Abstract
Guided by the extensive knowledge of CRISPR-Cas9 molecular mechanisms, protein engineering can be an effective method in improving CRISPR-Cas9 toward desired traits different from those of their natural forms. Here, we describe a directed protein evolution method that enables selection of catalytically enhanced CRISPR-Cas9 variants (CECas9) by targeting a shortened protospacer within a toxic gene. We demonstrate the effectiveness of this method with a previously characterized Type II-C Cas9 from Acidothermus cellulolyticus (AceCas9) and show by enzyme kinetics an up to fourfold improvement of the in vitro catalytic efficiency by AceCECas9. We further evolved the more widely used Streptococcus pyogenes Cas9 (SpyCas9) and demonstrated a noticeable improvement in the SpyCECas9-facilitated homology directed repair–based gene insertion in human colon cancer cells.
The CRISPR-Cas9 system has found widespread applications in genome manipulations due to its simplicity and effectiveness. Significant efforts in enzyme engineering have been made to improve the CRISPR-Cas9 systems beyond their natural power with additional functionalities such as DNA modification, transcriptional regulation, and high target selectivity.1–10 Relatively less attention, however, has been paid to improving the catalytic efficiency of CRISPR-Cas9. Increased catalytic efficiency may be desired in applications where the currently available CRISPR-Cas9 tools are either ineffective4,11–14 or of low efficiency such as with type II-C Cas915–18 or in non-mammals.19,20
Functional CRISPR-Cas9 enzymes are composed of a Cas9 protein, a CRISPR RNA (crRNA), and a trans-activating crRNA (tracrRNA). The crRNA and tracrRNA can often be replaced by a chimeric single-guide RNA (sgRNA).21The sgRNA or crRNA contains a complementary region that base pairs with the target DNA (protospacer). Cas9 possesses multiple domains: a nucleic acid recognition domain (REC), a protospacer adjacent motif (PAM)-interacting domain (PID), a RuvC nuclease domain, and a HNH nuclease domain (Fig. 1A). Cas9 searches for and initially locates a PAM by its PID, which leads to unwinding of the protospacer and formation of the DNA–sgRNA heteroduplex or R-loop. Finally, the HNH domain cleaves the target strand, while the RuvC domain cleaves the non-target strand.22–24 Previous biochemical and biophysical studies on Streptococcus pyogenes Cas9 (SpyCas9) showed that it is essentially a single turnover enzyme with the rate-limiting step at DNA binding25 or R-loop formation.26 Importantly, DNA cleavage efficiency scales with sampling of the HNH domain conformation; a correct conformation enhances the cleavage activity of the RuvC domain.27 This allostery between HNH and RuvC is believed to result from the hinge comprising two helices that connect RuvC-III to HNH.21,27–32 Mutations that disrupt one of the two hinge helices lowered the DNA cleavage activity of SpyCas9.27 These detailed mechanistic studies suggest that the allosteric hinge is also a potential target for engineering catalytically enhanced CRISPR-Cas9 (CECas9) variants.33 However, protein engineering by rational design to select for CECas9 variants would be laborious and have limited chances of successes.
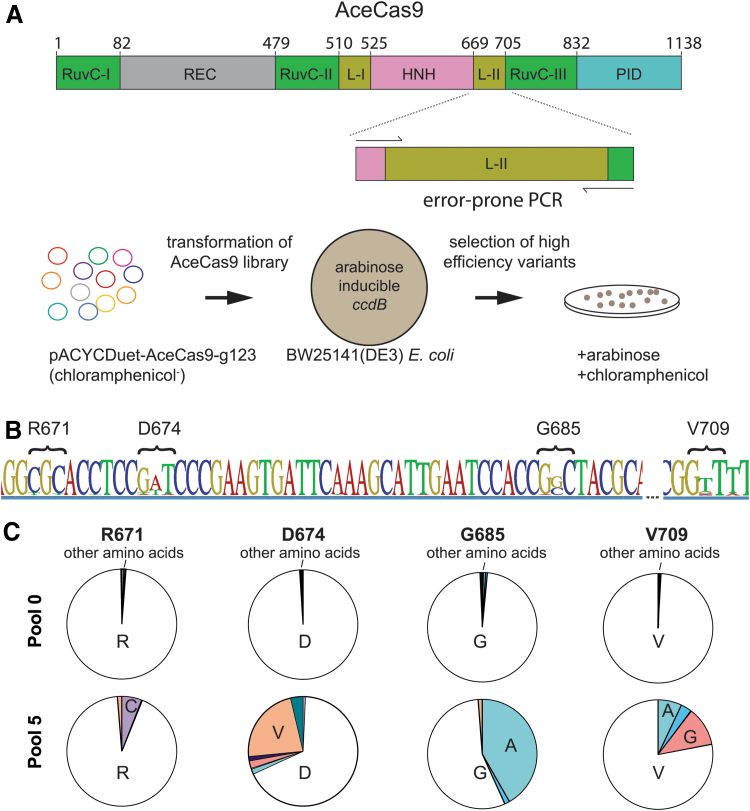
FIG. 1.
