Abstract
Background
Reduction of COL4A3, one of the six isoforms of collagen 4, in asthmatic airways results in increased inflammation and angiogenesis, implicating it as a central part of asthma pathogenesis. However, to date, the path underlying these diminished COL4A3 levels has been elusive. This study investigated a possible mechanism underlying the reduction of COL4A3 expression.
Methods
Bronchial biopsies of 76 patients with asthma and 83 controls were subjected to RNA-sequencing and DNA methylation bead arrays to identify expression and methylation changes. The binding of ZNF263 was analysed by chromatin-immunoprecipitation sequencing coupled with quantitative (q)PCR. Effects of ZNF263 silencing, using small interfering RNA, on the COL4A3 expression were studied using qPCR.
Results
COL4A3 expression was significantly reduced in bronchial biopsies compared to healthy controls, whereas DNA methylation levels at cg11797365 were increased. COL4A3 expression levels were significantly low in asthmatics without inhaled corticosteroid (ICS) use, whereas the expression was not statistically different between asthmatics using ICS and controls. Methylation levels at cg11797365 in vitro were increased upon consecutive rhinovirus infections.
Conclusion
Our data indicate an epigenetic modification as a contributing factor for the loss of COL4A3 expression in asthmatic airway epithelium.
Short abstract
An epigenetic modification interrupts ZNF263 binding, which may contribute to the loss of COL4A3 expression in asthmatic airway epithelium https://bit.ly/39cZbyn
Background
Asthma is one of the most common lung inflammatory diseases, which significantly impairs health throughout life, with approximately >300 million individuals worldwide affected [1]. The pathogenic mechanisms underlying the disease are multifactorial and still not completely understood [2].
Recent evidence suggests an increasing role of epigenetics in asthma, particularly DNA methylation [3]. DNA methylation regulates gene expression by addition of a methyl group to the cytosine [4]. Along with genetic susceptibility, exposure to allergens and tobacco smoke, repetitive human rhinovirus (HRV) infections are considered major contributors to asthma development [5–7]. Airway constriction along with loss of breath, wheezing, excess mucus production and airway remodelling are considered to be main characteristics of asthma [8, 9]. Airway epithelium plays an important role by balancing immune responses to all the various environmental and viral triggers [10]. The airway epithelium plays a critical role for inhaler therapies such as inhaled corticosteroids (ICS) [11].
ICS are considered to be the most effective treatment for asthma to date. They are also known to reduce the asthmatic airway inflammation [12]. Prolonged airway inflammation, one of the key asthmatic features can result in several changes to the extracellular matrix (ECM) and can lead to pathological tissue remodelling [13]. The ECM is the noncellular component of the lung, which includes myriad structural components such as glycoproteins, proteoglycans and proteins, including collagens secreted by ECM-embedded cells [14, 15].
Type IV collagen, a principal component of the basement membrane, is one of the abundant nonfibrillar collagens in the lung [16]. There are six genes that encode for six distinct α-chains of type IV collagen (α1–6 encoded by COL4A1-6, respectively). These α-chains form heterotrimers, which further assemble into complex collagen networks [17, 18]. COL4A3, like all collagen type IV isoforms, is known to be deposited as a heterotrimer in the ECM [19, 20]. Collagen type IV genes are located adjacently, orientated in a head-to-head fashion and are transcriptionally linked in pairs, i.e. COL4A1/COL4A2, COL4A3/COL4A4 and COL4A5/COL4A6 [21].
In the airways of high-risk wheezing asthmatic patients, collagen type IV deposition is decreased [22]. In particular, one of the six isoforms (COL4A3) protein levels are reduced 18-fold in the lung tissue of asthmatics [23–25]. However, the mechanism underlying reduced protein levels of COL4A3 remains elusive. Hence, in this report, we analysed whether reduced protein expression is a result of decreased levels of COL4A3 gene expression. Additionally, we have investigated whether an epigenetic modification is a contributing cause of the impairment of COL4A3 expression in asthma.
Methods
Cohort
Bronchial epithelial cells biopsy cohort
Bronchial biopsies from the right lower lobe between generations three and six were collected from 76 adult patients with a prior doctor's diagnosis of asthma [26, 27] and from 83 healthy adult control subjects who were derived from the Normal Values of Inflammatory Variables from Healthy Subjects study [28]. Clinical control of asthma was assessed using Asthma Quality of Life Questionnaire (AQLQ) score [29]. All study protocols were approved by the local medical ethics committees. The ethics approval reference number is METc 2004/271. All subjects provided written informed consent.
Cell culture
Bronchial epithelial cells-2B cell culture
Flasks were coated with fibronectin, collagen type I and bovine serum albumin. Human bronchial epithelial cells (BEAS-2B) were cultured in bronchial epithelial cell growth medium (BEGM) medium without gentamycin (GA)-1000 (Lonza, Switzerland) in pre-coated flasks.
Normal human bronchial epithelial cell culture
Normal human bronchial epithelial cells (NHBE) were purchased from Lonza (Switzerland) and cultured in BEGM medium without GA-1000.
HeLA cell culture
Human cervical carcinoma cells (HeLA) were grown in Earle's modified Eagle's medium (EMEM) supplemented with 2 mm l-glutamine (Lonza Group, Germany) and 10% heat-inactivated fetal bovine serum (FBS) (ThermoFisher, USA). No antibiotics were added.
HRV consecutive infections
BEAS-2Bs were seeded at 2×106 per T25 flask (Sarstedt, Germany) and cultured until 80% confluent. Cells were then infected with HRV-16, at multiplicity of infection (MOI) of 0, 5 for 24 h. Obtained infected cells were further split into two parts with 80% of cells transferred to RLT buffer (Qiagen, Hilden, Germany) for further analysis and 20% was re-seeded for following infections. This procedure was repeated for up to five times with a time interval of 72 h between each rhinovirus infection (RVI). Samples were analysed after first, third and fifth infection (supplementary figure 1).
Small interfering RNA transfections
Cells were cultured at 35 000 cells per well in 24-well tissue culture plate with EMEM medium supplemented with 10% FBS before the day of transfection. 25 nM small interfering (si)RNA (ON-TARGET; Horizon Discoveries, USA) against ZNF263 and nontarget siRNA (Horizon Discoveries, USA) as control were delivered via transfection by means of lipofectamine 2000 (Invitrogen, Germany). Manufacturer's recommendations were followed. Cells were harvested 18 h post-transfection.
