Abstract
Lung adenocarcinoma (LUAD), the most common histological type of non-small cell lung cancer, is one of the most malignant and deadly diseases. Current treatments for advanced LUAD patients are far from ideal and require further improvements. Here, we utilized a systematic integrative analysis of LUAD microRNA sequencing (miRNA-seq) and RNA-seq data from The Cancer Genome Atlas (TCGA) to identify clinically relevant tumor suppressor miRNAs. Three miRNA candidates (miR-195-5p, miR-101-3p, and miR-338-5p) were identified based on their differential expressions, survival significance levels, correlations with targets, and an additive effect on survival among them. We further evaluated mimics of the three miRNAs to determine their therapeutic potential in inhibiting cancer progression. The results showed not only that each of the miRNA mimics alone but also the three miRNA mimics in combination were efficient at inhibiting tumor growth and progression with equal final concentrations, meaning that the three miRNA mimics in combination were more effective than the single miRNA mimics. Moreover, the combined miRNA mimics provided significant therapeutic effects in terms of reduced tumor volume and metastasis nodules in lung tumor animal models. Hence, our findings show the potential of using the three miRNAs in combination to treat LUAD patients with poor survival outcomes.
Keywords: lung adenocarcinoma, miRNA, therapy, nucleotide drug, miRNA mimics, additive effect
Graphical abstract
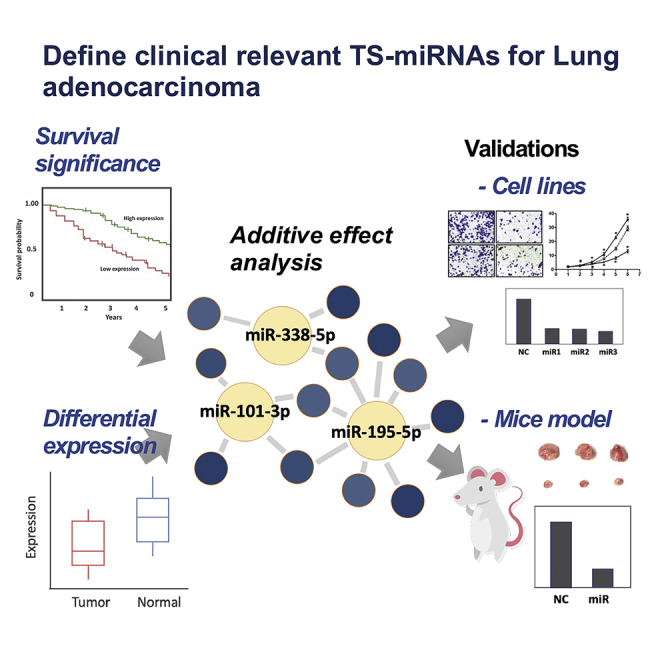
Utilizing an integrative bioinformatics approach, we identified three miRNA candidates that can serve in diagnostic and therapeutic roles for patients with lung adenocarcinoma. The experimental results prove that not only these single miRNA candidates but also the three miRNAs in combination were efficient at inhibiting tumor growth and progression.
Introduction
Lung adenocarcinoma (LUAD) is one of the most malignant and deadly diseases and also the most common histological subtype of non-small cell lung cancer (NSCLC).1,2 Notably, LUAD patients only survive for an average of 14 months after being diagnosed.3 Current standard treatment options for advanced lung cancer include combination chemotherapy with platinum-based agents, EGFR tyrosine kinase inhibitors, chemotherapy plus anti-angiogenesis or anti-EGFR antibodies, ALK inhibitors for ALK translocation, and immune checkpoint inhibitors. However, current treatments only benefit a small proportion of patients. Therefore, more in-depth studies to develop more promising therapeutics for lung cancer patients require the immediate attention of relevant researchers.
miRNAs, which are small and non-coding sequences of RNA (around 19–23 nucleotides in length), are highly conserved in modulating post-translational modifications, which effectively repress the multiple target genes through imperfect sequence bases, with complementarity, between miRNA and its targets.4 The role of a given miRNA in cancer can be either oncogenic, with such a miRNA termed an oncomiR, or tumor suppressing, with such a miRNA termed a tumor suppressor (TS) miRNA.1 Abnormal miRNA expression in various cancers, including lung cancer, has been demonstrated to involve tumor development, tumor progression, and events leading to treatment resistance.5,6 For instance, the signature of four miRNAs—miR-29c, miR-124, miR-200c, and miR-424—is considered by the European Lung Cancer Working Party (ELCWP) to effectively predict survival or drug response to cisplatin and vinorelbine in lung cancer patients.7
There are several ongoing pre-clinical or clinical trials using miRNAs as therapeutics to treat lung cancers. The drug application of antagomir or miRNA mimics depends on the roles of miRNA (oncomiR/TS miRNA). Patients with highly expressed oncomiRs are suitable for antagomir treatment; however, in many cases, it is bothersome to measure the miRNA expression for each patient. Mimics of TS miRNA can inhibit cancers, so several miRNAs have been delivered to suppress tumor cells; for example, let-7 can be used to negatively mediate oncogenic KRAS in NSCLC.8 A well-known miRNA mimic currently is MRX34, a miRNA-34a mimic, which was applied for the downregulation of miRNA-34 cancers in one phase I trial with NSCLC patients.9 Also, miR-16 mimics targeting EGFR were applied in a phase I trial, with the results indicating that these mimics are relatively safe for patients due to highly specific delivery by minicells.10,11 Even though miRNA therapeutics have shown encouraging outcomes in treating several cancers, there are still challenges in identifying efficacious therapeutic candidates and developing a safe and efficient delivery system for future medicines.12
In the present study, we utilized a systematic integrative analysis of LUAD miRNA-seq and RNA-seq data from The Cancer Genome Atlas (TCGA), and three miRNA candidates were filtered out for experimental validations. Not only these single miRNA candidates but also the three miRNAs in combination were efficient at inhibiting tumor growth and progression.