Selection of catalytically enhanced Type II-C Cas9 from Acidothermus cellulolyticus (AceCas9). (A) Top: Domain organization of AceCas9 with amplified view of linker II (L-II) with primers used for error-prone polymerase chain reaction primers. Bottom: Directed protein evolution selection strategy. pACYC-AceCas9-g123 and pACYC-AceCas9-g119 co-express AceCas9 and the single-guide RNA with 24mer and 20mer guide, respectively. (B) Sequence logo of consensus sequence from next-generation sequencing analysis of the final pool. The height of each letter is proportional to the observed frequency of each nucleotide in the alignment column. The locations with highest deviation are marked by their corresponding amino acid positions. (C) Pie charts illustrate enrichment of amino acids at the four highest frequency sites. Pool 0 reflects enrichment values in the starting library, whereas pool 5 reflects those from the last round of survival colonies. Single letters for amino acids are used. Color images are available online.
We previously described a toxicity-based bacterial survival assay that is amenable for a library-based directed protein evolution selection.34,35 In this approach, a library of plasmids encoding a collection of Cas9 variants is transformed into bacterial cells harboring a plasmid expressing the toxic protein ccdB under the induction by arabinose. In the presence of arabinose, cells transformed with the Cas9 variants capable of cleaving the ccdB-encoding plasmid would grow, whereas those containing the variants incapable of cleaving the ccdB-encoding plasmid would not. Thus, the survival cell colonies would allow identification of the Cas9 variants that cleave the ccdB-encoding plasmid. To identify CECas9 variants, we need to program the ccdB-encoding plasmid with a protospacer that is not cleaved by the wild-type Cas9.
We first applied the protein directed evolution method to a Type II-C Cas9 from Acidothermus cellulolyticus (AceCas9). Due to its thermostability and recently discovered sensitivity to DNA methylation,36,37 AceCas9 has been used for gene editing in model thermophilic bacteria for biofuel processing38 and is anticipated to have expanded application in epigenetics. AceCas9 was previously shown to be most efficient in cleaving protospacers with 24 nt of spacer complementarity.37 The rate of single-turnover cleavage of the 24 nt protospacer is fourfold that of the 20 nt protospacer on supercoiled plasmid DNA. In bacterial cells, while AceCas9 targeting a 24 nt protospacer causes near 100% survival in the ccdB-based cell survival assay, that targeting a 20 nt protospacer has a rate of survival of <0.2% (Supplementary Fig. S1). Therefore, in a typical transformation experiment, no surviving colonies would be observed when the wild-type AceCas9 is introduced to electrocompetent Escherichia coli cells harboring the ccdB plasmid with a 20 nt protospacer.35,37 We took advantage of this selection feature in identifying AceCECas9. We hypothesized that the allosteric hinge can be engineered to allow faster HNH conformational sampling, which leads to faster RuvC cleavage, resulting in more efficient formation of double-strand breaks. To this end, we generated a library of the hinge variants through error-prone polymerase chain reaction (PCR; Fig. 1A).
In contrast to negligible growth of cells containing the ccdB plasmid transformed with a 20 nt protospacer wild-type AceCas9, those transformed with the hinge library showed significant growth. The survival colonies were then pooled, and their plasmid DNA was extracted for a second round of selection. The selection cycle was repeated until the rate of survival per microgram of DNA no longer increased. The DNA pool from each selection cycle was subjected to next-generation sequencing (NGS), and plasmid DNA from 20 individual colonies of the final cycle were also isolated for Sanger sequencing. We identified the survival variants via single nucleotide polymorphism (SNP), calling on the assembled contigs of each DNA pool (Fig. 1B and C). The most deviations occur at the nucleotide positions corresponding to amino acid Arg671, Asp674, Gly685, and Val709(Fig. 1B and C). These survival variants encode AceCECas9 R671C, D674V, G685A, and V709A/G. The findings were supported by Sanger sequencing of individual colonies, which showed 11/20 to be G685A, 6/20 to be V709A/G, and 3/20 to be D674V. We mapped these amino acid positions onto our previously constructed AceCas9 structure model (Supplementary Fig. S2). Arg671, Asp674, and Gly685 are located downstream of the helix following the RuvC domain (helix 1; AceCas9, like other type II-C Cas9s, lacks the second helix following helix 1). Surprisingly, V709 is located outside the predicted hinge region on the β-strand directly packing against hinge helix 1 (Supplementary Fig. S2), suggesting that structural elements interacting with the hinge helices also impact the allosteric regulation.