Genome-wide analysis of DNA methylation and RNA expression
DNA and RNA were isolated from bronchial biopsies using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen). DNA and RNA concentrations were quantified using the FLUOstar Omega-system (BMG Labtech, Ortenberg, Germany). RNA quality was assessed by Agilent RNA6000 Nano Chip (Agilent, USA) on an Agilent 2100 Bioanalyzer. Only samples with a RNA integrity number >8 were accepted for further analysis by RNA-sequencing. Analysis of DNA methylation and RNA-sequencing was carried out at the Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel (Kiel, Germany), following the manufacturer's protocols. The genome-wide analysis of loci-specific DNA methylation was performed using 500 ng bisulfite-converted genomic DNA (EZ DNA methylation Kit; Zymo Research, Germany) and the HumanMethylation450 BeadChip kit (WG-314-1003; Illumina, Germany). β-values were Illumina-normalised (Partek Genomic Suite; Partek, USA) and categorised according to the GSE40699 and as described by Du et al. [30]. Any β-value ≤0.2 was considered to be fully unmethylated. β-values between 0.2 and 0.6 were considered to be partially methylated.
For RNA sequencing, 1000 ng total RNA was processed using HiSeq PE Cluster Kit v4, cBot (PE-401-4001; Illumina, USA), the HiSeq SBS Kit v4 (FC-401-4003; Illumina, USA) and the HiSeq2500 System (Illumina, USA). Strand-specific sequencing with 2×125 bp resulted in FASTQ data files further processed with Partek Flow. Average read quality (Phred) was set at >20 to optimise alignment. Alignment was performed using GSNAP (v3, 31.03.2013). All data are expressed as reads per kilobase per million mapped reads.
Pyrosequencing
The outcome of DNA methylation quantification, cg11797365, was verified by pyrosequencing. 200 ng DNA was converted into bisulfite DNA using EpiTect Bisulfite kit (Qiagen). Converted bisulfite DNA was analysed further using Q24 PyroMark PCR Kit 200 (Qiagen), PyroMark Gold Reagents 5×24 Kit (Qiagen) and PyroMark System (Qiagen). Primer pairs used for pyrosequencing are listed in supplementary table 1.
Chromatin-immunoprecipitation DNA-sequencing
Chromatin-immunoprecipitation sequencing (ChIP-seq) was performed by Active Motif (Carlsbad, USA). Concisely, the zinc finger protein 263 (ZNF263) ChIP-quantitative (q)PCR assay was performed using 30 μg BEAS-2B and NHBE chromatin and 5 μL antibody against ZNF263 (Novus Biologicals, Germany). Differential ZNF263 quantification results were verified using qPCR on three validation sites (COL4A3, AHDC1, AP1S3) around the region of interest and a positive control pair (ANO7). Untr4, an untranscribed genomic region on human chromosome 4 is used as a negative control primer pair. The primers used are listed in supplementary table 2.
qPCR
As mentioned earlier, total RNA was isolated following manufacturer's protocol and cDNA was synthesised using Superscript VILO cDNA synthesis kit (Invitrogen). Pre-designed prime-time qPCR assays for ZNF263 (Hs.PT.58.19342521), COL4A3 (Hs.PT.249834) and GAPDH (Hs.PT.39a.22214836) were used (IDT Technologies, the Netherlands). qPCR was performed using Taq-Man PCR master mix (Life Technologies, Carlsbad, USA) on a Bio-Rad Applied Biosystems 7900 HT Fast real-time PCR system (Thermo-Fisher Scientific, Waltham, USA). We used the 2−ΔΔct method to quantify the relative RNA expression level using glyceraldehyde 3-phosphate dehydrogenase as an endogenous reference. Three independent experiments were conducted with a minimum of three replicates per experiment.
Data analysis
The datasets of DNA methylation and RNA expression were normalised before analysis. For both datasets, the software package Partek Genomic Suite was used to detect differentially methylated CpGs as well as differentially expressed RNA. Resulting CpG sites were checked for known single nucleotide polymorphisms (SNPs). Statistical analysis was performed using JMP13 (SAS, USA) and Prism 6 (GraphPad, USA). Data were statistically analysed with a nonparametric Kruskal–Wallis with Dunn's post-test and Mann–Whitney U-test where appropriate.
Results
Study population
The cohort consisted of 159 subjects, of whom 83 were healthy. Among healthy controls, 38 (45.8%) subjects were female. Mean (range) age of controls was 46 (18–73) years. 76 subjects of the cohort had doctor-diagnosed asthma, of whom 38 (50%) patients were female. Among asthmatic patients, 50 were ICS users and 26 were not using ICS (table 1). Patients in the ICS group were receiving beclomethasone equivalent to 800 µg·day−1 (200–1000 µg·day−1).
TABLE 1.
Bronchial epithelial cell biopsy cohort
| Controls | Asthma | ||
| ICS | No ICS | ||
| Subjects n | 83 | 50 | 26 |
| Age# years | 46 | 48.6** | 46.3 |
| Female# % | 45.8 | 54 | 42.3 |
| FEV1# % pred | 100 | 82**** | 83**** |
| BMI kg·m−2 | 23.6 | 27*** | 27*** |
| ICS dose µg·day−1 | NA | 800 (200–1000) | NA |
FEV1: forced expiratory volume in 1 s; BMI: body mass index; ICS: inhaled corticosteroids (dose equivalent to beclomethasone); NA: not applicable. #: mean values. **: p<0.01, ***: p<0.001, ****: p<0.0001, asthma versus controls.
Control of asthma was assessed using the AQLQ in 76 patients, of whom 15 (32.8%) were well controlled, six (7.8%) were totally controlled and 28 (36.8%) were not controlled; the asthma control status of 15 patients was not available and two patients were not evaluated.
