Abstract
Glucose metabolism reprogramming is one of the hallmarks of cancer cells, although functional and regulatory mechanisms of long noncoding RNA (lncRNA) in the contribution of glucose metabolism in lung adenocarcinoma (LUAD) remain incompletely understood. The aim of this study was to uncover the role of GAS6-AS1 in the regulation of progression and glucose metabolism in LUAD. We discovered that overexpression of GAS6-AS1 suppressed tumor progression of LUAD both in vitro and in vivo. Metabolism-related assays revealed that GAS6-AS1 inhibited glucose metabolism reprogramming. Mechanically, GAS6-AS1 was found to repress the expression of glucose transporter GLUT1, a key regulator of glucose metabolism. Ectopic expression of GLUT1 restored the inhibition effect of GAS6-AS1 on cancer progression and glucose metabolism reprogramming. Further investigation identified that GAS6-AS1 directly interacted with transcription factor E2F1 and suppressed E2F1-mediated transcription of GLUT1, and GAS6-AS1 was downregulated in LUAD tissues and correlated with clinicopathological characteristics and survival of patients. Taken together, our results identified GAS6-AS1 as a novel tumor suppressor in LUAD and unraveled its underlying molecular mechanism in reprogramming glucose metabolism. GAS6-AS1 potentially may serve as a prognostic marker and therapeutic target in LUAD.
Keywords: GAS6-AS1, lung adenocarcinoma, glucose metabolism, E2F1, GLUT1
Graphical abstract
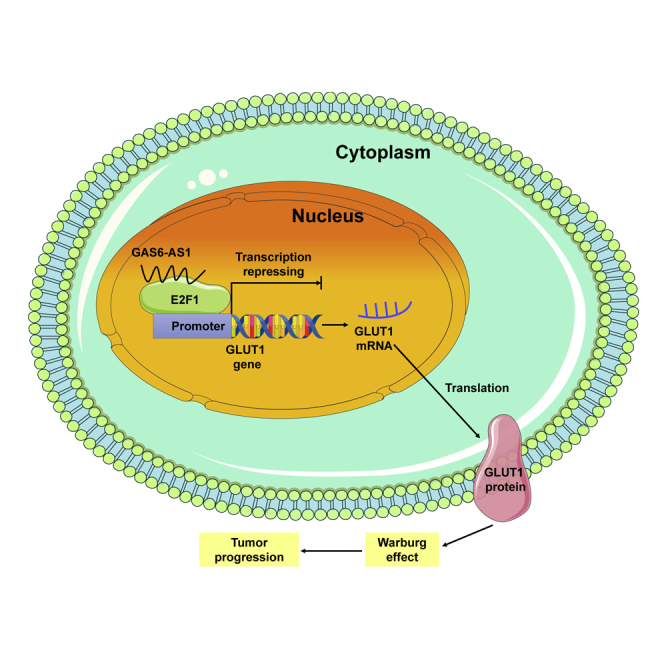
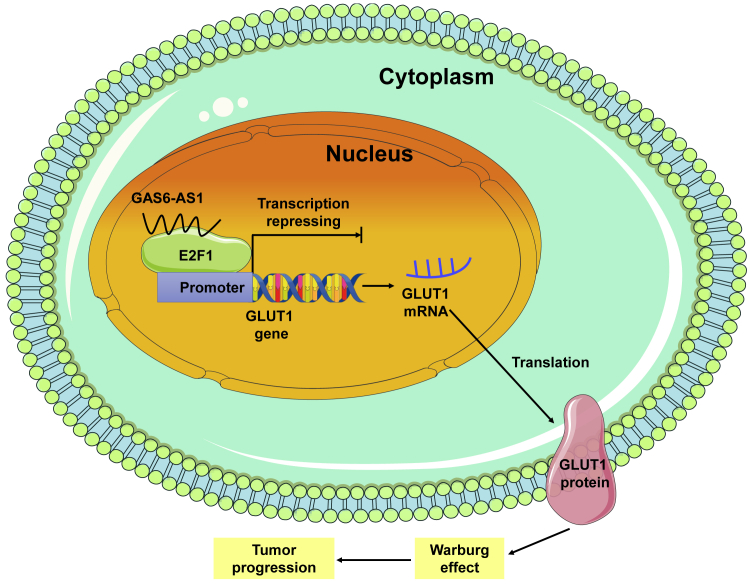
In this study, we identified lncRNA GAS6-AS1 as a suppressor in tumor progression and glucose metabolism reprogramming of LUAD. GAS6-AS1 exerted its biological role through binding with E2F1 and suppressing E2F1-mediated transcription of glucose transporter GLUT1.
Introduction
The rapid proliferation and growth of solid tumors lead to a shortage of energy supply and oxygen deprivation. Glucose is the major energy source to support tumor growth and can also provide a carbon source for biosynthetic reactions.1 Cancer cells exhibit dramatic aberration of glucose metabolism, displaying activation of glycolysis despite the presence of oxygen, also known as the “Warburg effect.”2 In glycolysis, cancer cells produce adenosine triphosphate (ATP) through pyruvate transformation to lactic acid in the cytosol, being characterized by more uptake of glucose, more production of lactate and pyruvate, and an extracellular acidic pH.3 Recently, glucose metabolism reprogramming has been recognized as a hallmark of cancer, and enhanced glycolytic effect has been proven to support the survival and growth of cancer cells.4 Given the crucial role played by glucose metabolism in cancer biology, it is necessary to identify key molecules or pathways that reprogram this process.
Long noncoding RNAs (lncRNAs) represent a large class of transcripts > 200 nucleotides with no protein coding capability.5 Current studies have uncovered the vital roles of lncRNAs in transcription, genomic imprinting, messenger RNA degradation, translation, protein kinetics, and function as RNA decoys or scaffolds,6 and increasing evidence demonstrates that lncRNAs function in the pathogenesis of cancers with diverse mechanisms.7,8 However, the potential role and mechanism of lncRNAs in the regulation of cancer metabolism remain largely unknown.
Lung cancer is the most common malignancy and the leading cause of cancer-related death worldwide.9 The two main groups of lung cancer are small-cell lung cancer (SCLC, 15%–20% cases) and non-small cell lung cancer (NSCLC, 80%–85% cases). Lung adenocarcinoma (LUAD) represents the most common histological subtype of NSCLC, accounting for ∼40%.10 Multiple studies have confirmed the enhancement of glycolysis in LUAD,11, 12, 13 but the specific mechanisms of glucose metabolism reprogramming in LUAD need to be further teased out. The aim of this study was to probe metabolism-related lncRNAs in LUAD and to study its functional mechanism. Here, we identified GAS6-AS1 as a metabolism-regulating lncRNA that was dysregulated in LUAD tissues and meanwhile in glucose-starved LUAD cells. Mechanistically, GAS6-AS1 exerted its effect on progression and glucose metabolism via repressing E2F1-mediated transcription of glucose transporter 1 (GLUT1). GLUT1, a pivotal rate-limiting element in the transport of glucose in malignancy cells, has been extensively documented to regulate glucose metabolism.14 Our results indicate that the GAS6-AS1/E2F1/GLUT1 axis may be a promising metabolism target for antitumor therapy.
Results
GAS6-AS1 suppressed tumor progression in LUAD
We analyzed a GEO dataset (GEO: GSE56843) and a GEPIA dataset to probe metabolism-related lncRNAs in LUAD. A microarray analysis of GEO: GSE56843 was performed to compare the gene expression profiles of A549 cells cultured with glucose and without glucose. A total of 4,245 dysregulated genes were detected in LUAD cells induced by glucose starvation (screening criterion: adj. p < 0.001, p < 0.001, |LogFC| > 1). The GEPIA dataset was searched to seek differentially expressed genes (screening criterion: |Log2FC| > 1, q < 0.01) and differential survival genes (screening criterion: group cutoff: median, p < 0.01, top 500) in LUAD. Finally, 13 genes were obtained by overlapping these results and two lncRNAs (LINC-PINT, GAS6-AS1) were included (Figure S1A). The expression profiles of 13 candidate genes in GEO: GSE56843 and GEPIA datasets are shown in Figures S1B and S1C. LINC-PINT has been reported to function as a tumor suppressor that exerts important regulatory roles in NSCLC progression by sponging miR-208a-3p/PDCD4.15 The role of GAS6-AS1 in lung cancer has not been identified, and we selected it as our research subject.
