother. 2010;54(8):3414–3426. doi: 10.1128/AAC.00381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Olofsson S.K., Geli P., Andersson D.I., Cars O. Pharmacodynamic model to describe the concentration-dependent selection of cefotaxime-resistant Escherichia coli. Antimicrob Agents Chemother. 2005;49(12):5081–5091. doi: 10.1128/AAC.49.12.5081-5091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Levin B.R., Lipsitch M., Perrot V., Schrag S., Antia R., Simonsen L. The population genetics of antibiotic resistance. Clin Infect Dis. 1997;24(Suppl 1):S9–S16. doi: 10.1093/clinids/24.supplement_1.s9. [DOI] [PubMed] [Google Scholar]
- 175.Blanquart F. Evolutionary epidemiology models to predict the dynamics of antibiotic resistance. Evol Appl. 2019;12(3):365–383. doi: 10.1111/eva.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Okeke I.N., Edelman R. Dissemination of antibiotic-resistant bacteria across geographic borders. Clin Infect Dis. 2001;33(3):364–369. doi: 10.1086/321877. [DOI] [PubMed] [Google Scholar]
- 177.Pehrsson E.C., Tsukayama P., Patel S., Mejia-Bautista M., Sosa-Soto G., Navarrete K.M. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533(7602):212–216. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Forsberg K.J., Reyes A., Wang B., Selleck E.M., Sommer M.O., Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337(6098):1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gupta S., Arango-Argoty G., Zhang L., Pruden A., Vikesland P. Identification of discriminatory antibiotic resistance genes among environmental resistomes using extremely randomized tree algorithm. Microbiome. 2019;7(1):123. doi: 10.1186/s40168-019-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Aarestrup F.M. Veterinary drug usage and antimicrobial resistance in bacteria of animal origin. Basic Clin Pharmacol Toxicol. 2005;96(4):271–281. doi: 10.1111/j.1742-7843.2005.pto960401.x. [DOI] [PubMed] [Google Scholar]
- 181.Aarestrup F.M., Seyfarth A.M., Emborg H.D., Pedersen K., Hendriksen R.S., Bager F. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob Agents Chemother. 2001;45(7):2054–2059. doi: 10.1128/AAC.45.7.2054-2059.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Aarestrup F.M., Bager F., Jensen N.E., Madsen M., Meyling A., Wegener H.C. Surveillance of antimicrobial resistance in bacteria isolated from food animals to antimicrobial growth promoters and related therapeutic agents in Denmark. Apmis. 1998;106(6):606–622. doi: 10.1111/j.1699-0463.1998.tb01391.x. [DOI] [PubMed] [Google Scholar]
- 183.Sun J., Liao X.P., D'Souza A.W., Boolchandani M., Li S.H., Cheng K. Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nat Commun. 2020;11(1):1427. doi: 10.1038/s41467-020-15222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mather A.E., Reid S.W., Maskell D.J., Parkhill J., Fookes M.C., Harris S.R. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science. 2013;341(6153):1514–1517. doi: 10.1126/science.1240578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hu Y., Yang X., Li J., Lv N., Liu F., Wu J. The Bacterial Mobile Resistome Transfer Network Connecting the Animal and Human Microbiomes. Appl Environ Microbiol. 2016;82(22):6672–6681. doi: 10.1128/AEM.01802-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.de Been M., Lanza V.F., de Toro M., Scharringa J., Dohmen W., Du Y. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 2014;10(12) doi: 10.1371/journal.pgen.1004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tamames J., Puente-Sanchez F. SqueezeMeta, A Highly Portable, Fully Automatic Metagenomic Analysis Pipeline. Front Microbiol. 2019;9:3349. doi: 10.3389/fmicb.2018.03349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Feldgarden M., Brover V., Haft D.H., Prasad A.B., Slotta D.J., Tolstoy I. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob Agents Chemother. 2019;63(11) doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lakin S.M., Dean C., Noyes N.R., Dettenwanger A., Ross A.S., Doster E. MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. 2017;45(D1):D574–D580. doi: 10.1093/nar/gkw1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Eddy S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23(1):205–211. [PubMed] [Google Scholar]