COL4A3 expression reduction in asthma is associated with use of ICS and increased levels of DNA methylation at cg11797365
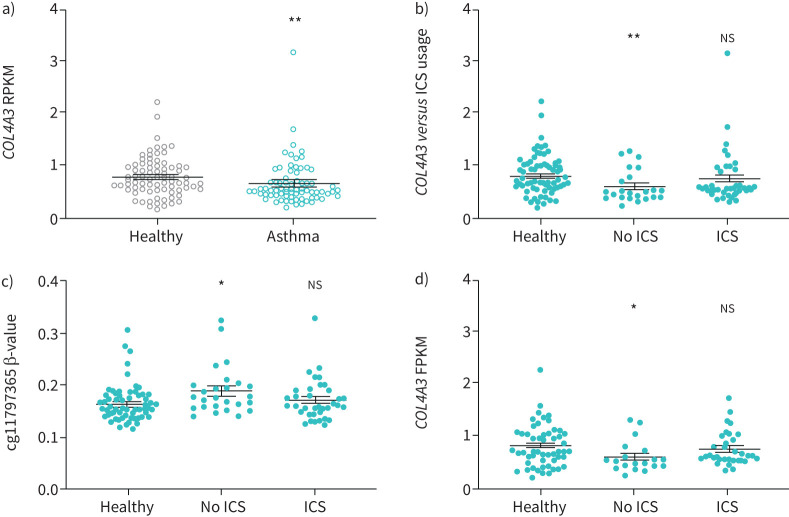
Aiming to investigate whether decrease of COL4A3 gene expression is causative for the previously found reduced COL4A3 protein levels [24], we analysed COL4A3 gene expression in bronchial biopsies (table 1) using RNA-seq. Overall expression levels of COL4A3 were significantly reduced in asthmatics (fragments per kilobase per million mapped reads (FPKM) 0.68±0.05, n=76) when compared to controls without respiratory disease (FPKM 0.81±0.04, n=83) (p<0.01; figure 1a). COL4A3 expression was found to be independent of sex (supplementary figure 2A), age, weight, atopy or use of ICS in a univariate analysis (supplementary table 3). Using a multivariant regression, COL4A3 expression correlated with diseases status and sex but not age, weight, atopy or use of ICS (supplementary table 4). Corticosteroids are known to have an effect on gene expression and glucocortico-response elements are present in the promotor region of COL4A3 [31, 32]. Furthermore, treatment with 100 nM dexamethasone in BEAS-2B cells in vitro for 24 h showed a significant ∼3.75-fold increase in COL4A3 transcripts compared to basal expression levels (supplementary figure 2B, p<0.01). Hence, we compared the COL4A3 expression levels in the cells of asthmatics from our bronchial epithelial cells biopsy cohort, either using ICS or not. COL4A3 expression levels were found to be significantly low in asthmatics without ICS use (figure 1b; p<0.01), whereas the expression was not found to be statistically different between asthmatic patients using ICS and controls.
FIGURE 1.
COL4A3 expression reduction in asthma is associated with use of inhaled corticosteroids (ICS) and increased levels of cg11797365 methylation. a) Expression of COL4A3 in controls (n=83) and asthmatics (n=76) (Mann–Whitney U-test, **: p<0.01). b) Expression of COL4A3 in controls (n=83), patients with asthma using ICS (n=50) and not using ICS (n=26) is significantly reduced in the latter (Kruskal–Wallis with Dunn's post-test, **: p<0.01 compared to controls). c) Methylation (β-value) of cg11797365 in asthmatic ICS users and non-users as compared to control subjects (Kruskal–Wallis with Dunn's post-test, *: p<0.05 compared to control subjects). d) COL4A3 expression in control and asthmatic subjects with low methylation (β-value <0.2); healthy controls n=59, patients with asthma using ICS n=33 and not using ICS n=19 (Kruskal–Wallis with Dunn's post-test *: p<0.05 compared to control subjects). Data are presented as mean±sem. RPKM: kilobase per million mapped reads; FPKM: fragments per kilobase per million mapped reads; ns: nonsignificant.
We reasoned that a possible mechanism for the decreased expression could be an altered gene regulation via SNPs or epigenetic modifications. The altered gene regulation was found not to be associated with any reported SNPs (data not shown). Hence, a comparative analysis of the DNA methylation sites in bronchial biopsies was performed. We identified one DNA methylation site, cg11797365, which is located in the first intron, having significantly higher methylation in asthmatic patients (table 2). This finding was independent of sex or age in our cohort (supplementary figures 2C and D).
TABLE 2.
Change of level of DNA methylation in bronchial biopsies isolated from bronchial epithelial cell biopsy cohort: asthma versus controls
| Current asthma | Control | p-value | |||
| Methylation β-value | Patients | Methylation β-value | Patients | ||
| cg27243623 | 0.455±0.073 | 70 | 0.479±0.068 | 70 | 0.041 |
| cg11797365# | 0.177±0.041 | 70 | 0.165±0.034 | 70 | 0.045 |
| cg04324308 | 0.081±0.031 | 70 | 0.075±0.009 | 70 | 0.088 |
| cg12349837 | 0.697±0.084 | 70 | 0.717±0.074 | 70 | 0.129 |
| cg19865525 | 0.084±0.027 | 70 | 0.087±0.027 | 70 | 0.388 |
| cg08280392 | 0.111±0.018 | 70 | 0.113±0.020 | 70 | 0.681 |
| cg22994281 | 0.072±0.013 | 70 | 0.072±0.011 | 70 | 0.929 |
Data are presented as mean±sd or n, unless otherwise stated. #: site with increased methylation levels in asthmatic patients.
Stratifying for ICS use, cg11797365 was significantly increased in asthmatic patients without ICS use as compared to controls (figure 1c; p<0.05). No significant difference was found between ICS users and healthy controls (figure 1c, supplementary table 5). To explore a possible regulatory function of cg11797365 on COL4A3 expression, we grouped subjects into low methylated and medium methylated levels at cg11797365 according to the β-value (<0.2; cut-off values described in the materials and methods section). Expression of COL4A3 remained significantly reduced in low-methylated asthmatics with no ICS use as compared to controls (figure 1d; p<0.05). In patients with medium methylated levels at cg11797365 (β-value ≥0.2), expression levels of COL4A3 did not change regardless of the ICS usage (supplementary figure 2E). This may suggest that medium methylation at cg11797365 alters the transcriptional response of COL4A3 to ICS in asthmatics.
