Abstract
Objective
To explore the neddylation pathway, found to be highly activated in various cancers, as a potential therapeutic target in endometrial carcinoma, one of the three most frequent malignant tumours in the female reproductive system.
Methods
Data from The Cancer Genome Atlas were analysed using online servers. Expression levels of key neddylation genes were validated by reverse-transcription polymerase chain reaction and western blots of tumour and adjacent tissues. Underlying mechanisms and the effects on cell activities of the neddylation pathway-specific inhibitor, MLN4924, were investigated in endometrial cancer cell lines.
Results
Key neddylation enzymes, ubiquitin conjugating enzyme E2 M (UBC12), ubiquitin conjugating enzyme E2 F (UBE2F), ring-box 1 (RBX1) and ring finger protein 7 (RBX2), were significantly overexpressed in endometrial carcinoma tissues versus normal tissues, but only UBE2F and RBX2 positively correlated with patient survival. MLN4924 significantly suppressed proliferation and colony formation in EC cells by inducing DNA re-replication, cell cycle arrest and apoptosis. Mechanism study revealed that MLN4924 induced the accumulation of cullin-RING ligase substrates in vitro.
Conclusions
The neddylation pathway was identified to play an important role in endometrial cancer. The neddylation specific inhibitor, MLN4924, may be a potential therapeutic drug for endometrial carcinoma.
Keywords: Endometrial cancer, neddylation, MLN4924, cell cycle arrest, apoptosis, cullin-RING ligase
Introduction
Endometrial cancer (EC) is one of the most common gynaecological malignancies, and the incidence rate of endometrial carcinoma is increasing worldwide.1,2 Surgery, chemotherapy, and radiotherapy are considered to be effective treatments for EC.3,4 The majority of patients with low grade, early stage endometrial cancer will have favourable survival with surgery resection, however, patients with more aggressive, high‐grade tumours, with tumour-cell spreading beyond the uterus, will progress quickly within one year and have a lower survival rate. 4 Neo-adjuvant chemotherapy for tumour reduction may enable surgical treatment in patients who were previously considered to have unresectable disease. Preoperative chemotherapy may also help identify patients with tumours that are sensitive to chemotherapy, who would be more likely to benefit from surgery, and it may enable a less aggressive surgery. Patients who received neo-adjuvant therapy have been shown to have improved or equivalent survival, maximal tumour debulking rates and reduced postoperative morbidity compared with patients who received primary surgery. 5 Therefore, it is urgent to identify the molecular mechanism underlying endometrial cancer and to develop new effective anti-endometrial cancer drugs.
Neddylation, a process of adding the ubiquitin-like protein NEDD8 to target proteins, is a recently discovered posttranslational modification that regulates subcellular localization, stability, conformation and functions of substrates. The neddylation process involves a three-step enzymatic cascade reaction, mediated by NEDD8-activating enzyme E1 (NAE, a heterodimer comprising NEDD8-activating enzyme E1 regulatory subunit [NAE1] and NEDD8-activating enzyme E1 catalytic subunit [UBA3]), NEDD8-conjugating E2 enzymes (ubiquitin conjugating enzyme E2 M [UBE2M/UBC12] or ubiquitin conjugating enzyme E2 F [UBE2F]), and NEDD8 E3 ligases (including ring-box 1 [RBX1/ROC1], ring finger protein 7 [RBX2/ROC2], and oncoprotein MDM2, etc).6–8 The most characteristic substrates of the neddylation system are the cullins, which control the degradation of approximately 20% of ubiquitinated cellular proteins.8,9 The neddylation pathway, including E1, E2, E3 and global neddylation of substrates, is reported to be hyperactivated in several cancer types, including lung cancer, liver cancer, and glioma, and is shown to be associated with overall patient survival. 10 MLN4924, a small molecular inhibitor of NAE, has been developed as a first-in-class neddylation inhibitor.11,12 Because of its significant anticancer efficacy in preclinical studies, MLN4924 has been advanced into phase I clinical trials for several solid tumours and haematologic malignancies.9,12 However, the gene expression status of the neddylation pathway, and the correlation with patient outcome, remain unclear. In addition, the effect and potential mechanism underlying neddylation targeting by MLN4924 in EC cells remains undetermined.
The aim of the present study was to investigate the neddylation pathway in EC, through analyses of The Cancer Genome Atlas (TCGA) datasets, gene expression and protein level assays of endometrial tumour and adjacent tissues, and subsequent investigation of the effects of MLN4924 on cellular activity in EC cell lines. 13
Patients and methods
Study population and clinical specimens
Endometrial adenocarcinoma tumour tissues and corresponding adjacent noncancerous tissues were obtained from patients with endometrial cancer who were being treated at Shanghai First Maternity and Infant Hospital between January 2013 and December 2017. The study was approved by the Ethics Committee of Shanghai First Maternity and Infant Hospital, Tongji University (approval date: 7 March 2018), and written informed consent was obtained from all patients providing tissue samples for the study. Samples were obtained from patients in the operating room, cut into small pieces and stored in liquid nitrogen before use. Samples then underwent total RNA extraction for real-time reverse transcription (RT)-polymerase chain reaction (PCR), or protein extraction and purification for western blots.
