Abstract
Background
Leprosy, a chronic infectious disease caused by Mycobacterium leprae, is often late- or misdiagnosed leading to irreversible disabilities. Blood transcriptomic biomarkers that prospectively predict those who progress to leprosy (progressors) would allow early diagnosis, better treatment outcomes and facilitate interventions aimed at stopping bacterial transmission. To identify potential risk signatures of leprosy, we collected whole blood of household contacts (HC, n=5,352) of leprosy patients, including individuals who were diagnosed with leprosy 4-61 months after sample collection.
Methods
We investigated differential gene expression (DGE) by RNA-Seq between progressors before presence of symptoms (n=40) and HC (n=40), as well as longitudinal DGE within each progressor. A prospective leprosy signature was identified using a machine learning approach (Random Forest) and validated using reverse transcription quantitative PCR (RT-qPCR).
Findings
Although no significant intra-individual longitudinal variation within leprosy progressors was identified, 1,613 genes were differentially expressed in progressors before diagnosis compared to HC. We identified a 13-gene prospective risk signature with an Area Under the Curve (AUC) of 95.2%. Validation of this RNA-Seq signature in an additional set of progressors (n=43) and HC (n=43) by RT-qPCR, resulted in a final 4-gene signature, designated RISK4LEP (MT-ND2, REX1BD, TPGS1, UBC) (AUC=86.4%).
Interpretation
This study identifies for the first time a prospective transcriptional risk signature in blood predicting development of leprosy 4 to 61 months before clinical diagnosis. Assessment of this signature in contacts of leprosy patients can function as an adjunct diagnostic tool to target implementation of interventions to restrain leprosy development.
Funding
This study was supported by R2STOP Research grant, the Order of Malta-Grants-for-Leprosy-Research, the Q.M. Gastmann-Wichers Foundation and the Leprosy Research Initiative (LRI) together with the Turing Foundation (ILEP# 702.02.73 and # 703.15.07).
Keywords: Leprosy, Diagnostics, RNA-Seq, Transcriptomics, Biomarker, Prediction
Research in context.
Evidence before this study
Leprosy, an infectious disease caused by Mycobacterium leprae that still poses a considerable health and economic threat in areas of several low and middle income countries. Unfortunately, leprosy is often late- or misdiagnosed leading to irreversible disabilities and deformities. Identifying individuals who are at risk of developing leprosy disease, before clinical symptoms arise, is crucial to reduce leprosy-associated disabilities as well as transmission of the bacterium.
Host transcriptomic biomarkers are intensely investigated as tools for diagnosis of tuberculosis, an infectious disease caused by the related Mycobacterium tuberculosis. In leprosy research, only a small amount of transcriptomic biomarker profiles have been identified as potential diagnostic tools for leprosy disease. However, the previously published biomarkers detect leprosy after occurrence of symptoms and involve invasive samples such as skin or nerve biopsies.
Added value of this study
In the present investigation, we collected venous blood of household contacts of leprosy patients from Bangladesh to identify a transcriptomic biomarker predicting leprosy development. This involved sampling at recruitment into the study before any clinical signs were present, and at diagnosis of leprosy. Importantly, we describe a 4-gene signature, RISK4LEP that can identify individuals who will develop leprosy 4–61 months prior to clinical diagnosis. This signature has potential for application in diagnostic tests for leprosy as it is based on unstimulated whole blood and only a low number of genes, thereby harbouring essential characteristics for rapid, user-friendly point-of-care tests.
Implications of all available evidence
This study demonstrated the potential of host whole blood transcriptomic biomarkers as tools for early diagnostics of leprosy and identified a prospective 4-gene signature, RISK4LEP, that can identify individuals who will develop leprosy allowing for prophylactic or early treatment. Since household contacts of leprosy patients present a higher risk of developing leprosy, application of RISK4LEP can guide health care workers to target implementation of prophylaxis.
Further longitudinal studies in other endemic areas are now required to validate the RISK4LEP signature.
Alt-text: Unlabelled box
1. Introduction
Leprosy, also known as Hansen's disease, is still a considerable health threat in pockets of several low- and middle-income countries worldwide. The annual number of new cases fluctuates around 200,000 people, reflecting a stable trend that has been observed during the last decade [1]. Affecting the skin and peripheral nerves, leprosy presents as a spectrum including several clinical forms paralleling immunity against Mycobacterium leprae, the pathogen causing leprosy [2]. On one pole of the immunopathological spectrum tuberculoid leprosy (TT) is situated, mainly characterized by low amount of bacteria and a cell-mediated immune response, and at the other pole lepromatous leprosy (LL) presenting high bacterial load, and a humoral response [3,4]. In between these polar forms patients present borderline leprosy (borderline tuberculoid [BT], borderline borderline [BB] and borderline lepromatous (BL]) [5].
Diagnosis still heavily relies on detection of clinical symptoms and early detection of leprosy represents a substantial hurdle in present-day leprosy health care. Besides, the reduced number of new cases has resulted in unfamiliarity of signs and symptoms of leprosy limiting suspicion and detection of leprosy. Only a small percentage (estimated 5%) of people exposed to M. leprae develop the disease [3]. In addition, leprosy displays a long incubation period (2 to >10 years) [6,7]. These factors contribute to limited awareness of the disease among both the public and healthcare providers, hampering the early detection of new cases, and are reinforced by the strong social stigma of leprosy. Detection delay not only results in frequent delay of treatment leading to irreversible disabilities, but also contributes to perpetuating transmission.
Leprosy is a multi-factorial disease influenced by the infectious agent (dose and frequency of exposure) but also by genetics [8], [9], [10], [11], [12], [13], [14], nutritional factors [15,16], living conditions [17,18] and individual characteristics (age, sex) [19,20]. Household contacts (HC) of leprosy patients are at highest risk [21], [22], [23], [24], and thus a recommendation for use of chemoprophylaxis as preventive treatment for contacts of leprosy patients was included in the WHO 2018 guidelines [25]. Given the low proportions of individuals actually developing leprosy after M. leprae exposure, biomarkers identifying who will develop disease would be very useful to target prophylactic measures.
In the past years, several studies have searched for biomarkers to (early) detect leprosy either based on the host immune response [26], [27], [28], [29], [30], [31], the pathogen [32], [33], [34], [35], [36], [37], or a combination of both [38], [39], [40], [41], [42], [43], [44], [45]. Molecular detection by identification of the repetitive element RLEP by (quantitative) PCR [33,46,47] as well as detection of anti-M. leprae phenolic glycolipid I (PGL-I) IgM in blood [28,29] are methods employed to assist leprosy diagnosis. Nevertheless, the sensitivity of these techniques to identify paucibacillary (PB) leprosy is not sufficient due to the low concentrations of bacilli in these patients [26,44,45]. On the other hand, PCR and anti-PGL-IgM, though useful to detect infection, are inadequate predictors of disease amongst HC of leprosy patients, as individuals remaining without disease may present positive PCR and/or PGL-I IgM [28,32,35,44,45]. In addition, combinations of other host proteins [24,27] have been shown to be useful to diagnose leprosy and detect M. leprae infection, but have not been studied prospectively yet.
Transcriptomic analysis of differential gene expression (DGE) represents an effective approach to identify new biomarkers for leprosy diagnosis [54]. RNA-Seq, a high-throughput and unbiased technique which includes the whole transcriptome instead of a selection of genes, has been successfully used to prospectively identify correlates of risk for leprosy reversal reaction [48], as well as for tuberculosis caused by the closely related bacteria Mycobacterium tuberculosis [49], [50], [51], [52].
The immune response during leprosy and leprosy reactions has also been investigated through transcriptomics [53], [54], [55], [56], [57], [58], [59], [60], [61], [62]. However, very few studies have employed transcriptomics to identify a biomarker risk signature for leprosy diagnosis: one study described that gene expression of LDR and CCL4 in nerve biopsies identified up to 80% of pure neural (PN) leprosy patients [63]. Likewise, a signature formed by four miRNA was identified using skin biopsies that could discriminate leprosy patients with 80% sensitivity and 91% specificity [53]. Although these transcriptomic biomarkers show potential, both are based on samples that require invasive techniques (nerve and skin biopsies) and were applied when clinical symptoms were already visible.
In contrast to previous work, this study aimed to identify a prospective biomarker signature that can predict development of leprosy. For this purpose, whole blood samples were collected from HC who were followed up for several years and re-sampled in case they developed leprosy. Transcriptomic differences were investigated between progressors and HC who remained without leprosy. Variation in gene expression of those individuals who developed leprosy was assessed between the timepoint before leprosy diagnosis and at onset of disease. A risk signature for leprosy development can guide post-exposure prophylactic strategies to avoid disease progression, reduce disability and contribute to stop M. leprae transmission.
2. Methods
2.1. Sample collection and study design
HC (n=5,352) of newly diagnosed leprosy patients were recruited and a first blood sample was collected from April 2013 to April 2018 as part of a field trial [28,[64], [65], [66]] in four districts in the northwest of Bangladesh (Nilphamari, Rangpur, Panchagarh and Thakurgaon). Patients and HC entered the study through the Rural Health Program of The Leprosy Mission International, Bangladesh, based at the Danish Bangladesh Leprosy Mission Hospital in Nilphamari, a referral hospital specialized in the detection and treatment of leprosy. The population of the four districts, which was around 7,000,000 at the start of intake, is mainly rural, but includes six main towns. The new case detection rate and the prevalence in the study area were 1.18 and 0.9 per 10,000 correspondingly [67].