In order to confirm that the enhanced AceCas9 was a direct consequence of mutations rather than other in vivo factors such as expression levels, we made individual AceCas9 mutants of G685A and V709A and tested both in-cell and in vitro cleavage assays. Not surprisingly, G685A and V709A had nearly 100% survival rate in E. coli cells harboring the ccdB plasmid with the 20 nt protospacer (Fig. 2A), while the wild-type AceCas9 would not survive (Fig. 2 and Supplementary Fig. S1). Consistently, the single-turnover rate of cleavage for both purified mutants are three- to fourfold that of the purified wild-type AceCas9 for either the 24 nt (24mer) or the 20 nt (20mer) protospacer (Fig. 2B).37 Thus, the significantly increased rate of cleavage allows AceCECas9 to eliminate the toxic ccdB plasmid effectively with a 20 nt protospacer, promoting cell survival. Finally, though AceCECas9 was selected using a specific 20 nt protospacer, in vitro cleavage with AceCECas9 programmed to target a site of the human EMX1 gene (NC_000002.12) also displayed accelerated cleavage in comparison with wild-type AceCas9 (Supplementary Fig. S3A), suggesting AceCECas9 is not specifically enhanced for a single protospacer.
FIG. 2.
Verification of the catalytically enhanced AceCas9 by survival assay and kinetic analysis. (A) Plasmids encoding the selected AceCas9 mutants were transformed into ccdB-harboring cells that are plated on plates with (Ara+) or without arabinose (Ara–). Images of the plates are shown with the corresponding survival rate plotted below. (B) Single-turnover DNA cleavage experiments and the resulting rates of the AceCECas9 with either the 20mer spacer or the 24mer spacer single-guide RNA (sgRNA). Molar excess of AceCas9–sgRNA complex were incubated with plasmid DNA for various times at 50°C before being resolved and visualized on agarose gels. Kinetic experiments were performed in triplicate (Supplementary Fig. S4).
In order to evaluate if AceCECas9 variants have increased tolerance of DNA-sgRNA mismatches, we assessed the survival rates of G685A and V709A in E. coli cells harboring the ccdB plasmids with mismatched protospacers at position −1 or −4. These mismatches were previously shown to prevent the wild-type AceCas9 from eliminating the ccdB plasmid, thus leading to death of the E. coli hosts.35 Similarly, no cell survived either the T(-1)G or the G(-4)A plasmid with G685A (Supplementary Fig. S3B and C), although a small percentage of cells survived both mismatches with V709A, suggesting that V709A has an elevated off-target activity on certain mismatched protospacers in comparison with the wild-type. The AceCECas9 in its current form may thus be applicable in cases where the enhanced catalytic activity outweighs the off-target effect. Further engineering efforts may combine strategies that increase specificity with those that enhance catalytic efficiency to yield an optimal gene-editing enzyme.
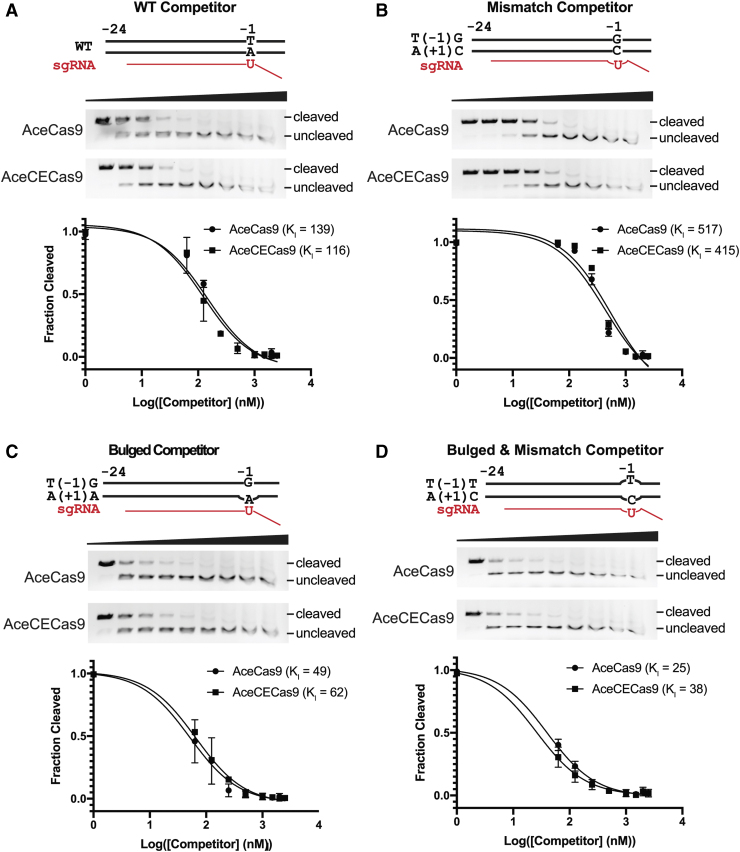