Cell lines, culture, and reagents
Human endometrial cancer cell lines, HHUA and AN3CA, were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in Gibco Dulbecco's Modified Eagle Medium (Invitrogen [ThermoFisher Scientific], Waltham MA, USA), containing 10% Gibco foetal bovine serum (FBS; Invitrogen [ThermoFisher Scientific]) and 1% penicillin–streptomycin solution, at 37 °C with 5% CO2. For in vitro studies into the effects of NAE inhibition, MLN4924 (Selleck Chemicals LLC; Houston, TX, USA) was dissolved in dimethyl sulfoxide (DMSO) and stored at –20 °C. 14
TCGA database
Publicly available data were extracted from the TCGA database (http://cancergenome.nih.gov/). Data were then analysed using the online web resources, UALCAN (http://ualcan.path.uab.edu/index.html), 15 Tumour IMmune Estimation Resource (TIMER), 16 and cBioPortal. 17
Real-time RT-PCR
Total RNA was isolated from endometrial adenocarcinoma or adjacent noncancerous tissue samples (50 mg each) using Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions, and treated with RNase-free DNase. Reverse transcription was performed using 2 µg total RNA per sample and the PrimeScript reverse transcription reagent kit (Takara, Japan) according to the manufacturer’s protocol. The resultant cDNA (50 ng per sample) was amplified by real-time PCR using Power SYBR Green PCR MasterMix (Applied Biosystems, Foster City, CA, USA) and the ABI 7900 thermocycler (Applied Biosystems) according to the manufacturer’s instructions, under the following cycling conditions: preliminary denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 30 s, and elongation at 72 °C for 30 s. PCR products were quantified using the cycle threshold (Ct) value, and target gene expression was calculated as 0.5Delta Ct (target gene Ct – β-actin Ct). Primer sequences for real-time PCR were as follows: human β-actin internal control: forward 5′-TGACGTGGACATCCGCAAA G-3′, reverse 5′-CTGGAAGGTGGACA GCGAGG-3′; human NAE1: forward 5′-AAGCACTTCACTACGCTTAGC-3′, reverse 5′-TGGAGTATGACTGTGGT CCTTT-3′; human UBA3: forward 5′-CGATCTGGACCCTTCACACAC-3′, reverse 5′-GCCAGCTCCAATGACTAG AAC-3′; human UBC12: forward 5′-ATGAGGGCTTCTACAAGAGTGG-3′, reverse 5′-ATTGTCTCACACTTCACCTT GG-3′; human UBE2F: forward 5′-GGTTTCTGTGAGAGACAAATTGC-3′, reverse 5′-TCTGGGGTTACTGTTAG CTGAA-3′; human RBX1: forward 5′-TT GTGGTTGATAACTGTGCCAT-3′, reverse 5′-GACGCCTGGTTAGCTTG ACAT-3′; and human RBX2: forward 5′-TGGAAGACGGAGAGGAAACCT-3′, reverse 5′-TCCCCAGACCACAACA CAG T-3′.
Cell cycle analysis using propidium iodide staining and fluorescence-activated cell-sorting
The HHUA and AN3CA cells were treated with 0.3 µM MLN 4924 or DMSO for 48 h, harvested and fixed in 70% ethanol at –20 °C overnight, and stained with propidium iodide (PI, 36 µg/ml; Sigma, St. Louis, MO, USA) containing RNase (10 µg/ml, Sigma) at 37 °C for 30 min. Cells were then analysed by CyAn™ ADP flow cytometry (Beckman Coulter, Fullerton, CA, USA) according to the manufacturer’s instructions, to determine the proportions of cells in subG1 and G2/M phases. Apoptosis was measured as the percentage of cells in the subG1 population, and data were analysed with ModFit LT software, version 3.0 (Verity Software House, Topsham, ME, USA).
Colony formation assay
The HHUA and AN3CA cells were seeded in triplicate into 6-well plates at 500 cells/well. After culture for 24 h at 37 °C/5% CO2, cells were treated with 0.3 µM MLN 4924 or DMSO for 48 h, then cultured in fresh drug-free media for a further 12 days. Cell colonies were fixed with 4% paraformaldehyde and stained with crystal violet, and colonies with more than 50 cells were counted. Mean ± SD was calculated from analysis of triplicate wells, and each experiment was performed three times.
Immunoblot assays
Total protein was extracted from endometrial adenocarcinoma or adjacent noncancerous tumour tissues (10 mg each) by homogenising in radioimmunoprecipitation assay buffer that contained protease and phosphatase inhibitors. Protein was also extracted from HHUA and AN3CA cells that had been seeded at 6 × 105 cells per 100-mm dish, cultured overnight and treated with 0, 0.1, 0.3 or 1.0 µM MLN4924 for 48 h. Total protein concentrations were measured using a Thermo Scientific™ Pierce™ BCA Protein Assay Kit (ThermoFisher Scientific). Total protein (50 µg per sample) was loaded into 4–20% sodium-dodecyl sulphate-polyacrylamide gradient gels for electrophoresis, and the separated proteins were transferred onto polyvinylidene fluoride membranes. Membranes were blocked at room temperature with 5% bovine serum albumin before incubating with the following primary antibodies at 4 °C overnight: rabbit anti-UBE2F and rabbit anti-RBX2 (1:1000 dilution, Millipore Sigma, Burlington, MA, USA), rabbit anti-CDT1 (DNA replication factor Cdt1), rat anti-ORC1 (origin recognition complex subunit 1), rabbit anti-p-IĸBα, rabbit anti-IĸBα, rabbit anti-p27, rabbit anti-p21, rabbit anti-p-H2AX, and rabbit anti-c-Caspase3 (1:1000 dilution; Cell Signaling Technology, Danvers, MA, USA), mouse anti-CUL5 (cullin 5; 1:500 dilution; Santa Cruz biotechnology, Santa Cruz, CA, USA), and rabbit anti-GAPDH loading control (1:1000 dilution; Cell Signaling Technology). Membranes were then washed three times (10 min each) in tris-buffered saline with 0.1% Tween 20 (TBST) before incubating with horse-radish peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG secondary antibodies (1:1000 dilution; Cell Signaling Technology) for 2 h at room temperature. Membranes were again washed three times in TBST and immunoreactive signals were visualized using an ECL Western Blotting detection reagent kit (cat No. 1705061; BioRad, Watford, UK) and ImageQuant LAS 4000 imaging system (GE Healthcare, Little Chalfont, UK).