HC were defined as those living in the same house, in a house on the same compound and sharing the same kitchen, or direct neighbours (first neighbours). Exclusion criteria included previous leprosy, refused informed consent, pregnant women, tuberculosis, children younger than 5 years, liver disease or jaundice and temporary residency in the study area [66]. Some HC in the study received BCG vaccination (n=657) after providing the first blood sample. Whole blood from HC was collected in PAXgene tubes at time of diagnosis of the index case (t=1) (Fig. 1). All contacts were followed up annually and checked for the absence of clinical signs and symptoms of leprosy. All individuals were followed up for 36 months or longer. Follow up is still ongoing. Contacts who were clinically diagnosed with leprosy within 4–61 months after recruitment were considered progressors (n=85). A second blood sample was collected from progressors at the time of leprosy diagnosis, before start with multidrug therapy (t=2) and bacteriological index (BI) was determined. Leprosy was diagnosed by a medical officer following the Rural Health Program guidelines in accordance to the National Leprosy Control Program [68]. Progressors who presented five or fewer skin lesions and BI 0 were classified as PB and those who presented more than five skin lesions were classified as MB [25].
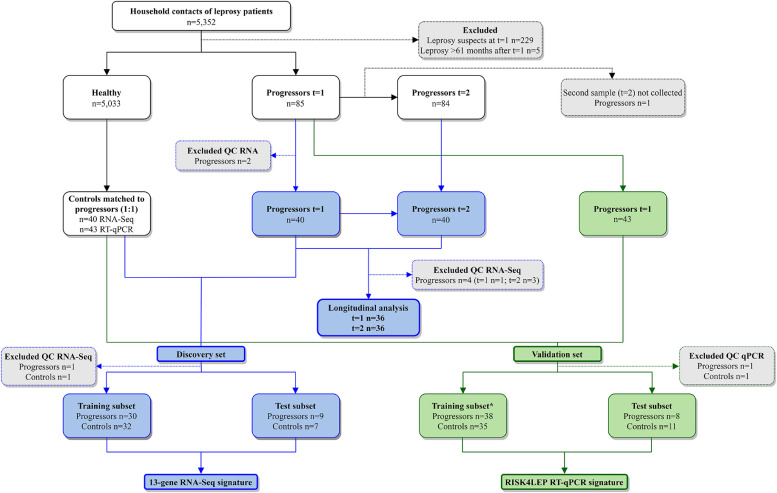
Fig. 1.
Study design to identify a transcriptomic signature associated with leprosy risk. In blue samples used in the discovery set (RNA-Seq) and in green samples used in the validation set (reverse transcription quantitative PCR (RT-qPCR)). Progressors are household contacts who developed leprosy within 4-61 months (Fig. S1) after recruitment. t=1 is the timepoint before disease and t=2 is the timepoint of leprosy diagnosis. Excluded QC (quality check) RNA refers to samples that did not meet RNA quality check for RNA-Seq (RNA integrity number [RIN] ≤ 6) and were not used for RT-qPCR (validation set). Excluded QC RNA-Seq refers to samples for which RNA-Seq data did not meet the quality requirements with respect to number and distribution of reads (Fig. S2). Excluded QC RT-qPCR were samples showing outlier Cycle threshold (Ct) values (>15) for the reference GAPDH gene (medians of two assays: 9.6 and 7.3). Training and test subsets were used in Random Forest to predict leprosy development. *RT-qPCR data of 8 samples (4 progressors and 4 HC controls) from the discovery set (RNA-Seq) were included in the training subset of the RT-qPCR Random Forest to improve the training of the model.
An initial discovery set was drawn from the cohort including 40 HC and 40 progressors who were diagnosed with leprosy 4–60 months after recruitment. To replicate and validate the results from the discovery set, a validation set was drawn later from the same cohort which included 43 HC and 43 progressors who were diagnosed with leprosy 4–61 months after recruitment. Subjects who developed leprosy > 61 months after recruitment (available only during the validation analysis) were excluded (n=5). The control HC group were optimally matched to the progressors by age, sex, date of recruitment, follow up time and BCG vaccination within the study (Table 1).
Table 1.
Cohort characterization.
|
Discovery set, RNA-Seq (n=80) | ||||||
|---|---|---|---|---|---|---|
| Group | Subjects | Sex | Age range (n) | RJ Classification | BI | Time to diagnosis (n) |
| Progressors | 40 | 26 females 14 males |
6-15 years (7) 16-30 years (9) 31-60 years (22) 61-70 years (2) |
37 BT 1 TT 1 I 1 PN |
34 BI-0 6 BI-und |
4-12 months (6) 13-24 months (10) 25-36 months (10) 37-48 months (7) 49-61 months (7) |
| HC | 40 | 27 females 13 males |
6-15 years (7) 16-30 years (8) 31-60 years (24) 61-70 years (1) |
- | - | |
| Validation set, RT-qPCR (n=86) | ||||||
| Group | Subjects | Sex | Age range (n) | RJ Classification | BI | Time to diagnosis (n) |
| Progressors | 43 | 23 females 20 males |
6-15 years (12) 16-30 years (12) 31-60 years (16) 61-70 years (3) |
40 BT 2 TT 1 I |
35 BI-0 8 BI-und |
4-12 months (7) 13-24 months (5) 25-36 months (9) 37-48 months (11) 49-61 months (11) |
| HC | 43 | 23 females 20 males |
6-15 years (12) 16-30 years (12) 31-60 years (16) 61-70 years (3) |
- | - | |
Group (leprosy progressors or household contact [HC] controls), number of individuals used for analyses, number of females and males, number of individuals in certain age range (at t=1), number of leprosy progressors according to Ridley-Jopling (RJ) classification (5), bacteriological index (BI) of progressors and time to diagnosis for progressors (time between the first sample before clinical diagnosis (t=1) and leprosy diagnosis (t=2)) are shown for the samples used in the RNA-Seq (discovery set) and the RT-qPCR (validation set) analyses. RT-qPCR: reverse transcription quantitative PCR. HC: Household contacts; BT: borderline tuberculoid leprosy; TT: tuberculoid leprosy; I: indeterminate leprosy; PN: pure neural leprosy; BI-und: bacteriological index undetermined as patient refused or was too young for skin slit smear and PB leprosy was diagnosed according to the number of lesions.
2.2. Ethics statement
This study was approved by the National Research Ethics Committee (BMRC/NREC/2016-2019/214) and followed the Helsinki Declaration (version Fortaleza, Brazil, October 2013). Participants were informed in the local language about the study objectives, the samples and their right to refuse to take part or withdraw without consequences for their treatment. All subjects gave written informed consent before enrolment and treatment was provided according to national guidelines [68].
2.3. RNA isolation, library preparation and sequencing
Blood was collected in PAXgene tubes (BD Biosciences, Franklin Lakes, NJ) in Bangladesh and sent on dry ice to Leiden University Medical Centre (The Netherlands) for analysis. RNA isolation from PAXgene tubes was automated using a QIAcube (Qiagen, Hilden, Germany) and PAXgene Blood RNA kits (Qiagen) according to the manufacturers’ protocol.
RNA concentrations were measured by Qubit RNA BR (Thermo Fisher Scientific, Waltham, MA) and integrity was determined by Fragment Analyzer (Agilent, Santa Clara, CA). Samples that passed the quality check (RNA integrity number [RIN] ≥ 6) were considered for RNA-Seq. RNA-Seq was performed by GenomeScan (Leiden, The Netherlands): libraries were prepared using NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA) including poly(A) enrichment. Additionally, globin reduction was performed using GLOBINclear kit (Thermo Fisher Scientific). Briefly, mRNA was isolated from total RNA using the oligo-dT magnetic beads. After fragmentation of the mRNA cDNA was synthesized. This was used for ligation with the sequencing adapters and PCR amplification of the resulting product. The quality and yield after sample preparation was measured by Fragment Analyzer. The size of the resulting products was consistent with the expected size distribution.
Clustering and sequencing were performed in a NovaSeq6000 System (Illumina, San Diego, CA) with a 2*150bp paired-end protocol in one single batch to avoid a batch effect. A concentration of 1.1 nM of DNA was used.
2.4. Gene expression quantitative PCR
Reverse transcription quantitative PCR (RT-qPCR) was performed using Biomark HD system (Fluidigm, South San Francisco, CA). Reverse Transcription Master Mix (Fluidigm) was used to convert 40 ng of RNA into cDNA following manufacturer's instructions. Prior to real-time amplification with the 48.48 Dynamic Array™ integrated fluidic circuit (IFC), a preamplification of 14 cycles was performed using Preamp Master Mix and TaqMan Assays (Table S1) according to manufacturer's instructions. Data were analysed using the software Real-Time PCR Analysis (v 4.5.2, Fluidigm).
2.5. RNA sequencing analysis
RNA-Seq files were processed using the opensource BIOWDL RNA-Seq pipeline v2.0 (https://github.com/biowdl/RNA-seq/tree/v2.0.0) developed at Leiden University Medical Centre. This pipeline performs FASTQ pre-processing (including quality control, quality trimming, and adapter clipping), RNA-Seq alignment and read quantification. FastQC was used for checking raw read QC. Adapter clipping was performed using Cutadapt (v2.4) with default settings. RNA-Seq reads’ alignment was performed using HISAT2 (v2.1.0) on GRCh38 reference genome analysis set. The gene read quantification was performed using HTSeq-count (v0.9.1) with setting “–stranded reverse”. The gene annotation used for quantification was Ensembl version 94. DGE and read normalization is explained in the next section, statistical analysis.
Functional analyses were performed using ClueGO plugin [69] to identify Gene Ontology (GO) terms and Ingenuity Pathway Analysis (IPA, Qiagen, Hildern, Germany) to establish canonical pathways.
2.6. Statistical analysis
Using RNA-Seq data, we performed DGE analysis to identify genes significantly differentially expressed between leprosy progressors at t=1 (n=40) and HC (n=40), and between progressors at t=1 and t=2 (n =40). We used an established R package, edgeR [70], executed according to their guidelines, and using raw counts normalized for library sizes with the Trimmed Mean of the M-values (TMM) method. The first comparison (t=1 vs HC) was evaluated in an unpaired design whereas the second comparison (t=1 vs t=2) was evaluated in a paired design because the samples were composed of 2 time points from the same individuals. We also modelled the difference of gene expression between the time points (t=1 vs t=2) as a linear function of the number of months which elapsed between the time points. Genes with false discovery rates below 0.05 (adjusted p-values < 0.05) were classified as differentially expressed.