Results
Identification of miRNA candidates by integrative bioinformatics approaches
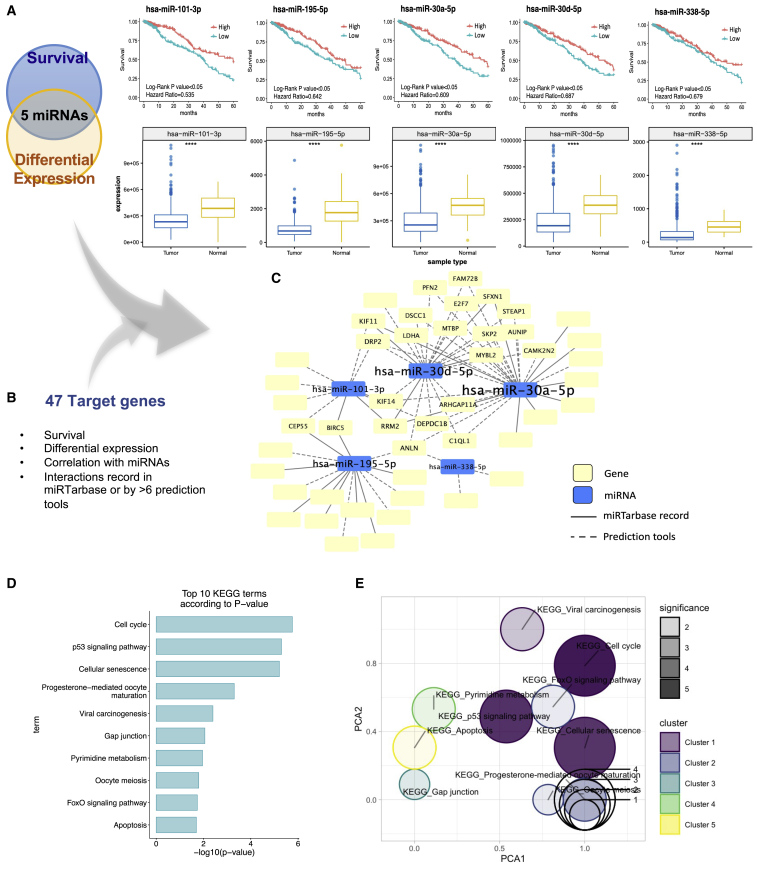
In order to develop efficacious miRNA-based treatments for LUAD patients, we sought to uncover differentially expressed and survival-relevant miRNAs with tumor-suppressing features using systematic analysis, as shown in Figure 1. First, to define clinically relevant miRNAs, two crucial features, differential expression and survival significance, were exploited to intensify the significance of the candidates (Figure 1A). Figure 1A shows the 5-year survival of five miRNA candidates with a statistically significant p value (log-rank p < 0.05). The expression levels of these miRNAs were significantly different between primary tumor (TP) and adjacent normal (NT) samples in TCGA LUAD dataset. The consistency of the direction between the fold change of expression (log2 fold change < −1) and the hazard ratio (HR) of survival (HR < 1) was also considered. Thus, five miRNA candidates (miR-195-5p, miR-30a-5p, miR-30d-5p, miR-101-3p, and miR-338-5p) were screened out.
Figure 1.
Defining clinically relevant miRNAs through bioinformatics integrative analysis
(A) Clinically relevant miRNA candidates were filtered out by both significance in terms of survival and differential expression in TCGA LUAD dataset. 5-year survival analysis of five miRNA candidates indicated that they were significant, with log-rank p < 0.05 and HR values < 1. The expression level differences of these five miRNA candidates between LUAD tumor and normal samples in TCGA are displayed. The asterisks (∗∗∗∗) denote statistical significance with p < 0.0001. (B) Target genes were defined by their survival significance levels, differential expression levels, correlations with the miRNA candidates, and interactions as indicated in the miRTarbase or by >6 prediction tools. (C) The network displays the interactions between the miRNA candidates and the target genes recorded in the miRTarbase (solid lines) or defined by the prediction tools (dashed lines). (D) The top 10 significant KEGG pathways of the 47 target genes are shown based on the p values of the functional annotation analysis results. (E) A gene set overrepresentation of significant KEGG pathways displays overlapped genes in the different pathways.
In identifying the target genes regulated by these miRNA candidates, we found that 349 genes exhibited survival significance, differential expression levels, and high correlations with the miRNA candidates. The application of more stringent criteria, which further considered higher-level evidence recorded in the miRTarbase or predicted by >6 tools (Figure 1B), yielded 47 target genes. Table S1 lists the target genes regulated by each miRNA candidate according to the network in Figure 1C, and Figure S1 also validated the relationships between the miRNA candidates and their targets with significant p value. Relatedly, Figure 1C shows the network of higher-evidence-level miRNA-gene interactions. This network clearly indicates important targets regulated by multiple miRNA candidates, meaning that several miRNAs contribute simultaneously to cancer progression. For instance, target genes such as BIRC5 and RRM2 are shown in the network, and these genes are regulated by more than one of our miRNA candidates, which are validated by previous studies. This observation indicates that those miRNA candidates simultaneously play crucial roles in cancer progression. To investigate the crucial functions of those target genes regulated by the miRNAs, the 47 genes were annotated, and a bar chart (Figure 1D) showing the top 10 significant functions of the KEGG pathway, which clearly shows that the most significant functions were “cell cycle” and “p53 signaling pathway” functions, was generated. Figure 1E visualizes the gene set overrepresentation, indicating the genes that simultaneously played roles in different functions. Obviously, most of the functions were highly associated with cell cycle regulation, as indicated by the shorter distances, while only a few functions were located at the top of the plot some distance from those cell cycle functions. More genes shown by the larger circles were found to play roles in more significant functions like the cell cycle functions, which are indicated by darker colors. Overall, the figure shows that most of the functions had high levels of association between them.