Increased DNA methylation at site cg11797365 interrupts ZNF263 binding, leading to reduced COL4A3 expression
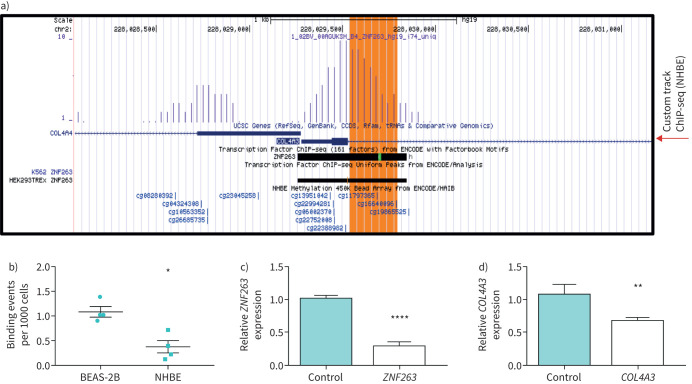
Through analysis of publicly available data (www.genome.ucsc.edu) we identified cg11797365 to be located in close proximity (five base pairs downstream towards the 3′-end) to the consensus motive TcCTCCc for a confirmed transcription factor, ZNF263 binding site in the first intron of the COL4A3 gene. ChIP-seq analysis carried out in NHBE cells confirmed the binding of ZNF263 in the first intron of the COL4A3 gene (figure 2a). This quantitative analysis revealed that ZNF263 bound less to the intronic COL4A3 region in NHBE cells which exhibited increased methylation of 10.7% at cg11797365 as compared to BEAS-2B with methylation of 6.4% (p<0.05; figure 2b, supplementary table 6). To prove the enhancing function of ZNF263, we analysed COL4A3 mRNA expression in HeLa cells transfected with siRNA targeting ZNF263. Silencing of ZNF263 was associated with a significant reduction of COL4A3 mRNA expression in siRNA-transfected HeLa cells (figure 2c and d). Our data indicate that a significant increase in DNA methylation levels at cg11797365 is present in asthmatics. Reduced COL4A3 expression in asthmatics is likely mediated by abrogated transcriptional control at cg11797365, possibly exerted via decreased binding of the transcription factor ZNF263. We have further analysed the impact of ICS on ZNF263 expression in our bronchial biopsies samples stratified for ICS use. No significant impact of ICS use on ZNF263 expression (supplementary figure S4) has been found, which suggests that the effect of ICS is not associated with ZNF263 expression in asthmatic patients.
FIGURE 2.
ZNF263 binding is interrupted in asthmatic patients due to increased methylation at site cg11797365 leading to a reduced COL4A3 expression. a) Detection hits of chromatin-immunoprecipitation sequencing (ChIP-seq) data of ZNF263 in COL4A3 region. Large peak corresponds to region of first exon and start of first intron. cg11797365 is indicated by the red arrow and corresponds with previous report on ZNF263 binding (ENCODE track). The methylation site cg11797365 is close to the consensus binding site (green highlight in ENCODE track) of ZNF263. Custom track from ChIP-seq data from normal human bronchial epithelial cells (NHBE) is shown as indicated. b) Quantitative ChIP-seq analysis of ZNF263 binding to genomic region COL4A3 (1. intron) of cg11797365 in human bronchial epithelial cells (BEAS-2B) and NHBEs. Normalised binding events (per 1000 cells), n=4 per cell line (BEAS-2B and NHBE; Lonza). c) Quantitative (q)PCR to quantify ZNF263 expression levels in HeLA cells transfected with small interfering (si)RNA against ZNF263 in comparison to cells transfected with a nontarget control. d) Expression of COL4A3 in HeLA cells transfected with siRNA against ZNF263 compared to cells transfected with a nontarget control. Δct values are normalised to GAPDH. *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001, Mann–Whitney U-test.
RVI induces increased DNA methylation levels at cg11797365 in vitro
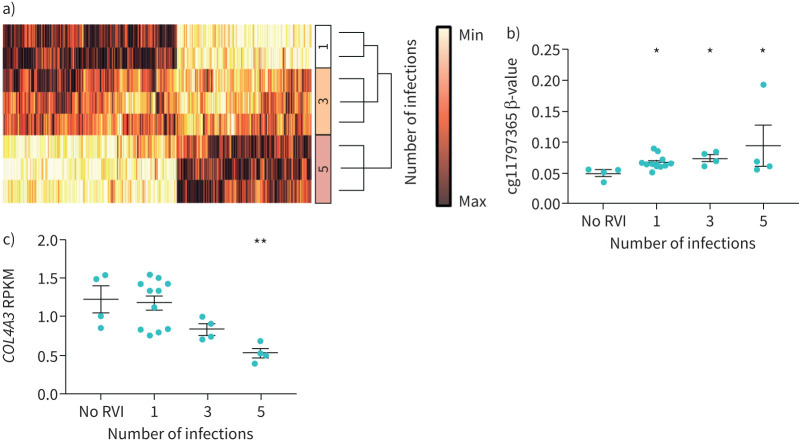
It was recently reported that RVI can induce DNA methylation changes in nasal epithelial cells in vitro [33, 34]. However, the consequences of consecutive RVIs on the COL4A3 DNA methylation levels are not fully understood. To outline the effects of RVIs on genome-wide methylation in epithelial cells, we infected BEAS-2B cells consecutively with HRV-16 and measured DNA methylation using human methylation 450k chips. Consecutive RVIs significantly altered levels of methylation in 33 826 CpGs (figure 3a; linear regression false discovery rate corrected p<0.01), out of which 17 methylation sites were found to be located on COL4A3. To understand the consequences of consecutive RVIs on COL4A3 methylation and expression, we quantified DNA methylation and gene expression levels of COL4A3 in BEAS-2B cells by pyrosequencing and qPCR, respectively. 15 out of the 17 DNA methylation sites from the 450k chips in COL4A3 were significantly associated (p<0.05) with either the number of infections, the viral titre (MOI) or the duration of the in vitro infection (table 3). Of the 15 methylation sites investigated, eight CpG sites were correlated with basal COL4A3 expression (table 4).
FIGURE 3.