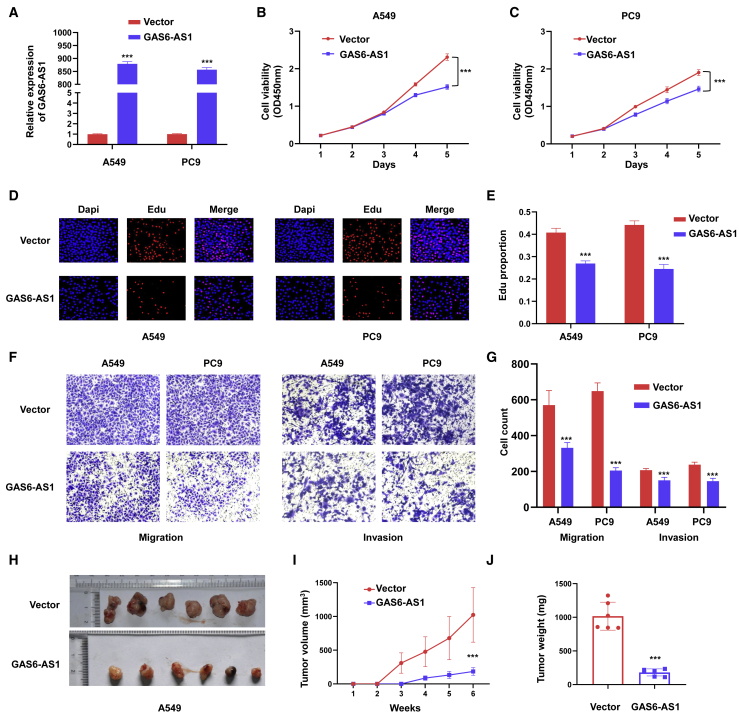
To elucidate the function of GAS6-AS1 in LUAD, plasmids were transfected into LUAD cells to overexpress GAS6-AS1. Confirmed by qRT-PCR, expression plasmids effectively manipulated the expression of GAS6-AS1 in A549 and PC9 cells (Figure 1A). Cell proliferation was analyzed with CCK8 and 5-ethynyl-2′-deoxyuridine (EdU) staining assays, and the results showed that cell proliferative capability was remarkably attenuated by overexpression of GAS6-AS1 (Figures 1B–1E). Transwell and Matrigel assays were used to test the migration and invasion ability of LUAD cells, respectively. The results indicated that ectopic expression of GAS6-AS1 inhibited the migration and invasion of A549 cells and PC9 cells (Figures 1F and 1G). In addition, knockdown of GAS6-AS1 promoted proliferation, migration, and invasion of A549 and PC9 cells (Figures S2A–S2E). A xenograft tumor model was used to evaluate the tumor-suppressive role of GAS6-AS1 in vivo. Empty vector (n = 6)- and GAS6-AS1 (n = 6)-transfected cells were injected subcutaneously in axilla of nude mice. Tumor nodules were resected 6 weeks after injection, and the volume and weight of tumors were recorded (Figure 1H). As shown in Figures 1I and 1J, tumors of the GAS6-AS1-transfected group grew slowly, with smaller size and lighter weight. These results suggested that GAS6-AS1 exerted tumor-suppressive roles in LUAD both in vitro and in vivo.
Figure 1.
GAS6-AS1 functioned as a tumor suppressor in LUAD
(A) GAS6-AS1 was effectively upregulated in A549 and PC9 cells after transfection of overexpression plasmid of GAS6-AS1. (B and C) CCK8 assays were used to detect cell proliferation of A549 cells (B) and PC9 cells (C) after ectopic expression of GAS6-AS1. (D and E) EdU assays were performed in A549 (D) and PC9 cells (E) after ectopic expression of GAS6-AS1. (F and G) Transwell (F) and Matrigel (G) assays were used to test the migration and invasion ability of LUAD cells after ectopic expression of GAS6-AS1, respectively. (H) Images of xenograft tumors derived from nude mice bearing A549 cells of different groups. (I) Tumor volume was measured every week after injection. (J) Tumor weight was measured after resection of xenograft tumors.
GAS6-AS1 impaired glucose metabolism reprogramming in LUAD cells
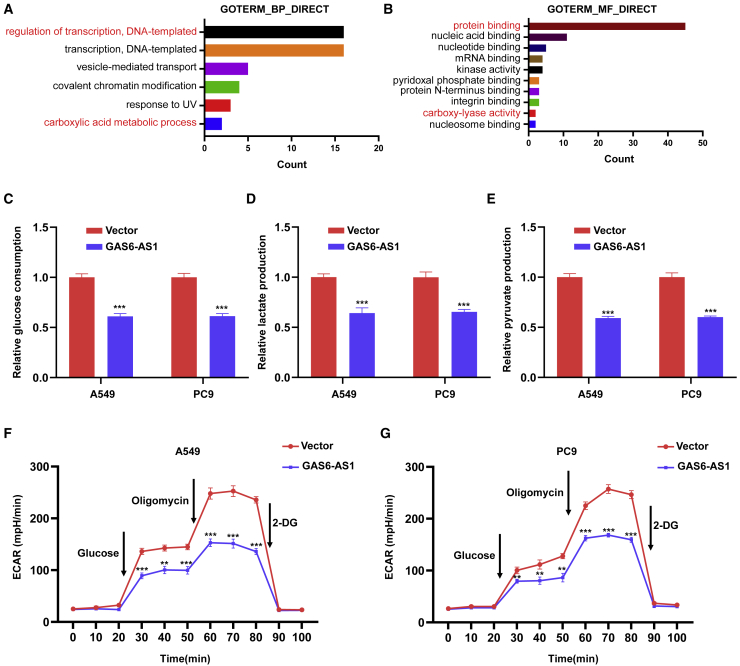
Consistently with the data in Figure S1B, GAS6-AS1 was downregulated by glucose starvation in A549 and PC9 cells (Figure S3A). Ectopic expression of GAS6-AS1 inhibited proliferation, migration, and invasion of LUAD cells in both glucose-sufficient and glucose-free conditions (Figures S3B–S3G). It was reported in 1988 that glucose starvation is required for insulin stimulation of glucose uptake and metabolism in cultured microvascular endothelial cells.16 Moreover, cell growth and survival depend on a delicate balance between energy production and synthesis of metabolites, and energy stress is closely linked to metabolic alterations.17 In tumor cells, glucose metabolism reprogramming will appear during energy stress to guarantee the energy supply and survival, manifested by cancer cells taking up more glucose and producing more lactate and pyruvate (Warburg effect).18 We performed Gene_Ontology enrichment analysis on genes highly co-expressed with GAS6-AS1, and the results suggested that these genes might participate in the biological processes of “regulation of transcription, DNA-templated” and “carboxylic acid metabolic process” (Figure 2A) and the molecular functions of “protein binding” and “carboxy-lyase activity” (Figure 2B). Based on these theories and predictions, we intended to study the role of GAS6-AS1 in glucose metabolism reprogramming of LUAD cells. It turned out that ectopic expression of GAS6-AS1 inhibited glucose consumption of A549 and PC9 cells (Figure 2C), and the production of lactate and pyruvate in LUAD cells were decreased after overexpression of GAS6-AS1 (Figures 2D and 2E). In addition, Seahorse analysis revealed decreased extracellular acidification rate (ECAR) in A549 and PC9 cells by ectopic expression of GAS6-AS1 (Figures 2F and 2G). Moreover, knockdown of GAS6-AS1 promoted glucose consumption and lactate and pyruvate production and increased ECAR of A549 and PC9 cells (Figures S4A–S4E). These results implied that GAS6-AS1 could impair glucose metabolism reprogramming of LUAD cells.
Figure 2.
GAS6-AS1 suppressed glucose metabolism reprogramming in LUAD cells
(A) Terms of biological process with statistical significance were presented in Gene_Ontology enrichment analysis. (B) Terms of molecular function with statistical significance were presented in Gene_Ontology enrichment analysis. (C) Detection of glucose consumption in LUAD cells after overexpression of GAS6-AS1. (D and E) Detection of lactate (D) and pyruvate (E) production in LUAD cells after overexpression of GAS6-AS1. (F and G) ECARs were measured by Seahorse XF in LUAD cells after overexpression of GAS6-AS1.