DGE of RT-qPCR data was measured using Mann-Whitney U test using the package stats (version 3.6.3) in RStudio (version 1.2.5033) and genes with p-value below 0.05 were considered significantly expressed.
2.7. Machine learning to predict leprosy progression
Random Forest, a machine learning approach [71], was applied to select gene features (chi-squared method) and design a model to predict leprosy progression using gene expression data from RNA-Seq and RT-qPCR. For this purpose, the package mlr (version 2.17.1) [72] was employed in RStudio (version 1.2.5033). Accuracy, sensitivity, specificity and Area Under the Curve (AUC) were also obtained using mlr.
The sample set for the RNA-Seq model (discovery set) included leprosy progressors at t=1 (n=40) and HC controls (n=40) (Fig. 1). After RNA-Seq quality check (read count and MDS plot), two samples were excluded and the rest were divided into training (80%, n=62) and test (20%, n=16) subsets. An independent sample set (validation set) with 43 progressors and 43 controls was used for the RT-qPCR model. Samples were also placed 80% in the training (n=65) and 20% in the test (n=19) subsets (two samples excluded due to RT-qPCR quality check). Eight samples (four progressors and four controls) from the discovery set were added to the training subset (total training subset=73) for the RT-qPCR model to improve training input.
Training subsets were employed to train the models using a leave-one-out cross-validation (LOOCV) approach and subsequently evaluated in the test subsets. Parameters were set to ntree 50-1,000, mtry 1-10, nodsize 10-50, 72 iterations and 5 iterations of cross-validation. For the RT-qPCR model mtry and iterations were 1-4 and 1,000 respectively.
Using RNA-Seq data, an initial feature selection was performed limiting the model to 8-20 features. After feature selection lncRNA and pseudogenes were discarded from the selection set and the model was retrained and re-evaluated using the final set of features (n=13). Gene expression in TMM-normalized counts per million mapped reads (CPM) of differentially expressed genes (n=1,613) were the input for the RNA-Seq Random Forest model (discovery set). ΔCts (Cycle threshold) of differentially expressed genes (Mann-Whitney U test, n=4) were used for the RT-qPCR model (validation set). ΔCts were calculated as the difference of Ct of target gene and Ct of the reference gene, where GAPDH (assay ID Hs99999905_m1) was the reference gene. Initially, two assays with different primers and probes of GAPDH (Hs99999905_m1 and Hs02786624_g1, Table S1) were included in the RT-qPCR. Mann-Whitney U test of Ct values showed that GAPDH from assay Hs02786624_g1 presented significant differences between groups, whilst Ct values obtained from assay ID Hs99999905_m1 did not differ significantly between groups (Table S2). Therefore, only Ct values from GAPDH assay ID Hs99999905_m1 were used to calculate the ΔCt values.
2.8. Role of the funding source
Funding sources had no role in the study design, data collection, data analyses, data interpretation, writing of the report and the decision to submit the manuscript for publication.
3. Results
3.1. Cohort characterization
Between 2013 and 2018, HC of leprosy patients (n=5,123) without any clinical signs and symptoms of leprosy were recruited in Bangladesh (Fig. 1) and whole blood was collected for RNA isolation. HC who were suspected to have leprosy at recruitment (n=229) were excluded from the study. Leprosy progressors were defined as HC who were clinically diagnosed with leprosy 4-61 months after recruitment (n=85, Fig. S1).
RNA quality of samples from progressors with two timepoints present (before disease [t=1] and at time of diagnosis [t=2]) was assessed and samples from progressors which passed the RNA-Seq quality check (RIN ≥ 6) of both timepoints (n=40) were further analysed by RNA-Seq (Fig. 1). An equal number of HC (n=40) who did not develop disease (controls) were matched to progressors by age, sex, time of sample recruitment, follow up time and BCG vaccination (Table 1, discovery set). A separate sample set from the same area in Bangladesh, including samples from progressors (n=43) before disease (t=1) and matched controls (n=43) were used for RT-qPCR (Table 1, validation set).
In the discovery set, BT leprosy was reported in 37 of the progressors, one presented TT leprosy, one indeterminate (I) and one PN (Table 1). Similarly, the progressors in the validation set included 40 BT, two TT and one I leprosy patients.
In the discovery set, two individuals in the progressors group and one in the control group (one paired control) received BCG vaccination after samples collection at t=1 as part of a field trial [64] in Bangladesh. None of the individuals in the validation set received BCG vaccination after sample collection.
3.2. Gene expression differences in blood can be observed between leprosy progressors and contacts up to 5 years before leprosy diagnosis
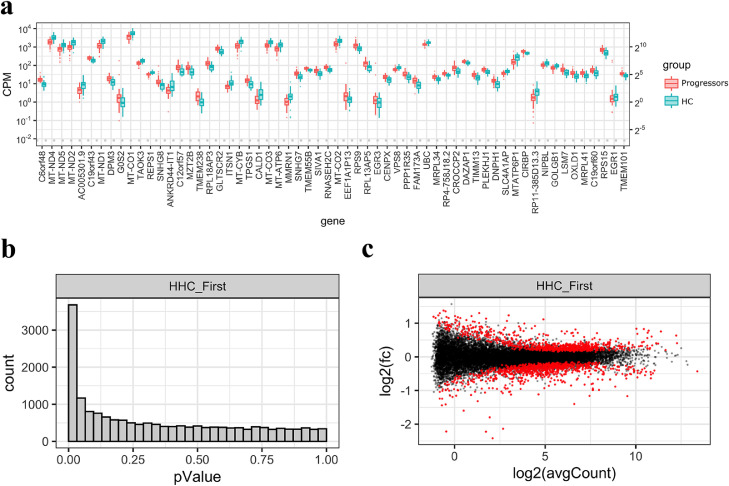
RNA-Seq gene expression data from blood of leprosy progressors (n=40) 4-61 months before diagnosis (t=1) was compared to HC who did not develop disease (n=40) (Fig. 2A). Initial quality analysis of the RNA-Seq revealed a low number of on-feature unique reads for two samples (one progressor at t=1 and one control, Fig. S2) which were subsequently excluded and thus 39 samples per group were considered for further analyses (Fig. 1). From the total of 17,435 genes, we identified 1,613 which were significantly differentially expressed (adjusted p-value < 0.05, Fig. 2B) between progressors and HC using an unpaired analysis with edgeR. From these, 836 were upregulated and 777 were downregulated in leprosy progressors compared to HC (Fig. 2C).
Fig. 2.
RNA-Seq differential gene expression analysis of leprosy progressors before clinical diagnosis and household contacts. RNA-Seq data of whole blood from leprosy progressors (n=39) 4-61 months before clinical diagnosis of leprosy (t=1 or First time point) was compared to control household contacts (HC/HHC, n=39), after exclusion of one sample per group due to low number of on-feature unique reads (Fig. S2). A two-group (unpaired samples) analysis was performed using edgeR (71) in R. a) Boxplot of Trimmed Mean of the M-values (TMM)-normalized counts per million mapped reads (CPM) per group of the most significantly differentially expressed genes. Y-axis shows CPM, expressed in power of 10 (left) or power of 2 (right). Progressors at t=1 are shown in red and HC controls in blue. b) Histogram of p-values. Number of genes (y-axis) with a given p-value (x-axis). c) MA plot showing log2 of fold change (FC) in gene expression (y-axis) and log2 of average CPM (x-axis) per gene. In red, genes significantly differentially expressed (adjusted p-value < 0.05) and in black, genes not differentially expressed. C6orf48 is also known as SNHG32 and C19orf60 as REX1BD.
Enriched GO terms and pathways were identified in upregulated and downregulated genes. Upregulated GO terms and canonical pathways included “cotranslational protein targeting to membrane”, “protein targeting to endoplasmic reticulum (ER)”, “protein localization to endoplasmic reticulum”, “eIF2 signalling”, “mammalian target of rapamycin (mTOR) signalling”, “regulation of eIF4 and p70S6K signalling” and “coronavirus pathogenesis pathway” (Table 2). Within the downregulated genes common GO terms and canonical pathways were “organelle organization”, “cellular component organization”, “clathrin-mediated endocytosis signalling”, “integrin signalling”, “Focal adhesion kinase (FAK) signalling” and “p70S6K signalling”.
Table 2.
Functional analysis of differentially expressed genes in blood of leprosy progressors.
|
Upregulated genes |
Downregulated genes |
||||
|---|---|---|---|---|---|
| GO terms | adj p-value | % associated genes | GO terms | adj p-value | % associated genes |
| SRP-dependent cotranslational protein targeting to membrane | 1.50E-33 | 41.51 | organelle organization | 1.33E-20 | 5.92 |
| cotranslational protein targeting to membrane | 1.05E-32 | 40.00 | cellular component organization | 1.50E-17 | 5.02 |
| protein targeting to ER | 5.23E-32 | 37.50 | regulation of cellular component organization | 7.92E-15 | 6.31 |
| establishment of protein localization to endoplasmic reticulum | 2.87E-31 | 36.29 | regulation of organelle organization | 5.61E-14 | 7.49 |
| protein localization to endoplasmic reticulum | 2.22E-30 | 31.79 | positive regulation of organelle organization | 6.43E-13 | 9.28 |
| Canonical pathway | adj p-value | % associated genes | Canonical pathway | adj p-value | % associated genes |
| eIF2 signalling | 4.29E-28 | 21.90 | clathrin-mediated endocytosis signalling | 3.57E-08 | 11.40 |
| mTOR signalling | 3.04E-13 | 14.80 | 14-3-3-mediated signalling | 6.72E-07 | 12.60 |
| regulation of eIF4 and p70S6K signalling | 1.70E-12 | 16.60 | integrin signalling | 8.44E-07 | 9.90 |
| coronavirus pathogenesis pathway | 1.63E-10 | 15.30 | FAK signalling | 2.92E-06 | 13.70 |
| oxidative phosphorylation | 1.04E-06 | 13.80 | p70S6K signalling | 4.13E-06 | 11.60 |
Top Gene Ontology (GO) terms identified by ClueGO (70) and canonical pathways identified by Ingenuity Pathway Analysis (Qiagen) from 836 upregulated and 777 downregulated genes in leprosy progressor before clinical diagnosis compared to household contacts who did not develop leprosy. P-values were adjusted for multiple testing with Bonferroni correction (adj p-value). Percentages of associated upregulated or downregulated genes from the pathway are shown.