Methylation levels at cg11797365 are increased, expression levels of COL4A3 are decreased after consecutive rhinovirus infections (RVI) in vitro. a) Hierarchical clustering heat map (Ward's minimum variance) of genome-wide methylation changes after consecutive numbers of infections with human rhinovirus-16 (p<0.01, false discovery rate adjusted linear regression p-values). All methylation levels are normalised. b) Methylation level at site cg11797365 increased significantly after RVI. Levels of methylation after each RVI were compared to no RVI (n=4; 1 RVI n=10, 3 RVI n=4, 5 RVI n=4). c) COL4A3 expression (reads per kilobase per million mapped reads (RPKM)) decreases after each RVI (no RVI n=4; 1 RVI n=10, 3 RVI n=4, 5 RVI n=4). Data are presented as mean±sem. *: p<0.05, **: p<0.01 by Kruskal–Wallis with Dunn's post-test.
TABLE 3.
Effects of rhinovirus infection on DNA methylation levels of different CpGs in the COL4A3 gene
| Form of infection | Log-worth# | p-value | |
| cg08280392 | DOI | 2.62 | 0.0024 |
| cg19865525 | DOI | 1.399 | 0.03987 |
| cg27243623 | DOI | 2.593 | 0.00255 |
| cg22994281 | DOI | 1.805 | 0.01566 |
| cg04324308 | DOI | 3.407 | 0.00039 |
| cg11797365¶ | DOI*NOI | 3.89 | 0.00013 |
| cg13951042 | DOI*NOI | 3.462 | 0.00035 |
| cg27243623 | DOI*NOI | 2.193 | 0.00641 |
| cg22994281 | DOI*NOI | 1.473 | 0.03368 |
| cg04324308 | DOI*NOI | 2.175 | 0.00668 |
| cg12349837 | MOI | 2.547 | 0.00284 |
| cg19865525 | MOI | 1.332 | 0.04655 |
| cg08280392 | MOI*NOI | 1.744 | 0.01802 |
| cg27243623 | MOI*NOI | 4.009 | 0.0001 |
| cg12349837 | NOI | 3.616 | 0.00024 |
DOI: duration of infection (4 h, 24 h, 48 h); NOI: number of consecutive infections (0, 1, 3, 5); MOI: multiplicity of infection (number of virus particles per cell); DOI*NOI, MOI*NOI: combination effect of DOI and NOI, MOI and NOI determining increase of methylation. #: log-worth: −log10(p-value); ¶: site with increased methylation levels in asthmatic patients .
TABLE 4.
Correlated basal COL4A3 expression and CpG sites
| Basal COL4A3 expression | |
| cg12349837 | −0.736 |
| cg11797365# | −0.662 |
| cg13951042 | −0.519 |
| cg08280392 | 0.542 |
| cg19865525 | 0.609 |
| cg27243623 | 0.685 |
| cg22994281 | 0.722 |
| cg04324308 | 0.812 |
#: target CpG site is associated with basal COL4A3 expression.
DNA methylation levels at cg11797365 significantly increased with successive numbers of infections (p<0.05; figure 3b), whereas RNA-seq analysis revealed a significantly decreased COL4A3 expression upon successive numbers of infections (figure 3c; five consecutive infections versus no infection, p<0.01). The expression level was also confirmed by qPCR (supplementary figure 4). These findings suggest that consecutive RVI in vitro increased methylation levels at cg11797635 within the COL4A3 gene, while being associated with decreased levels of gene expression in BEAS-2B cells.
COL4A3 expression and DNA methylation at cg11797365 are independently associated with asthma and correlate with lung function
To better understand the association of cg11797365 methylation and COL4A3 expression level with asthma, we combined our data with the dataset from Nicodemus-Johnson et al. [35] (GSE85568) to 207 matched DNA methylation and expression pairs. COL4A3 expression levels were z-transformed and β-values of DNA methylation levels were used without additional transformation. To compensate dataset inherent variations, a variable “dataset” was used in our logistic regression analysis. We did not find a direct association of gene expression and levels of DNA methylation in the combined dataset (p=0.832; supplementary table 7). Next, we performed a logistic regression analysis for both variables for asthma outcome. We included age, sex, β-values of cg11797365, z-values of COL4A3 expression and the dataset. Logistic regression returned a significant and independent association for age (p=0.0072), cg11797365 (p=0.0254), z-value (COL4A3) (p=0.0498) and dataset (p=0.001), but not for sex (p=0.0624) (supplementary table 8).
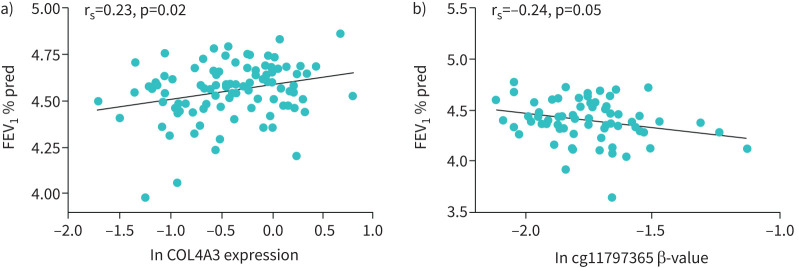
Additionally, better lung function (forced expiratory volume in 1 s (FEV1) % predicted) was associated with higher COL4A3 expression (Spearman correlation coefficient rs=0.23, p=0.02; figure 4c) in controls and asthmatic patients who did not receive ICS. However, no correlation was found in patients after ICS treatment (data not shown). Furthermore, higher levels of methylation at cg11797365 correlated with lower lung function (FEV1 % pred) in asthmatics regardless of ICS use (Spearman correlation coefficient rs=−0.24, p=0.05; figure 4d). These results may suggest that COL4A3 expression and DNA methylation are independently associated with asthma.
FIGURE 4.
Association of COL4A3 expression and methylation level at cg11797365 with lung function. a) Spearman correlation plot of logarithmic transformed (natural logarithm) forced expiratory volume in 1 s (FEV1) % predicted and COL4A3 expression. Correlation includes controls and asthmatic patients without inhaled corticosteroids. rs=0.23, p=0.02, n=101. Line indicates correlation. b) Spearman correlation plot of logarithmic transformed (natural logarithm) FEV1 % predicted and methylation levels at cg11797365 (β-value, age- and sex-adjusted). Correlation includes patients with asthma with and without ICS. rs=−0.24, p=0.05, n=70. Line indicates correlation.
