Abstract
Alternating Hemiplegia of Childhood (AHC) is a devastating autosomal dominant disorder caused by ATP1A3 mutations, resulting in severe hemiplegia and dystonia spells, ataxia, debilitating disabilities, and premature death. Here, we determine the effects of delivering an extra copy of the normal gene in a mouse model carrying the most common mutation causing AHC in humans, the D801N mutation. We used an adeno-associated virus serotype 9 (AAV9) vector expressing the human ATP1A3 gene under the control of a human Synapsin promoter. We first demonstrated that intracerebroventricular (ICV) injection of this vector in wild-type mice on postnatal day 10 (P10) results in increases in ouabain-sensitive ATPase activity and in expression of reporter genes in targeted brain regions. We then tested this vector in mutant mice. Simultaneous intracisterna magna and bilateral ICV injections of this vector at P10 resulted, at P40, in reduction of inducible hemiplegia spells, improvement in balance beam test performance, and prolonged survival of treated mutant mice up to P70. Our study demonstrates, as a proof of concept, that gene therapy can induce favorable effects in a disease caused by a mutation of the gene of a protein that is, at the same time, an ATPase enzyme, a pump, and a signal transduction factor.
Keywords: AAV9, Na+/K+-ATPase, Mashl+/− mice, alternating hemiplegia of childhood
Introduction
ATP1A3-related disorders resulting from mutations in the ATP1A3 gene represent a new group of disorders, the most common of which is Alternating Hemiplegia of Childhood (AHC).1,2 AHC is a devastating neurological disorder that manifests disabling episodes of hemiplegia and dystonia, ataxia, severe developmental impairment, and premature death.3–8 Approximately 80% of all AHC cases are caused by heterozygous ATP1A3 mutations, with the D801N mutation accounting for 40% of AHC patients.9–11 Currently, the only available therapy is the calcium channel blocker flunarizine, which causes partial reduction in hemiplegia spells, but has no effect on the other usually very severe manifestations of AHC.12–14 Thus, there is need for the development of therapies for this disease.
Our group has developed two knock-in mouse models of AHC: (1) The first carries the D801N mutation (Mashlool, Atp1a3Mashl/+, Mashl+/−), which faithfully reproduces the human condition and (2) the other carries the E815K mutation, the second most common mutation in humans, which is present in 25% of AHC patients and which results in the most severe form of the disease.15–17 The prohibitively high mortality exhibited by the E815K mouse model, as compared with the lower mortality of the D801N model, precluded it from this study. Advances made in reversing the phenotype in the D801N mutant mouse model would still be applicable to the more severe E815K mutation.15–17
The gene therapy approach to treat AHC and other genetic ATP1A3-related diseases is supported by studies demonstrating the rescue of behavioral abnormalities in the mouse model carrying the I810N Atp1a3 mutation by injection of a bacterial artificial chromosome (BAC) containing the wild-type (WT) Atp1a3 gene into pronuclei of fertilized oocytes.18 In this model of Atp1a3 mutation, Clapcote and associates showed that there is a 36–42% reduction of ouabain-sensitive Na+, K+-ATPase activity in whole brain homogenates as compared with WT; similarly, in our D801N Mashlool model, we found a 44% reduction of ouabain-sensitive Na+, K+-ATPase activity in hippocampal samples as compared with WT mice.18,19 These reductions were even more pronounced in cell cultures expressing such mutations.10,18 Due to the limited benefits and feasibility of the BAC-mediated rescue approach to be translated to the human condition, we pursued gene therapy-mediated rescue of the AHC phenotype.18,20
Adeno-associated virus (AAV) mediated gene therapy is a promising mode of therapy for rare and severe neurogenetic disorders, in mouse models and in humans.7,21–24 Gene therapy that will prove to be effective in the D801N model can be used as a therapy for AHC patients with other mutations and for neurological disorders that are secondary to ATP1A3-related ATPase deficiency such as Alzheimer's and Parkinson's diseases as ATP1A3 dysfunction has been implicated to contribute to cell death in these disorders.25–28 In this study, we investigated the effects of AAV-mediated gene therapy in the D801N mutant mouse model. We chose to study the rescue effects of an adeno-associated virus serotype 9 (AAV9) due to its high transduction efficiency into neurons, the site of ATP1A3 gene expression; in addition, AAV9 has a safety profile that enabled its approval as a vector for gene therapy for spinal muscular atrophy.7,29–36
Materials and Methods
Mice
D801N mutant mice, as generated and as previously shown to faithfully reproduce the human condition, were used in this study.16,17 In all experiments, sex-matched littermates were used. The experimenters were blinded to the genotype of mice, and to the groups of animals that received the test and control vectors; all experiments were analyzed in a blinded manner. All procedures were approved by the Duke University Institutional Animal Care and Use Committee.
Test and control vectors
AAV vectors with the ATP1A3 transgene (test vector) and lacking transgene (control) were generated by the Duke University Department of Neurobiology Viral Vector Core facility. The test vector (Fig. 1), AAV9-hSyn-ATP1A3-Myc-FLAG-p2A-mCherry-SV40polyA, contained FLAG-tag at the 3′ terminal of ATP1A3 coding sequence with a P2A cleavage site inserted between mCherry and FLAG-tag DNA coding regions. The control vector consisted of AAV9-hSyn-mCherry.
Figure 1.