GAS6-AS1 exerted tumor-suppressive roles via regulating expression of glucose transporter GLUT1
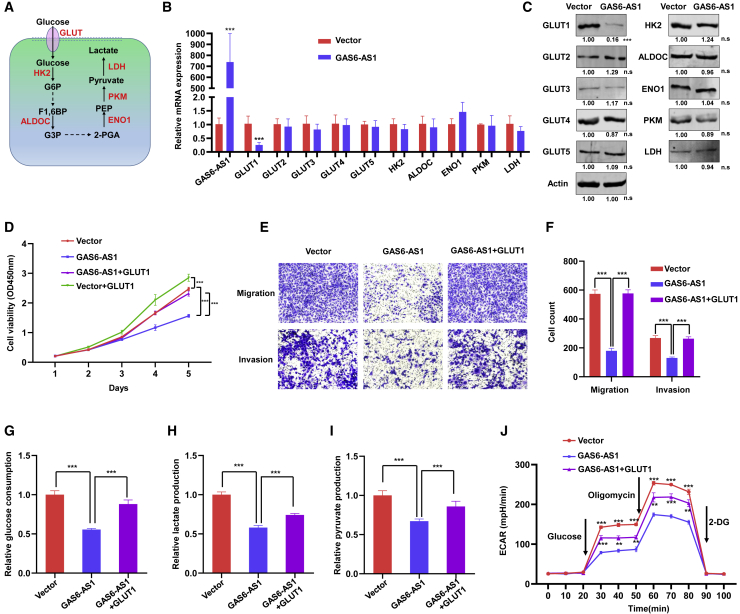
Glucose metabolism reprogramming of cancer cells is driven by a number of specific enzymes, including GLUT1–5 (glucose transporters 1–5, SLC2A1–5), HK2 (hexokinase 2), ALDOC (aldolase C), ENO1 (enolase 1), PKM (pyruvate kinase), and LDH (lactate dehydrogenase) (Figure 3A).19 Real-time RT-PCR and western blotting were performed to explore the influence of GAS6-AS1 on these metabolism-related enzymes. Results showed that the mRNA and protein levels of GLUT1 (also known as SLC2A1) were significantly downregulated after overexpression of GAS6-AS1 (Figures 3B and 3C). GLUT1 belongs to the glucose transporter family and regulates glucose transport across the cell membrane.20 Increasing studies indicate that GLUT1 participates in tumor progression and energy metabolism reprogramming in multiple cancer types.21, 22, 23 To verify whether the function of GAS6-AS1 was dependent on GLUT1, we performed rescue experiments by transfecting expression plasmids of GLUT1 into GAS6-AS1-overexpressing A549 cells. CCK8, Transwell, and Matrigel assays revealed that ectopic expression of GLUT1 (Figure S5A) restored the inhibition effect of GAS6-AS1 on proliferation, migration, and invasion (Figures 3D–3F). Furthermore, glucose consumption, lactate production, pyruvate production, and ECAR were detected. Consistently, ectopic expression of GLUT1 restored the inhibition effect of GAS6-AS1 on glucose metabolism reprogramming in A549 cells (Figures 3G–3J). These results suggested that increased GLUT1 expression dysregulated the GAS6-AS1-mediated effects on tumor suppression and glucose metabolism reprogramming.
Figure 3.
GAS6-AS1 inhibited progression and glucose metabolism reprogramming of LUAD cells via downregulating GLUT1
(A) Diagram of key enzymes that play vital roles in glucose metabolism reprogramming. (B) The mRNA expression of metabolism-related genes was measured by qRT-PCR after overexpression of GAS6-AS1 in A549 cells. (C) The protein expression of metabolism-related enzymes was measured by western blotting after overexpression of GAS6-AS1 in A549 cells. (D) Ectopic expression of GLUT1 restored the inhibition effect of GAS6-AS1 on proliferation in A549 cells. (E and F) Ectopic expression of GLUT1 restored the inhibition effect of GAS6-AS1 on migration and invasion in A549 cells. (G–J) Glucose consumption (G), lactate production (H), pyruvate production (I), and ECAR (J) were measured, and results indicated that ectopic expression of GLUT1 restored the inhibition effect of GAS6-AS1 on glucose metabolism reprogramming in A549 cells.
GAS6-AS1 interacted with E2F1
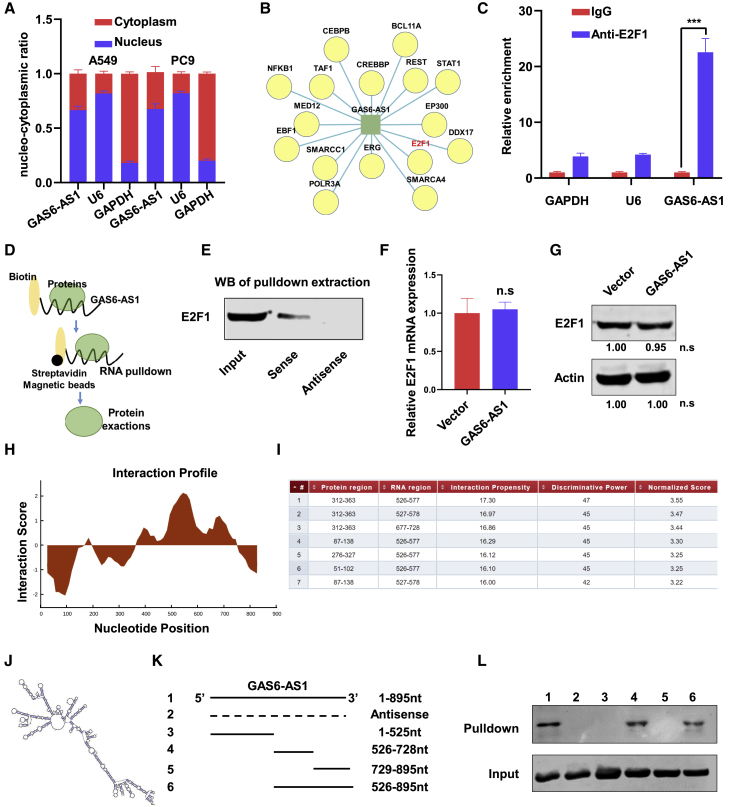
As the subcellular localization of lncRNAs is crucial for their functional biology, nucleo-cytoplasmic separation assays were performed, and the results showed that GAS6-AS1 was mainly located in the nucleus (Figure 4A). The nucleus is the place for transcription, and our previous analysis indicated that GAS6-AS1 might participate in the biological process of “regulation of transcription, DNA-templated” (Figure 2A). In recent years, multiple lncRNAs were found to regulate the transcription of target genes via binding with specific transcription factors.24, 25, 26 Through a search of the LncRNA Modulator Atlas in Pan-cancer (LncMAP) datasets, a cluster of transcription factors were predicted to interact with GAS6-AS1 (Figure 4B). Among these transcription factors, we noticed that E2F1 had been reported to enhance glycolysis and reprogram energy metabolism in cancer cells,27 and Albert Tauler and colleagues also found that E2F1 induced mTOR complex 1 activation to regulate glucose metabolism.28 Moreover, transcription chromatin immunoprecipitation sequencing (TF ChIP-seq) data of ChIPBase (http://rna.sysu.edu.cn/chipbase/) showed that GLUT1 (SLC2A1) gene was predicted as a direct target of E2F1 in HeLa-S3 cells (Figure S6A). We suspected that lncRNA GAS6-AS1 might regulate the expression of GLUT1 via interacting with transcription factor E2F1. To validate our hypothesis, RNA immunoprecipitation (RIP) assay was performed in A549 cells, and the results indicated E2F1 recruitment of the RNA fragment of GAS6-AS1 (Figure 4C). Western blot analysis of proteins extracted from GAS6-AS1 pull-down assay revealed that E2F1 protein specifically binds to the sense sequence of GAS6-AS1 but not to the antisense control (Figures 4D and 4E). Additionally, the mRNA and protein expression of E2F1 were not altered after overexpression of GAS6-AS1 (Figures 4F and 4G). Furthermore, we used catRAPID tools to analyze the interaction region of GAS6-AS1 with E2F1 (Figure 4H), and the results showed that E2F1 might bind to nucleotides 526–728 of GAS6-AS1 with high propensities (Figure 4I). Meanwhile, the secondary structure of GAS6-AS1 was predicted in the online database AnnoLnc (Figure 4J). We next constructed a series of GAS6-AS1 truncations to map its binding fragment with E2F1 (Figure 4K). Verified by RNA pull-down assays, a fragment of GAS6-AS1 (526–728 nt) was responsible for its interaction with E2F1 (Figure 4L). These data indicated that GAS6-AS1 specifically bound to transcription factor E2F1.
Figure 4.