Discussion
To the best of our knowledge, our study is the first to analyse COL4A3 expression levels and report a possible role of altered DNA methylation site which is located in COL4A3. Since our first report of an 18-fold reduction of COL4A3 expression in asthmatic airways, the fate of COL4A3 has remained elusive [24]. Loss of COL4A3 in asthmatic airway sections has been linked to the pathogenesis of asthma by aggravating inflammation, mucus production, angiogenesis, goblet-cell hyperplasia, neutrophil migration and increased bronchial hyperreactivity [23, 25, 36]. Here we shed light on decreased mRNA expression of COL4A3 and an increased intragenic DNA methylation in asthmatics. Furthermore, our study also demonstrates the association of ICS treatment with COL4A3 expression and DNA methylation levels at cg11797365. We explored the loss of COL4A3 protein expression as a consequence of decreased COL4A3 gene expression.
In our study, expression of COL4A3 in bronchial biopsies from asthma patients was found to be significantly reduced when compared to controls. We identified an intronic DNA methylation site, cg11797365, to be significantly increased in asthmatics. Our analysis suggests an association between increased levels of DNA methylation at cg11797365 and decreased COL4A3 expression. Furthermore, in our cohort stratified for different levels of DNA methylation at cg11797365 (low and medium levels), usage of ICS resulted in increased levels of COL4A3 in asthmatics with low levels of DNA methylation, whereas expression levels of COL4A3 were found to be almost similar irrespective of the use of ICS in asthmatics with medium levels of DNA methylation. This may suggest that medium methylation levels at cg11797365 might alter the transcriptional response to ICS in asthmatics.
The second aim of our study was to observe if DNA methylation level and the gene expression are associated with asthma. It has been shown that increased methylation level at cg11797365 is correlated with decreased expression level of COL4A3 [37]. In addition, Nicodemus-Johnson et al. [35] reported a wide-ranging set of Illumina DNA-methylation array (GSE85568) and RNA-seq data (GSE85567) generated from freshly isolated asthmatic and healthy endobronchial airway epithelial cells. In addition, their comprehensive dataset indicated that cg11797365 was significantly increased in asthmatics and the pathway analysis revealed that type IV collagen gene family was associated with a “tumour necrosis factor remodelling-centred endotype”. This endotype was strongly associated with asthma severity, ICS use and bronchoalveolar lavage eosinophil counts. This is in line with current literature in which COL4A3 is important in reducing aggravated airway inflammation (i.e. eosinophilic, neutrophilic) and goblet cell hyperplasia, as well as airway smooth muscle cell ECM remodelling [25, 36]. Mechanistically, we found that an increase in DNA methylation levels at cg11797365 was responsible for a decreased binding of the expression enhancer ZNF263. Frietze et al. [38] identified ZNF263 to have repressive functions in promotor regions, but did not fully exclude the possibility of an additional enhancer function. Furthermore, ZNF263 exhibited an enhancing effect on members of the “cellular component organisation and biogenesis” family, to which type IV collagens belong [38]. We observed that silencing ZNF263 upon siRNA transfection was associated with a significant reduction of COL4A3 mRNA expression. This is supportive of our hypothesis that the reduced levels of COL4A3 expression is a consequence of decreased binding of ZNF263. Our results imply that epigenetic modifications in COL4A3 may have a detrimental impact on ECM homeostasis in the lung. This may suggest that DNA methylation and gene expression to be independent variables for asthma prediction, yet the additional mechanistic factors regulating COL4A3 expression require further research.
RVIs have been shown to affect epigenetic markers such as DNA methylations in asthma-relevant genes, especially the host immune responsive genes to HRVs such as (HLA-B)-associated transcript 3 (BAT3), neuraminidase 1 (NEU-1) and Mothers against decapentaplegic homologue 3 (SMAD3), a susceptible signalling intermediator activated by transforming growth factor-β in asthma [33, 39]. These findings suggest that RVIs influence DNA methylation and are considered an additional risk factor for asthma patients. Consistently, we found that consecutive RVIs in BEAS-2Bs led to an increased methylation at cg11797365, while COL4A3 expression is reduced, suggesting that persistent RVIs may contribute to the severity, and potentially the progression of asthma by affecting abundance of protein levels of COL4A3 in bronchial airways. However, further work is warranted to understand how consecutive RVIs can lead to an increased DNA methylation at site cg11797365 in COL4A3 in human airway epithelium.
Impairment of lung function is a key feature of asthma [40]. We postulated whether DNA methylation at cg11797365 and/or COL4A3 expression levels are associated with lung function in patients from our cohort. We found that higher levels of COL4A3 expression in controls and asthmatics who are not receiving ICS was associated with better lung function. Furthermore, higher levels of DNA methylation at cg11797365 correlated with lower lung function in asthmatics regardless of their ICS use. These results may suggest that COL4A3 expression and methylation levels at cg11797365 are both implicated in lung function in healthy and asthmatics. However, further studies are required to validate our findings in a larger cohort.
The major limitation of our study is an insufficient number of medium-methylated subjects (n=16) in our cohort, when compared to the number of low-methylated subjects (n=111). Investigating more subjects with medium methylation in the future to be comparable with low methylated subjects would be necessary.
BEAS-2B cells have low DNA methylation levels as compared to NHBEs. Additionally, BEAS-2Bs have the ability to maintain their expansion capabilities even with high number of passages. Thus BEAS-2Bs proved the best fit for our experiments in order to study DNA-methylation increase after consecutive RVIs. Unfortunately, we were not successful in achieving comparable levels of DNA methylation at cg11797365 as seen in our human data. Lastly, we cannot exclude the possibility of carry-over of RV16 in our consecutive infections experiment, as the viral titre was not analysed in the supernatant. Yet, we detected no mRNA and protein expression of interleukin (IL)-6 and IL-8, chemokines which are known to be specifically induced by RVIs, in our re-cultured but noninfected cells [33, 41]. Although RV16 carry-over cannot be completely eliminated, the impact of any such carry-over would be minimal.