In order to identify the molecular basis for the enhanced catalytic efficiency, we performed DNA binding competition experiments to learn if AceCECas9 enhances DNA binding, DNA unwinding, or HNH conformational sampling. To a typical DNA cleavage assay containing 6 nM plasmid target and 250 nM AceCas9 (or AceCECas9), double-stranded DNA oligo competitors were added at increasing concentrations. The fraction of cleavage was then quantified and plotted against the logarithm of competitor concentrations. The plot was fitted using a standard competition binding model to yield an apparent competitor binding constant, KI. The wild-type protospacer DNA oligo has nearly identical KI for both AceCas9 and AceCECas9 (Fig. 3A). The T(-1)G protospacer DNA oligo is a worse competitor than the wild-type but again with equal KI for both AceCas9 and AceCECas9, suggesting that AceCECas9 does not enhance, in comparison with AceCas9, DNA binding for either the wild-type or mismatched protospacer (Fig. 3A). Finally, AceCas9 and AceCECas9 showed similar binding constants to two bubbled DNA oligos (either complementary or not to sgRNA; Fig. 3B), indicating that the DNA unwinding was not altered in AceCECas9. These results confirm that the enhanced catalytic efficiency of AceCECas9 is through a process other than DNA binding and unwinding, most likely HNH domain sampling.
FIG. 3.
DNA binding competition assay results. The DNA oligo substrates used are schematically shown as wild-type (WT), T(-1)G/A(+1)C (dsDNA with mismatch to sgRNA at position +1), T(-1)T/A(+1)C (bulged DNA oligo at position +1 that mismatches with sgRNA), and non-target strand bulge (bulged DNA oligo at position at +1 that matches sgRNA). For each competition binding experiment, the cleavage gel image and fraction of cleavage versus competitor concentration at logarithmic scale are shown. Solid curves are fitted theoretical curves and fitted KI values are indicated. (A) Competition binding result for the wild-type or T(-1)G oligo with AceCas9 (WT) and the V709A variant. (B) Competition binding result for the bulged oligos with AceCas9 (WT) and the V709A variant. Experiments performed in triplicate (Supplementary Fig. S5). Color images are available online.
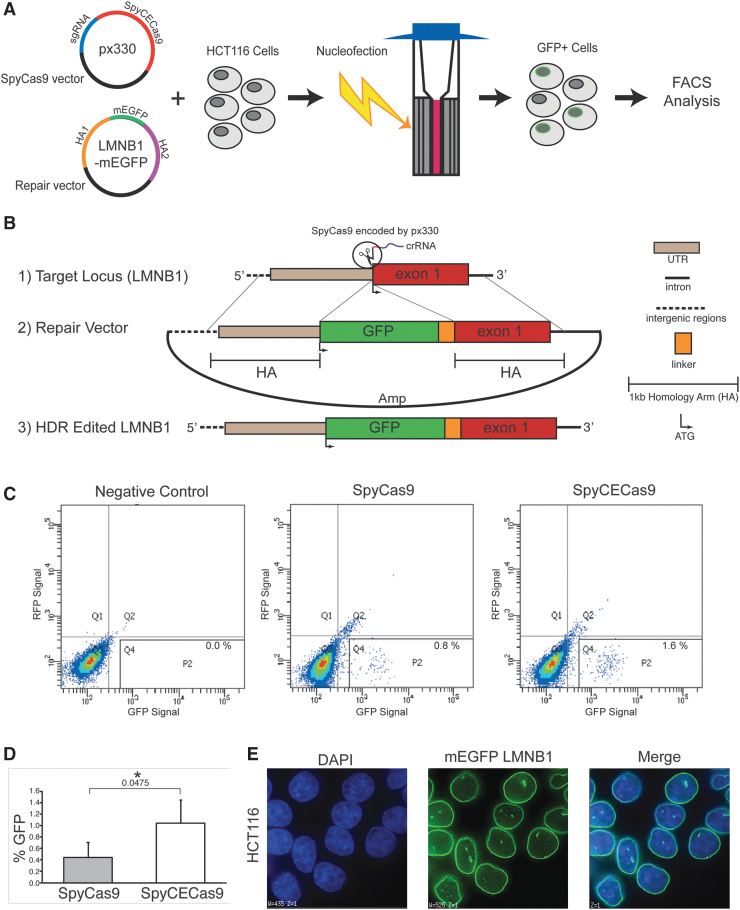
Finally, we applied the same directed evolution strategy to the more widely used SpyCas9. Similar to AceCas9, we systematically reduced the base pairs in the protospacer complementary to the guide RNA and determined that 17 bp slowed the wild-type SpyCas9 sufficiently to obstruct survival. We then generated a DNA library that encodes SpyCas9 hinge variants (with guide RNA) and transformed it to the cells harboring the ccdB plasmid with a 17 bp protospacer. Unlike the wild-type SpyCas9 transformation that resulted in no cell survival, the library transformation resulted in significant growth on arabinose-containing plates, and after iterative rounds of evolution, a single variant, K918N, emerged (Supplementary Fig. S6). We termed the K918N variant “catalytically enhanced SpyCas9” (SpyCECas9). To determine if the SpyCECas9 improves gene-editing applications relative to the SpyCas9, we employed both to achieve homology directed repair (HDR)-based insertion of the green fluorescent protein (GFP) gene39 in the LMNB1 gene within HCT116 cells (Fig. 4 and Supplementary Fig. S7). As shown in Figure 4C–E, co-transfection of the SpyCECas9-encoding plasmid with the HDR template into HTC116 consistently resulted in a twofold increase in GFP incorporation versus that of SpyCas9, likely due to its enhanced cleavage of the target DNA.