GAS6-AS1 interacted with transcription factor E2F1
(A) RNA expression was measured by qRT-PCR after nuclear and cytosolic separation. U6 small nuclear RNA (U6) was used as a nucleus marker, and glyceraldehyde-phosphate dehydrogenase (GAPDH) was used as a cytosol marker. (B) Transcription factors that are predicted to interact with GAS6-AS1 by LncMAP. (C) RIP experiments were performed in A549 cells with E2F1 antibody, and IgG was used as an internal reference. (D) Schematic diagram of RNA pull-down assay. (E) Lysates from A549 cells were used for RNA pull-down with biotinylated GAS6-AS1 transcript or antisense transcript. Western blotting demonstrated the specific association of GAS6-AS1 with E2F1. (F and G) qRT-PCR (F) and western blotting (G) were performed to detect the mRNA and protein expression of E2F1 after overexpression of GAS6-AS1. (H and I) The predicted interaction profile (H) and binding region (I) of GAS6-AS1 with E2F1 are shown. (J) The secondary structure of GAS6-AS1 in AnnoLnc is presented. (K) A series of GAS6-AS1 deletion mapping fragments were constructed, and RNA pull-down assays were performed. (L) Western blotting analysis was performed with the proteins retrieved from pull-down assays, and input was used as a positive control.
GAS6-AS1 suppressed E2F1-mediated transcription of GLUT1
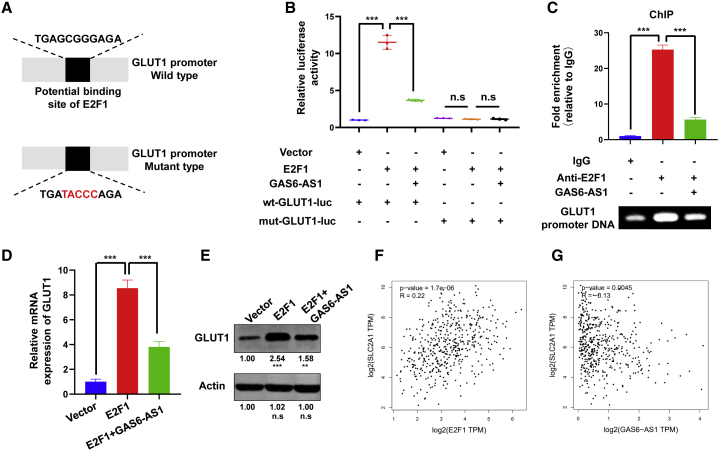
Transcription factors regulated the expression of target genes through binding with specific DNA sequences. Predicted by the JASPAR database, we found that E2F1 potentially bound to the promoter region of GLUT1 (Figure S6B). To prove that GLUT1 is a transcriptional target of E2F1 and the regulation can be repressed by GAS6-AS1, luciferase vectors consisting of wild-type (WT) or mutant (mut) GLUT1 promoters were constructed and transfected into A549 cells (Figure 5A). The luciferase assay revealed that overexpression of E2F1 (Figure S5B) stimulated the WT GLUT1 promoter activity, as indicated by increased luciferase activity, but had no effect on mut-type GLUT1 promoter activity. Moreover, GAS6-AS1 suppressed the increase of luciferase activity induced by E2F1 (Figure 5B). ChIP assay was also performed, and the results suggested that E2F1 was bound to GLUT1 promoter and the interaction was repressed by GAS6-AS1 (Figure 5C). In addition, the enforced expression of E2F1 significantly upregulated the mRNA and protein levels of GLUT1, which could also be restored by GAS6-AS1 (Figures 5D and 5E). These results suggested that E2F1 directly bound to promoter region of GLUT1 to activate its transcription and that GAS6-AS1 could inhibit the process. We checked the expression levels of GLUT1 and E2F1 in GEPIA and GSE56843 datasets. Results indicated that GLUT1 and E2F1 were both upregulated in LUAD tissues (Figures S5C and S5D), whereas the expression of GLUT1 and E2F1 were not altered under glucose-free conditions in LUAD cells (Figure S5E). In addition, by searching the GEPIA dataset, we found that the expression of E2F1 was positively correlated with GLUT1 in LUAD, whereas GAS6-AS1 was negatively correlated with GLUT1 (Figures 5F and 5G).
Figure 5.
GAS6-AS1 repressed E2F1-mediated transcription of GLUT1
(A) The potential binding site of E2F1 with GLUT1 promoter was shown, and the wild-type or mutant-type luciferase vectors were constructed. (B) Luciferase activity was assayed in A549 cells transfected with luciferase vectors (wild type or mutant type) and meantime co-transfected with expression plasmids (empty vectors, E2F1 expression plasmids, or GAS6-AS1 expression plasmids). (C) ChIP experiments of E2F1 (IgG as an internal control) were performed, and the co-precipitated DNA was subjected to PCR amplification with primers specific to GLUT1 promoter region. (D and E) The level of GLUT1 under ectopic expression of E2F1 or GAS6-AS1 was detected by qRT-PCR (D) and western blotting (E). (F and G) The correlation between E2F1 (F) and GAS6-AS1 (G) with GLUT1 in LUAD tissues is presented.
GAS6-AS1 was clinically relevant in LUAD
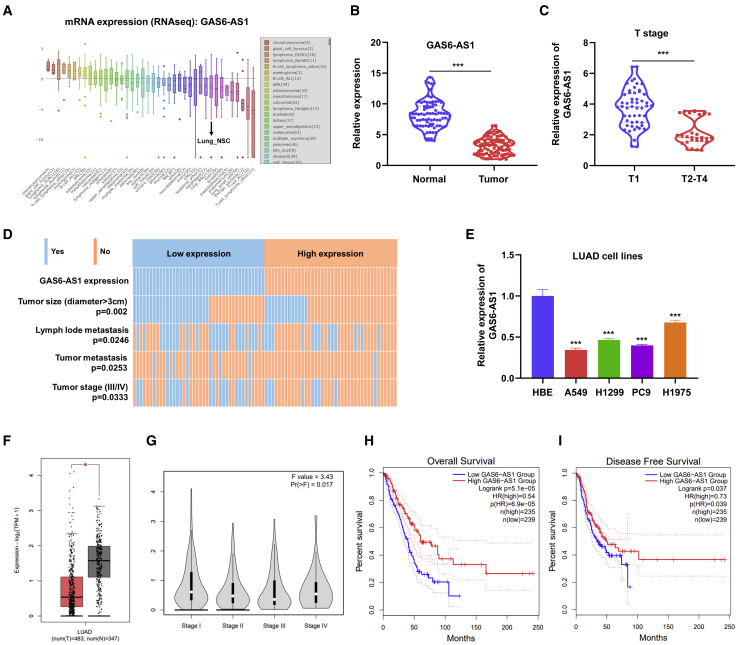
To confirm the expression pattern of GAS6-AS1 in LUAD, we first searched the CCLE database, and the results showed that the expression of GAS6-AS1 in NSCLC cells (Lung_NSC) was at a relatively low level (Figure 6A). Furthermore, we performed qRT-PCR assay on 80 paired LUAD tissues and adjacent normal tissues and found that GAS6-AS1 was significantly lowly expressed in LUAD tissues (Figure 6B). Moreover, patients of advanced T stages (T2–T4) exerted lower expression of GAS6-AS1 than patients of early T stages (T1) (Figure 6C). Additionally, we divided the 80 LUAD samples into a GAS6-AS1 low expression group (40 cases) and a high expression group (40 cases) based on median GAS6-AS1 levels to investigate the clinical significance of GAS6-AS1 in LUAD. Results suggested that the expression of GAS6-AS1 correlated with tumor size, lymph node metastasis, cancer distant metastasis and TNM stage of LUAD tissues (Figure 6D; Table S1). The expression of GAS6-AS1 in common LUAD cell lines was also detected by qRT-PCR, and we found that GAS6-AS1 was lowly expressed in 4 LUAD cell lines compared with normal human bronchial epithelial (HBE) cells (Figure 6E). Furthermore, the GEPIA dataset was explored to analyze the clinical correlation of GAS6-AS1 in LUAD. Results showed that GAS6-AS1 was lowly expressed in LUAD tissues (Figure 6F), and the expression of GAS6-AS1 in LUAD correlated with tumor stage (Figure 6G), overall survival (Figure 6H), and disease-free survival (Figure 6I) of patients. Collectively, these results indicated that GAS6-AS1 was downregulated in LUAD tissues and correlated with tumor stages and survival of patients.