Cell proliferation assay
The HHUA and AN3CA cells were seeded in triplicate into 96-well plates at 1 × 103 cells/well, and then treated with 0.3 µM MLN4924 or DMSO for 0, 24, 48, 72, 96 or 120 h. Cell viability was determined by Cell Counting Kit-8 assay (CCK8; Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions.
Statistical analyses
Data are presented as mean ± SD, and were analysed using SPSS software, version 17.0 (SPSS Inc., Chicago, IL, USA). Parameters between two groups were compared with Student’s t-test, and two-way analysis of variance was used to compare more than two groups. Survival was analysed by the Kaplan–Meier method. A P value < 0.05 was considered to be statistically significant, and three significance levels are reported: P < 0.05, P < 0 .01, and P < 0.001.
Results
Expression of neddylation genes in cancers
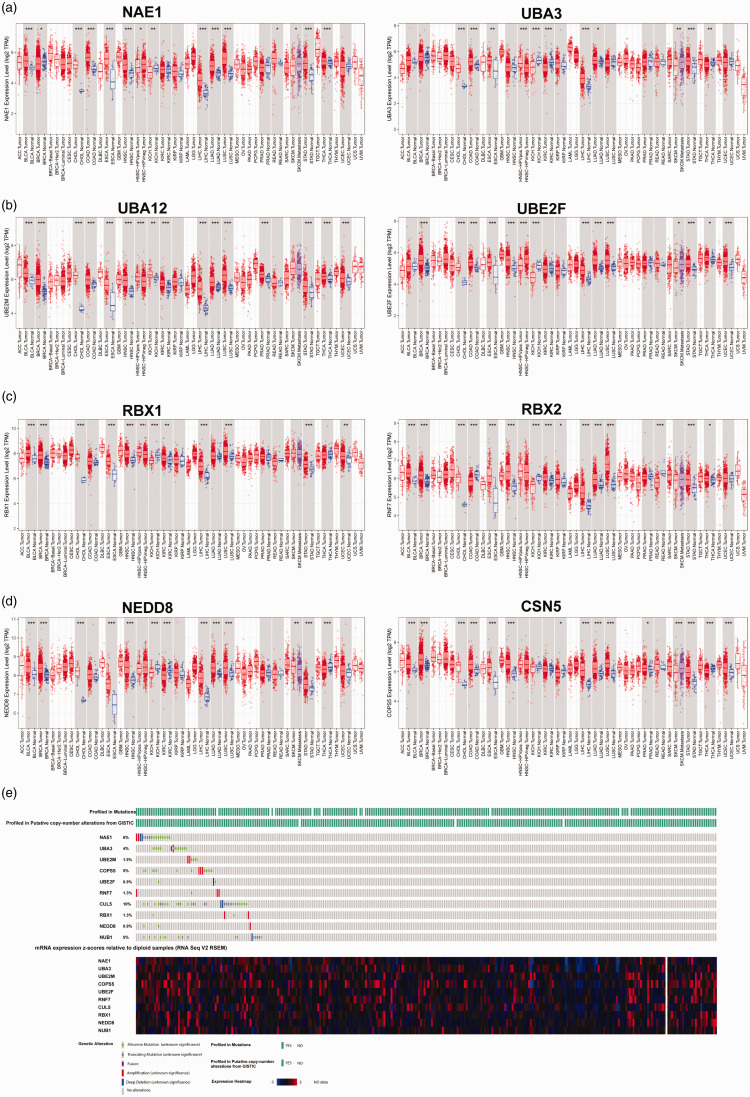
Expression of key genes involved in the neddylation pathway, including E1, E2, and E3 enzymes, NEDD8 ubiquitin like modifier (NEDD8), and COP9 signalosome subunit 5 (COPS5 or CSN5), a key gene for de-neddylation of the cullins, 18 was analysed using the online server TIMER, and was found to vary between cancer types (Figure 1a–d). Compared with normal tissue, NAE1 was overexpressed in bladder urothelial carcinoma, cholangiocarcinoma, oesophageal carcinoma, head and neck squamous cell carcinoma, liver hepatocellular carcinoma, lung adenocarcinoma (LUAD), lung squamous cell carcinoma, skin cutaneous melanoma, and stomach adenocarcinoma; and downregulated in breast invasive carcinoma and kidney chromophobe. UBA3 was overexpressed in cholangiocarcinoma, colon adenocarcinoma, oesophageal carcinoma, liver hepatocellular carcinoma, LUAD, and stomach adenocarcinoma; and downregulated in kidney chromophobe, kidney renal clear cell carcinoma and thyroid carcinoma. Similar variability was observed for the other key genes in different cancer types (Figure 1).
Figure 1.
Expression and somatic variations of E1, E2 and E3 enzymes of neddylation in samples from The Cancer Genome Atlas (TCGA): (a) NEDD8-activating enzyme E1 regulatory subunit (NAE1) and NEDD8-activating enzyme E1 catalytic subunit (UBA3) expression; (b) ubiquitin conjugating enzyme E2 M (UBC12) and ubiquitin conjugating enzyme E2 F (UBE2F) expression; (c) ring-box 1 (RBX1) and ring finger protein 7 (RBX2) expression; (d) NEDD8 and COP9 signalosome subunit 5 (CSN5) expression; and (e) somatic variations in neddylation genes. BLCA, bladder urothelial carcinoma; CHOL, cholangiocarcinoma; ESCA, oesophageal carcinoma; HNSC, head and neck squamous cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; BRCA, breast invasive carcinoma; KICH, kidney chromophobe; COAD, colon adenocarcinoma; KIRC, kidney renal clear cell carcinoma; and THCA, thyroid carcinoma. *P < 0.05, **P < 0.01, ***P < 0.001.