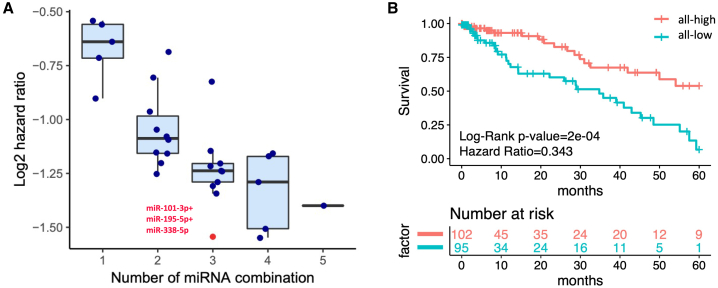
To advance miRNA candidates that could potentially be developed into promising treatments for LUAD patients, we further examined whether there was an additive effect in terms of reducing the risks among LUAD patients by increasing the number of miRNAs combined in different combinations for a survival analysis of the five miRNA candidates. The HR values of the different miRNA combinations gradually declined when more miRNA candidates were included (Figure 2A). These phenomena illustrate that there were, in fact, additive effects among the different miRNA candidates. Accordingly, a combination of three of the miRNAs (miR-195-5p, miR-101-3p, and miR-338-5p) with roughly the lowest HR value (0.343) was selected as the combination used for the following analysis. Moreover, survival analysis of this three-miRNA combination demonstrated that the patients with all-high expression levels of the three miRNAs in the combination had significantly better survival times than patients with all-low expression levels of the three miRNAs in the combination (Figure 2B). That suggests that the three-miRNA combination is correlated with good prognosis in LUAD patients. Thus, the three miRNAs and their combination were selected as our preferred candidates for the experimental validations.
Figure 2.
Additive survival analysis of five miRNA candidates
(A) The HR values of each of the miRNA combinations were calculated by additive survival analysis and are shown in a boxplot with dot plots overlaid. The x axis indicates the number of miRNAs combined; the y axis indicates the log2 transformation of the HR. (B) Survival analysis of the combination of the 3 miRNAs shows a significant difference between the all-high and all-low expression groups, with a log-rank p < 0.05.
In vitro assays for validation of 3 TS miRNA candidates
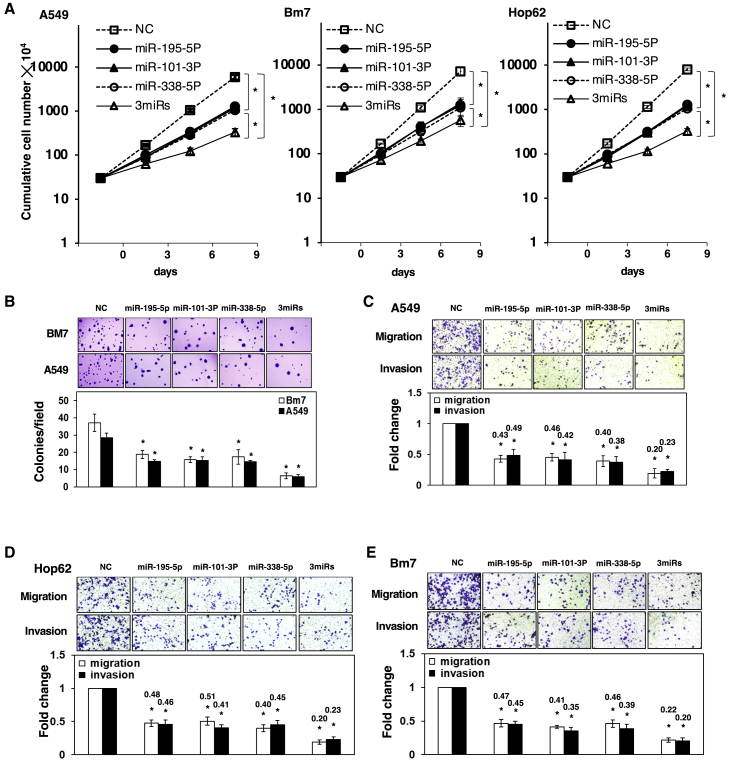
To certify that the roles of the three miRNA candidates were correlated with good outcomes in LUAD, we investigated the functional analysis of the three miRNAs in several lung cancer cell lines (namely, the A549, Bm7, and Hop62 cell lines). The expression of miRNA mimics was first examined with time (Figure S2). Compared to the negative control group, each singly transfected miRNA resulted in 5-fold decreases in the number of A549 cells, with p < 0.05 (precisely 5.88 × 107 in control to 1.27 × 107/1.18 × 107/1.06 × 107 in miR-195-5p/miR-101-3p/miR-338-5p transfected, respectively); however, the three miRNAs in combination shows 17 times reduction in the number of cells (3.31 × 106), with p < 0.05 by day 9 (Figure 3A). By using equal total amounts of miRNA mimics of each miRNA for each group, we found that the cell number following transfection with the three miRNA mimics combined (20/3 + 20/3 + 20/3 = 20 nM) was significantly reduced in comparison with transfection with the individual miRNA mimics (each at 20 nM). We also observed similar inhibition effects on Bm7 and Hop62 cells. Also, in comparisons with the negative control group, colony-formation assay results indicated that decreases of the colony numbers reached around 50% (p < 0.05) with overexpression of each of the individual miRNA mimics, and almost 75% (p < 0.05) with overexpression of the 3 miRNA mimics in combination (Figure 3B). The cell viability was consistently and obviously reduced over time with overexpression of the miRNAs according to the 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazolium bromide (MTT) assays (Figure S3). Furthermore, cell migration and invasion assays showed significant declines in the cell numbers in individual miRNA plates, with decreases of more than 50% with p < 0.05, yet 2-fold greater effects (that is, decreases to less than 25% of negative control levels) for the 3 miRNAs in combination, as shown in Figures 3C–3E. These results clearly indicate that cancer growth and progression are more vulnerable to treatment with the three miRNAs in combination than to treatment with each individual miRNA.
Figure 3.
Overexpression of miR-195-5p, miR-101-3p, or miR-338-5p mimics reduced the growth and tumor progression levels of A549, Bm7, and Hop62 cells in vitro
(A) The transfected cells were seeded and counted using the trypan blue exclusion method at the indicated time intervals. The results demonstrated that the overexpression of the miRNA mimics attenuated the growth of the lung cancer cells. (B) The overexpression of the miRNA mimics decreased the colony numbers of A549 and Bm7 cells in soft agar, as determined by in vitro colony formation assays. The overexpression of the miRNA mimics suppressed the in vitro migration and invasion abilities of A549 (C), Hop62 (D), and Bm7 (E) cells. A scrambled oligonucleotide was used as negative control (NC). All the cells were individually transfected with the miRNA mimics at a final concentration of 20 nM. In the combined setup (3miRs), the cells were transfected with all three of the miRNA mimics at one-third concentrations. The mean values obtained from three independent experiments are depicted. The error bars depict the standard error of each mean. The asterisks (∗) indicate statistically significant differences (p < 0.05) between the experimental and control groups. 3miRs is denoted as the combination of three miRNAs (miR-195-5p, miR-101-3p, and miR-338-5p).