- 191.Boulund F., Johnning A., Pereira M.B., Larsson D.G., Kristiansson E. A novel method to discover fluoroquinolone antibiotic resistance (qnr) genes in fragmented nucleotide sequences. BMC Genomics. 2012;13:695. doi: 10.1186/1471-2164-13-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Allen H.K., Cloud-Hansen K.A., Wolinski J.M., Guan C., Greene S., Lu S. Resident microbiota of the gypsy moth midgut harbors antibiotic resistance determinants. DNA Cell Biol. 2009 doi: 10.1089/dna.2008.0812. [DOI] [PubMed] [Google Scholar]
- 193.D'Costa V.M., McGrann K.M., Hughes D.W., Wright G.D. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 194.Fitzpatrick D., Walsh F. Antibiotic resistance genes across a wide variety of metagenomes. FEMS Microbiol Ecol. 2016;92(2) doi: 10.1093/femsec/fiv168. [DOI] [PubMed] [Google Scholar]
- 195.Mahnert A., Moissl-Eichinger C., Zojer M., Bogumil D., Mizrahi I., Rattei T. Man-made microbial resistances in built environments. Nat Commun. 2019;10(1):968. doi: 10.1038/s41467-019-08864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Clemente J., Pehrsson E., Blaser M., Sandhu K., Gao Z., Wang B. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1(3) doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bhullar K., Waglechner N., Pawlowski A., Koteva K., Banks E.D., Johnston M.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.D'Costa V.M., King C.E., Kalan L., Morar M., Sung W.W., Schwarz C. Antibiotic resistance is ancient. Nature. 2011;477(7365):457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 199.Allen H.K., Moe L.A., Rodbumrer J., Gaarder A., Handelsman J. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 2009;3(2):243–251. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 200.Manaia C.M. Assessing the Risk of Antibiotic Resistance Transmission from the Environment to Humans: Non-Direct Proportionality between Abundance and Risk. Trends Microbiol. 2017;25(3):173–181. doi: 10.1016/j.tim.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 201.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264(5157):375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 202.Gibson M.K., Forsberg K.J., Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2014 doi: 10.1038/ismej.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Forsberg K.J., Patel S., Gibson M.K., Lauber C.L., Knight R., Fierer N. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509(7502):612–616. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lira F., Vaz-Moreira I., Tamames J., Manaia C.M., Martínez J.L. Metagenomic analysis of an urban resistome before and after wastewater treatment. Sci Rep. 2020;10(1):8174. doi: 10.1038/s41598-020-65031-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cox G., Wright G.D. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol. 2013;303(6–7):287–292. doi: 10.1016/j.ijmm.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 206.Martinez J.L. The antibiotic resistome: challenge and opportunity for therapeutic intervention. Fut Med Chem. 2012;4:347–359. doi: 10.4155/fmc.12.2. [DOI] [PubMed] [Google Scholar]
- 207.Moura A., Soares M., Pereira C., Leitao N., Henriques I., Correia A. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009;25(8):1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 208.Leplae R., Lima-Mendez G., Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements, update 2010. Nucl Acids Res. 2010;38(Database issue):D57–D61. doi: 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lai S., Jia L., Subramanian B., Pan S., Zhang J., Dong Y. mMGE: a database for human metagenomic extrachromosomal mobile genetic elements. Nucleic Acids Res. 2021;49(D1):D783–D791. doi: 10.1093/nar/gkaa869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Garmendia L., Hernandez A., Sanchez M.B., Martinez J.L. Metagenomics and antibiotics. Clin Microbiol Infect : Off Publ the Eur Soc Clin Microbiol Infect Dis. 2012;18(Suppl 4):27–31. doi: 10.1111/j.1469-0691.2012.03868.x. [DOI] [PubMed] [Google Scholar]