Expression and somatic variations of neddylation genes in EC tissues and association with patient survival
Expression of two E1 enzyme subunits, NAE1 and UBA3, was analysed in EC versus normal non-tumour tissues using TIMER, and no differences in expression were found (Figure 1a). Expression of the E2 enzymes, UBC12 and UBE2F, was also analysed using TIMER, and both were found to be significantly overexpressed in EC tumour tissues compared with normal non-tumour tissues (P < 0.05; Figure 1b). As cullin family proteins are well-studied neddylation substrates that can be neddylated by RBX1 (Cul1-4) or RBX2 (Cul5),8,19 the expression of these two E3 enzymes was also analysed in the TGCA EC dataset. RBX1, but not RBX2 (RNF7), was found to be significantly overexpressed in EC tumour tissues compared with normal non-tumour tissues (P < 0.05; Figure 1c). In addition, the key deneddylation gene, CSN5, was overexpressed in EC versus normal tissue (P < 0.05; Figure 1d), which may indicate the deneddylation activating status in EC.
Using the cBioPortal online server, the somatic mutation profile of various NEDD8 associated genes in TCGA EC samples was analysed, and revealed the following somatic mutation frequency rates: NEA1, 6%; UBA3, 4%; UBE2M, 1.9%; CSN5, 5%; UBE2F, 0.9%; RBX2, 1.3%; RBX1, 1.3%; and NEDD8, 0.9% (Figure 1e). Most of the diploid samples were observed to express neddylation genes.
Endometrial adenocarcinoma and adjacent noncancerous tissues were collected from a total of 71 patients (15 aged >50 years, and 56 aged ≤50 years). World Health Organization tumour classifications were as follows: grade I tumour, 17 patients; grade II tumour, 28 patients; and grade III tumour, 26 patients. Tumour mRNA was extracted from 41 samples for RT-qPCR, and 30 samples were used for western blot analysis.
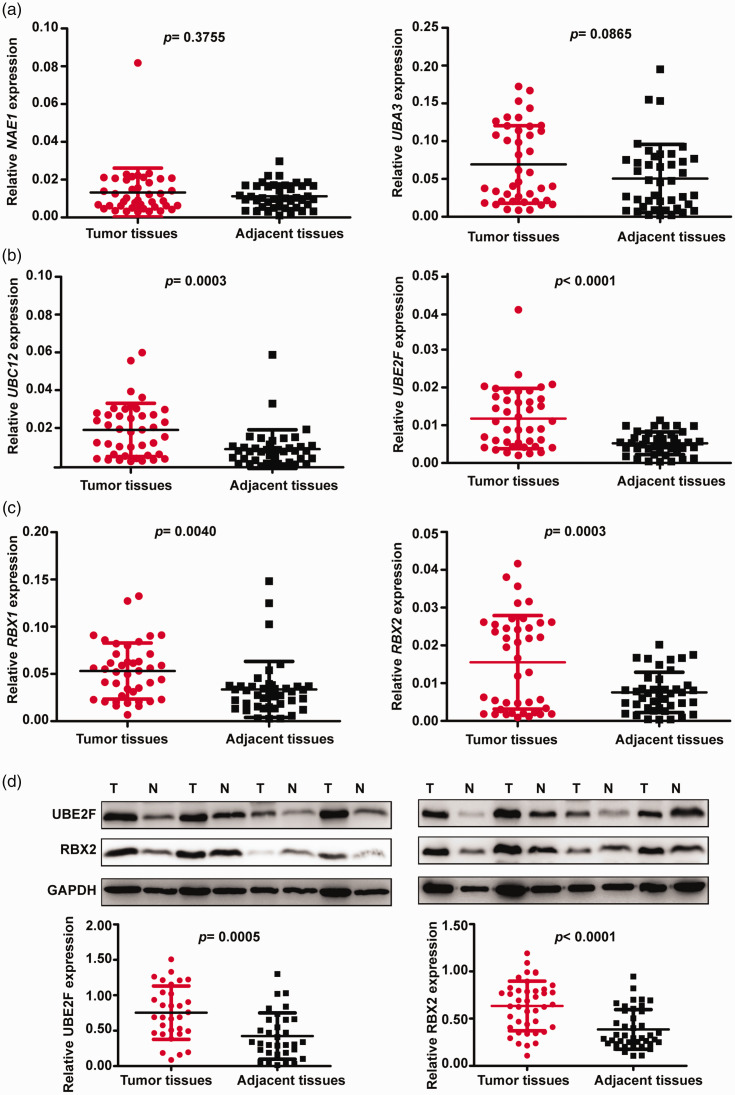
Expression levels of E1, E2 and E3 genes in EC and adjacent tissues from 41 patients were investigated by RT-PCR (Figure 2). Consistent with TCGA data, no differences in expression of NAE1 and UBA3 were observed between tumour tissues and adjacent tissues (Figure 2a), but expression of E2 and E3 genes were significantly elevated in EC versus normal adjacent tissues (P < 0.01; Figure 2b and 2c).
Figure 2.