Figure 6.
GAS6-AS1 was downregulated in LUAD tissues and correlated with clinical characteristics of LUAD patients
(A) The expression of GAS6-AS1 in different cancer cells is shown from CCLE datasets. (B) GAS6-AS1 was downregulated in 80 LUAD tissues compared with paired adjacent normal tissues. (C) Patients of advanced T stages exerted lower expression of GAS6-AS1. (D) Expression of GAS6-AS1 correlated with tumor size, lymph node metastasis, cancer distant metastasis, and TNM stage of LUAD. (E) GAS6-AS1 was lowly expressed in LUAD cells (A549, H1299, PC9, H1965) compared with HBE cells. (F) GAS6-AS1 was lowly expressed in LUAD tissues in the GEPIA dataset. (G–I) The expression of GAS6-AS1 in LUAD correlated with tumor stage (G), overall survival (H), and disease free (I) survival of patients in the GEPIA dataset.
Discussion
Increased aerobic glycolysis even in the abundance of oxygen, or the “Warburg effect,” has been recognized as an emerging hallmark of cancer. As distinguished from healthy cells, cancer cells exert glucose metabolism remodeling, characterized by consumption of glucose and excretion of lactate at a significantly higher rate and synthesis of ATP by producing lactic acid as the end product.29 These metabolic changes benefit cancer cells in multiple aspects. The switch to oxygen-independent glycolysis makes cancer cells more resistant to hypoxia conditions associated with tumor growth, and the glycolytic intermediates fuel several biosynthetic pathways that produce de novo nucleotides, lipids, amino acids, and NADPH. Moreover, the increased production of lactic acid lowers the pH of the extracellular microenvironment and supports the invasion and metastasis of cancer cells.30 The pivotal molecular mechanisms for the initiation and development of metabolic remodeling still remain unclear. Targeting cancer cells from the point of glucose metabolism might provide attractive and effective therapeutic approaches.
Lung cancer is the leading cause of cancer-related death, with limited curative treatment options, and extensive studies have revealed the reprogramming of glucose metabolism in lung cancer.31,32 The alterations of oncogenes and tumor suppressor genes, leading to activation or inhibition of signaling pathways and transcriptional networks, were reported to participate in the glucose metabolism reprogramming of lung cancer.33 However, the potential involvement of lncRNAs is poorly defined in glucose metabolism reprogramming of lung cancer. In our study, utilizing publicly available lncRNA expression profiling data in A549 cells under glucose starvation and integrating analyses of GEPIA data, we screened the metabolism-related lncRNA GAS6-AS1 in LUAD. GAS6-AS1, which is located at chromosome 13q34, is an antisense transcript of GAS6. Previous research revealed that low expression of GAS6-AS1 predicted a poor prognosis in patients with NSCLC.34 In our study, we further found that the expression of GAS6-AS1 was frequently downregulated in LUAD tissues, and clinical analysis reveals that downregulation of GAS6-AS1 correlated with advanced tumor stage and poor prognosis of LUAD patients. Moreover, ectopic expression of GAS6-AS1 inhibited tumor growth and glucose metabolism remodeling in LUAD cells.
GAS6-AS1 has been reported to facilitate the progression of breast cancer by targeting the miR-324-3p/SETD1A axis,35 and in gastric cancer and hepatocellular carcinoma GAS6-AS1 was identified to promote oncogenicity.36, 37 This suggested that the roles of GAS6-AS1 in the development of diverse cancer types were different.
The reprogramming of cancer metabolism is driven by several key enzymes. GLUTs are membrane proteins that transport glucose from the capillaries into cells and mediate the first step for cellular glucose usage. Five main classical GLUTs (GLUT1, GLUT2, GLUT3, GLUT4, GLUT5) with different affinities for glucose have been reported to function in glucose uptake for glycolysis of cancers.38 The phosphorylation of glucose, a crucial step in cellular metabolism, is catalyzed by HK2, and HK2 is considered the hub in the regulation of cancer cell glycolysis.39 ALDOC is a glycolytic enzyme that catalyzes the reversible aldol cleavage of fructose-1,6-biphosphate (F1,6BP) to glyceraldehyde-3-phosphate (G3P).40 ENO1 transforms 2-phospho-d-glycerate (2-PGA) into phosphoenolpyruvate (PEP) during glycolysis,41 followed by PKM transferring of a phosphoryl group from PEP to ADP, generating ATP and pyruvate.42 And LDH is responsible for the conversion of l-lactate and NAD to pyruvate and NADH in the final step of anaerobic glycolysis.43 We performed qRT-PCR and western blotting to detect the influence of GAS6-AS1 on these enzymes, and the results indicated that GAS6-AS1 remarkably decreased the expression of GLUT1 at both mRNA and protein levels. GLUT1 (also known as solute carrier family 2 member 1, SLC2A1) is a key rate-limiting factor in the transport of glucose in cancer cells and plays a critical role in glycolysis and tumor progression in multiple cancer types.44, 45, 46 Numerous lncRNAs have been proved to regulate GLUT1 in various ways. HOX transcript antisense RNA (HOTAIR) influences glucose metabolism through GLUT1 upregulation via mammalian target of rapamycin signaling.47 lncRNA-p23154 promotes glycolysis in oral squamous cell carcinoma by binding to the promoter region of miR-378a-3p, which represses GLUT1 expression by targeting to its 3′ UTR directly.48 SLC2A1-AS1 regulates aerobic glycolysis and progression in hepatocellular carcinoma by competitively binding to STAT3 and resulting in inactivation of the FOXM1/GLUT1 axis.49 Consistent with these studies, we showed that GAS6-AS1 exerted its role in tumor progression and glucose metabolism in LUAD by downregulating GLUT1.
Mechanistic investigations revealed that GAS6-AS1 could bind to transcription factor E2F1 but not affect E2F1 expression. Previous studies have shown that E2F1 coactivated by KDM4A regulates the pyruvate dehydrogenase kinase-dependent metabolic switch between mitochondrial oxidation and glycolysis.50 In addition, E2F1 was reported to significantly enhance glucose uptake and lactate production in bladder and prostate cancer cell lines.27 lncRNAs have been identified to function as master regulators for gene expression in many ways, and one is to interact with transcription factor and thus transcriptionally regulate downstream genes.51 For instance, lncRNA CAIF directly binds to p53 protein and blocks p53-mediated myocardin transcription in cardiomyocyte autophagy,52 and PAXIP1-AS1 facilitates cell invasion and angiogenesis of glioma by recruiting transcription factor ETS1 to upregulate KIF14 expression.53 Likewise, our results suggested that GAS6-AS1 integrated with E2F1 and suppressed E2F1-mediated transcription of GLUT1. The protein function domains of E2F1 (the conserved domains that E2F1 binds with DNA promoter to function as a transcription factor) were protein regions 206–299 and 129–192 (Figure S5F), and in our study, GAS6-AS1 was predicted to bind with E2F1 in protein regions 312–363, 87–138, 276–327, and 51–102 (Figure 4I). This indicated that they shared some common binding motif (region 129–138 and region 276–299). We speculated that GAS6-AS1 decreased the interaction of E2F1 with GLUT1 promotor by competitive binding.
Taken together, our results identified lncRNA GAS6-AS1 as a suppressor in tumor progression and glucose metabolism reprogramming of LUAD. The downregulation of GAS6-AS1 correlated with tumor stage and overall survival of LUAD patients, and GAS6-AS1 exerted its biological role through binding with E2F1 and suppressing the E2F1-mediated transcription of glucose transporter GLUT1 (Figure 7). Therefore, GAS6-AS1 may serve as a potential diagnostic and prognostic biomarker for LUAD patients, and targeting the GAS6-AS1/E2F1/GLUT1 axis may present new perspectives in the prevention or treatment of LUAD.
Figure 7.
Schematic illustration of GAS6-AS1 in inhibition of glycolytic metabolism and progression of LUAD
Materials and methods
Bioinformatics
GEO microarray dataset GEO: GSE56843 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56843) and a GEPIA dataset (http://gepia2.cancer-pku.cn/#index)54 were used to screen metabolism-related lncRNAs in LUAD. Gene clusters (Table S2) highly co-expressed with GAS6-AS1 (Pearson correlation coefficient ≥ 0.35) were submitted to DAVID Bioinformatics Resources 6.8 (http://david.abcc.ncifcrf.gov/)55 for Gene_Ontology enrichment analysis. GO terms with statistical significance (p < 0.05) were presented. LncMAP56 was browsed to seek for transcription factors potentially interacting with GAS6-AS1. The secondary structure of GAS6-AS1 was predicted in AnnoLnc (http://annolnc.gao-lab.org/),57 and catRAPID (http://s.tartaglialab.com/page/catrapid_group)58 was explored to estimate the binding propensity of GAS6-AS1 with E2F1. The JASPAR database (http://jaspardev.genereg.net/)59 was used to evaluate the transcription factor binding profiles with gene promoters. The CCLE database (https://portals.broadinstitute.org/ccle/about) was searched to verify the expression of GAS6-AS1 in different cancer cell lines.