- 211.Muniesa M., Colomer-Lluch M., Jofre J. Potential impact of environmental bacteriophages in spreading antibiotic resistance genes. Fut Microbiol. 2013;8(6):739–751. doi: 10.2217/fmb.13.32. [DOI] [PubMed] [Google Scholar]
- 212.Hadjadj L., Baron S.A., Diene S.M., Rolain J.M. How to discover new antibiotic resistance genes? Exp Rev Mol Diagn. 2019;19(4):349–362. doi: 10.1080/14737159.2019.1592678. [DOI] [PubMed] [Google Scholar]
- 213.Ruppé E., Ghozlane A., Tap J., Pons N., Alvarez A.-S., Maziers N. Prediction of the intestinal resistome by a three-dimensional structure-based method. Nat Microbiol. 2019;4(1):112–123. doi: 10.1038/s41564-018-0292-6. [DOI] [PubMed] [Google Scholar]
- 214.Ma D., Cook D.N., Alberti M., Pon N.G., Nikaido H., Hearst J.E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16(1):45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 215.Hachler H., Cohen S.P., Levy S.B. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1991;173(17):5532–5538. doi: 10.1128/jb.173.17.5532-5538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shen L.L., Kohlbrenner W.E., Weigl D., Baranowski J. Mechanism of quinolone inhibition of DNA gyrase. Appearance of unique norfloxacin binding sites in enzyme-DNA complexes. J Biol Chem. 1989;264(5):2973–2978. [PubMed] [Google Scholar]
- 217.Yoshida H., Bogaki M., Nakamura M., Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34(6):1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Li Y., Xu Z., Han W., Cao H., Umarov R., Yan A. HMD-ARG: hierarchical multi-task deep learning for annotating antibiotic resistance genes. Microbiome. 2021;9(1):40. doi: 10.1186/s40168-021-01002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Frank J.A., Pan Y., Tooming-Klunderud A., Eijsink V.G., McHardy A.C., Nederbragt A.J. Improved metagenome assemblies and taxonomic binning using long-read circular consensus sequence data. Sci Rep. 2016;6:25373. doi: 10.1038/srep25373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rhoads A., Au K.F. PacBio Sequencing and Its Applications. Genom Proteom Bioinf. 2015;13(5):278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Deamer D., Akeson M., Branton D. Three decades of nanopore sequencing. Nat Biotechnol. 2016;34(5):518–524. doi: 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.de Koning W., Miladi M., Hiltemann S., Heikema A., Hays J.P., Flemming S. NanoGalaxy: Nanopore long-read sequencing data analysis in Galaxy. GigaScience. 2020;9(10) doi: 10.1093/gigascience/giaa105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Martin C., Stebbins B., Ajmani A., Comendul A., Hamner S., Hasan N.A. Nanopore-based metagenomics analysis reveals prevalence of mobile antibiotic and heavy metal resistome in wastewater. Ecotoxicology (London, England) 2021 doi: 10.1007/s10646-020-02342-w. [DOI] [PubMed] [Google Scholar]
- 224.Urban L., Holzer A., Baronas J.J., Hall M.B., Braeuninger-Weimer P., Scherm M.J. Freshwater monitoring by nanopore sequencing. eLife. 2021:10. doi: 10.7554/eLife.61504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ma L., Li B., Jiang X.T., Wang Y.L., Xia Y., Li A.D. Catalogue of antibiotic resistome and host-tracking in drinking water deciphered by a large scale survey. Microbiome. 2017;5(1):154. doi: 10.1186/s40168-017-0369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Narciso-da-Rocha C., Rocha J., Vaz-Moreira I., Lira F., Tamames J., Henriques I. Bacterial lineages putatively associated with the dissemination of antibiotic resistance genes in a full-scale urban wastewater treatment plant. Environ Int. 2018;118:179–188. doi: 10.1016/j.envint.2018.05.040. [DOI] [PubMed] [Google Scholar]