Taken together, our data provide evidence that epigenetic changes in COL4A3 may lead to a change in the expression of COL4A3 in the bronchial airways of asthmatic patients. Usage of ICS leads to increase in COL4A3 expression levels; however, this effect was only observed in asthma patients with low DNA methylation levels at cg11797365, while ICS usage had no effect on COL4A3 expression levels in patients with medium levels of DNA methylation at cg11797365. We postulate that ZNF263 binds to COL4A3 and acts as a gene expression enhancer, while DNA methylation at cg11797365 in COL4A3 prevents the binding and reduces the gene expression. Additionally, our findings may suggest that risk factors like RVIs might modify the DNA-methylation of COL4A3, which leads to worse disease outcome (e.g. lung function). Our findings suggest that patients with a DNA methylation level similar to healthy controls would respond to ICS and benefit. When medium levels of DNA methylation at cg11797365 are present, our data suggest that ICS treatment did not modulate COL4A3 expression, and the efficacy of ICS was found to be less than in patients with low levels of DNA methylation at cg11797365. However, further work is necessary to validate our findings in a larger cohort.
Collectively, based on our data, COL4A3 expression and DNA methylation are both associated with asthma and COL4A3 can be recognised as an important molecular player in asthma.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary figures 00802-2020.supplement_figures (940.1KB, pdf)
Supplementary methods and tables 00802-2020.SUPPLEMENTARY_METHODS_TABLES (128KB, pdf)
Acknowledgements
The authors would like to thank Nadine Weissheimer, Detlev Schult-Badusche and Elvira Ehlers-Jeske (Division of Paediatric Pneumology and Allergology, University Children's Hospital, Lübeck, Germany) for their excellent technical support.
Footnotes
This article has supplementary material available from openres.ersjournals.com
Author contributions: S.S.P. Nemani, M. Weckmann, M. Pech and M.V. Kopp designed the experiments, performed analysis and wrote the manuscript. C.J. Vermeulen and A. Faiz together with M. Weckmann analysed the bronchial epithelial cell biopsy cohort data and analysed the chromatin immunoprecipitation sequencing data. B.G.G. Oliver provided rhinovirus-16 and edited the manuscript. M. van den Berge and J.K. Burgess extensively reviewed and edited the manuscript. M. Weckmann analysed data and edited the manuscript. All authors approved the final version of the manuscript.
Conflict of interest: S.S.P. Nemani has nothing to disclose.
Conflict of interest: C.J. Vermeulen has nothing to disclose.
Conflict of interest: M. Pech has nothing to disclose.
Conflict of interest: A. Faiz has nothing to disclose.
Conflict of interest: B.G.G. Oliver has nothing to disclose.
Conflict of interest: M. van den Berge reports grants paid to his university by GlaxoSmithKline, AstraZeneca and Genentech, outside the submitted work.
Conflict of interest: J.K. Burgess reports grants from the National Health and Medical Research Council, Australia; the University of Groningen; and the European Union, all during the conduct of the study.
Conflict of interest: M.V. Kopp reports grants from the Federal Ministry of Research and Education (BMBF) during the conduct of the study; and personal fees from ALK-Abello, Allergopharma, Boehringer Ingelheim, Chiesi, Glaxo, Infectopharm, Meda, Sanofi-Aventis, Leti Pharma, Novartis and Vertex, outside the submitted work.
Conflict of interest: M. Weckmann reports grants from the BMBF, University of Lübeck and German Academic Exchange Service during the conduct of the study.
Support statement: This work was supported by the National Health and Medical Research Council (NHMRC), Australia. J.K. Burgess was supported by a NHMRC R. Douglas Wright Fellowship (number 402835), a NHMRC Career Development Fellowship (1032695) and a Rosalind Franklin Fellowship (University of Groningen, The Netherlands, and The European Union). Markus Weckmann was supported by a young investigator award (E42-2012, University of Lübeck), a junior research cluster programme (JC01-2016, University of Lübeck), the German Centre for Lung Research (DZL, Federal Ministry of Education and Research) and a Group of Eight/German Academic Exchange Service grant (56266000). Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Nunes C, Pereira AM, Morais-Almeida M. Asthma costs and social impact. Asthma Res Pract 2017; 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomsen SF. Genetics of asthma: an introduction for the clinician. Eur Clin Respir J 2015; 2: 24643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang IV, Schwartz DA. Epigenetic mechanisms and the development of asthma. J Allergy Clin Immunol 2012; 130: 1243–1255. doi: 10.1016/j.jaci.2012.07.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baccarelli A, Rusconi F, Bollati V, et al. Nasal cell DNA methylation, inflammation, lung function and wheezing in children with asthma. Epigenomics 2012; 4: 91–100. doi: 10.2217/epi.11.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sly PD, Kusel M, Holt PG. Do early-life viral infections cause asthma? J Allergy Clin Immunol 2010; 125: 1202–1205. doi: 10.1016/j.jaci.2010.01.024 [DOI] [PubMed] [Google Scholar]
- 6.Holgate ST. Pathogenesis of asthma. Clin Exp Allergy 2008; 38: 872–897. doi: 10.1111/j.1365-2222.2008.02971.x [DOI] [PubMed] [Google Scholar]
- 7.Engelmann I, Mordacq C, Gosset P, et al. Rhinovirus and asthma: reinfection, not persistence. Am J Respir Crit Care Med 2013; 188: 1165–1167. doi: 10.1164/rccm.201303-0585LE [DOI] [PubMed] [Google Scholar]
- 8.Hudon Thibeault AA, Laprise C. Cell-specific DNA methylation signatures in asthma. Genes 2019; 10: 932. doi: 10.3390/genes10110932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh ER, Stokes K, August A. The role of eosinophils in allergic airway inflammation. Discov Med 2010; 9: 357–362. [PubMed] [Google Scholar]
- 10.Lambrecht BN, Hammad H. The airway epithelium in asthma. Nat Med 2012; 18: 684–692. doi: 10.1038/nm.2737 [DOI] [PubMed] [Google Scholar]