Mouse model validations of 3 miRNA candidates
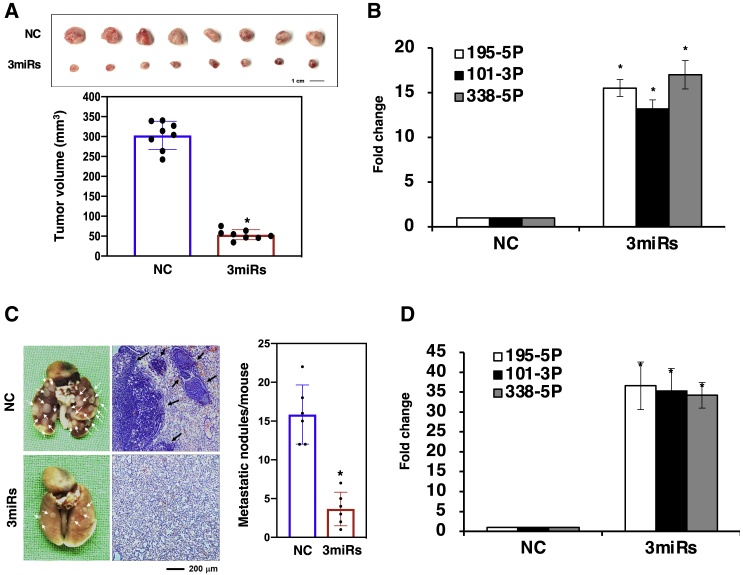
To confirm the efficacy of the miRNA candidates, we further performed animal experiments. Since we discovered that the effectiveness of the miRNAs in combination was greater than that of any single miRNA, the three-miRNA combination was further evaluated in the mouse experiments. We subcutaneously injected Bm7 human lung cancer cells with control or the three-miRNA combination into nude mice and monitored the sizes of the resulting tumors. The mice were injected with control or miRNA-transfected cells, and the tumor-bearing mice were sacrificed at the indicated time. The mice injected with three-miRNA-combination-transfected cells had reduced tumor volumes of more than 80% (p < 0.05) compared to the negative control group mice at the indicated time, suggesting a much slower tumor growth rate (Figure 4A). The quantitative expression levels of miR-195-5p, miR-101-3p, and miR-338-5p in the subcutaneous tumors showed an over 8-fold increase compared to those in the negative control mice (Figure 4B). In addition, the negative control group and those injected with the miRNA-transfected cells had the respective cells injected via the tail vein to examine their metastatic ability, which was analyzed based on the number of nodules in the lungs. We evaluated the effects of the three miRNAs in combination in lung tumor metastasis animal models and found that the metastatic nodules in the lungs were significantly decreased by one-fourth fold in comparison to the nodules in the negative-control-injected mice (Figure 4C). According to qPCR, the miR-195-5p, miR-101-3p, and miR-338-5p expression levels in the metastatic nodules of the mice injected with the three-miRNA-combination-transfected cells were 30 times higher than those in nodules of the negative control group (Figure 4D). Taken together, these results indicated that the three miRNAs in combination provide a TS function to reduce cancer growth and metastases.
Figure 4.
The combined overexpression of the miRNA mimics suppressed the tumor growth and metastasis of Bm7 cells in vivo
(A) As determined by xenograft assays, the combined overexpression of the miRNA mimics in Bm7 cells decreased the tumor volume. Following transfection with three miRNA mimics, namely, miR-195-5p, miR-101-3p, and miR-338-5p, or with the scrambled oligonucleotide (which served as an NC), the viable Bm7-combination cells and the Bm7-NC cells were subcutaneously injected into nude mice (n = 8 per group). At 30 days after implantation, the mice were sacrificed, and the tumor volume of the xenografts was measured. (B) The subcutaneous tumors were excised in order to detect the expression levels of miR-195-5p, miR-101-3p, and miR-338-5p through quantitative real-time PCR. Bar, 1.0 cm. (C) The combined overexpression of the miRNA mimics reduced the in vivo metastatic activity of the Bm7 cells. The metastatic nodules in the lungs were measured by injecting non-obese diabetic severe-combined immunodeficiency mice (n = 6 per group) with the viable Bm7-combination cells or the Bm7-NC cells through the tail vein injection method. After 15 weeks, the mice were sacrificed, and the metastatic nodules in the lungs were counted by gross and microscopic examination (left and middle images). (D) The metastatic nodules were excised in order to detect the expression levels of miR-195-5p, miR-101-3p, and miR-338-5p through quantitative real-time PCR. Bar, 200 μm. The results are expressed as the mean ± standard error of the mean. The asterisks (∗) indicate statistically significant differences (p < 0.05) between the experimental and control cells. 3miRs is denoted as the combination of three miRNAs (miR-195-5p, miR-101-3p, and miR-338-5p).
Discussion
In this study, a systematic integrative approach was applied to identify clinically relevant miRNAs that might have a TS function, and the results showed that three miRNAs (miR-195-5p, miR-101-3p, and miR-338-5p) in combination showed the greatest effect, with that result shown not merely through bioinformatics methods but also through biological assays of cell system and animal models. The three miRNA mimics provided strong TS effects in terms of reducing cancer growth and cancer metastases and thus could be of potential benefit in lung cancer treatment strategies.