- 227.Nielsen H.B., Almeida M., Juncker A.S., Rasmussen S., Li J., Sunagawa S. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32(8):822–828. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- 228.Imelfort M., Parks D., Woodcroft B.J., Dennis P., Hugenholtz P., Tyson G.W. GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ. 2014;2 doi: 10.7717/peerj.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Alneberg J., Bjarnason B.S., de Bruijn I., Schirmer M., Quick J., Ijaz U.Z. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11(11):1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 230.Kelley D.R., Salzberg S.L. Clustering metagenomic sequences with interpolated Markov models. BMC Bioinf. 2010;11:544. doi: 10.1186/1471-2105-11-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Strous M., Kraft B., Bisdorf R., Tegetmeyer H.E. The binning of metagenomic contigs for microbial physiology of mixed cultures. Front Microbiol. 2012;3:410. doi: 10.3389/fmicb.2012.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kang D.D., Froula J., Egan R., Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3 doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wu Y.W., Simmons B.A., Singer S.W. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32(4):605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 234.Tully B.J., Graham E.D., Heidelberg J.F. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci Data. 2018;5 doi: 10.1038/sdata.2017.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Plaza Oñate F., Le Chatelier E., Almeida M., Cervino A.C.L., Gauthier F., Magoulès F. MSPminer: abundance-based reconstitution of microbial pan-genomes from shotgun metagenomic data. Bioinformatics. 2019;35(9):1544–1552. doi: 10.1093/bioinformatics/bty830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Maguire F., Jia B., Gray K.L., Lau W.Y.V., Beiko R.G., Brinkman F.S.L. Metagenome-assembled genome binning methods with short reads disproportionately fail for plasmids and genomic Islands. Microb Genomics. 2020;6(10) doi: 10.1099/mgen.0.000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Spencer S.J., Tamminen M.V., Preheim S.P., Guo M.T., Briggs A.W., Brito I.L. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 2016;10(2):427–436. doi: 10.1038/ismej.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Beitel C.W., Froenicke L., Lang J.M., Korf I.F., Michelmore R.W., Eisen J.A. Strain- and plasmid-level deconvolution of a synthetic metagenome by sequencing proximity ligation products. PeerJ. 2014;2 doi: 10.7717/peerj.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lanza V.F., Baquero F., Martinez J.L., Ramos-Ruiz R., Gonzalez-Zorn B., Andremont A. In-depth resistome analysis by targeted metagenomics. Microbiome. 2018;6(1):11. doi: 10.1186/s40168-017-0387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Demetrius L. Thermodynamics and evolution. J Theor Biol. 2000;206(1):1–16. doi: 10.1006/jtbi.2000.2106. [DOI] [PubMed] [Google Scholar]
- 241.Giuditta A. Natural computing and biological evolution: a new paradigm. Riv Biol. 2008;101(1):119–128. [PubMed] [Google Scholar]
- 242.Gardner T.S., Hawkins K. Synthetic biology: evolution or revolution? A co-founder's perspective. Curr Opin Chem Biol. 2013;17(6):871–877. doi: 10.1016/j.cbpa.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 243.Baquero F., Levin B.R. Proximate and ultimate causes of the bactericidal action of antibiotics. Nat Rev Microbiol. 2020;1–10 doi: 10.1038/s41579-020-00443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Okasha S. Oxford University Press; Oxford: 2006. Evolution and the levels of selection. [Google Scholar]
- 245.Baquero F. Genetic hyper-codes and multidimensional Darwinism: replication modes and codes in evolutionary individuals of the bacterial world. In: Trueba G., editor. Why does Evolution Matter? The Importance of Understanding Evolution. Newcastle upon Tyne. Cambridge Scholar Publishing; United Kingdom: 2014. pp. 165–180. [Google Scholar]
- 246.Jiménez M.J.P., Jiménez Á.R., Caparrini F.S. Complexity classes in models of cellular computing with membranes. Nat Comput. 2003;2(3):265–285. [Google Scholar]
- 247.Păun G. Computing with Membranes. J Comput Syst Sci. 2000;61(1):108–143. [Google Scholar]
- 248.Martinez J.L., Baquero F. Emergence and spread of antibiotic resistance: setting a parameter space. Ups J Med Sci. 2014;119(2):68–77. doi: 10.3109/03009734.2014.901444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Baquero F., Campos M., Llorens C., Sempere J.M. A Model of Antibiotic Resistance Evolution Dynamics Through P Systems with Active Membranes and Communication Rules. In: Graciani C., Riscos-Núñez A., Păun G., Rozenberg G., Salomaa A., editors. Enjoying Natural Computing. Lecture Notes in Computer Science. Springer, Cham; Switzerland: 2018. pp. 33–44. [Google Scholar]