3.3. Gene expression in whole blood does not vary during leprosy development
To identify biomarkers indicative of disease development, we studied longitudinal variation of gene expression in leprosy progressors between 4-61 months before diagnosis and at time of diagnosis. Since the quality check for the RNA-Seq data of one sample at t=1 and three samples at t=2 failed due to low amount of aligned on-feature unique reads or the sample was an outlier (multidimensional scaling (MDS) plot) (Fig. S2 and S3), these samples were excluded from the analysis with their paired sample (Fig. 1). Thus, for a total of 36 progressors longitudinal comparison was feasible. Surprisingly, a paired DGE analysis showed no genes that were significantly differentially expressed (adjusted p-value < 0.05, edgeR) between timepoint of diagnosis compared to the timepoint before diagnosis (Fig. S4), indicating that gene expression in blood does not vary intra-individually between the pre-clinical (no symptoms) and clinical (symptoms visible) phases of leprosy. Similarly, in a separate gene expression analysis we did not find any gene to display significant changes of expression level proportional to the number of months which elapsed between the two sample collection moments.
3.4. Machine learning identifies gene expression signature predicting leprosy
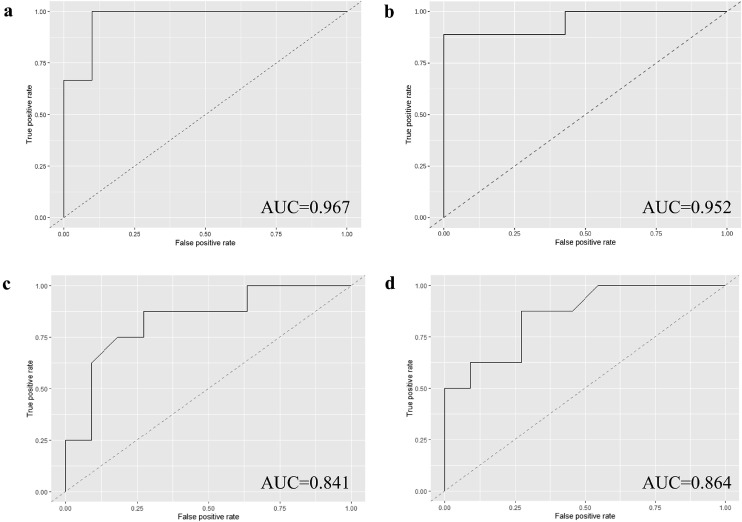
Next, a machine learning model was applied to select a subset of genes that optimized prediction of risk of leprosy development amongst HC. Random Forest was performed splitting the samples into training (80%, n=62) and test (20%, n=16) subsets, followed by a LOOCV approach and limiting the model to 8 to 20 features/genes. TMM-normalized CPM of genes differentially expressed (n=1,613) between progressors and HC in RNA-Seq of whole blood were used as input. The model (Table S3, 19-gene RNA-Seq) which included 19 genes (Table 3, Fig. S5), showed a strong predictive potential for leprosy with an accuracy of 87.5% (sensitivity 100.0%, specificity 80.0%) and AUC of 96.7% (Fig. 3A, Table S4). This set of genes contained protein coding genes but also long non-coding (lnc)RNA and pseudogenes.
Table 3.
Gene selection using a machine learning approach.
| Gene name | Ensembl ID | Type of RNA |
|---|---|---|
| SNHG32orC6orf48 | ENSG00000204387 | ncRNA, small nuclear RNA |
| MT-ND4 | ENSG00000198886 | protein coding |
| MT-ND5 | ENSG00000198786 | protein coding |
| MT-ND2 | ENSG00000198763 | protein coding |
| lnc-IL17RA-36 or AC005301.9 | ENSG00000283633 | lncRNA |
| MT-CO1 | ENSG00000198804 | protein coding |
| TAOK3 | ENSG00000135090 | protein coding |
| REPS1 | ENSG00000135597 | protein coding |
| MT-CYB | ENSG00000198727 | protein coding |
| TPGS1 | ENSG00000141933 | protein coding |
| MMRN1 | ENSG00000138722 | protein coding |
| UBC | ENSG00000150991 | protein coding |
| MTATP6P1 | ENSG00000248527 | pseudogene |
| RP11-385D13.4 | ENSG00000266538 | lncRNA |
| REX1BDorC19orf60 | ENSG00000006015 | protein coding |
| CCDC85B | ENSG00000175602 | protein coding |
| HCG4P12 | ENSG00000225864 | pseudogene |
| RNU6-238P | ENSG00000200183 | pseudogene |
| AC009303.2 | ENSG00000279227 | lncRNA |
Genes identified by Random Forest to predict leprosy progression amongst household contacts of leprosy patients. In bold genes that were included in the final RNA-Seq signature and tested by reverse transcription quantitative PCR (RT-qPCR). Underlined the genes present in the final RT-qPCR RISK4LEP signature.
Fig. 3.
AUC of leprosy risk RNA-Seq and RT-qPCR signatures in blood. Area Under the Curve (AUC) of risk signatures in whole blood to prospectively predict leprosy progressors within household contacts (HC). The models were built using Random Forest, were trained with 80% of the sample sets and evaluated in 20%. a) AUC of RNA-Seq 19-gene signature where 8 to 20 features/genes were automatically selected by the model from a total of 1,613 features. b) AUC of RNA-Seq 13-gene signature based on the 19-gene signature but excluding pseudogenes and long non-coding (lnc)RNA (n=6). c) AUC of reverse transcription quantitative PCR (RT-qPCR) 13-gene signature selected in the RNA-Seq signature. d) AUC of RT-qPCR 4-gene signature RISK4LEP (final RT-qPCR signature) where only genes significantly differentially expressed in the RT-qPCR were selected.
To validate the signature in an independent sample set, we aimed at selecting a set of genes with commercially available probes for RT-qPCR. For this reason, the lncRNA and pseudogenes were excluded (n=6). A new model (Table S3, 13-gene RNA-Seq) was re-trained and re-evaluated in the reduced 13-gene signature (Table 3, SNHG32/C6orf48, MT-ND4, MT-ND5, MT-ND2, MT-CO1, TAOK3, REPS1, MT-CYB, TPGS1, MMRN1, UBC, REX1BD/C19orf60, CCDC85B) and showed an accuracy of 87.5% with a sensitivity of 88.9%, specificity of 85.7% and AUC of 95.2% (Fig. 3B, Table S4). It is of note that five of these 13 genes (MT-ND2, MT-ND4, MT-ND5, MT-CO1, and MT-CYB) are mitochondrial genes involved in oxidative phosphorylation, and are all down-regulated in leprosy progressors.
In addition, we evaluated whether using genes from previously described tuberculosis risk signatures could also predict leprosy. For this purpose, a Random Forest was performed with genes from the Sweeney3 (GBP5, DUSP3, KLF2) [73], the Suliman2 (ANKRD22, OSBPL10) [50] or the RISK6 (GBP2, FCGR1B, SERPING1, TUBGCP6, TRMT2A, SDR39U1) [51] signatures as input. However, the tuberculosis risk signatures showed poor or moderate performance to predict leprosy with AUCs of 51.6%, 58.7% and 78.3% respectively (Table S4). Thus, the Sweeney3 and Suliman2 signatures resemble an algorithm that predicts leprosy randomly. The RISK6 signature, although presenting a reasonably good prediction of leprosy, showed lower performance compared to our novel 19-gene (AUC=96.7%) and 13-gene RNA-Seq (AUC=95.2%) signatures.
3.5. Validation of a leprosy predictive biomarker signature
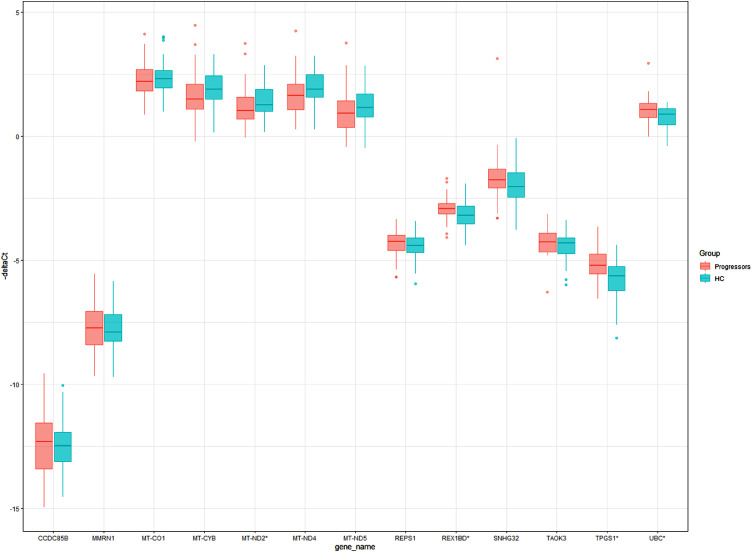
The 13-gene RNA-Seq signature was validated by RT-qPCR in an independent set of subjects. Gene expression of the 13 genes and a reference gene (GAPDH_m1) were tested using Biomark HD system (Fluidigm), a high-throughput RT-qPCR. Validation was performed on a separate set which included 43 leprosy progressors at t=1 and 43 HC controls as well as four progressors (at t=1) and four controls from the discovery set that were included to improve training of the model. Two outlier samples (one progressor and one control) presenting Cts of the reference gene >15 (median 7.3 GAPDH) were excluded from the analysis (Fig. S6).