The TS roles of the three miRNA candidates in lung cancer have been described in many previous studies, and our experimental results also showed that these miRNAs indeed serve as TS miRNAs.13, 14, 15 Lu et al.13 described the value of miR-101-3p as a prognostic marker, which they determined by examining survival probabilities based on its expression level. Other researchers showed that the expression of miR-101-3p contributes to cancer progression and metastasis in lung cancers.14 Meanwhile, a recent study indicated that miR-195-5p can serve as a risk biomarker of lung cancer.16 In still another study, miR-338-5p was found to be downregulated in lung cancer, with its targets’ functions mainly being verified as related to cell cycle regulation.17 Here, we further demonstrated the clinical importance of miR-338-5p to survival. In addition, additive effects among these three miRNAs exist, although those effect had not yet been discovered in previous research. We also validated the additive effects of the combination of these three miRNAs through in vitro and in vivo experiments. Those data indicated that the three miRNAs can be prognostic biomarkers and could serve as potential targets of miRNA-based drug development. The network of miRNA-gene interactions produced in the present study (Figure 1C) is a clear demonstration of significant miRNA-gene relations, with that network revealing several oncogenic targets governed by multiple miRNA candidates. Among them, RRM2 and BIRC5 are crucial oncogenes previously identified as playing roles in multiple cancers.18, 19, 20
The functional enrichment analysis conducted in this study (Figures 1D and 1E) indicated that the most significant functions affected by the candidate miRNAs were related to cell cycle regulation. As expected, several aberrant miRNAs have been reported to inhibit targets involved the cell cycle and proliferation.17,21 For instance, the miR-34 family has been reported to influence p53 signaling in a manner that results in cell cycle arrest.21 Among the three candidate miRNAs identified in this study, miR-338-5p was shown to be associated with cell cycle progression in a previous study.17 Therefore, we further confirmed the functions of the associated target genes to verify that those vital target genes were included in the integrative bioinformatics analysis. Three hundred forty-nine target genes were first identified as having differential expression levels and survival significance, with most of those genes mainly involved in the cell cycle with significant p values, as shown in Figure S4A. Moreover, 25 target genes regulated by the final three miRNAs (miR-195-5p, miR-101-3p, and miR-338-5p) were also functionally annotated (Figure S4B). Three bar plots (Figure 1D; Figure S4) indeed demonstrated that the top 10 significant functions of the major target genes governed by our miRNA candidates were highly similar. The p53 signaling pathway was also identified as the crucial function of the targets for those miRNAs. In a previous study, Liu et al.22 showed that several miRNAs mediate the function of p53 signaling to promote tumorigenesis.
Previously, we established a synergistic survival analysis method, which is aimed at considering how the expression levels of two genes concurrently affect survival.23 We further developed an additive effect on survival analysis to examine more than two candidates in combination. The method of additive effect to identify the most promising miRNA candidates is based on the purpose of seeking the fewest candidates with the most effectiveness. For this reason, we determine the most significant change in HR values among all potential combinations (Figure 2A). By applying this additive survival analysis, we examined three miRNAs in combination to identify the lowest HR for the TS role. The biological experiments also exhibited the additive effects of the three miRNAs in combination (Figures 3 and 4). In multiple functional assays, the results showed that the overexpression of the miRNAs in combination more effectively suppressed cancer cell growth and progression than each miRNA alone, even though the final concentrations of each miRNA and the miRNAs in combination transfected were the same (20 nM). Our mouse experiments also illustrated the great difference between the miRNAs in combination when transfected into cells versus a negative control transfected into cells in terms of tumor volumes and metastasis nodules in mice. These experiments indicated the evident additive effects among the miRNA candidates. Even though many studies have shown that miRNA cocktails have a concurrent or double effect on the particular functions of tumorigenesis, such as the effect of a cocktail of let-7B and miR-34a on cell invasion or the effect of a cocktail of miR-143 and miR-145 on apoptosis,24 we have demonstrated that the three miRNAs identified in this study effectively affect cell proliferation, tumor growth, and metastasis. In addition, we researched the potential of miRNA drugs with other therapeutic agents. To validate the efficiency of combinatorial therapy over the use of traditional therapeutic agents, we have performed cytotoxicity assays to measure whether a synergistic effect of the microRNAs and chemotherapy drug cisplatin in three lung cancer cell lines. As shown in Figure S5, a combination of three TS miRNAs and cisplatin synergistically reduced the cell survival of lung cancer cells (combination index < 1). That indicates the efficiency of combinatorial therapy of miRNAs and cisplatin.
A novel approach has been constructed to define multiple miRNA combinations with the lowest HR, though there are few limitations of this approach. First, the limitation may exist due to lack of validation by other cohort studies. The miRNA combinations were calculated based on the survival time, which outcome might be slightly different in another cohort. Also, the best combination of multiple miRNAs was chosen by the analyzers as shown in Figure 2, which could lead to different conclusions depending on the analyzers. However, our miRNA candidates are still reliable, because multiple functional assays successfully verify the efficacy of miRNA candidates.
Diagnostics and therapeutics are commonly inseparable and involve reactive procedures that thoroughly depend on the personal symptoms, family history, laboratory and imaging diagnosis, and current medical care of patients. This has progressively resulted in a concept called “theranostics,” which aims at providing safer and more effective patient-centered care by considering the association between diagnosis and therapeutics in order to improve the quality of medical care.25 According to the theranostics approach, our miRNA candidates have potential for use in both diagnosis and therapy. The expression levels of the three miRNAs combined can effectively distinguish cancer patients in terms of survival, which makes the combination of the three miRNAs an ideal biomarker for uncovering patients likely to have poor survival outcomes. For this group of patients, mimics of the three miRNAs can also serve as therapeutics. Thus, our study shows the potential of using the three miRNAs in combination to treat lung cancer patients likely to have poor survival outcomes.
Materials and methods
Data collection and preparation
Data were first obtained from our databases, the DriverDB23,26,27 and YM50028, 29, 30 databases. In brief, TCGA level 3 miRNA-seq and RNA-seq LUAD data were also collected from TCGA data portal: https://portal.gdc.cancer.gov/. The relevant clinical dataset of LUAD patients was likewise downloaded from TCGA website. The preparation and processing of the miRNA-seq and RNA-seq data were addressed by in-house Perl and R scripts, the details of which are documented in previous publications.26,28 Thereby, 515 tumor and 59 normal samples of RNA-seq data were included, while 513 tumor and 46 normal samples of miRNA-seq data were used for the following analyses.
Identification of differentially expressed miRNAs and genes
To identify prognosis-related miRNAs, we examined the differentially expressed miRNAs (DEmiRNAs) and genes (DEgenes) so as to distinguish the significant differences between TP samples and adjacent NT samples. An R package, DEseq (Version 1.28.0),31 was applied. The criteria to filter the miRNA candidates had adjusted p <0.05 and log2 fold change values <−1, which was effective in identifying tumor versus normal samples for the tumor suppressor role. Likewise, the filtering for the gene candidates had adjusted p <0.05 and log2 fold change values >1 for the oncogenic role. Also, to filter out the candidates with extremely low expression levels, we further selected the mean of normalized counts (baseMean) >1 for miRNAs and >10 for genes.