Vertical scatter plots showing expression of E1, E2 and E3 neddylation enzymes in endometrial cancer (EC) tissues and adjacent normal tissues: (a) reverse transcription (RT)-polymerase chain reaction (PCR) results showing NEDD8-activating enzyme E1 regulatory subunit (NAE1) and NEDD8-activating enzyme E1 catalytic subunit (UBA3) expression; (b) RT-PCR results showing ubiquitin conjugating enzyme E2 M (UBC12) and ubiquitin conjugating enzyme E2 F (UBE2F) expression; (c) RT-PCR results showing ring-box 1 (RBX1) and ring finger protein 7 (RBX2) expression; and (d) western blot analysis showing representative immunoblot panels and relative UBE2F and RBX2 protein levels. T, tumour tissue; N, normal adjacent tissue; GAPDH, loading control. Data presented as mean ± SD.
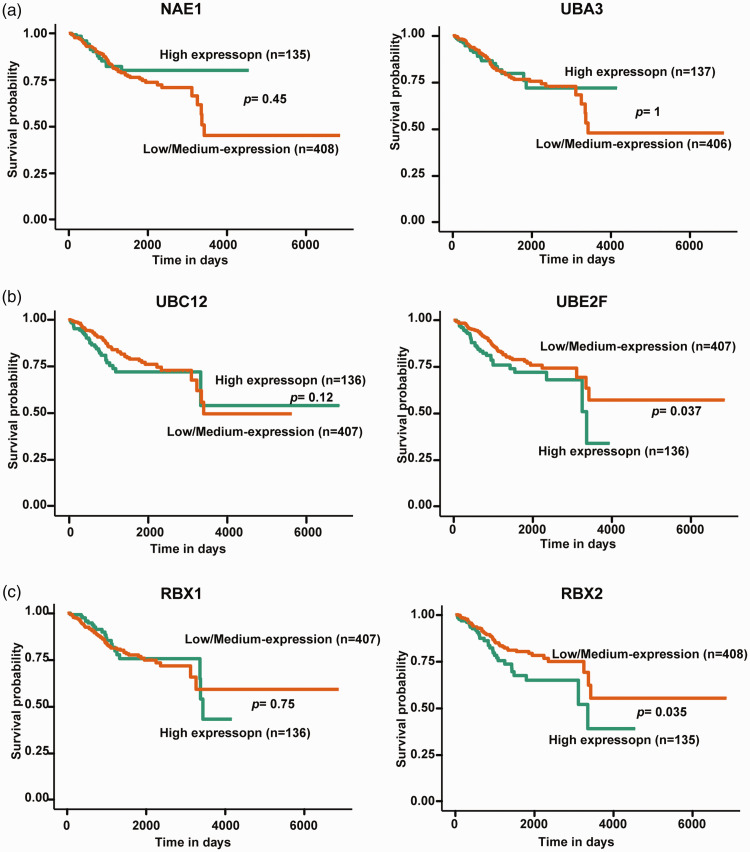
Further western blot analyses of relative protein levels in tissue samples from 30 patients showed that UBE2F and RBX2 proteins were significantly elevated in EC tumour tissues compared with normal adjacent tissue (P < 0.001; Figure 2d). Survival analysis of TCGA data revealed that only UBE2F and RBX2 overexpression were indicators of poor prognosis in patients with EC (Figure 3).
Figure 3.
Kaplan–Meier curves showing the correlation between neddylation enzyme expression and survival of patients with endometrial cancer (EC) from The Cancer Genome Atlas dataset: (a) E1 enzymes NEDD8-activating enzyme E1 regulatory subunit (NAE1) and NEDD8-activating enzyme E1 catalytic subunit (UBA3); (b) E2 enzymes ubiquitin conjugating enzyme E2 M (UBC12) and ubiquitin conjugating enzyme E2 F (UBE2F); and (c) E3 enzymes ring-box 1 (RBX1) and ring finger protein 7 (RBX2). Kaplan–Meier curves were analysed using the UALCAN online server.
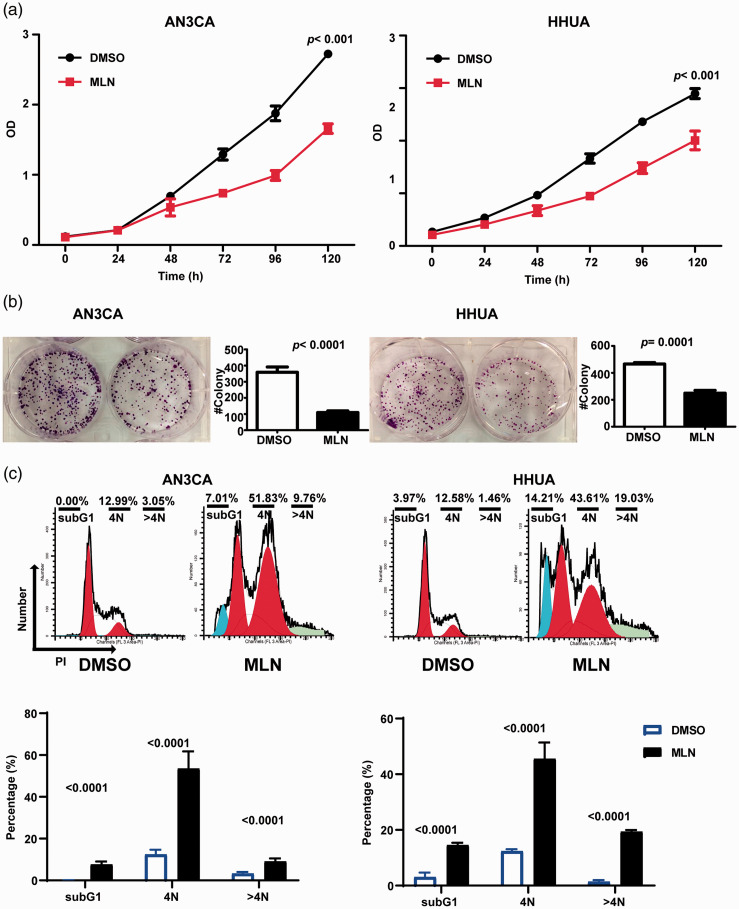
MLN4924 suppresses proliferation and cloning of EC cells
A specific inhibitor of neddylation, MLN4924, was investigated for its in vitro effects in EC cells. HHUA and AN3CA cell lines treated with 0.3 µM MLN4924 over 120 h showed significantly reduced proliferation versus cells treated with DMSO only (P < 0.001; Figure 4a). In addition, the colony forming ability of both cell lines was significantly reduced after 12 days by treatment with 0.3 µM MLN 4924 for 48 h versus treatment with DMSO (P ≤ 0.0001; Figure 4b).