Significantly differentially expressed genes were determined using ΔCts (Ct of target gene – Ct of reference gene). Four genes, MT-ND2, REX1BD, TPGS1 and UBC (Table 4), presented significant differential expression (p-values 0.0483, 0.0101, 0.0004 and 0.0060 respectively, Mann-Whitney U test) between leprosy progressors and controls (Fig. 4).
Table 4.
RT-qPCR ΔCts of the 13-gene signature in leprosy progressors and household contacts.
| Gene | p-value | ΔCt progressors | ΔCt HC | ΔΔCt | FC | Log2FC |
|---|---|---|---|---|---|---|
| CCDC85B | 0.840901 | 12.32 | 12.48 | -0.16 | 1.12 | 0.16 |
| MMRN1 | 0.654911 | 7.73 | 7.88 | -0.15 | 1.11 | 0.15 |
| MT-CO1 | 0.594361 | -2.22 | -2.31 | 0.09 | 0.94 | -0.09 |
| MT-CYB | 0.054951 | -1.50 | -1.90 | 0.40 | 0.76 | -0.40 |
| MT-ND2 | 0.048303 | -1.03 | -1.26 | 0.23 | 0.85 | -0.23 |
| MT-ND4 | 0.062337 | -1.64 | -1.90 | 0.26 | 0.84 | -0.26 |
| MT-ND5 | 0.159386 | -0.94 | -1.17 | 0.24 | 0.85 | -0.24 |
| REPS1 | 0.298712 | 4.24 | 4.40 | -0.16 | 1.12 | 0.16 |
| REX1BD | 0.010086 | 2.92 | 3.18 | -0.27 | 1.20 | 0.27 |
| SNHG32 | 0.238032 | 1.77 | 2.03 | -0.26 | 1.20 | 0.26 |
| TAOK3 | 0.178822 | 4.26 | 4.30 | -0.04 | 1.03 | 0.04 |
| TPGS1 | 0.000448 | 5.20 | 5.62 | -0.42 | 1.34 | 0.42 |
| UBC | 0.005958 | -1.07 | -0.89 | -0.18 | 1.13 | 0.18 |
P-values of Mann-Whitney U test of reverse transcription quantitative PCR (RT-qPCR) ΔCts (Cycle threshold (Ct) of target gene – Ct of reference gene) between leprosy progressors (n=47) and household contact (HC) controls (n=47). In bold genes significantly differentially expressed (p-value <0.05). Median of ΔCts per group, ΔΔCt (median ΔCt progressors – median ΔCt HC), Fold Change (FC, 2–∆∆Ct) for progressors and log2 of Fold Change (log2FC).
Fig. 4.
Boxplot showing -ΔCts of 13 genes. Boxplot of -ΔCts (-(Cycle threshold (Ct) target gene – Ct reference gene, GAPDH)) obtained by reverse transcription quantitative PCR (RT-qPCR) in whole blood. Genes identified in the RNA-Seq signature (n=13) are shown. Leprosy progressors before clinical diagnosis of leprosy are shown in red (t=1, n=47) and household contact (HC) controls in blue (n=47). *Genes significantly differentially expressed between the two groups using Mann-Whitney U test (MT-ND2, REX1BD, TPGS1 and UBC).
An RT-qPCR model to predict leprosy risk and validate the RNA-Seq signature was established by Random Forest using ΔCts as input. Samples from the discovery set (n=8) were only used in the training subset which included 80% of samples (n=73). Thus, the model was evaluated in a separate subset from the validation set consisting of 20% (n=19) of the sample set. A Random Forest model including the 13 genes (Table S3, 13-gene RT-qPCR) showed an AUC of 84.1% (Fig. 3C, Table S4) and accuracy of 73.7% (sensitivity 87.5%, specificity 63.6%). Slightly improved predictive potential for leprosy was observed if Random Forest was performed using only the four genes significantly differentially expressed (Table S3, 4-gene RT-qPCR RISK4LEP), showing an AUC of 86.4% (Fig. 3D, Table S4), accuracy of 79.0%, sensitivity of 87.5% and specificity of 72.7%. From these four genes, REX1BD, TPGS1 and UBC were upregulated, whilst MT-ND2 was downregulated in leprosy progressors before clinical diagnosis compared to HC (Fig. 4). This is in line with the RNA-Seq results in the discovery set, except for UBC (Fig. 2). However, removing UBC from the signature (3-gene signature) and addition of the following gene with lowest p-value (MT-CYB) in the 3-gene signature led to decreased performances (Table S4). Addition of demographic variables (sex, age and Ridley-Jopling [5] classification of the index leprosy contact) into the 4-gene or a reduced 2-gene (TPGS1 and UBC) signature did not improve the performance either (Table S4). Therefore, the 4-gene RT-qPCR risk signature, which we named RISK4LEP, is preferred to predict leprosy development in HC due to the improved performance and the lower number of genes required (Table 3).
4. Discussion
Leprosy diagnosis is often ascertained after the occurrence of clinical symptoms, which may already coincide with the presence of irreversible tissue damage. Early diagnosis and prompt treatment are critical to reduce leprosy-associated disabilities and block M. leprae transmission. However, a sensitive diagnostic test with potential to predict the development of leprosy is not available.
To identify a transcriptomic risk signature for leprosy, this study investigated gene expression differences by RNA-Seq between HC of leprosy patients in Bangladesh who later developed leprosy and those who remained without clinical sign and symptoms. Initially a 13-gene signature that could predict leprosy development was identified using Random Forest, a machine learning approach. Subsequently, the signature was adapted and validated in a separate set by RT-qPCR. Validation of the signature in a new sample set (validation set) showed that reducing the signature to four genes improved prediction of leprosy in this sample set. The RISK4LEP signature allowed discrimination of leprosy progressors with a sensitivity of 87.5%, a specificity of 72.3% and an AUC of 86.4%. This 4-gene signature identified leprosy progressors amongst individuals exposed to leprosy bacilli from 4 to 61 months before clinical diagnosis, thus representing the first transcriptomic risk signature to prospectively predict leprosy progressors at an asymptomatic stage. This signature is unique for leprosy and does not overlap with known tuberculosis risk signatures. Since leprosy has a long incubation time and low disease prevalence, more than 5,000 samples had to be collected during 8 years to obtain samples of 85 individuals before and at disease onset. As such, this is the first study of its kind in leprosy research.
The RISK4LEP predictive signature is composed by four genes: MT-ND2, REX1BD, TPGS1 and UBC. MT-ND2 encodes a subunit (core subunit 2) of the mitochondrial NADH:Ubiquinone Oxidoreductase [74]. MT-ND2 together with MT-ND6 are the essential subunits forming the mitochondrial membrane respiratory chain NADH dehydrogenase which plays a critical role in oxidative phosphorylation. One of the functions of mitochondrial reactive oxygen species resulting from oxidative phosphorylation is to regulate immunity. MT-ND2 is under-expressed in leprosy progressors, hence presenting a disadvantage to successful elimination of M. leprae [75]. Little is known of REX1BD and TPGS1: REX1BD encodes the Required For Excision 1-B Domain Containing Protein and TPGS1, Tubulin Polyglutamylase Complex Subunit 1, is a gene related to microtubule binding and tubulin-glutamic acid ligase activity that may act in the targeting of the tubulin polyglutamylase complex [74]. UBC is one of the four genes encoding the human ubiquitin involved in several pathways such as protein degradations, DNA repair, cell cycle regulation, kinase modification, endocytosis and regulation of other cell signalling. It has been previously reported as having a high degree of connectivity in a protein-protein interaction network with differentially expressed genes in patients with active tuberculosis [76]. The ubiquitin system is involved in the innate immune response in tuberculosis and has been suggested as a potential target for host-directed therapy, indicating that UBC might play a role in the innate response against M. leprae as well. Moreover, variants in the regulation regions of the PRKN gene (previously known as PARK2), which is part of the ubiquitin system, have been associated with susceptibility to leprosy [14]. PRKN codes a ubiquitin ligase that is essential for autophagy of mycobacteria and damaged mitochondria [77,78].
Our data show that differences in gene expression could be observed up to 61 months before the disease manifests (Fig. S1). In contrast, intra-individual expression remains stable in individuals between the pre-symptomatic phase and time of diagnosis. This indicates that differences in expression of some genes in blood of leprosy progressors precede appearance of symptoms.
Further exploration of the pathways that could be responsible for the observed differences in gene expression between leprosy progressors and controls showed that genes overexpressed in leprosy progressors are involved in translation pathways and cotranslation of membrane and ER proteins. EIF2, eIF4 and p70S6K signalling pathways, overexpressed in leprosy progressors, are downstream pathways to the mTOR pathway which also displayed higher levels in progressors and regulates protein translation, gene expression, metabolic processes, immune receptor signalling and migratory activity [79,80]. Likewise, in several other diseases such as cancer, type 2 diabetes, rheumatoid arthritis and viral infections the mTOR pathway is deregulated [79,81]. In general, antigen recognition activates the mTOR signalling pathway as a result of which naïve CD4+ T-cells differentiate into Th1, Th2 and Th17 [80]. This process may thus lead to a higher expression of mRNA related to Th1, Th2 and Th17 in leprosy patients compared to healthy individuals as previously observed [82]. Interestingly, upregulation of the coronavirus pathogenesis pathway was also observed in leprosy progressors. This could be caused by activation of the inflammatory and autophagy regulation pathways in individuals infected with coronaviruses as well as BT leprosy patients [83,84].
Downregulated gene expression in leprosy progressors was observed and occurred in organelle and cellular component organization pathways as well as integrin and FAK signalling pathways. FAK is a tyrosine kinase downstream of integrin growth factor. Nuclear FAK regulates transcription of inflammatory signalling, immune escape, angiogenesis and p53 [85]. Moreover, overexpression of FAK has been linked to advanced cancer and metastasis [85,86]. Although FAK inhibitors are currently being tested for use in cancer treatment, the FAK signalling pathway has never been studied in leprosy. Hence the significance of under-expression in leprosy progressors before diagnosis as observed in this study requires further investigation.