Survival analysis and the additive effect on survival of miRNAs
To define clinically relevant candidates, another R package, Survival (version 2.41-3),32 was used to calculate the Cox regression (or Cox proportional hazards) model between two pre-defined groups, which provided survival estimations in the presence of time-to-event datasets. We stratified cancer patients according to the medians of miRNA expression levels. Significant survival-relevant miRNA and gene candidates could then be identified with log-rank p <0.05 and HR values <1 for miRNAs (HR values > 1 for genes). In this study, we applied 5-year survival for the following analyses.
To determine miRNA candidates with a significant additive effect on survival, we constructed an analytic model to evaluate the additive effect between co-expressed miRNAs. We first calculated every survival estimation of any combination of miRNA candidates, in no particular order. For each miRNA combination, we stratified patients by the medians of expression levels, and patients were classified into an all-high or all-low group; the all-high group consisted of the patients who had very high expression of all the miRNAs in the combination, and vice versa. Then, survival estimations were computed based on the patients in the all-high and all-low groups. Significant combinations were defined with log-rank p <0.05 and HR values <1. Thus, survival estimations could be compared by applying the differences among the HR values of any of the miRNA combinations.
Compiling target genes of miRNA candidates and functional enrichment analysis
To compile the information regarding miRNA-gene interactions, three analytic steps were adopted. First, the correlations between the RNA expression and miRNA expression levels were calculated, including three types of correlation: Pearson, Spearman, and Kendall correlations. Those genes and miRNAs for which one of the three types of correlations was <−0.3 were taken into account. Then, in order to consider more substantial evidence regarding interactions between miRNAs and genes, the miRTarbase data: http://miRTarBase.cuhk.edu.cn/ were downloaded and applied to show the interactions as networks.33 The interactions were not only recorded in the miRTarbase used; 12 more prediction tools were applied to investigate the relations between the miRNA candidates and target genes, as detailed in a previous publication.30 The interactions predicted by more than 6 tools were taken as higher-level evidence.
Functional enrichment analysis of target genes was also performed as detailed in our previous publications,26,27 for which KEGG pathway annotations were used with adjusted p <0.05. The top 10 significant KEGG terms were extracted from the annotation results and regarded as the most critical functions regulated by the miRNA candidates. Moreover, gene set overrepresentation analysis was performed by using another R package, GSOAP,34 which explores overlapped gene sets in multiple functions.
Cell culture and transfection
Human lung adenocarcinoma A549 and Hop62 cell lines were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (PS; Gibco). The Bm7 cell line, a lung adenocarcinoma cell line with brain metastatic ability,35 was cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium with 10% FBS. The various cells were maintained at 37°C in a humidified incubator containing 5% CO2. The A549, Bm7, and Hop62 cells were then transfected with miRNA mimics (hsa-miR-195-5p, hsa-miR-101-3p, and hsa-miR-338-5p) or a scrambled oligonucleotide as a negative control (Ambion) at a final concentration of 20 nM using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. For detection of mature miRNAs, total RNA was reverse transcribed into complementary DNA using MultiScribe Reverse Transcriptase (Thermo Fisher Scientific) and the specific primers designed for miR-195-5p, miR-101-3p, and miR-338-5p (Applied Biosystems). The expression levels of miR-195-5p, miR-101-3p, and miR-338-5p were determined on the ABI StepOnePlus system with quantitative real-time PCR using TaqMan Universal PCR Master Mix and TaqMan MicroRNA Assay (Applied Biosystems). The miRNA levels were normalized to the expression of RNU48 small nuclear RNA. For miRNA target gene expression analysis, sequence of primers used in the quantitative real-time PCR experiment are shown in Table S2; the relative mRNA levels were normalized to the expression of 18S rRNA.
Cell growth and viability assays
For the evaluation of cell growth, the transfected cells (3 × 105) were seeded into 6-well plates and then counted using the trypan blue exclusion method at the time indicated, as described previously.36 For the detection of cell viability, the transfected cells (1 × 104) were seeded into 24-well plates. After incubation for 24, 48, or 72 h, an MTT assay (Sigma-Aldrich) was used to assess cell viability with a microplate ELISA reader (TECAN Infinite 200).36
Soft-agar colony-formation assay
The transfected cells (5 × 103) were used for the assay of anchorage-independent growth in soft agar, as described previously.36,37 Then, the cells were stained, and colonies were counted from 10 random fields under a microscope.
Migration and invasion assays
Migration and invasion ability assays (3 × 104 cells for the migration assay and 5 × 104 cells for the invasion assay) were performed in 24-well plates using Millicell tissue culture plate well inserts (Millipore, Bedford, MA, USA) for 12 h and BD BioCoat Matrigel Invasion Chambers (Becton Dickson, Mountain View, CA, USA) for 20 h, respectively, as described previously.36,38
Xenografted tumorigenicity assay in nude mice
Five-week-old male mice (BALB/c nu/nu, National Science Council Animal Center, Taipei, Taiwan) were subcutaneously injected with 3 × 106 viable transfected Bm7 cells, in a total volume of 100 μL PBS, into both hind limbs. Thirty days after implantation, the mice were sacrificed, and the tumor volume (V) of the xenografts determined by measuring the length (L), width (W), and depth (D) of the tumors with a measuring caliper and using the formula: V = (L × W × D × π)/6; the average value was calculated.38,39 The expression levels of miR-195-5p, miR-101-3p, and miR-338-5p in the excised tumor samples were detected using miRNA quantitative real-time PCR, as previously described.36,40 All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) of China Medical University.
In vivo tail vein metastasis assay
As described previously,36,41 the transfected cells (1 × 106) were injected into 6-week-old male non-obese diabetic severe-combined immunodeficiency mice (NOD-SCID mice) by tail vein injection. After 15 weeks, the mice were sacrificed, and the metastatic lung nodules were counted by gross and microscopic examination. The expression levels of miR-195-5p, miR-101-3p, and miR-338-5p in excised lung tumor samples were detected by miRNA quantitative real-time PCR.