- 250.Campos M., Llorens C., Sempere J.M., Futami R., Rodriguez I., Carrasco P. A membrane computing simulator of trans-hierarchical antibiotic resistance evolution dynamics in nested ecological compartments (ARES) Biol Direct. 2015;10:41. doi: 10.1186/s13062-015-0070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Campos M., San Millán Á., Sempere J.M., Lanza V.F., Coque T.M., Llorens C. Simulating the Influence of Conjugative-Plasmid Kinetic Values on the Multilevel Dynamics of Antimicrobial Resistance in a Membrane Computing Model. Antimicrob Agents Chemother. 2020;64(8):e00593–e620. doi: 10.1128/AAC.00593-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Campos M., Capilla R., Naya F., Futami R., Coque T., Moya A. Simulating Multilevel Dynamics of Antimicrobial Resistance in a Membrane Computing Model. MBio. 2019;10(1) doi: 10.1128/mBio.02460-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Olivares J., Alvarez-Ortega C., Linares J.F., Rojo F., Köhler T., Martínez J.L. Overproduction of the multidrug efflux pump MexEF-OprN does not impair Pseudomonas aeruginosa fitness in competition tests, but produces specific changes in bacterial regulatory networks. Environ Microbiol. 2012;14:1968–1981. doi: 10.1111/j.1462-2920.2012.02727.x. [DOI] [PubMed] [Google Scholar]
- 254.Olivares J., Álvarez-Ortega C., Martínez J.L. Metabolic compensation of fitness costs associated with overexpression of the multidrug efflux pump MexEF-OprN in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58:3904–3913. doi: 10.1128/AAC.00121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Alcalde-Rico M., Olivares-Pacheco J., Alvarez-Ortega C., Camara M., Martinez J.L. Role of the Multidrug Resistance Efflux Pump MexCD-OprJ in the Pseudomonas aeruginosa Quorum Sensing Response. Front Microbiol. 2018;9:2752. doi: 10.3389/fmicb.2018.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Alcalde-Rico M, Olivares-Pacheco J, Halliday N, Cámara M, Martínez JL. The analysis of the role of MexAB-OprM on quorum sensing homeostasis shows that the apparent redundancy of Pseudomonas <em>aeruginosa</em> multidrug efflux pumps allows keeping the robustness and the plasticity of this intercellular signaling network. bioRxiv. 2020:2020.03.10.986737.
- 257.Freihofer P., Akbergenov R., Teo Y., Juskeviciene R., Andersson D.I., Bottger E.C. Nonmutational compensation of the fitness cost of antibiotic resistance in mycobacteria by overexpression of tlyA rRNA methylase. RNA. 2016;22(12):1836–1843. doi: 10.1261/rna.057257.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Baquero F., Martinez J.L. Interventions on Metabolism: Making Antibiotic-Susceptible Bacteria. mBio. 2017;8(6) doi: 10.1128/mBio.01950-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Banerjee D., Raghunathan A. Constraints-based analysis identifies NAD+ recycling through metabolic reprogramming in antibiotic resistant Chromobacterium violaceum. PLoS One. 2019;14(1) doi: 10.1371/journal.pone.0210008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhu Y., Czauderna T., Zhao J., Klapperstueck M., Maifiah M.H.M., Han M.L. Genome-scale metabolic modeling of responses to polymyxins in Pseudomonas aeruginosa. GigaScience. 2018;7(4) doi: 10.1093/gigascience/giy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.O'Brien E.J., Monk J.M., Palsson B.O. Using Genome-scale Models to Predict Biological Capabilities. Cell. 2015;161(5):971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Bosi E., Monk J.M., Aziz R.K., Fondi M., Nizet V., Palsson B.O. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. PNAS. 2016;113(26):E3801–E3809. doi: 10.1073/pnas.1523199113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Kim H.U., Kim S.Y., Jeong H., Kim T.Y., Kim J.J., Choy H.E. Integrative genome-scale metabolic analysis of Vibrio vulnificus for drug targeting and discovery. Mol Syst Biol. 2011;7:460. doi: 10.1038/msb.