Patient specimens
Eighty paired LUAD and adjacent non-tumor tissue samples were obtained from patients undergoing operations in the Department of Thoracic Surgery, Jiangsu Cancer Hospital, from 2015 to 2018. None of the patients was previously treated with radiotherapy, chemotherapy, or immunotherapy. All LUAD tissues were confirmed by the histopathological reports. The study was approved by the ethics committee of Jiangsu Cancer Hospital, and written informed consent was obtained from all participants.
Cell culture
HBE and LUAD cell lines (A549, H1299, PC9, and H1975) were purchased from Shanghai Institutes for Biological Science (Shanghai, China). HBE and H1975 were cultivated in DMEM (KeyGen, Nanjing, China), and A549, H1299, and PC9 cells were cultivated in RPMI-1640 (KeyGen, Nanjing, China), all supplemented with 10% fetal bovine serum (FBS) (KeyGen, Nanjing, China) and cultured in a standard 37°C incubator. For glucose starvation experiments, cells were cultured in medium with different concentrations of glucose.
RNA extraction and qRT-PCR
RNA was extracted from tissue samples or cultured cells with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The PrimeScript RT Master Mix Perfect Real Time Kit (Takara, Nanjing, China) was used to synthesize cDNA, and qRT-PCR was performed with SYBR Select Master Mix (Takara, Nanjing, China). The expression levels of mRNA were normalized to ACTIN as reference genes. Primers used in this study are shown in Table S3.
Transfection
Human GAS6-AS1, GLUT1, and E2F1 cDNA were PCR-amplified and then cloned into the expression vector pcDND3.1 (RiboBio, Guangzhou, China). Transfection was carried out with Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, and empty vector plasmid was used as negative control. For experiments in vivo, GAS6-AS1 expression plasmids were packaged with pPACKH1 Lentivector Packaging Plasmid Mix (System Biosciences, Palo Alto, CA, USA) and were introduced into 293T cells. The liquid supernatant was transected into A549 cells, and stable transfected cells were positively selected with puromycin (Thermo Fisher Scientific, Shanghai, China) for 4–6 weeks.
Cell proliferation assays
Cell proliferation assays were performed 24 h after transfection. For CCK8 viability assay, cells were seeded at a density of 2,000 cells/well in a 96-well plate. 10% CCK8 (Beyotime, Suzhou, China) was added to the well, and the absorbance at 450 nm was measured every 24 h. For EdU assay, 8,000 cells/100 μL were plated in 96-well plates. One hundred microliters of 50 μM EdU solution (EdU Apollo488 In Vitro Imaging Kit, RiboBio, Guangzhou, China) was incubated with cells. EdU was stained with red fluorescence, and images were photographed by fluorescence microscope to count the proportion of proliferating cells.
Migration and invasion assays
The migration and invasion assays were conducted with Transwell assay inserts (8 μm PET, 24-well Millicell) and Matrigel-coated inserts (BD Biosciences, Bedford, MA, USA), respectively. The upper chambers of the inserts were filled with 200 μL of serum-free medium, and the lower chambers were filled with 800 μL of medium containing 10% FBS. A total of 5 × 104 cells were added to the upper chambers and incubated for 24 h (migration) or 48 h (invasion). The migrated or invaded cells in the lower chambers were stained with crystal violet and counted.
Tumor xenograft experiment
The procedures for animal care and use were approved by Nanjing Medical University Animal Care Committee. Ten female nude mice (4–6 weeks old) were purchased from Nanjing Medical University School of Medicine’s accredited animal facility and randomly divided into two groups. A549 cells stably expressing GAS6-AS1 or empty vector were injected subcutaneously in axilla of the mice. The volume of xenografts was measured every week with calipers ([length × width2]/2). All mice were euthanized 6 weeks after injection, and tumor xenografts were harvested and weighed.
Glucose uptake
Glucose uptake was measured with the Glucose Uptake Assay Kit (Abcam, Cambridge, UK). Cells were seeded in a 96-well plate at a density of 2,000 cells/well and were starved in 100 μL of serum-free medium overnight to increase glucose uptake. 2-Deoxyglucose (2-DG) was added to cells and incubated for 20 min at 37°C. Exogenous 2-DG was removed by washing cells with PBS, and cells were lysed with extraction buffer. The uptake of 2-DG was measured by colorimetric assay, following the manufacturer’s guidelines.
Lactate and pyruvate measurement
Lactate and pyruvate levels of cell supernatant were measured with an l-Lactate Assay Kit (Abcam, Cambridge, UK) and a Pyruvate Assay Kit (Abcam, Cambridge, UK), respectively, according to the manufacturer’s instructions. The final results were normalized by total protein concentration.
ECAR
The Seahorse XF 24 Extracellular Flux Analyzer (Agilent, Beijing, China) was used to monitor ECAR of cells. Briefly, cells were seeded in a XF 24-well plate at a density of 1 × 104 per well and allowed to attach overnight. After serum starvation for 24 h, ECAR was detected with a Seahorse XF Glycolysis Stress Test kit (Agilent, Beijing, China) according to the manufacturer’s instructions. The measurements were normalized to mitochondrial DNA.
Western blotting
Cells were lysed in radio immunoprecipitation assay buffer (KeyGen, Nanjing, China) containing protease inhibitor PMSF (KeyGen, Nanjing, China) and analyzed for total protein concentration. Western blotting was obtained utilizing 20–40 μg of lysate protein according to the standard protocol. The following antibodies were used in this study: GLUT1 (Abcam, ab652, 1:1,000 dilution), GLUT2 (Abcam, ab54460, 1:1,000 dilution), GLUT3 (Abcam, ab41525, 1:1,000 dilution), GLUT4 (Abcam, ab654, 1:1,000 dilution), GLUT5 (Abcam, ab36057, 1:1,000 dilution), HK2 (Cell Signaling Technology, 2867, 1:1,000 dilution), ALDOC (Abcam, ab87122, 1:1,000 dilution), ENO1 (Proteintech, 11204-1-AP, 1:1,000 dilution), PKM (Abcam, ab38237, 1:1,000 dilution), LDH (Cell Signaling Technology, 3582, 1:1,000 dilution), ACTIN (Abcam, ab15265, 1:1,000 dilution), and E2F1 (Santa Cruz Biotechnology, sc-251, 1:1,000 dilution).
Subcellular fractionation analysis
Subcellular isolation of RNA was performed with a PARIS Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The isolated RNA was purified and further analyzed by qRT-PCR. glyceraldehyde-phosphate dehydrogenase and U6 small nuclear RNA were used as cytoplasmic and nuclear controls, respectively.
RIP
RIP assay was carried out with a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. Cells were lysed in the presence of protease and RNase inhibitors. Magnetic beads were then pre-incubated with IgG (Santa Cruz Biotechnology, sc-2025) or anti-E2F1 (Santa Cruz Biotechnology, sc-251). The prepared cell lysates underwent immunoprecipitation at 4°C overnight, and the co-precipitated RNAs were detected by qRT-PCR.
Biotin-RNA pull-down assay
The mMESSAGE mMACHINE T7 Transcription Kit (Invitrogen, Carlsbad, CA, USA) was used for translation assays in vitro. Then, the targeted RNA was biotin-labeled with desthiobiotinylation using the Pierce RNA 3′ End Desthiobiotinylation Kit (Invitrogen, California, USA). Cells were collected and lysed by protein lysis buffer, and streptavidin magnetic beads were used to capture the biotin-labeled RNA probe. RNA pull-down assays were performed with the Pierce Magnetic RNA-Protein Pull-Down Kit (Invitrogen, California, USA) according to the manufacturer’s instructions. The pull-down extractions were eluted and subjected to western blotting analysis.