Statistical analysis for in vivo and in vitro experiments
Unless otherwise noted, each sample was assayed in triplicate. Statistical analyses were performed using the Student’s t test for a simple comparison of two groups. Differences were considered statistically significant if the p was <0.05.
Acknowledgments
This research was funded by the Ministry of Science and Technology, Taiwan (MOST 108-2314-B-039-060, MOST 108-2622-E-039-005-CC2, MOST 109-2622-E-039-004-CC2, MOST 109-2628-E-039-001-MY3, MOST 109-2327-B-039-002, MOST 108-2628-B-039-003, MOST 109-2628-B-039-006, 109-2314-B-182-078-MY3, and 109-2628-B-182-008), China Medical University, Taiwan (CMU108-Z-02), China Medical University Hospital, Taiwan (DMR-109-055, DMR-109-223, DMR-110-072, and DMR-109-206), and Chang Gung Memorial Hospital at Linkou (CMRPD1J0321 and CMRPD1H0473). The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Author contributions
Conceptualization, W.C. and Y.S.; validation, K.H., C.L., L.L., and P.P.; formal analysis, S.L.; data curation, K.H., M.W., and S.W.; writing – original draft preparation, S.L.; writing – review & editing, Y.L., F.C., and H.W.; visualization, S.L.; supervision, W.C. and Y.S.; project administration, S.L.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.04.020.
Contributor Information
Yuh-Pyng Sher, Email: ypsher@gmail.com.
Wei-Chung Cheng, Email: cwc0702@gmail.com.
Supplemental information
References
- 1.Zhang Y., Yang Q., Wang S. MicroRNAs: a new key in lung cancer. Cancer Chemother. Pharmacol. 2014;74:1105–1111. doi: 10.1007/s00280-014-2559-9. [DOI] [PubMed] [Google Scholar]
- 2.Barta J.A., Powell C.A., Wisnivesky J.P. Global Epidemiology of Lung Cancer. Ann. Glob. Health. 2019;85:8. doi: 10.5334/aogh.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balzer B.W.R., Loo C., Lewis C.R., Trahair T.N., Anazodo A.C. Adenocarcinoma of the Lung in Childhood and Adolescence: A Systematic Review. J. Thorac. Oncol. 2018;13:1832–1841. doi: 10.1016/j.jtho.2018.08.2020. [DOI] [PubMed] [Google Scholar]
- 4.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Macfarlane L.A., Murphy P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li G., Fang J., Wang Y., Wang H., Sun C.C. MiRNA-based Therapeutic Strategy in Lung Cancer. Curr. Pharm. Des. 2018;23:6011–6018. doi: 10.2174/1381612823666170725141954. [DOI] [PubMed] [Google Scholar]
- 7.Berghmans T., Ameye L., Willems L., Paesmans M., Mascaux C., Lafitte J.J., Meert A.P., Scherpereel A., Cortot A.B., Cstoth I., European Lung Cancer Working Party Identification of microRNA-based signatures for response and survival for non-small cell lung cancer treated with cisplatin-vinorelbine A ELCWP prospective study. Lung Cancer. 2013;82:340–345. doi: 10.1016/j.lungcan.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 8.Trang P., Medina P.P., Wiggins J.F., Ruffino L., Kelnar K., Omotola M., Homer R., Brown D., Bader A.G., Weidhaas J.B., Slack F.J. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouchie A. First microRNA mimic enters clinic. Nat. Biotechnol. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 10.van Zandwijk N., Pavlakis N., Kao S.C., Linton A., Boyer M.J., Clarke S., Huynh Y., Chrzanowska A., Fulham M.J., Bailey D.L. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 11.Wu S.G., Chang T.H., Liu Y.N., Shih J.Y. MicroRNA in Lung Cancer Metastasis. Cancers (Basel) 2019;11:265. doi: 10.3390/cancers11020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 13.Lu H.M., Yi W.W., Ma Y.S., Wu W., Yu F., Fan H.W., Lv Z.W., Yang H.Q., Chang Z.Y., Zhang C. Prognostic implications of decreased microRNA-101-3p expression in patients with non-small cell lung cancer. Oncol. Lett. 2018;16:7048–7056. doi: 10.3892/ol.2018.9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X., He X., Liu Y., Zhang H., Chen H., Guo S., Liang Y. MiR-101-3p inhibits the growth and metastasis of non-small cell lung cancer through blocking PI3K/AKT signal pathway by targeting MALAT-1. Biomed. Pharmacother. 2017;93:1065–1073. doi: 10.1016/j.biopha.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Zheng J., Xu T., Chen F., Zhang Y. MiRNA-195-5p Functions as a Tumor Suppressor and a Predictive of Poor Prognosis in Non-small Cell Lung Cancer by Directly Targeting CIAPIN1. Pathol. Oncol. Res. 2019;25:1181–1190. doi: 10.1007/s12253-018-0552-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L., Feng T., Zhang W., Gao S., Wang R., Lv W., Zhu T., Yu H., Qian B. MicroRNA Biomarker hsa-miR-195-5p for Detecting the Risk of Lung Cancer. Int. J. Genomics. 2020;2020:7415909. doi: 10.1155/2020/7415909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu N., Yong S., Kim H.K., Choi Y.L., Jung Y., Kim D., Seo J., Lee Y.E., Baek D., Lee J. Identification of tumor suppressor miRNAs by integrative miRNA and mRNA sequencing of matched tumor-normal samples in lung adenocarcinoma. Mol. Oncol. 2019;13:1356–1368. doi: 10.1002/1878-0261.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H., Zhang H., Du Y., Gu X. Prognostic significance of BRCA1, ERCC1, RRM1, and RRM2 in patients with advanced non-small cell lung cancer receiving chemotherapy. Tumour Biol. 2014;35:12679–12688. doi: 10.1007/s13277-014-2592-7. [DOI] [PubMed] [Google Scholar]