FIG. 4.
Gene insertion facilitated by the wild-type and the catalytically enhanced Streptococcus pyogenes Cas9 (SpyCas9) in HCT116 cells. (A) Cartoon illustration depicting the genome insertion process. The px330 vector co-expressing either the wild-type SpyCas9 or the SpyCECas9 with the sgRNA targeting the LMNB1 gene is co-transfected with the repair vector containing mEGFP flanked by 1 kb homology arms (HAs). Each transfection is performed with the Lonza SE cell line 4-D nucleofector solution kit by electroporating one million HCT116 cells in an electro-cuvette. Cells are plated and cultured for 48 h post transfection (positive cells are depicted as gray with a green ring around the nuclear lamina). The HCT116 cells that successfully achieved homology directed repair (HDR) by incorporation of the green fluorescent protein (GFP) are detected, sorted, collected via flow cytometry, and expanded for imaging. (B) Detailed scheme of the CRISPR-Cas9-mediated GFP tagging strategy. (1) The crRNA (purple) co-expressing with the wild-type SpyCas9 (px330, Addgene #42230) or the catalytically enhanced (CE) SpyCECas9 on a modified px330 vector targets a site upstream of exon 1 (red) of the LMNB1 gene. (2) A repair vector containing 1 kb HAs flanking the GFP-encoding sequence fused with a linker sequence (AICSDP-10:LMNB1-mEGFP, Addgene #87422). (3) Successfully edited target locus following SpyCas9 cleavage and HDR contains an in-frame insertion of the GFP after the start codon of LMNB1 exon 1. (C) The observed GFP signal of the wild-type and the catalytically enhanced SpyCas9. Flow cytometry plots displaying GFP fluorescence intensity (x-axis) versus RFP fluorescence intensity (y-axis; to control for auto-fluorescence) following 48 h post transfection for the wild-type (SpyCas9) and catalytically enhanced (SpyCECas9). GFP+ cells fall within the P2 gate. The negative control is obtained from cells transfected with the repair vector and SpyCas9 without a sgRNA. Experiments were conducted in triplicate (Supplementary Figure S7). (D) Bar graph displaying the averages of the quadruplicate measurement for both SpyCas9 and SpyCECas9, and error bars displaying standard deviation. Statistical significance was determined by an unpaired two-tailed t-test (*p ≤ 0.05, exact value shown below the asterisk). (E) Representative fluorescence microscopy images of GFP-tagged LMNB1 in HCT116 cells obtained on a DeltaVision (GE Life Sciences) microscope. The cell nuclei were stained with DAPI. Color images are available online.
In order to ensure that SpyCECas9 did not result in elevated off-target cleavage in the HDR-mediated gene insertion, we analyzed the target and the top two predicted off-target sites in the HTC116 genome40 via sequencing (Supplementary Fig. S8). We show that the target site indeed contains the incorporated gene sequence encoding GFP, while no indel is detected in the two predicted off-target sites (Supplementary Fig. S8).
Our results present the first engineered Cas9s with increased catalytic efficiency through a directed protein evolution method. We demonstrated the effectiveness of this method on AceCas9 that yielded enhanced DNA cleavage in bacterial cells. We then used the same strategy to produce SpyCECas9 that doubled the efficiency of the SpyCas9-mediated HDR in human colon cancer cells where the overall level of HDR-mediated gene insertion is typically only a few percent. These catalytically enhanced Cas9s can be applied in such areas where high catalytic efficiency is needed.