Luciferase reporter assay
GLUT1 promoter sequences (−2,000 bp upstream of start codon) containing E2F1 putative binding site (WT) and mut site (mut type) were designed. The target fragments were inserted into firefly luciferase plasmids by restriction endonuclease digestion using SpeI and Hind III and ligated with T4 DNA ligase. Overexpression constructs (empty vectors, E2F1, or GAS6-AS1) were co-transfected with WT-GLUT1-luc and mut-GLUT1-luc, respectively, and a plasmid carrying Renilla luciferase was used as an internal reference. The cells were harvested to detect luciferase activity with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) 24 h after transfection. The ratio of firefly luciferase to Renilla activity was calculated to measure luciferase activity.
ChIP
ChIP was performed with a ChIP assay kit (Beyotime, Shanghai, China) according to the manufacturer’s protocol. Briefly, A549 cells were cross-linked in 4% paraformaldehyde, and DNA was sheared by sonication to yield soluble chromatin. Cell lysates were incubated with protein A/G beads coated with anti-E2F1 (Santa Cruz Biotechnology, sc-251) or anti-mouse IgG (Santa Cruz Biotechnology, sc-2025). Immunoprecipitation was conducted at 4°C overnight. Cross-linked DNA was eluted from the protein-DNA complex and further subjected to qRT-PCR.
Statistical analysis
Statistical analysis was performed with SPSS 22.0 software and GraphPad Prism 8.0. All values are presented as means ± standard deviation (SD). Student’s t test was used to determine statistical differences between two groups. The chi-square test was used to analyze the relationship between GAS6-AS1 expression and clinicopathological characteristics. All the repetitive experiments were repeated three times,and p < 0.05 was considered statistically significant (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0. 001; n.s., no significance).
Declarations
Ethics approval and consent to participate
The study was approved by the ethics committee of Jiangsu Cancer Hospital, and written informed consent was obtained from all participants.
Consent for publication
Consent for publication has been obtained from all authors.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
The authors thank NanJing XinJia Medical Technology Co. Ltd for providing technical platform support. This research was supported by National Natural Science Foundation for Youth of China (no. 81902354); the National Natural Science Foundation of China (81672869); Jiangsu Provincial Medical Outstanding Talent (to L.X.); Jiangsu Provincial Medical Youth Talent (QNRC2016657 to B.R.); the talents program of Jiangsu Cancer Hospital (YC201814); the “333” talent project (BRA2019325); Science and Technology program of Xu Zhou(no. KC18037); and Xuzhou Clinical Technology Key Research Project (2019GG021).
Author contributions
L.X., Y.S., and B.R. designed and supervised the study. J.L., H.W., and L.W. performed most of the experiments and wrote the manuscript. G.W. was responsible for statistical analysis. Y.Y. helped to conduct experiments in vivo. K.X. and X.L. performed some mechanistic experiments and edited the manuscript. All authors read and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.04.022.
Contributor Information
Lin Xu, Email: xulin83@njmu.edu.cn.
Yi Shen, Email: dryishen@nju.edu.cn.
Binhui Ren, Email: renbinhui@jszlyy.com.cn.
Supplemental information
References
- 1.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantor J.R., Sabatini D.M. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boroughs L.K., DeBerardinis R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khorkova O., Hsiao J., Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv. Drug Deliv. Rev. 2015;87:15–24. doi: 10.1016/j.addr.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charles Richard J.L., Eichhorn P.J.A. Platforms for Investigating LncRNA Functions. SLAS Technol. 2018;23:493–506. doi: 10.1177/2472630318780639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhan A., Soleimani M., Mandal S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J., Zhang X., Chen W., Hu X., Li J., Liu C. Regulatory roles of long noncoding RNAs implicated in cancer hallmarks. Int. J. Cancer. 2020;146:906–916. doi: 10.1002/ijc.32277. [DOI] [PubMed] [Google Scholar]
- 9.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 10.Herbst R.S., Heymach J.V., Lippman S.M. Lung cancer. N. Engl. J. Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan T.W., Lane A.N., Higashi R.M., Farag M.A., Gao H., Bousamra M., Miller D.M. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol. Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerr E.M., Martins C.P. Metabolic rewiring in mutant Kras lung cancer. FEBS J. 2018;285:28–41. doi: 10.1111/febs.14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jozwiak P., Lipinska A. [The role of glucose transporter 1 (GLUT1) in the diagnosis and therapy of tumors] Postepy Hig. Med. Dosw. 2012;66:165–174. doi: 10.5604/17322693.988242. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L., Hu J., Li J., Yang Q., Hao M., Bu L. Long noncoding RNA LINC-PINT inhibits non-small cell lung cancer progression through sponging miR-218-5p/PDCD4. Artif. Cells Nanomed. Biotechnol. 2019;47:1595–1602. doi: 10.1080/21691401.2019.1605371. [DOI] [PubMed] [Google Scholar]
- 16.Gerritsen M.E., Burke T.M., Allen L.A. Glucose starvation is required for insulin stimulation of glucose uptake and metabolism in cultured microvascular endothelial cells. Microvasc. Res. 1988;35:153–166. doi: 10.1016/0026-2862(88)90059-3. [DOI] [PubMed] [Google Scholar]
- 17.Bezawork-Geleta A., Wen H., Dong L., Yan B., Vider J., Boukalova S., Krobova L., Vanova K., Zobalova R., Sobol M. Alternative assembly of respiratory complex II connects energy stress to metabolic checkpoints. Nat. Commun. 2018;9:2221. doi: 10.1038/s41467-018-04603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., Huang Q., Long X., Guo X., Sun X., Jin X., Li Z., Ren T., Yuan P., Huang X. Mitochondrial elongation-mediated glucose metabolism reprogramming is essential for tumour cell survival during energy stress. Oncogene. 2017;36:4901–4912. doi: 10.1038/onc.2017.98. [DOI] [PubMed] [Google Scholar]
- 19.Doherty J.R., Cleveland J.L. Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng D., Xu C., Sun P., Wu J., Yan C., Hu M., Yan N. Crystal structure of the human glucose transporter GLUT1. Nature. 2014;510:121–125. doi: 10.1038/nature13306. [DOI] [PubMed] [Google Scholar]
- 21.Xiao H., Wang J., Yan W., Cui Y., Chen Z., Gao X., Wen X., Chen J. GLUT1 regulates cell glycolysis and proliferation in prostate cancer. Prostate. 2018;78:86–94. doi: 10.1002/pros.23448. [DOI] [PubMed] [Google Scholar]
- 22.Chen J., Cao L., Li Z., Li Y. SIRT1 promotes GLUT1 expression and bladder cancer progression via regulation of glucose uptake. Hum. Cell. 2019;32:193–201. doi: 10.1007/s13577-019-00237-5. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y.X., Feng J.Y., Sun M.M., Liu B.W., Yang G., Bu Y.N., Zhao M., Wang T.J., Zhang W.Y., Yuan H.F., Zhang X.D. Aspirin inhibits the proliferation of hepatoma cells through controlling GLUT1-mediated glucose metabolism. Acta Pharmacol. Sin. 2019;40:122–132. doi: 10.1038/s41401-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang E., He X., Zhang C., Su J., Lu X., Si X., Chen J., Yin D., Han L., De W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154. doi: 10.1186/s13059-018-1523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng Z., Wang J., Shan B., Li B., Peng W., Dong Y., Shi W., Zhao W., He D., Duan M. The long noncoding RNA LINC00312 induces lung adenocarcinoma migration and vasculogenic mimicry through directly binding YBX1. Mol. Cancer. 2018;17:167. doi: 10.1186/s12943-018-0920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang H., Li T., Qu Y., Wang X., Li B., Song J., Sun X., Tang Y., Wan J., Yu Y. Long non-coding RNA SNHG15 interacts with and stabilizes transcription factor Slug and promotes colon cancer progression. Cancer Lett. 2018;425:78–87. doi: 10.1016/j.canlet.2018.03.038. [DOI] [PubMed] [Google Scholar]