Recently, Leal-Calvo and Moraes performed a comprehensive reanalysis of nine publicly available microarrays of leprosy patients from variable origin. The authors found DGE in skin development processes including genes such as AQP3, AKR1C3, CYP27B1, LTB, VDR and keratinocyte biology with CSTA, DSG1, KRT14, KRT5, PKP1 and IVL [87]. None of the genes identified by that study were, however, found in our analysis. This could be due to the fact that this reanalysis mainly investigated DGE between different leprosy types (BT vs LL), ENL reaction and LL or LL and healthy controls, whereas our study included mostly BT leprosy patients and HC. Moreover, RNA was obtained from skin biopsies or cell cultures instead of whole blood. Consequently, comparison of their results with the present work is limited and while possible biomarker genes were identified in the microarray reanalysis, application for diagnosis would be restricted to patients with visible symptoms as well as requiring more invasive samples (skin biopsies or cell culture). In contrast, the prospective 4-gene signature RISK4LEP identified in this study is measured in whole blood.
Similarly, other transcriptomic studies described leprosy biomarkers associated with leprosy but after occurrence of symptoms and using skin biopsies [53,63,88,89]. Serrano-Coll and colleagues showed that RT-qPCR of Oct-6 identified multibacillary (MB) patients (n=30) in Colombia with an AUC of 83.0% (89]. However, S-100 immunohistochemistry alone showed a better AUC (96.0%). Pinto et al. investigated the expression of non-coding RNAs in leprosy patients (5 TT and 6 LL) from Brazil [89] and found five P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs) that classified leprosy patients with an AUC of 90.0%. Jorge et al. established a non-coding RNA signature consisting of four miRNA, that discriminated leprosy patients (6 LL and 6 TT; AUC=87.3%) in Brazil [53]. Furthermore, Guerreiro and colleagues using nerve biopsies of PN leprosy patients (n=28) identified a transcriptomic signature based on a classification tree including LDR and CCL4 which could ascertain 80% of PN leprosy patients [63]. Although CCL4 and LDR were not significantly differentially expressed in our study, we also found lower expression levels of mitochondrial genes involved in the oxidative phosphorylation pathway in blood of leprosy progressors, in line with their findings in M. leprae-infected Schwann cells and nerve biopsies of Brazilian leprosy patients. This reduction may be caused by down-regulation of mitochondrial genes by mycobacteria during M. leprae infection to inhibit apoptosis and promote intracellular bacterial survival [90].
We found moderate prediction (AUC=78.3%) of leprosy when the RISK6 genes (GBP2, FCGR1B, SERPING1, TUBGCP6, TRMT2A, SDR39U1) were used as input in the Random Forest with RNA-Seq data. This is likely due to similarities in the immune response to mycobacteria in leprosy and tuberculosis patients [91]. In line with this, FCGR1A and GBP genes were previously found to be upregulated in leprosy patients or during leprosy reactions in and outside Bangladesh [48,54,92].
It has been previously reported that RNA profiles in blood of leprosy patients are different from those derived from skin [62]. Although transcriptomic analysis in skin of leprosy patients provide deeper insight into leprosy pathogenesis, the aim of this study was to identify leprosy predictive biomarkers, preferably measurable in rapid diagnostic tests. Thus, whole blood is a preferred biosample because it can be collected relatively easily and translated into field-friendly tests applying fingerstick blood [51].
In summary, the RISK4LEP signature described here, offers potential for the development of a point of care test allowing the identification of leprosy progression among HC in blood years before symptom development. Since the present study was performed in Bangladesh, additional, longitudinal studies are required to determine whether this signature predicts leprosy progression in endemic populations from different origins. Moreover, since the majority of the Bangladeshi patients who developed leprosy during this study developed BT leprosy, similar studies will also need to provide information on the performance of the signature to predict occurrence of LL types. It is tempting to speculate that this signature could identify early forms of BT leprosy, thus preventing the more severe LL types from developing.
Nevertheless, the novel RISK4LEP signature predicts development of (BT) leprosy up to 61 months before clinical diagnosis. Such signatures, when properly validated in other populations as well, can be applied for targeted preventive treatment and reduction of M. leprae transmission among HC.
Contributors
Conceptualization: AG, MT, JHR
Data collection: JCR, MK, SS, ASC, KA
Experiments: MT, SE
Data Curation: MT, JCR, AH
Data Analysis: MT, SK, HM, JW
Funding Acquisition: AG, JHR
Supervision: AG
Writing original draft: MT
Writing – Review & Editing: MT, AG
All authors reviewed, discussed, and agreed with manuscript.
Data sharing statement
Sequence data have been submitted to NCBI Gene Expression Omnibus (GEO) under accession number GSE163498.
RNA-Seq files were processed using the open source BIOWDL RNA-Seq pipeline v2.0 (https://github.com/biowdl/RNA-seq/tree/v2.0.0) developed at Leiden University Medical Centre (The Netherlands).
Declaration of Competing Interest
The authors declare no conflicts of interests.
Acknowledgements
The authors gratefully acknowledge all patients and control participants in Bangladesh and would like to thank Suzanne van Veen and Mariëlle Haks for assistance and advice on the Biomark HD system (Fluidigm).
This study was supported by an R2STOP Research grant from effect:hope, Canada and the Mission to End Leprosy, Ireland; the Order of Malta-Grants-for-Leprosy-Research (MALTALEP); the Q.M. Gastmann-Wichers Foundation; the Leprosy Research Initiative (LRI) together with the Turing Foundation (ILEP; 702.02.73 and 703.15.07).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103379.
Appendix. Supplementary materials
References
- 1.World Health Organization. Global leprosy update moving towards a leprosy-free world. Weekly Epidemiol Record. 2018;2019(94):389–412. (35/36) [Google Scholar]
- 2.Scollard D.M. International textbook of leprosy. 2016. Pathogenesis and pathology of leprosy.www.internationaltextbookofleprosy.org In: Scollard DM, Gillis TP, editors. 18 Septemberposting date. [Google Scholar]
- 3.Scollard D.M., Adams L.B., Gillis T.P., Krahenbuhl J.L., Truman R.W., Williams D.L. The continuing challenges of leprosy. Clin Microbiol Rev. 2006;19(2):338–381. doi: 10.1128/CMR.19.2.338-381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar B., Uprety S., Dogra S. International textbook of leprosy. 2017. Clinical diagnosis of leprosy.www.internationaltextbookofleprosy.org In: Scollard DM, Gills TP, editors. 9 Februaryposting date. [Google Scholar]
- 5.Ridley D.S., Jopling W.H. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. 1966;34(3):255–273. [PubMed] [Google Scholar]
- 6.Richardus J.H., Ignotti E., Smith W.C.S. International textbook of leprosy. 2016. Epidemiology of leprosy.www.internationaltextbookofleprosy.org In: Scollard DM, Gillis TP, editors. 18 Septemberposting date. [Google Scholar]
- 7.Walker S.L., Withington S.G., Lockwood D.N.J. Manson's tropical infectious diseases. 23rd Ed. W.B. Saunders; London: 2014. 41 - Leprosy. In: Farrar J, Hotez PJ, Junghanss T, Kang G, Lalloo D, White NJ, editors. 506-18.e1. [Google Scholar]
- 8.Uaska Sartori P.V., Penna G.O., Bührer-Sékula S., Pontes M.A.A., Gonçalves H.S., Cruz R. Human genetic susceptibility of leprosy recurrence. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-58079-3. 1284- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang F.R., Huang W., Chen S.M., Sun L.D., Liu H., Li Y. Genomewide association study of leprosy. N Engl J Med. 2009;361(27):2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 10.Cardoso C.C., Pereira A.C., de Sales Marques C., MO Moraes. Leprosy susceptibility: genetic variations regulate innate and adaptive immunity, and disease outcome. Future Microbiol. 2011;6(5):533–549. doi: 10.2217/fmb.11.39. [DOI] [PubMed] [Google Scholar]
- 11.Alter A., Grant A., Abel L., Alcais A., Schurr E. Leprosy as a genetic disease. Mamm Genome. 2011;22(1-2):19–31. doi: 10.1007/s00335-010-9287-1. [DOI] [PubMed] [Google Scholar]
- 12.Wang D., Xu L., Lv L., Su L.Y., Fan Y., Zhang D.F. Association of the LRRK2 genetic polymorphisms with leprosy in Han Chinese from Southwest China. Genes Immun. 2015;16(2):112–119. doi: 10.1038/gene.2014.72. [DOI] [PubMed] [Google Scholar]
- 13.Sales-Marques C., Cardoso C.C., Alvarado-Arnez L.E., Illaramendi X., Sales A.M., Hacker M.A. Genetic polymorphisms of the IL6 and NOD2 genes are risk factors for inflammatory reactions in leprosy. PLoS NeglTrop Dis. 2017;11(7) doi: 10.1371/journal.pntd.0005754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mira M.T., Alcais A., Nguyen V.T., Moraes M.O., Di Flumeri C., Vu H.T. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427(6975):636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 15.Dwivedi V.P., Banerjee A., Das I., Saha A., Dutta M., Bhardwaj B. Diet and nutrition: an important risk factor in leprosy. Microb Pathog. 2019;137 doi: 10.1016/j.micpath.2019.103714. [DOI] [PubMed] [Google Scholar]