Methods
Plasmid and library construction
The arabinose-inducible ccdB-encoding plasmid (p11-LacY-wtx1) was a gift from Dr. David Edgell (University of Western Ontario). The target sequence was inserted downstream of the ccdB gene using restriction enzymes. Point mutations were made via Q5 site-directed mutagenesis (NEB).
Genes encoding AceCas9 and the sgRNA were inserted into the pACYCDuet vector (Novagen 71147) to form pACYCDuet-Ac9-g119 (20mer) or pACYCDuet-Ac9-g123 (24mer), respectively, as previously described.37 The hinge library was created by amplifying the predicted hinge region with Gibson assembly primers via error-prone PCR (Ser663- Gly712) at an average rate of one mutation per 100 bp and inserting the PCR products into the pACYCDuet-Ac9-g119 vector. The resulting DNA was transformed into DH5α cells. About 60,000 colonies were obtained after overnight growth on media containing chloramphenicol. The colonies were pooled, grown for 2 h, and then collected for extraction of plasmid DNA.
In vivo selection
The survival assay was carried out as previously described.34,35,37 Briefly, electrocompetent E. coli BW25141 containing the modified p11-LacY-wtx1 (ccdB-encoding) plasmid were transformed with 100–200 ng of AceCas9 library plasmids. Cells were then recovered in pre-warmed Super Optimal Broth for 30 min with shaking at 37°C before 0.05 mM IPTG was added, and recovery continued for an additional 60 min. Transformants were then plated on LB agar plates containing either chloramphenicol or chloramphenicol and 10 mM arabinose, and incubated at 37°C for 16–20 h. Colonies were counted manually, and survival rates were calculated by dividing the colony forming units of arabinose-containing plates by that of the chloramphenicol only plates.
NGS
The hinge region of each survival pool was PCR amplified with primers containing Illumina indexes. Adaptors were added to these PCR products, and then the pooled libraries were subjected to 300 cycle single-end sequencing using MiSEQ (Illumina). Data were analyzed using Geneious v9.0.5 NGS analysis tools.
Protein expression and purification
AceCas9 and mutant variants were expressed and purified as previously described.37 Briefly, harvested cells in a lysis buffer were lysed via sonication. Clarified lysate was loaded onto an Ni-NTA agarose (Qiagen) column. Elutant was further purified via a HiTrap Heparin-HP (GE Healthcare) column and then a size exclusion column (HiLoad 26/60 Superdex 200 increase; GE Healthcare) before concentration and storage at −80°C.
In vitro transcription and purification of sgRNA
DNA oligos coding for a T7 promoter followed by the sgRNA were annealed in 10 × PCR buffer by heating in boiling water for 5 min followed by slow cooling. In vitro transcription was performed by adding 1 μM of this dsDNA to 50 mM Tris, pH 8.0, 40 mM MgCl2, 10 mM dithiothreitol (DTT), 2 mM Spermindine, 5 mM ATP, UTP, CTP, and GTP, pH 8.0, 0.1% Triton X-100, and 0.15 mg/mL of homemade T7 RNA polymerase, and incubating at 37°C overnight. The reaction was quenched the following morning by adding 50 mM EDTA. The sgRNA was then extracted via phenol/chloroform pH 4.5 extraction and purified via 10% denaturing polyacrylamide gel electrophoresis.
Plasmid cleavage assay
Plasmid cleavage assays were performed as previously described.34,35,37 Briefly, 500 nM pre-annealed AceCas9:sgRNA complex was incubated with 6 nM plasmid DNA in cleavage buffer (20 mM Tris, pH 7.5, 150 mM KCl, 2 mM DTT, 10 mM MgCl2, 5% glycerol) at 50°C for a given time. Reactions were then stopped by adding 5 × stop buffer (25 mM Tris, pH 7.5, 250 mM EDTA, 1% sodium dodecyl sulfate, 0.05% w/v bromophenol blue, 30% glycerol) and resolved on a 1 × TBE 0.5% agarose gel containing ethidium bromide. Gels were imaged using the ChemiDoc XRS System (Bio-Rad).
Single turnover kinetics
Plasmid kinetics assays were performed in a similar manner as the plasmid cleavage assays, but all reagents (DNA, RNP complex, and Eppendorf tubes) were kept ice-cold before reactions were initiated. After initiation of reaction, samples were taken from a 50°C water bath in triplicate at each time point and placed on ice. Then, ice-cold stop buffer was quickly added. Gels were analyzed by Image Lab v5.2.1 build 11 (Bio-Rad) and fitted to an exponential plateau curve in Prism v8.1.1 to extract kcleavage:
where Ff and Fi represent the final and initial fraction of cleavage, respectively.