Figure 4.
MLN4924 suppressed proliferation and colony formation of endometrial cancer cells in vitro: (a) cell counting kit-8 assay showing reduced proliferation of HHUA and AN3CA cells treated with 0.3 µM MLN4924. Data presented as mean ± SD of three replicates; (b) representative images of crystal violet-stained cells, and colony forming data showing reduced colony forming ability, after 12 days in culture, of HHUA and AN3CA cells treated with 0.3 µM MLN4924 for 48 h. Data presented as mean ± SD of three independent experiments; (c) Representative flow cytometry images and data showing MLN4924-induced cell cycle arrest and apoptosis of HHUA and AN3CA cells treated with 0.3 µM MLN4924 or DMSO for 48 h. The percentages of cells in G2/M and subG1 phases were determined. Data presented as mean ± SD of three replicates. MLN, MLN4924; OD, optical density; PI, propidium iodide.
MLN4924 induces cell cycle arrest and apoptosis of EC cells
The anticancer effects of neddylation inhibition by MLN4924 are reported to be attributable to induction of the DNA damage response, cell cycle arrest and apoptosis in cancer cells.6,20 PI staining and flow cytometry showed that MLN4924 significantly induced G2/M cell cycle arrest (P < 0.0001; Figure 4c), and the subG1 population was found to be increased, indicating apoptosis. 12 Furthermore, the population of cells with DNA > 2N was increased (P < 0.0001; Figure 4c).
MLN4924 induces cullin-RING ligase substrate accumulation
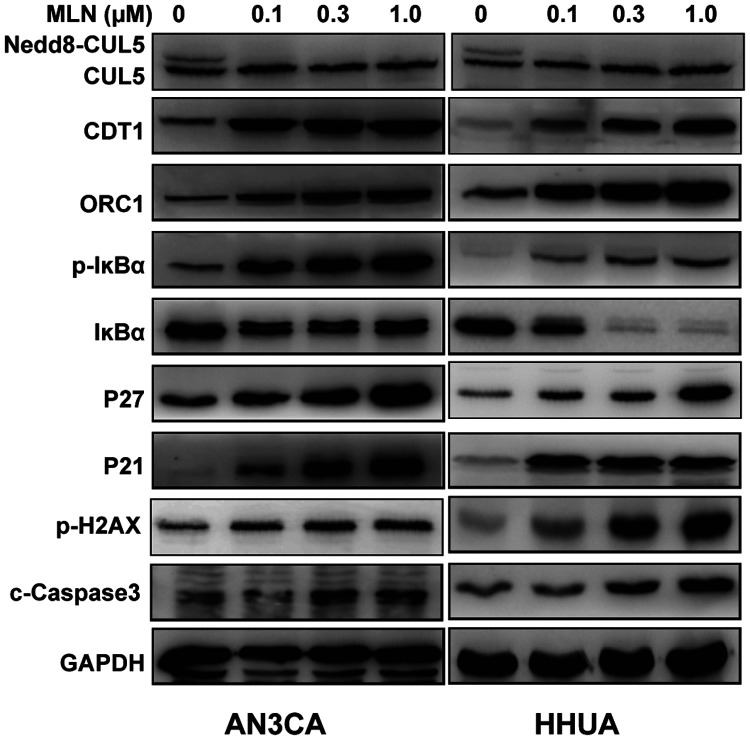
Cullin family members are the most studied neddylation substrates, and cullin-RING ligases (CRLs) are dysregulated in many types of cancers, leading to accelerated degradation of tumour suppressors and promotion of tumorigenesis and tumour progression. 21 Western blot analyses of HHUA and AN3CA cell lines treated with 0, 0.1, 0.3 or 1.0 µM MLN4924 over 48 h revealed that MLN4924 inactivated neddylation modification of cullin 5, and induced the accumulation of CRL substrates, including CDT1 and ORC1, p27 and p21, and IκBα, a key protein of the nuclear factor (NF)-κB signalling pathway (Figure 5). As MLN4924 was found to induce subG1 as well as DNA replication stress, the DNA damage and/or cell apoptosis marker p-H2AX, and c-Caspase3 was also analysed in the MLN4924 treated EC cells. As shown in Figure 5, p-H2AX and c-Caspase3 were increased upon MLN4924 treatment.
Figure 5.
Representative western blot panels showing relative protein levels in HHUA and AN3CA cells in response to treatment with 0–1.0 µM MLN4924 for 48 h. MLN4924 was shown to inhibit cullin 5 neddylation and induce cullin-RING ligase substrate accumulation. CUL5, cullin 5; CDT1, DNA replication factor Cdt1; ORC1, origin recognition complex subunit 1; P21, cyclin dependent kinase inhibitor 1A; P27, cyclin dependent kinase inhibitor 1B; IκBα, nuclear factor κB inhibitor alpha; p-IκBα, phosphorylated-IκBα; P-H2AX, phosphorylated Histone H2A.X; c-Caspase3, cleaved-Caspase-3; and GADPH loading control.