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Krueger A.S., Munck C., Dantas G., Church G.M., Galagan J., Lehar J. Simulating Serial-Target Antibacterial Drug Synergies Using Flux Balance Analysis. PLoS One. 2016;11(1) doi: 10.1371/journal.pone.0147651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Aziz R.K., Monk J.M., Lewis R.M., In Loh S., Mishra A., Abhay Nagle A. Systems biology-guided identification of synthetic lethal gene pairs and its potential use to discover antibiotic combinations. Sci Rep. 2015;5:16025. doi: 10.1038/srep16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Bordbar A., Monk J.M., King Z.A., Palsson B.O. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15(2):107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- 267.Hussein M., Han M.L., Zhu Y., Zhou Q., Lin Y.W., Hancock R.E.W. Metabolomics Study of the Synergistic Killing of Polymyxin B in Combination with Amikacin against Polymyxin-Susceptible and -Resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2019;64(1) doi: 10.1128/AAC.01587-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Thiele I., Palsson B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5(1):93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Norsigian C.J., Fang X., Seif Y., Monk J.M., Palsson B.O. A workflow for generating multi-strain genome-scale metabolic models of prokaryotes. Nat Protoc. 2020;15(1):1–14. doi: 10.1038/s41596-019-0254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Agren R., Liu L., Shoaie S., Vongsangnak W., Nookaew I., Nielsen J. The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLoS Comput Biol. 2013;9(3) doi: 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Dias O., Rocha M., Ferreira E.C., Rocha I. Reconstructing High-Quality Large-Scale Metabolic Models with merlin. Methods Mol Biol. 2018;1716:1–36. doi: 10.1007/978-1-4939-7528-0_1. [DOI] [PubMed] [Google Scholar]
- 272.Henry C.S., DeJongh M., Best A.A., Frybarger P.M., Linsay B., Stevens R.L. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28(9):977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 273.Karp P.D., Latendresse M., Paley S.M., Krummenacker M., Ong Q.D., Billington R. Pathway Tools version 19.0 update: software for pathway/genome informatics and systems biology. Brief Bioinform. 2016;17(5):877–890. doi: 10.1093/bib/bbv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Pitkanen E., Jouhten P., Hou J., Syed M.F., Blomberg P., Kludas J. Comparative genome-scale reconstruction of gapless metabolic networks for present and ancestral species. PLoS Comput Biol. 2014;10(2) doi: 10.1371/journal.pcbi.1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Machado D., Andrejev S., Tramontano M., Patil K.R. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018;46(15):7542–7553. doi: 10.1093/nar/gky537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Merigueti T.C., Carneiro M.W., Carvalho-Assef A.P.D., Silva-Jr F.P., da Silva F.A.B. FindTargetsWEB: A User-Friendly Tool for Identification of Potential Therapeutic Targets in Metabolic Networks of Bacteria. Front Genet. 2019;10:633. doi: 10.3389/fgene.2019.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Biggs M.B., Papin J.A. Novel multiscale modeling tool applied to Pseudomonas aeruginosa biofilm formation. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0078011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Vital-Lopez F.G., Reifman J., Wallqvist A. Biofilm Formation Mechanisms of Pseudomonas aeruginosa Predicted via Genome-Scale Kinetic Models of Bacterial Metabolism. PLoS Comput Biol. 2015;11(10) doi: 10.1371/journal.pcbi.1004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Bartell J.A., Blazier A.S., Yen P., Thogersen J.C., Jelsbak L., Goldberg J.B. Reconstruction of the metabolic network of Pseudomonas aeruginosa to interrogate virulence factor synthesis. Nat Commun. 2017;8:14631. doi: 10.1038/ncomms14631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Oberhardt M.A., Goldberg J.B., Hogardt M., Papin J.A. Metabolic network analysis of Pseudomonas aeruginosa during chronic cystic fibrosis lung infection. J Bacteriol. 