- 27.Wu M., Seto E., Zhang J. E2F1 enhances glycolysis through suppressing Sirt6 transcription in cancer cells. Oncotarget. 2015;6:11252–11263. doi: 10.18632/oncotarget.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almacellas E., Pelletier J., Manzano A., Gentilella A., Ambrosio S., Mauvezin C., Tauler A. Phosphofructokinases Axis Controls Glucose-Dependent mTORC1 Activation Driven by E2F1. iScience. 2019;20:434–448. doi: 10.1016/j.isci.2019.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anastasiou D. Tumour microenvironment factors shaping the cancer metabolism landscape. Br. J. Cancer. 2017;116:277–286. doi: 10.1038/bjc.2016.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jóźwiak P., Forma E., Bryś M., Krześlak A. O-GlcNAcylation and Metabolic Reprograming in Cancer. Front. Endocrinol. (Lausanne) 2014;5:145. doi: 10.3389/fendo.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamoto Y., Zasadny K.R., Minn H., Wahl R.L. Reproducibility of common semi-quantitative parameters for evaluating lung cancer glucose metabolism with positron emission tomography using 2-deoxy-2-[18F]fluoro-D-glucose. Mol. Imaging Biol. 2002;4:171–178. doi: 10.1016/s1536-1632(01)00004-x. [DOI] [PubMed] [Google Scholar]
- 32.Momcilovic M., Bailey S.T., Lee J.T., Zamilpa C., Jones A., Abdelhady G., Mansfield J., Francis K.P., Shackelford D.B. Utilizing 18F-FDG PET/CT Imaging and Quantitative Histology to Measure Dynamic Changes in the Glucose Metabolism in Mouse Models of Lung Cancer. J. Vis. Exp. 2018;137:57167. doi: 10.3791/57167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanhove K., Graulus G.J., Mesotten L., Thomeer M., Derveaux E., Noben J.P., Guedens W., Adriaensens P. The Metabolic Landscape of Lung Cancer: New Insights in a Disturbed Glucose Metabolism. Front. Oncol. 2019;9:1215. doi: 10.3389/fonc.2019.01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han L., Kong R., Yin D.D., Zhang E.B., Xu T.P., De W., Shu Y.Q. Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Med. Oncol. 2013;30:694. doi: 10.1007/s12032-013-0694-5. [DOI] [PubMed] [Google Scholar]
- 35.Zhang P., Dong Q., Zhu H., Li S., Shi L., Chen X. Long non-coding antisense RNA GAS6-AS1 supports gastric cancer progression via increasing GAS6 expression. Gene. 2019;696:1–9. doi: 10.1016/j.gene.2018.12.079. [DOI] [PubMed] [Google Scholar]
- 36.Ai J., Sun J., Zhou G., Zhu T., Jing L. Long non-coding RNA GAS6-AS1 acts as a ceRNA for microRNA-585, thereby increasing EIF5A2 expression and facilitating hepatocellular carcinoma oncogenicity. Cell Cycle. 2020;19:742–757. doi: 10.1080/15384101.2020.1729323. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Ancey P.B., Contat C., Meylan E. Glucose transporters in cancer - from tumor cells to the tumor microenvironment. FEBS J. 2018;285:2926–2943. doi: 10.1111/febs.14577. [DOI] [PubMed] [Google Scholar]
- 38.Smith T.A. Mammalian hexokinases and their abnormal expression in cancer. Br. J. Biomed. Sci. 2000;57:170–178. [PubMed] [Google Scholar]
- 39.Buono P., Cassano S., Alfieri A., Mancini A., Salvatore F. Human aldolase C gene expression is regulated by adenosine 3¢,5¢-cyclic monophosphate (cAMP) in PC12 cells. Gene. 2002;291:115–121. doi: 10.1016/s0378-1119(02)00586-3. [DOI] [PubMed] [Google Scholar]
- 40.Chen J.M., Chiu S.C., Chen K.C., Huang Y.J., Liao Y.A., Yu C.R. Enolase 1 differentially contributes to cell transformation in lung cancer but not in esophageal cancer. Oncol. Lett. 2020;19:3189–3196. doi: 10.3892/ol.2020.11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong Y., Lei Q.Y., Zhao S., Guan K.L. Regulation of glycolysis and gluconeogenesis by acetylation of PKM and PEPCK. Cold Spring Harb. Symp. Quant. Biol. 2011;76:285–289. doi: 10.1101/sqb.2011.76.010942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeung C., Gibson A.E., Issaq S.H., Oshima N., Baumgart J.T., Edessa L.D., Rai G., Urban D.J., Johnson M.S., Benavides G.A. Targeting Glycolysis through Inhibition of Lactate Dehydrogenase Impairs Tumor Growth in Preclinical Models of Ewing Sarcoma. Cancer Res. 2019;79:5060–5073. doi: 10.1158/0008-5472.CAN-19-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koh Y.W., Park S.Y., Hyun S.H., Lee S.J. Associations Between PET Textural Features and GLUT1 Expression, and the Prognostic Significance of Textural Features in Lung Adenocarcinoma. Anticancer Res. 2018;38:1067–1071. doi: 10.21873/anticanres.12324. [DOI] [PubMed] [Google Scholar]
- 44.Khabaz M.N., Qureshi I.A., Al-Maghrabi J.A. GLUT 1 expression is a supportive mean in predicting prognosis and survival estimates of endometrial carcinoma. Ginekol. Pol. 2019;90:582–588. doi: 10.5603/GP.2019.0102. [DOI] [PubMed] [Google Scholar]
- 45.Sawayama H., Ogata Y., Ishimoto T., Mima K., Hiyoshi Y., Iwatsuki M., Baba Y., Miyamoto Y., Yoshida N., Baba H. Glucose transporter 1 regulates the proliferation and cisplatin sensitivity of esophageal cancer. Cancer Sci. 2019;110:1705–1714. doi: 10.1111/cas.13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei S., Fan Q., Yang L., Zhang X., Ma Y., Zong Z., Hua X., Su D., Sun H., Li H., Liu Z. Promotion of glycolysis by HOTAIR through GLUT1 upregulation via mTOR signaling. Oncol. Rep. 2017;38:1902–1908. doi: 10.3892/or.2017.5840. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y., Zhang X., Wang Z., Hu Q., Wu J., Li Y., Ren X., Wu T., Tao X., Chen X. LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating Glut1-mediated glycolysis. Cancer Lett. 2018;434:172–183. doi: 10.1016/j.canlet.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 48.Shang R., Wang M., Dai B., Du J., Wang J., Liu Z., Qu S., Yang X., Liu J., Xia C. Long noncoding RNA SLC2A1-AS1 regulates aerobic glycolysis and progression in hepatocellular carcinoma via inhibiting the STAT3/FOXM1/GLUT1 pathway. Mol. Oncol. 2020;14:1381–1396. doi: 10.1002/1878-0261.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L.Y., Hung C.L., Chen Y.R., Yang J.C., Wang J., Campbell M., Izumiya Y., Chen H.W., Wang W.C., Ann D.K., Kung H.J. KDM4A Coactivates E2F1 to Regulate the PDK-Dependent Metabolic Switch between Mitochondrial Oxidation and Glycolysis. Cell Rep. 2016;16:3016–3027. doi: 10.1016/j.celrep.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Long Y., Wang X., Youmans D.T., Cech T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017;3:eaao2110. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu C.Y., Zhang Y.H., Li R.B., Zhou L.Y., An T., Zhang R.C., Zhai M., Huang Y., Yan K.W., Dong Y.H. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat. Commun. 2018;9:29. doi: 10.1038/s41467-017-02280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu H., Zhao G., Zhang Y., Jiang H., Wang W., Zhao D., Yu H., Qi L. Long non-coding RNA PAXIP1-AS1 facilitates cell invasion and angiogenesis of glioma by recruiting transcription factor ETS1 to upregulate KIF14 expression. J. Exp. Clin. Cancer Res. 2019;38:486. doi: 10.1186/s13046-019-1474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y., Li L., Wang Z., Pan T., Sahni N., Jin X., Wang G., Li J., Zheng X., Zhang Y. LncMAP: Pan-cancer atlas of long noncoding RNA-mediated transcriptional network perturbations. Nucleic Acids Res. 2018;46:1113–1123. doi: 10.1093/nar/gkx1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou M., Tang X., Tian F., Shi F., Liu F., Gao G. AnnoLnc: a web server for systematically annotating novel human lncRNAs. BMC Genomics. 2016;17:931. doi: 10.1186/s12864-016-3287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agostini F., Zanzoni A., Klus P., Marchese D., Cirillo D., Tartaglia G.G. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics. 2013;29:2928–2930. doi: 10.1093/bioinformatics/btt495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathelier A., Fornes O., Arenillas D.J., Chen C.Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44(D1):D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li S., Jia H., Zhang Z., Wu D. LncRNA GAS6-AS1 facilitates the progression of breast cancer by targeting the miR-324-3p/SETD1A axis to activate the PI3K/AKT pathway. Eur. J. Cell Biol. 2020;99:151124. doi: 10.1016/j.ejcb.2020.151124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.