- 16.Feenstra S.G., Nahar Q., Pahan D., Oskam L., Richardus J.H. Recent food shortage is associated with leprosy disease in Bangladesh: a case-control study. PLoS NeglTrop Dis. 2011;5(5):e1029. doi: 10.1371/journal.pntd.0001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serrano-Coll H., Mora H.R., Beltrán J.C., Duthie M.S., Cardona-Castro N. Social and environmental conditions related to Mycobacterium leprae infection in children and adolescents from three leprosy endemic regions of Colombia. BMC Infect Dis. 2019;19(1):520. doi: 10.1186/s12879-019-4120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerr-Pontes L.R.S., Montenegro A.C.D., Barreto M.L., Werneck G.L., Feldmeier H. Inequality and leprosy in Northeast Brazil: an ecological study. Int J Epidemiol. 2004;33(2):262–269. doi: 10.1093/ije/dyh002. [DOI] [PubMed] [Google Scholar]
- 19.Vieira M.C.A., Nery J.S., Paixão E.S., Freitas de Andrade K.V., Oliveira Penna G., Teixeira M.G. Leprosy in children under 15 years of age in Brazil: A systematic review of the literature. PLoS NeglTrop Dis. 2018;12(10) doi: 10.1371/journal.pntd.0006788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar R., Pradhan S. Leprosy and women. Int J Women's Dermatol. 2016;2(4):117–121. doi: 10.1016/j.ijwd.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakker M.I., Hatta M., Kwenang A., Van Mosseveld P., Faber W.R., Klatser P.R. Risk factors for developing leprosy–a population-based cohort study in Indonesia. Lepr Rev. 2006;77(1):48–61. [PubMed] [Google Scholar]
- 22.Goulart I.M., Bernardes Souza D.O., Marques C.R., Pimenta V.L., Goncalves M.A., Goulart L.R. Risk and protective factors for leprosy development determined by epidemiological surveillance of household contacts. Clin Vaccine Immunol. 2008;15(1):101–105. doi: 10.1128/CVI.00372-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sales A.M., Ponce de Leon A., Duppre N.C., Hacker M.A., Nery J.A., Sarno E.N. Leprosy among patient contacts: a multilevel study of risk factors. PLoS Negl Trop Dis. 2011;5(3):e1013. doi: 10.1371/journal.pntd.0001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quilter E.E.V., Butlin C.R., Singh S., Alam K., Lockwood D.N.J. Patients with skin smear positive leprosy in Bangladesh are the main risk factor for leprosy development: 21-year follow-up in the household contact study (COCOA) PLoS NeglTrop Dis. 2020;14(10) doi: 10.1371/journal.pntd.0008687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization . Guidelines for the diagnosis, treatment and prevention of leprosy. World Health Organization Regional Office for South-East Asia; 2018. Regional office for South-East Asia. [Google Scholar]
- 26.van Hooij A., van den Eeden S., Richardus R., Tjon Kon Fat E., Wilson L., Franken K. Application of new host biomarker profiles in quantitative point-of-care tests facilitates leprosy diagnosis in the field. EBioMedicine. 2019;47:301–308. doi: 10.1016/j.ebiom.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Hooij A., Tjon Kon Fat E.M., Batista da Silva M., Carvalho Bouth R., Cunha Messias A.C., Gobbo A.R. Evaluation of Immunodiagnostic Tests for Leprosy in Brazil, China and Ethiopia. Sci Rep. 2018;8(1):17920. doi: 10.1038/s41598-018-36323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Hooij A., Tjon Kon Fat E.M., Richardus R., van den Eeden S.J., Wilson L., de Dood C.J. Quantitative lateral flow strip assays as user-friendly tools to detect biomarker profiles for leprosy. Sci Reports. 2016;6:34260. doi: 10.1038/srep34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Hooij A., Tjon Kon Fat E.M., van den Eeden S.J.F., Wilson L., Batista da Silva M., Salgado C.G. Field-friendly serological tests for determination of M. leprae-specific antibodies. Sci Rep. 2017;7(1):8868. doi: 10.1038/s41598-017-07803-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X., You Y.G., Yuan Y.H., Yuan L.C., Wen Y. Host immune responses induced by specific Mycobacterium leprae antigens in an overnight whole-blood assay correlate with the diagnosis of paucibacillary leprosy patients in China. PLoS NeglTrop Dis. 2019;13(4) doi: 10.1371/journal.pntd.0007318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Queiroz E.A., Medeiros N.I., Mattos R.T., Carvalho A.P.M., Rodrigues-Alves M.L., Dutra W.O. Immunological biomarkers of subclinical infection in household contacts of leprosy patients. Immunobiology. 2019;224(4):518–525. doi: 10.1016/j.imbio.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Gama R.S., Gomides T.A.R., Gama C.F.M., Moreira S.J.M., de Neves Manta F.S., de Oliveira L.B.P. High frequency of M. leprae DNA detection in asymptomatic household contacts. BMC Infectious Diseases. 2018;18(1):153. doi: 10.1186/s12879-018-3056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbieri R.R., Manta F.S.N., Moreira S.J.M., Sales A.M., Nery J.A.C., Nascimento L.P.R. Quantitative polymerase chain reaction in paucibacillary leprosy diagnosis: A follow-up study. PLoS NeglTrop Dis. 2019;13(3) doi: 10.1371/journal.pntd.0007147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez A.N., Ribeiro-Alves M., Sarno E.N., Moraes M.O. Evaluation of qPCR-based assays for leprosy diagnosis directly in clinical specimens. PLoS NeglTrop Dis. 2011;5(10):e1354. doi: 10.1371/journal.pntd.0001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manta F.S.N., Barbieri R.R., Moreira S.J.M., Santos P.T.S., Nery J.A.C., Duppre N.C. Quantitative PCR for leprosy diagnosis and monitoring in household contacts: A follow-up study, 2011-2018. Sci Rep. 2019;9(1):16675. doi: 10.1038/s41598-019-52640-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carvalho R.S., Foschiani I.M., Costa M., Marta S.N., da Cunha Lopes Virmond M. Early detection of M. leprae by qPCR in untreated patients and their contacts: results for nasal swab and palate mucosa scraping. Eur J Clin Microbiol Infect Dis. 2018;37(10):1863–1867. doi: 10.1007/s10096-018-3320-9. [DOI] [PubMed] [Google Scholar]
- 37.Pathak V.K., Singh I., Turankar R.P., Lavania M., Ahuja M., Singh V. Utility of multiplex PCR for early diagnosis and household contact surveillance for leprosy. Diagn Microbiol Infect Dis. 2019;95(3) doi: 10.1016/j.diagmicrobio.2019.06.007. 114855- [DOI] [PubMed] [Google Scholar]
- 38.Gama R.S., Souza M.L.M., Sarno E.N., Moraes M.O., Goncalves A., Stefani M.M.A. A novel integrated molecular and serological analysis method to predict new cases of leprosy amongst household contacts. PLoS NeglTrop Dis. 2019;13(6) doi: 10.1371/journal.pntd.0007400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Beers S.M., Izumi S., Madjid B., Maeda Y., Day R., Klatser P.R. An epidemiological study of leprosy infection by serology and polymerase chain reaction. Int J Leprosy Other Mycobacterial Dis. 1994;62(1):1–9. [PubMed] [Google Scholar]
- 40.Bakker M.I., Hatta M., Kwenang A., Van Mosseveld P., Faber W.R., Klatser P.R. Risk factors for developing leprosy–a population-based cohort study in Indonesia. Lepr Rev. 2006;77(1):48–61. [PubMed] [Google Scholar]
- 41.Martins ACdC, Miranda A., Oliveira MLW-d-Rd, Bührer-Sékula S., Martinez A. Nasal mucosa study of leprosy contacts with positive serology for the phenolic glycolipid 1 antigen. Braz J Otorhinolaryngol. 2010;76(5):579–587. doi: 10.1590/S1808-86942010000500008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cardona-Castro N., Beltran-Alzate J.C., Manrique-Hernandez R. Survey to identify Mycobacterium leprae-infected household contacts of patients from prevalent regions of leprosy in Colombia. Mem Inst Oswaldo Cruz. 2008;103(4):332–336. doi: 10.1590/s0074-02762008000400003. [DOI] [PubMed] [Google Scholar]
- 43.Santos D.F.D., Mendonca M.R., Antunes D.E., Sabino E.F.P., Pereira R.C., Goulart L.R. Molecular, immunological and neurophysiological evaluations for early diagnosis of neural impairment in seropositive leprosy household contacts. PLoS NeglTrop Dis. 2018;12(5) doi: 10.1371/journal.pntd.0006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tió-Coma M., Avanzi C., Verhard E.M., Pierneef L., van Hooij A., Benjak A. Genomic characterization of mycobacterium leprae to explore transmission patterns identifies new subtype in Bangladesh. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Hooij A., Tió-Coma M., Verhard E.M., Khatun M., Alam K., Tjon Kon Fat E. Household contacts of leprosy patients in endemic areas display a specific innate immunity profile. Front Immunol. 2020;11(1811) doi: 10.3389/fimmu.2020.01811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donoghue H.D., Holton J., Spigelman M. PCR primers that can detect low levels of Mycobacterium leprae DNA. J Med Microbiol. 2001;50(2):177–182. doi: 10.1099/0022-1317-50-2-177. [DOI] [PubMed] [Google Scholar]