2010;192(20):5534–5548. doi: 10.1128/JB.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Moyne O., Castelli F., Bicout D.J., Boccard J., Camara B., Cournoyer B. Metabotypes of Pseudomonas aeruginosa Correlate with Antibiotic Resistance. Virulence and Clinical Outcome in Cystic Fibrosis Chronic Infections. Metabolites. 2021;11(2) doi: 10.3390/metabo11020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Rêgo A.M., Alves da Silva D., Ferreira N.V., de Pina L.C., Evaristo J.A.M., Caprini Evaristo G.P. Metabolic profiles of multidrug resistant and extensively drug resistant Mycobacterium tuberculosis unveiled by metabolomics. Tuberculosis (Edinburgh, Scotland) 2020;126:102043. doi: 10.1016/j.tube.2020.102043. [DOI] [PubMed] [Google Scholar]
- 283.Vestergaard M., Nohr-Meldgaard K., Bojer M.S., Krogsgard Nielsen C., Meyer R.L., Slavetinsky C. Inhibition of the ATP Synthase Eliminates the Intrinsic Resistance of Staphylococcus aureus towards Polymyxins. mBio. 2017;8(5) doi: 10.1128/mBio.01114-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Jiang M., Kuang S.F., Lai S.S., Zhang S., Yang J., Peng B. Na(+)-NQR Confers Aminoglycoside Resistance via the Regulation of l-Alanine Metabolism. mBio. 2020;11(6) doi: 10.1128/mBio.02086-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Su Y.B., Kuang S.F., Peng X.X., Li H. The depressed P cycle contributes to the acquisition of ampicillin resistance in Edwardsiella piscicida. J Proteomics. 2020;212 doi: 10.1016/j.jprot.2019.103562. [DOI] [PubMed] [Google Scholar]
- 286.Gardner S.G., Marshall D.D., Daum R.S., Powers R., Somerville G.A. Metabolic Mitigation of Staphylococcus aureus Vancomycin Intermediate-Level Susceptibility. Antimicrob Agents Chemother. 2018;62(1) doi: 10.1128/AAC.01608-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Zampieri M., Zimmermann M., Claassen M., Sauer U. Nontargeted Metabolomics Reveals the Multilevel Response to Antibiotic Perturbations. Cell Rep. 2017;19(6):1214–1228. doi: 10.1016/j.celrep.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 288.Ye J.Z., Lin X.M., Cheng Z.X., Su Y.B., Li W.X., Ali F.M. Identification and efficacy of glycine, serine and threonine metabolism in potentiating kanamycin-mediated killing of Edwardsiella piscicida. J Proteomics. 2018;183:34–44. doi: 10.1016/j.jprot.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 289.Kim T.Y., Kim H.U., Lee S.Y. Metabolite-centric approaches for the discovery of antibacterials using genome-scale metabolic networks. Metab Eng. 2010;12(2):105–111. doi: 10.1016/j.ymben.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 290.Kauffman K.J., Prakash P., Edwards J.S. Advances in flux balance analysis. Curr Opin Biotechnol. 2003;14(5):491–496. doi: 10.1016/j.copbio.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 291.Mazumdar V., Snitkin E.S., Amar S., Segre D. Metabolic network model of a human oral pathogen. J Bacteriol. 2009;191(1):74–90. doi: 10.1128/JB.01123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Mahadevan R., Schilling C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng. 2003;5(4):264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 293.Price N.D., Schellenberger J., Palsson B.O. Uniform sampling of steady-state flux spaces: means to design experiments and to interpret enzymopathies. Biophys J. 2004;87(4):2172–2186. doi: 10.1529/biophysj.104.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Charusanti P., Chauhan S., McAteer K., Lerman J.A., Hyduke D.R., Motin V.L. An experimentally-supported genome-scale metabolic network reconstruction for Yersinia pestis CO92. BMC Syst Biol. 2011;5:163. doi: 10.1186/1752-0509-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Hartman H.B., Fell D.A., Rossell S., Jensen P.R., Woodward M.J., Thorndahl L. Identification of potential drug targets in Salmonella enterica sv. Typhimurium using metabolic modelling and experimental validation. Microbiology (Reading) 2014;160(Pt 6):1252–1266. doi: 10.1099/mic.0.076091-0. [DOI] [PubMed] [Google Scholar]