DNA binding competition assays
Oligonucleotides were annealed by heating in a 95°C water bath followed by slow cooling. Serial dilutions of oligos were made from 25 to 0.063 μM, and added to 6 nM plasmid DNA. The plasmid/oligo mixture was then added to annealed RNP complex (250 nM AceCas9 [or AceCECas9] and 250 nM sgRNA) and allowed to react for 15 min at 50°C. Reactions were stopped with stop buffer and resolved on agarose gels before being visualized and quantified as before. The inhibitor binding constant, KI, is extracted by fitting fraction of cleavage to a typical competitive binding model:
where [competitor] is the concentration of the competitor and Fi and Ff are the initial and final fraction of cleavage, respectively.
CRISPR-mediated HDR assay in HCT116 cells
HCT116 cells were maintained in McCoy's 5A media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C with 5% CO2. The plasmids encoding the wild-type SpyCas9 (px330, Addgene #42230) or its catalytically enhanced variant, SpyCECas9 (modified px330), targeting the LMNB1 gene (GGGGTCGCAGTCGCCATGGC; 2 μg) and an LMNB1-mEGFP containing repair vector (Addgene plasmid #87422) (3 μg) were nucleofected into HCT116 cells grown to 70–80% confluency using the Lonza SE cell line 4-D Nucleofector solution kit with pulse code EN113.39 Forty-eight hours post nucleofection, cells were evaluated for the presence of GFP using FACSCanto.
Supplementary Material
Acknowledgments
We thank D. Edgell for providing cells and plasmids for the in vivo study; B. Washburn, C. Pye, and K. Poduch of the FSU Molecular Cloning Facility for the cloning experiments and discussion; S. Miller and A. Brown of the FSU Sequencing facility for assistance with NGS library preparation and sequencing; and D.Q. Mesa for assistance in enzymatic assays.
Author Disclosure Statement
The authors declare that they have no conflict of interest.
Funding Information
This work was supported by NIH grants R01 GM099604 to H.L. and R01 GM083337 to D.M.G.
Supplementary Material
References
- 1. Kleinstiver BP, Pattanayak V, Prew MS, et al. . High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016;529:490–495. DOI: 10.1038/nature.16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rees HA, Komor AC, Yeh W-H, et al. . Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun 2017;8:15790. DOI: 10.1038/ncomms15790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Slaymaker IM, Gao L, Zetsche B, et al. . Rationally engineered Cas9 nucleases with improved specificity. Science 2016;351:84–88. DOI: 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen JS, Dagdas YS, Kleinstiver BP, et al. . Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017;550:407–410. DOI: 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaudelli NM, Komor AC, Rees HA, et al. . Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017;551:464–471. DOI: 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 2014;32:577–582. DOI: 10.1038/nbt.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu JH, Miller SM, Geurts MH, et al. . Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018;556:57–63. DOI: 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li C, Zong Y, Wang Y, et al. . Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 2018;19:59. DOI: 10.1186/s13059-018-1443-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee JK, Jeong E, Lee J, et al. . Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun 2018;9:3048. DOI: 10.1038/s41467-018-05477-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Didovyk A, Borek B, Tsimring L, et al. . Transcriptional regulation with CRISPR-Cas9: principles, advances, and applications. Curr Opin Biotechnol 2016;40:177–184. DOI: 10.1016/j.copbio.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doench JG, Fusi N, Sullender M, et al. . Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184–191. DOI: 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doench JG, Hartenian E, Graham DB, et al. . Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 2014;32:1262–1267. DOI: 10.1038/nbt.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graf R, Li X, Chu VT, et al. . sgRNA sequence motifs blocking efficient CRISPR/Cas9-mediated gene editing. Cell Rep 2019;26:1098–1103.e3. DOI: 10.1016/j.celrep.2019.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gagnon JA, Valen E, Thyme SB, et al. . Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 2014;9:e98186. DOI: 10.1371/journale.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee CM, Cradick TJ, Bao G. The Neisseria meningitidis CRISPR-Cas9 system enables specific genome editing in mammalian cells. Mol Ther 2016;24:645–654. DOI: 10.1038/mt.2016.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harrington LB, Paez-Espino D, Staahl BT, et al. . A thermostable Cas9 with increased lifetime in human plasma. Nat Commun 2017;8:1424. DOI: 10.1038/s41467-017-01408-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma E, Harrington LB, O'Connell MR, et al. . Single-stranded DNA cleavage by divergent CRISPR-Cas9 enzymes. Mol Cell 2015;60:398–407. DOI: 10.1016/j.molcel.