Discussion
Endometrial carcinoma is one of the most frequently diagnosed gynaecological malignancies, characterized by high rates of recurrence and poor prognosis. 22 Neddylation, a post-translational modification that conjugates the ubiquitin-like protein NEDD8 to substrate proteins, is an important biochemical process that regulates protein function. 20 The neddylation pathway is reported to be overactivated in multiple cancer types, including liver, lung and colon cancer, and its overactivation indicates poor prognosis. 23 In the present study, the expression profiles of key neddylation genes were analysed in TCGA samples using an online server.
To the best of our knowledge, the present study is the first one to investigate the expression profiles of neddylation genes in endometrial carcinoma. The expression of E1, E2 and E3 neddylation enzymes was analysed in sample data from the TCGA database and in a cohort of samples obtained for the present investigation. No differences were found in E1 enzyme mRNA levels between endometrial carcinoma tissues and adjacent normal tissues; E2 enzymes, including UBE2M and UBE2F, were found to be elevated, as were E3 enzymes, including RBX1 and RBX2, however, only UBE2F and RBX2 overexpression were demonstrated to predict poor prognosis in patients with endometrial carcinoma. The neddylation enzyme, NAE1, is reported to be elevated in lung cancer. 6 The present results showed that expression levels of E1 genes in EC were not elevated, but E2 gene expression was elevated in EC, which was not consistent with the report in lung cancer. 6 We surmise that the expression of neddylation pathway genes may be tissue type-dependent, and different enzymes are elevated to support the overactivation of neddylation in different cancers.
Ubiquitin and ubiquitin-like proteins are directed to targets by cascades of E1, E2, and E3 enzymes. CRLs are one of the largest families of multiple subunit ubiquitin E3 ligases, containing one cullin protein (CUL1, -2, -3, -4, or -5) and one RING protein (RBX1 or RBX2). 24 UBC12–RBX1 and UBE2F–RBX2 pairs are reported to specifically regulate the neddylation of cullins 1–4 and CUL5, respectively. 19 In the present research, UBE2F and RBX2 expression was found to be elevated in TCGA data, and their overexpression was associated with a poor prognosis, suggesting that the UBE2F–RBX2 axis may play an important role in endometrial carcinoma development. Thus, the expression of UBE2F and RBX2 was examined in a cohort of endometrial carcinoma tissues and adjacent normal tissues, and revealed that expression of UBE2F and RBX2 was significantly higher in endometrial carcinoma tissues at both the protein and mRNA levels, which was consistent with the TCGA data.
Inhibition of protein neddylation has been described as an attractive anticancer strategy. 14 High throughput screening identified N6-benzyl adenosine as an inhibitor of NAE, and further medicinal chemistry investigations resulted in the discovery of MLN4924. 21 Preclinical studies have revealed that MLN4924 has a potent activity, with well-tolerated toxicity against a range of solid tumours and haematological malignancies. Furthermore, combinations of MLN4924 with chemoradiotherapy have been shown to increase antitumour activity in AML and solid tumour cell lines, and in xenograft models.25–28 For example, inhibition of neddylation with MLN4924 overcame platinum resistance in in vitro models of platinum-resistant ovarian cancer. 29 However, the effect of MLN4924 on endometrial carcinoma is unreported. In the present study, treatment of endometrial carcinoma cells with MLN4924 was found to effectively induce G2/M cell cycle arrest, cell apoptosis and DNA duplication, and was shown to inhibit proliferation and colony formation. Thus, whether MLN4924 can enhance the radiotherapy effects during treatment of endometrial cancer deserves further study.
The best-known neddylation substrates are the cullin family members, and neddylation inhibition by MLN4924 has been shown to exert significant anticancer effects mainly through inactivation of CRLs and accumulation of critical CRL substrates, thus triggering multiple cellular responses, leading to the induction of cell cycle arrest, apoptosis, senescence and autophagy in a cell-type dependent manner. 20 MLN4924 is reported to cause accumulation of the CRL substrates CDT1 and ORC1 to trigger DNA re-replication stress and subsequent DNA damage, leading to cell cycle arrest and cell death in cancer cells.11,20 MLN4924 has also been shown to induce apoptosis via a time-dependent stabilization of the CRL substrate p-IκBα, to inactivate NF-κB in haematological malignancies.26,30 In the present study, MLN4924 indeed induced the accumulation of CDT1, ORC1 and p-IκBα, which may result in cell cycle defects and apoptosis in endometrial carcinoma cells. In addition, MLN4924 was shown to induce p27 and p21 accumulation, which is a previously reported mechanism of MLN4924.31–33
The results of the present study may be limited by several factors, for example, the expression of key neddylation genes was not validated using tumour tissue arrays, which contain large numbers of patients’ samples; and survival values associated with key neddylation genes were not validated in a separate cohort of samples. Further studies should focus on the identification of substrates for neddylation or the neddylation-related signalling pathway, that may play a role in EC tumorigenesis and progression.
In conclusion, the results of the present study indicated that the neddylation pathway is activated in EC, particularly the UBE2F–RBX2–CUL5 axis, and that expression levels of key molecules within this pathway are upregulated. In addition, MLN4924 treatment significantly suppressed EC cell growth in vitro. MLN4924 may suppress EC cell proliferation by inducing the accumulation of CRL substrates, such as CDT1, ORC1, p27, p-IκBα and p21, which may result in cell cycle arrest, DNA damage, and cell apoptosis.
Footnotes
Author contributions: HRL developed the hypothesis, conducted the experiments and drafted the manuscript. HRL, QB and XL contributed to data analysis and interpretation.