- 11.Yuksel H, Turkeli A. Airway epithelial barrier dysfunction in the pathogenesis and prognosis of respiratory tract diseases in childhood and adulthood. Tissue Barriers 2017; 5: e1367458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes PJ. Inhaled corticosteroids. Pharmaceuticals 2010; 3: 514–540. doi: 10.3390/ph3030514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito JT, Lourenço JD, Righetti RF, et al. Extracellular matrix component remodelling in respiratory diseases: what has been found in clinical and experimental studies? Cells 2019; 8: 342. doi: 10.3390/cells8040342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci 2010; 123: 4195–4200. doi: 10.1242/jcs.023820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White ES. Lung extracellular matrix and fibroblast function. Ann Am Thorac Soc 2015; 12: S30–S33. doi: 10.1513/AnnalsATS.201406-240MG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunsmore SE. Treatment of COPD: a matrix perspective. Int J Chron Obstruct Pulmon Dis 2008; 3: 113–122. doi: 10.2147/COPD.S1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heidet L, Arrondel C, Forestier L, et al. Structure of the human type IV collagen gene COL4A3 and mutations in autosomal Alport syndrome. J Am Soc Nephrol 2001; 12: 97–106. [DOI] [PubMed] [Google Scholar]
- 18.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer 2003; 3: 422–433. doi: 10.1038/nrc1094 [DOI] [PubMed] [Google Scholar]
- 19.Faiz A, Tjin G, Harkness L, et al. The expression and activity of cathepsins D, H and K in asthmatic airways. PLoS One 2013; 8: e57245. doi: 10.1371/journal.pone.0057245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudson BG, Tryggvason K, Sundaramoorthy M, et al. Alport's syndrome, Goodpasture's syndrome, and type IV collagen. N Engl J Med 2003; 348: 2543–2556. [DOI] [PubMed] [Google Scholar]
- 21.Li YY, Yu H, Guo Z-M, et al. Systematic analysis of head-to-head gene organization: evolutionary conservation and potential biological relevance. PLoS Comput Biol 2006; 2: e74. doi: 10.1371/journal.pcbi.0020074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson PRA, Burgess JK, Underwood PA, et al. Extracellular matrix proteins modulate asthmatic airway smooth muscle cell proliferation via an autocrine mechanism. J Allergy Clin Immunol 2004; 113: 690–696. doi: 10.1016/j.jaci.2003.12.312 [DOI] [PubMed] [Google Scholar]
- 23.Van der Velden J, Harkness LM, Barker DM, et al. The effects of tumstatin on vascularity, airway inflammation and lung function in an experimental sheep model of chronic asthma. Sci Rep 2016; 6: 26309. doi: 10.1038/srep26309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgess JK, Boustany S, Moir LM, et al. Reduction of tumstatin in asthmatic airways contributes to angiogenesis, inflammation, and hyperresponsiveness. Am J Respir Crit Care Med 2010; 181: 106–115. doi: 10.1164/rccm.200904-0631OC [DOI] [PubMed] [Google Scholar]
- 25.Harkness LM, Weckmann M, Kopp M, et al. Tumstatin regulates the angiogenic and inflammatory potential of airway smooth muscle extracellular matrix. J Cell Mol Med 2017; 21: 3288–3297. doi: 10.1111/jcmm.13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broekema M, Volbeda F, Timens W, et al. Airway eosinophilia in remission and progression of asthma: accumulation with a fast decline of FEV1. Respir Med 2010; 104: 1254–1262. doi: 10.1016/j.rmed.2010.03.030 [DOI] [PubMed] [Google Scholar]
- 27.Broekema M, Timens W, Vonk JM, et al. Persisting remodelling and less airway wall eosinophil activation in complete remission of asthma. Am J Respir Crit Care Med 2011; 183: 310–316. doi: 10.1164/rccm.201003-0494OC [DOI] [PubMed] [Google Scholar]
- 28.Boudewijn IM, Postma DS, Telenga ED, et al. Effects of ageing and smoking on pulmonary computed tomography scans using parametric response mapping. Eur Respir J 2015; 46: 1193–1196. doi: 10.1183/09031936.00009415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volbeda F, Broekema M, Lodewijk ME, et al. Clinical control of asthma associates with measures of airway inflammation. Thorax 2013; 68: 19–24. doi: 10.1136/thoraxjnl-2012-201861 [DOI] [PubMed] [Google Scholar]
- 30.Du P, Zhang X, Huang C-C, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010; 11: 587. doi: 10.1186/1471-2105-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Berge M, Steiling K, Timens W, et al. Airway gene expression in COPD is dynamic with inhaled corticosteroid treatment and reflects biological pathways associated with disease activity. Thorax 2014; 69: 14–23. doi: 10.1136/thoraxjnl-2012-202878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy TE, Pauli F, Sprouse RO, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res 2009; 19: 2163–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pech M, Weckmann M, König IR, et al. Rhinovirus infections change DNA methylation and mRNA expression in children with asthma. PLoS One 2018; 13: e0205275. doi: 10.1371/journal.pone.0205275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McErlean P, Favoreto S, Costa FF, et al. Human rhinovirus infection causes different DNA methylation changes in nasal epithelial cells from healthy and asthmatic subjects. BMC Med Genomics 2014; 7: 37. doi: 10.1186/1755-8794-7-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicodemus-Johnson J, Myers RA, Sakabe NJ, et al. DNA methylation in lung cells is associated with asthma endotypes and genetic risk. JCI Insight 2016; 1: e90151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nissen G, Hollaender H, Tang FSM, et al. Tumstatin fragment selectively inhibits neutrophil infiltration in experimental asthma exacerbation. Clin Exp Allergy 2018; 48: 1483–1493. doi: 10.1111/cea.13236 [DOI] [PubMed] [Google Scholar]
- 37.Kulis M, Heath S, Bibikova M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet 2012; 44: 1236–1242. doi: 10.1038/ng.2443 [DOI] [PubMed] [Google Scholar]
- 38.Frietze S, Lan X, Jin VX, et al. Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J Biol Chem 2010; 285: 1393–1403. doi: 10.1074/jbc.M109.063032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lund RJ, Osmala M, Malonzo M, et al. Atopic asthma after rhinovirus-induced wheezing is associated with DNA methylation change in the SMAD3 gene promoter. Allergy 2018; 73: 1735–1740. doi: 10.1111/all.13473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dijkstra A, Vonk JM, Jongepier H, et al. Lung function decline in asthma: association with inhaled corticosteroids, smoking and sex. Thorax 2006; 61: 105–110. doi: 10.1136/thx.2004.039271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chun YH, Park JY, Lee H, et al. Rhinovirus-infected epithelial cells produce more IL-8 and RANTES compared with other respiratory viruses. Allergy Asthma Immunol Res 2013; 5: 216–223. doi: 10.4168/aair.2013.5.4.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary figures 00802-2020.supplement_figures (940.1KB, pdf)
Supplementary methods and tables 00802-2020.SUPPLEMENTARY_METHODS_TABLES (128KB, pdf)