- 296.Chavali A.K., D'Auria K.M., Hewlett E.L., Pearson R.D., Papin J.A. A metabolic network approach for the identification and prioritization of antimicrobial drug targets. Trends Microbiol. 2012;20(3):113–123. doi: 10.1016/j.tim.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Campos A.I., Zampieri M. Metabolomics-Driven Exploration of the Chemical Drug Space to Predict Combination Antimicrobial Therapies. Molecular cell. 2019;74(6) doi: 10.1016/j.molcel.2019.04.001. 1291-303.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Papin J.A., Reed J.L., Palsson B.O. Hierarchical thinking in network biology: the unbiased modularization of biochemical networks. Trends Biochem Sci. 2004;29(12):641–647. doi: 10.1016/j.tibs.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 299.Jamshidi N., Palsson B.O. Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst Biol. 2007;1:26. doi: 10.1186/1752-0509-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.McConville M.J., de Souza D., Saunders E., Likic V.A., Naderer T. Living in a phagolysosome; metabolism of Leishmania amastigotes. Trends Parasitol. 2007;23(8):368–375. doi: 10.1016/j.pt.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 301.Schwegmann A., Brombacher F. Host-directed drug targeting of factors hijacked by pathogens. Sci Signal. 2008;1(29):re8. doi: 10.1126/scisignal.129re8. [DOI] [PubMed] [Google Scholar]
- 302.Blount K.F., Wang J.X., Lim J., Sudarsan N., Breaker R.R. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol. 2007;3(1):44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 303.Laxminarayan R., Van Boeckel T., Frost I., Kariuki S., Khan E.A., Limmathurotsakul D. The Lancet Infectious Diseases Commission on antimicrobial resistance: 6 years later. Lancet Infect Dis. 2020;20(4):e51–e60. doi: 10.1016/S1473-3099(20)30003-7. [DOI] [PubMed] [Google Scholar]
- 304.Robinson T.P., Bu D.P., Carrique-Mas J., Fevre E.M., Gilbert M., Grace D. Antibiotic resistance is the quintessential One Health issue. Trans R Soc Trop Med Hyg. 2016;110(7):377–380. doi: 10.1093/trstmh/trw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Sanz-Garcia F., Hernando-Amado S., Martinez J.L. Mutation-driven evolution of Pseudomonas aeruginosa in the presence of either ceftazidime or ceftazidime/avibactam. Antimicrob Agents Chemother. 2018 doi: 10.1128/AAC.01379-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Blanco P., Corona F., Martinez J.L. Involvement of the RND efflux pump transporter SmeH in the acquisition of resistance to ceftazidime in Stenotrophomonas maltophilia. Sci Rep. 2019;9(1):4917. doi: 10.1038/s41598-019-41308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Blanco P., Corona F., Martinez J.L. Mechanisms and phenotypic consequences of acquisition of tigecycline resistance by Stenotrophomonas maltophilia. J Antimicrob Chemother. 2019 doi: 10.1093/jac/dkz326. [DOI] [PubMed] [Google Scholar]
- 308.Winsor G.L., Griffiths E.J., Lo R., Dhillon B.K., Shay J.A., Brinkman F.S. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44(D1):D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Asnicar F., Thomas A.M., Beghini F., Mengoni C., Manara S., Manghi P. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat Commun. 2020;11(1):2500. doi: 10.1038/s41467-020-16366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Brown C.L., Keenum I.M., Dai D., Zhang L., Vikesland P.J., Pruden A. Critical evaluation of short, long, and hybrid assembly for contextual analysis of antibiotic resistance genes in complex environmental metagenomes. Sci Rep. 2021;11(1):3753. doi: 10.1038/s41598-021-83081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Borderes M., Gasc C., Prestat E., Galvão Ferrarini M., Vinga S., Boucinha L. A comprehensive evaluation of binning methods to recover human gut microbial species from a non-redundant reference gene catalog. NAR Genom Bioinform. 2021;3(1) doi: 10.1093/nargab/lqab009. lqab009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Beaulaurier J., Zhu S., Deikus G., Mogno I., Zhang X.S., Davis-Richardson A. Metagenomic binning and association of plasmids with bacterial host genomes using DNA methylation. Nat Biotechnol. 2018;36(1):61–69. doi: 10.1038/nbt.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]