- 19.Sun H., Yang B., Zhang H., Song J., Zhang Y., Xing J., Yang Z., Wei C., Xu T., Yu Z. RRM2 is a potential prognostic biomarker with functional significance in glioma. Int. J. Biol. Sci. 2019;15:533–543. doi: 10.7150/ijbs.30114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu X., Zhang Y., Wu B., Kurie J.M., Pertsemlidis A. The miR-195 Axis Regulates Chemoresistance through TUBB and Lung Cancer Progression through BIRC5. Mol. Ther. Oncolytics. 2019;14:288–298. doi: 10.1016/j.omto.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J., Zhang C., Zhao Y., Feng Z. MicroRNA Control of p53. J. Cell. Biochem. 2017;118:7–14. doi: 10.1002/jcb.25609. [DOI] [PubMed] [Google Scholar]
- 23.Liu S.H., Shen P.C., Chen C.Y., Hsu A.N., Cho Y.C., Lai Y.L., Chen F.H., Li C.Y., Wang S.C., Chen M. DriverDBv3: a multi-omics database for cancer driver gene research. Nucleic Acids Res. 2020;48(D1):D863–D870. doi: 10.1093/nar/gkz964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miroshnichenko S., Patutina O. Enhanced Inhibition of Tumorigenesis Using Combinations of miRNA-Targeted Therapeutics. Front. Pharmacol. 2019;10:488. doi: 10.3389/fphar.2019.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeelani S., Reddy R.C., Maheswaran T., Asokan G.S., Dany A., Anand B. Theranostics: A treasured tailor for tomorrow. J. Pharm. Bioallied Sci. 2014;6(Suppl 1):S6–S8. doi: 10.4103/0975-7406.137249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng W.C., Chung I.F., Chen C.Y., Sun H.J., Fen J.J., Tang W.C., Chang T.Y., Wong T.T., Wang H.W. DriverDB: an exome sequencing database for cancer driver gene identification. Nucleic Acids Res. 2014;42:D1048–D1054. doi: 10.1093/nar/gkt1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung I.F., Chen C.Y., Su S.C., Li C.Y., Wu K.J., Wang H.W., Cheng W.C. DriverDBv2: a database for human cancer driver gene research. Nucleic Acids Res. 2016;44(D1):D975–D979. doi: 10.1093/nar/gkv1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng W.C., Chung I.F., Huang T.S., Chang S.T., Sun H.J., Tsai C.F., Liang M.L., Wong T.T., Wang H.W. YM500: a small RNA sequencing (smRNA-seq) database for microRNA research. Nucleic Acids Res. 2013;41:D285–D294. doi: 10.1093/nar/gks1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng W.C., Chung I.F., Tsai C.F., Huang T.S., Chen C.Y., Wang S.C., Chang T.Y., Sun H.J., Chao J.Y., Cheng C.C. YM500v2: a small RNA sequencing (smRNA-seq) database for human cancer miRNome research. Nucleic Acids Res. 2015;43:D862–D867. doi: 10.1093/nar/gku1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung I.F., Chang S.J., Chen C.Y., Liu S.H., Li C.Y., Chan C.H., Shih C.C., Cheng W.C. YM500v3: a database for small RNA sequencing in human cancer research. Nucleic Acids Res. 2017;45(D1):D925–D931. doi: 10.1093/nar/gkw1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Therneau T.M., Lumley T. Package ‘survival’. R Top. Doc. 2015;128:112. [Google Scholar]
- 33.Chou C.H., Shrestha S., Yang C.D., Chang N.W., Lin Y.L., Liao K.W., Huang W.C., Sun T.H., Tu S.J., Lee W.H. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tokar T., Pastrello C., Jurisica I. GSOAP: a tool for visualization of gene set over-representation analysis. Bioinformatics. 2020;36:2923–2925. doi: 10.1093/bioinformatics/btaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin C.Y., Chen H.J., Huang C.C., Lai L.C., Lu T.P., Tseng G.C., Kuo T.T., Kuok Q.Y., Hsu J.L., Sung S.Y. ADAM9 promotes lung cancer metastases to brain by a plasminogen activator-based pathway. Cancer Res. 2014;74:5229–5243. doi: 10.1158/0008-5472.CAN-13-2995. [DOI] [PubMed] [Google Scholar]
- 36.Hsu K.W., Wang A.M., Ping Y.H., Huang K.H., Huang T.T., Lee H.C., Lo S.S., Chi C.W., Yeh T.S. Downregulation of tumor suppressor MBP-1 by microRNA-363 in gastric carcinogenesis. Carcinogenesis. 2014;35:208–217. doi: 10.1093/carcin/bgt285. [DOI] [PubMed] [Google Scholar]
- 37.Hsu K.W., Hsieh R.H., Lee Y.H., Chao C.H., Wu K.J., Tseng M.J., Yeh T.S. The activated Notch1 receptor cooperates with alpha-enolase and MBP-1 in modulating c-myc activity. Mol. Cell. Biol. 2008;28:4829–4842. doi: 10.1128/MCB.00175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu K.W., Hsieh R.H., Wu C.W., Chi C.W., Lee Y.H., Kuo M.L., Wu K.J., Yeh T.S. MBP-1 suppresses growth and metastasis of gastric cancer cells through COX-2. Mol. Biol. Cell. 2009;20:5127–5137. doi: 10.1091/mbc.E09-05-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao W.R., Hsieh R.H., Hsu K.W., Wu M.Z., Tseng M.J., Mai R.T., Wu Lee Y.H., Yeh T.S. The CBF1-independent Notch1 signal pathway activates human c-myc expression partially via transcription factor YY1. Carcinogenesis. 2007;28:1867–1876. doi: 10.1093/carcin/bgm092. [DOI] [PubMed] [Google Scholar]
- 40.Wang A.M., Huang T.T., Hsu K.W., Huang K.H., Fang W.L., Yang M.H., Lo S.S., Chi C.W., Lin J.J., Yeh T.S. Yin Yang 1 is a target of microRNA-34 family and contributes to gastric carcinogenesis. Oncotarget. 2014;5:5002–5016. doi: 10.18632/oncotarget.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu K.W., Hsieh R.H., Huang K.H., Fen-Yau Li A., Chi C.W., Wang T.Y., Tseng M.J., Wu K.J., Yeh T.S. Activation of the Notch1/STAT3/Twist signaling axis promotes gastric cancer progression. Carcinogenesis. 2012;33:1459–1467. doi: 10.1093/carcin/bgs165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.