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mir A, Edraki A, Lee J, et al. . Type II-C CRISPR-Cas9 biology, mechanism, and application. ACS Chem Biol 2018;13:357–365. DOI: 10.1021/acschembio.7b00855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mushtaq M, Sakina A, Wani SH, et al. . Harnessing genome editing techniques to engineer disease resistance in plants. Front Plant Sci 2019;10:550. DOI: 10.3389/fpls.2019.00550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jaganathan D, Ramasamy K, Sellamuthu G, et al. . CRISPR for crop improvement: an update review. Front Plant Sci 2018;9:985. DOI: 10.3389/fpls.2018.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 2017;46:505–529. DOI: 10.1146/annurev-biophys-062215-010822 [DOI] [PubMed] [Google Scholar]
- 22. Sternberg SH, Redding S, Jinek M, et al. . DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014;507:62–67. DOI: 10.1038/nature13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 2013;82:237–266. DOI: 10.1146/annurev-biochem-072911-172315 [DOI] [PubMed] [Google Scholar]
- 24. Singh D, Sternberg SH, Fei JY, et al. . Real-time observation of DNA recognition by the RNA-guided endonuclease Cas9 using single-molecule FRET. Biophys J 2015;108:26a. DOI: 10.1038/ncomms12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raper AT, Stephenson AA, Suo Z. Functional insights revealed by the kinetic mechanism of CRISPR/Cas9. J Am Chem Soc 2018;140:2971–2984. DOI: 10.1021/jacs.7b13047 [DOI] [PubMed] [Google Scholar]
- 26. Gong SZ, Yu HH, Johnson KA, et al. . DNA unwinding is the primary determinant of CRISPR-Cas9 specificity. Biophys J 2018;114:251a. DOI: 10.1016/j.celrep.2017.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sternberg SH, LaFrance B, Kaplan M, et al. . Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 2015;527:110–113. DOI: 10.1038/nature15544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang F, Taylor DW, Chen JS, et al. . Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 2016;351:867–871. DOI: 10.1126/science.aad8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palermo G, Chen JS, Ricci CG, et al. . Key role of the REC lobe during CRISPR-Cas9 activation by “sensing,” “regulating,” and “locking” the catalytic HNH domain. Q Rev Biophys 2018;51:e91. DOI: 10.1017/S0033583518000070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jackson RN, van Erp PB, Sternberg SH, et al. . Conformational regulation of CRISPR-associated nucleases. Curr Opin Microbiol 2017;37:110–119. DOI: 10.1016/j.mib.2017.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zuo Z, Liu J. Structure and dynamics of Cas9 HNH domain catalytic state. Sci Rep 2017;7:17271. DOI: 10.1038/s41598-017-17578-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palermo G, Ricci CG, Fernando A, et al. . Protospacer adjacent motif-induced allostery activates CRISPR-Cas9. J Am Chem Soc 2017;139:16028–16031. DOI: 10.1021/jacs.7b05313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zeng Y, Cui Y, Zhang Y, et al. . The initiation, propagation and dynamics of CRISPR-SpyCas9 R-loop complex. Nucleic Acids Res 2018;46:350–361. DOI: 10.1093/nar/gkx1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hand TH, Das A, Li H. Directed evolution studies of a thermophilic Type II-C Cas9. Methods Enzymol 2019;616:265–288. DOI: 10.1016/bs.mie.2018.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hand TH, et al. . Phosphate lock residues of Acidothermus cellulolyticus Cas9 are critical to its substrate specificity. ACS Synth Biol 2018;7:2908–2917. DOI: 10.1021/acssynbio.8b00455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Das A, Hand TH, Smith CL, et al. . The molecular basis for recognition of 5′-NNNCC-3′ PAM and its methylation state by Acidothermus cellulolyticus Cas9. Nat Commun 2020;11:6346. DOI: 10.1038/s41467-020-20204-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsui TKM, Hand TH, Duboy EC, et al. . The impact of DNA topology and guide length on target selection by a cytosine-specific Cas9. ACS Synth Biol 2017;6:1103–1113. DOI: 10.1021/acssynbio.7b00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walker JE, Lanahan AA, Zheng T, et al. . Development of both type I-B and type II CRISPR/Cas genome editing systems in the cellulolytic bacterium Clostridium thermocellum. Metab Eng Commun 2020;10:e00116. DOI: 10.1016/j.mec.2019.e00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roberts B, Haupt A, Tucker A, et al. . Systematic gene tagging using CRISPR/Cas9 in human stem cells to illuminate cell organization. Mol Biol Cell 2017;28:2854–2874. DOI: 10.1091/mbc.E17-03-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alkan F, Wenzel A, Anthon C, et al. . CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol 2018;19:177. DOI: 10.1186/s13059-018-1534-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.