- 47.Martinez A.N., Lahiri R., Pittman T.L., Scollard D., Truman R., Moraes M.O. Molecular determination of Mycobacterium leprae viability by use of real-time PCR. J Clin Microbiol. 2009;47(7):2124–2130. doi: 10.1128/JCM.00512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teles R.M.B., Lu J., Tió-Coma M., Goulart I.M.B., Banu S., Hagge D. Identification of a systemic interferon-γ inducible antimicrobial gene signature in leprosy patients undergoing reversal reaction. PLoS NeglTrop Dis. 2019;13(10) doi: 10.1371/journal.pntd.0007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zak D.E., Penn-Nicholson A., Scriba T.J., Thompson E., Suliman S., Amon L.M. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387(10035):2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suliman S., Thompson E., Sutherland J., Weiner Rd J., Ota M.O.C., Shankar S. Four-gene pan-African blood signature predicts progression to tuberculosis. Am J Respiratory Crit Care Med. 2018;197(9):1198–1208. doi: 10.1164/rccm.201711-2340OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Penn-Nicholson A., Mbandi S.K., Thompson E., Mendelsohn S.C., Suliman S., Chegou N.N. RISK6, a 6-gene transcriptomic signature of TB disease risk, diagnosis and treatment response. Sci Rep. 2020;10(1):8629. doi: 10.1038/s41598-020-65043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner C.T., Gupta R.K., Tsaliki E., Roe J.K., Mondal P., Nyawo G.R. Blood transcriptional biomarkers for active pulmonary tuberculosis in a high-burden setting: a prospective, observational, diagnostic accuracy study. Lancet Respir Med. 2020;8(4):407–419. doi: 10.1016/S2213-2600(19)30469-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jorge K., Souza R.P., Assis M.T.A., Araujo M.G., Locati M., Jesus A.M.R. Characterization of MicroRNA expression profiles and identification of potential biomarkers in leprosy. J Clin Microbiol. 2017;55(5):1516–1525. doi: 10.1128/JCM.02408-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tió-Coma M., van Hooij A., Bobosha K., van der Ploeg-van Schip J.J., Banu S., Khadge S. Whole blood RNA signatures in leprosy patients identify reversal reactions before clinical onset: a prospective, multicenter study. Sci Rep. 2019;9(1):17931. doi: 10.1038/s41598-019-54213-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Montoya D.J., Andrade P., Silva B.J.A., Teles R.M.B., Ma F., Bryson B. Dual RNA-Seq of human leprosy lesions identifies bacterial determinants linked to host immune response. Cell Rep. 2019;26(13):3574–3585. doi: 10.1016/j.celrep.2019.02.109. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andrade P.R., Mehta M., Lu J., Teles R.M.B., Montoya D., Scumpia P.O. The cell fate regulator NUPR1 is induced by mycobacterium leprae via type I interferon in human leprosy. PLoS NeglTrop Dis. 2019;13(7) doi: 10.1371/journal.pntd.0007589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inkeles M.S., Teles R.M., Pouldar D., Andrade P.R., Madigan C.A., Lopez D. Cell-type deconvolution with immune pathways identifies gene networks of host defense and immunopathology in leprosy. JCI Insight. 2016;1(15):e88843. doi: 10.1172/jci.insight.88843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Realegeno S., Kelly-Scumpia K.M., Dang A.T., Lu J., Teles R., Liu P.T. S100A12 is part of the antimicrobial network against mycobacterium leprae in human macrophages. PLoS Pathog. 2016;12(6) doi: 10.1371/journal.ppat.1005705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orlova M., Cobat A., Huong N.T., Ba N.N., Van T.N., Spencer J. Gene set signature of reversal reaction type I in leprosy patients. PLoS Genet. 2013;9(7) doi: 10.1371/journal.pgen.1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guerreiro L.T., Robottom-Ferreira A.B., Ribeiro-Alves M., Toledo-Pinto T.G., Rosa B.T., Rosa P.S. Gene expression profiling specifies chemokine, mitochondrial and lipid metabolism signatures in leprosy. PLoS One. 2013;8(6):e64748. doi: 10.1371/journal.pone.0064748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bleharski J.R., Li H., Meinken C., Graeber T.G., Ochoa M.T., Yamamura M. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science. 2003;301(5639):1527–1530. doi: 10.1126/science.1087785. [DOI] [PubMed] [Google Scholar]
- 62.Salgado C.G., Pinto P., Bouth R.C., Gobbo A.R., Messias A.C.C., Sandoval T.V. miRNome expression analysis reveals new players on leprosy immune physiopathology. Front Immunol. 2018;9(463) doi: 10.3389/fimmu.2018.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guerreiro L.T., Robottom-Ferreira A.B., Ribeiro-Alves M., Toledo-Pinto T.G., Rosa Brito T., Rosa P.S. Gene expression profiling specifies chemokine, mitochondrial and lipid metabolism signatures in leprosy. PLoS One. 2013;8(6):e64748. doi: 10.1371/journal.pone.0064748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardus R., Alam K., Kundu K., Chandra Roy J., Zafar T., Chowdhury A.S. Effectiveness of single-dose rifampicin after BCG vaccination to prevent leprosy in close contacts of patients with newly diagnosed leprosy: a cluster randomized controlled trial. Int J Infect Dis. 2019;88:65–72. doi: 10.1016/j.ijid.2019.08.035. [DOI] [PubMed] [Google Scholar]
- 65.Richardus R., van Hooij A., van den Eeden S.J.F., Wilson L., Alam K., Richardus J.H. BCG and adverse events in the context of leprosy. Front Immunol. 2018;9:629. doi: 10.3389/fimmu.2018.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richardus R.A., Alam K., Pahan D., Feenstra S.G., Geluk A., Richardus J.H. The combined effect of chemoprophylaxis with single dose rifampicin and immunoprophylaxis with BCG to prevent leprosy in contacts of newly diagnosed leprosy cases: a cluster randomized controlled trial (MALTALEP study) BMC Infect Dis. 2013;13:456. doi: 10.1186/1471-2334-13-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yearly district activity report 2018. Internal communication: rural health program, the leprosy mission international Bangladesh.
- 68.National Leprosy Elimination Programme . 4th ed. Dhaka: Bangladesh National Leprosy Elimination Programme Directorate General of Health Services; 2014. National guidelines and technical manual on leprosy. [Google Scholar]
- 69.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 72.Lang M., Kotthaus H., Marwedel P., Weihs C., Rahnenführer J., Bischl B. Automatic model selection for high-dimensional survival analysis. J Statist Comput Simulation. 2015;85(1):62–76. [Google Scholar]
- 73.Sweeney T.E., Braviak L., Tato C.M., Khatri P. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir Med. 2016;4(3):213–224. doi: 10.1016/S2213-2600(16)00048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S. The GeneCards Suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformat. 2016;54 doi: 10.1002/cpbi.5. 1.30.1-1.3. [DOI] [PubMed] [Google Scholar]
- 75.Vanlalhruaii Tonsing M., Vanlalbiakdiki Sailo C., Zothansanga Chhakchhuak L., Chhakchhuak Z., Pandit B. Analysis of variants in mitochondrial genome and their putative pathogenicity in tuberculosis patients from Mizoram. North East India. Mitochondrion. 2020;54:21–25. doi: 10.1016/j.mito.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 76.Wang Z., Arat S., Magid-Slav M., Brown J.R. Meta-analysis of human gene expression in response to Mycobacterium tuberculosis infection reveals potential therapeutic targets. BMC Syst Biol. 2018;12(1) doi: 10.1186/s12918-017-0524-z. 3- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manzanillo P.S., Ayres J.S., Watson R.O., Collins A.C., Souza G., Rae C.S. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501(7468):512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scarffe L.A., Stevens D.A., Dawson V.L., Dawson T.M. Parkin and PINK1: much more than mitophagy. Trends Neurosci. 2014;37(6):315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Sage V., Cinti A., Amorim R., Mouland A.J. Adapting the stress response: viral subversion of the mTOR signaling pathway. Viruses. 2016;8(6):152. doi: 10.3390/v8060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12(5):325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laplante M., Sabatini D.M. mTOR signaling at a glance. J Cell Sci. 2009;122(20):3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Azevedo M.C.S., Marques H., Binelli L.S., Malange M.S.V., Devides A.C., Silva E.A. Simultaneous analysis of multiple T helper subsets in leprosy reveals distinct patterns of Th1, Th2, Th17 and Tregs markers expression in clinical forms and reactional events. Med Microbiol Immunol. 2017;206(6):429–439. doi: 10.1007/s00430-017-0519-9. [DOI] [PubMed] [Google Scholar]
- 83.Vardhana S.A., Wolchok J.D. The many faces of the anti-COVID immune response. J Exp Med. 2020;217(6) doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168. doi: 10.1016/j.cell.2020.08.017. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou J., Yi Q., Tang L. The roles of nuclear focal adhesion kinase (FAK) on Cancer: a focused review. J Exp Clin Cancer Res. 2019;38(1):250. doi: 10.1186/s13046-019-1265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yoon H., Dehart J.P., Murphy J.M., Lim S.T. Understanding the roles of FAK in cancer: inhibitors, genetic models, and new insights. J Histochem Cytochem. 2015;63(2):114–128. doi: 10.1369/0022155414561498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leal-Calvo T., MO Moraes. Reanalysis and integration of public microarray datasets reveals novel host genes modulated in leprosy. Mol Genet Genomics. 2020 doi: 10.1007/s00438-020-01705-6. [DOI] [PubMed] [Google Scholar]
- 88.Serrano-Coll H., Salazar-Peláez L.M., Mesa-Betancourt F., Cardona-Castro N. Oct-6 transcriptional factor a possible biomarker for leprosy diagnosis. Diagn Microbiol Infect Dis. 2020;99(2) doi: 10.1016/j.diagmicrobio.2020.115232. [DOI] [PubMed] [Google Scholar]
- 89.Pinto P., da Silva M.B., Moreira F.C., Bouth R.C., Gobbo A.R., Sandoval T.V. Leprosy piRnome: exploring new possibilities for an old disease. Sci Rep. 2020;10(1):12648. doi: 10.1038/s41598-020-69355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dubey R. Assuming the role of mitochondria in mycobacterial infection. Int J Mycobacteriol. 2016;5(4):379–383. doi: 10.1016/j.ijmyco.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 91.Modlin R.L., Bloom B.R. TB or not TB: that is no longer the question. Sci Transl Med. 2013;5(213) doi: 10.1126/scitranslmed.3007402. 213sr6. [DOI] [PubMed] [Google Scholar]
- 92.Geluk A., van Meijgaarden K.E., Wilson L., Bobosha K., vdP-vS J.J., van den Eeden S.J. Longitudinal immune responses and gene expression profiles in type 1 leprosy reactions. J Clin Immunol. 2014;34(2):245–255. doi: 10.1007/s10875-013-9979-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.