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by a grant to HRL from the Scientific Research Project (Type A) of Shanghai First Maternity and Infant Hospital (2017).
ORCID iD: Huanrong Liu https://orcid.org/0000-0001-6501-2668
References
- 1.Li BL, Lu W, Qu JJ, et al. Loss of exosomal miR-148b from cancer-associated fibroblasts promotes endometrial cancer cell invasion and cancer metastasis. J Cell Physiol 2019; 234: 2943–2953. [DOI] [PubMed] [Google Scholar]
- 2.Sarink D, Wilkens LR, White KK, et al. Racial/ethnic differences in anthropometric and hormone-related factors and endometrial cancer risk: the Multiethnic Cohort Study. Br J Cancer 2021; 124: 1724–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morice P, Leary A, Creutzberg C, et al. Endometrial cancer. Lancet 2016; 387: 1094–1108. [DOI] [PubMed] [Google Scholar]
- 4.Fleming GF, Brunetto VL, Cella D, et al. Phase III trial of doxorubicin plus cisplatin with or without paclitaxel plus filgrastim in advanced endometrial carcinoma: a Gynecologic Oncology Group Study. J Clin Oncol 2004; 22: 2159–2166. [DOI] [PubMed] [Google Scholar]
- 5.Rabinovich A. Neo-adjuvant chemotherapy for advanced stage endometrial carcinoma: a glimmer of hope in select patients. Arch Gynecol Obstet 2016; 293: 47–53. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Wang M, Yu G, et al. Overactivated neddylation pathway as a therapeutic target in lung cancer. J Natl Cancer Inst 2014; 106: dju083. [DOI] [PubMed] [Google Scholar]
- 7.Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects' review series. EMBO Rep 2008; 9: 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enchev RI, Schulman BA, Peter M. Protein neddylation: beyond cullin-RING ligases. Nat Rev Mol Cell Biol 2015; 16: 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Kang J, Zhang W, et al. Validation of NEDD8-conjugating enzyme UBC12 as a new therapeutic target in lung cancer. EBioMedicine 2019; 45: 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia X, Li C, Li L, et al. Neddylation inactivation facilitates FOXO3a nuclear export to suppress estrogen receptor transcription and improve fulvestrant sensitivity. Clin Cancer Res 2019; 25: 3658–3672. [DOI] [PubMed] [Google Scholar]
- 11.Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009; 458: 732–736. [DOI] [PubMed] [Google Scholar]
- 12.Luo Z, Yu G, Lee HW, et al. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res 2012; 72: 3360–3371. [DOI] [PubMed] [Google Scholar]
- 13.Gai W, Peng Z, Liu CH, et al. Advances in cancer treatment by targeting the neddylation pathway. Front Cell Dev Biol 2021; 9: 653882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Q, Yu GY, Shi JY, et al. Neddylation pathway is up-regulated in human intrahepatic cholangiocarcinoma and serves as a potential therapeutic target. Oncotarget 2014; 5: 7820–7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017; 19: 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017; 77: e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson IR, Irwin MS, Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell 2011; 19: 168–176. [DOI] [PubMed] [Google Scholar]
- 19.Huang DT, Ayrault O, Hunt HW, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell 2009; 33: 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L, Zhang W, Sun Y, et al. Protein neddylation and its alterations in human cancers for targeted therapy. Cell Signal 2018; 44: 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Q, Jiang Y, Sun Y. Anticancer drug discovery by targeting cullin neddylation. Acta Pharm Sin B 2020; 10: 746–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu J, Liu B, Li B, et al. TRIB3 suppresses proliferation and invasion and promotes apoptosis of endometrial cancer cells by regulating the AKT signaling pathway. Onco Targets Ther 2019; 12: 2235–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng Y, Iv YS, Pan QH, et al. An overactive neddylation pathway serves as a therapeutic target and MLN4924 enhances the anticancer activity of cisplatin in pancreatic cancer. Oncol Lett 2019; 18: 2724–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y, Morgan MA, Sun Y. Targeting neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid Redox Signal 2014; 21: 2383–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swords RT, Kelly KR, Smith PG, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood 2010; 115: 3796–3800. [DOI] [PubMed] [Google Scholar]
- 26.Milhollen MA, Traore T, Adams-Duffy J, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-κB-dependent lymphoma. Blood 2010; 116: 1515–1523. [DOI] [PubMed] [Google Scholar]
- 27.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res 2009; 15: 3912–3916. [DOI] [PubMed] [Google Scholar]
- 28.Abidi N, Xirodimas DP. Regulation of cancer-related pathways by protein NEDDylation and strategies for the use of NEDD8 inhibitors in the clinic. Endocr Relat Cancer 2015; 22: T55–T70. [DOI] [PubMed] [Google Scholar]
- 29.Jazaeri AA, Shibata E, Park J, et al. Overcoming platinum resistance in preclinical models of ovarian cancer using the neddylation inhibitor MLN4924. Mol Cancer Ther 2013; 12: 1958–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godbersen JC, Humphries LA, Danilova OV, et al. The Nedd8-activating enzyme inhibitor MLN4924 thwarts microenvironment-driven NF-κB activation and induces apoptosis in chronic lymphocytic leukemia B cells. Clin Cancer Res 2014; 20: 1576–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao WT, Wu JF, Yu GY, et al. Suppression of tumor angiogenesis by targeting the protein neddylation pathway. Cell Death Dis 2014; 5: e1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia L, Li H, Sun Y. Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia 2011; 13: 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Hou D, Luo Z, et al. The novel protective role of P27 in MLN4924-treated gastric cancer cells. Cell Death Dis 2015; 6: e1867. [DOI] [PMC free article] [PubMed] [Google Scholar]