AAV9 vector design: schematic diagram of the test vector with transgene (4.891 kb including ITRs). AAV9, adeno-associated virus serotype 9; ITRs, inverted terminal repeats. Color images are available online.
Design and cloning of the ATP1A3 gene into AAV vector backbone: ATP1A3 (NM_152296) Human Tagged ORF Clone obtained from OriGene was amplified by polymerase chain reaction (PCR). The PCR product was digested with BamHI-KpnI restriction enzymes and cloned upstream to mCherry ORF into the pAAV backbone carrying the human synapsin (hSyn) promoter, WPRE, and SV40 poly-A signal. The plasmid (pBK828) sequence was verified by Sanger sequencing. pBK292 expressing mCherry-WPRE-SV40-poly-A was used as control. mCherry was included in the design of the vector for two reasons: (1) It was included in the control vector to demonstrate transduction of that vector. (2) It was included in the test vector so that this would be as close as possible to the control vector. In addition, because of the cleavage site between mCherry and FLAG, mCherry would be cleaved away from FLAG-ATP1A3. This would allow detection of any neurons that may have vector transduction with expression of mCherry but not of the fusion, FLAG-ATP1A3, protein.
To generate AAV vectors, triple-plasmid protocol of production was used as previously described.37 The expression plasmid carrying the hSyn-ATP1A3-mCherry transgene was supplemented with the helper plasmid, pAdDeltaF6 (gift from Dr. Jim Wilson, plasmid No. 112867; Addgene) and with the packaging plasmid expressing rep2; cap9 (pAAV2/9; gift from Dr. Jim Wilson, plasmid No. 112865; Addgene). The plasmids were transfected into 293T cells (ATCC® CRL-3216™), and replicated viral vectors were collected and purified by CsCl gradient. The purified vector was subjected to dialysis followed by assessing physical titer by real-time PCR and functional titer by transduction into SH-SY5Y cells (ATCC CRL-2266™).
Vector injection procedures
Postnatal day 10 (P10) mice were pre-anesthetized in an induction chamber with 3% isoflurane and then transferred to a stereotaxic frame. Isoflurane, 1.5–2%, was supplied through a facemask throughout the duration of the surgery.16,17 A Hamilton syringe filled with test or control vector was inserted through the frontal plate (lateral to the metopic suture and rostral to the coronal suture) into the right or left ventricle of the mouse to deliver the viral vector.38 For intra-cisterna maga (ICM) injections, we first exposed the posterior dura mater. We then similarly injected the vector through the dura mater overlying the cerebellum. Initial pilot experiments to establish the technique consisted of ICM injection of Trypan blue 0.4% to demonstrate staining of the cerebellum by using our technique.
Age of testing
Mice were injected at P10. This corresponds to early infancy in humans, the age at which AHC symptoms typically appear.6,13 They underwent behavioral testing starting at P40, corresponding to pubescence in humans, the age at which all symptoms of AHC have been fully manifested for years.6,13,14 Behavioral testing was again repeated, in adulthood, at P70 and mice were sacrificed for carrying out further western blot analyses.
ATPase enzyme activity assays
Mice were injected with viral vectors at P10 and decapitated at P40 after anesthesia with urethane (1,000–2,000 mg/kg; intraperitoneal [IP] injection). The head was cooled in liquid nitrogen and then the different brain regions were dissected in a cold (4°C) assay buffer. The hippocampi, cortex, basal ganglia/thalamus, brainstem, and cerebellum were dissected, and they were stored at −80°C for use in ATPase assays. Tissues were homogenized by using 30 mM Tris-HCl (pH = 7.4).36,39,40
Protein concentration was measured by using the Bradford method (Coomassie Protein Assay Kit; Thermo Fischer, Waltham, MA), and ATPase activity assay using 10 μg protein was performed by following the instructions from the assay kit manufacturer (MAK113; Sigma-Aldrich, St. Louis, MO). The amount of inorganic phosphate (Pi) released from the added ATP was determined colorimetrically and quantitated by using a phosphate standard curve. Ouabain (3 mM) was used to determine ouabain-sensitive ATPase activity from the total ATPase activity. The ATPase assay coefficient of variation was consistently below 5%, usually in the 1–2% range, and correlation coefficients of the standard curves were consistently >0.98.
Immunohistochemistry and reporter gene expression
Mice were anesthetized with urethane (1,000–2,000 mg/kg; IP injection) and perfused transcardially with cold phosphate buffered saline (1 × PBS) followed by 4% paraformaldehyde (PFA) solution, as previously described.16,17 The brains were extracted and postfixed in 4% PFA at 4°C overnight. After postfixing, brains were cryoprotected in 30% sucrose solution in PBS at 4°C until the brains sank to the bottom. Frozen cryostat sections (40 μm) from the whole brain were collected in PBS containing 0.01% sodium azide and stored at 4°C until use for immunohistochemistry.
To evaluate reporter gene expression, the selected brain sections were mounted on superfrost+slides and air-dried. Sections were then washed with 1 × PBS three times and incubated in NGS 10%, Triton X-100 0.1%, bovine serum albumin (BSA) 1% for 2 h at room temperature. Slices were washed three times with 1 × PBS and incubated with NGS 5%, Triton X-100 0.1%, BSA 1%, Anti-FLAG-Tag M2 antibody (1:1,000, Cat. No. F3165; Sigma-Aldrich) in 1 × PBS at 4°C on a plate shaker overnight. Sections were washed three times with 1 × PBS after incubation with secondary antibody goat-anti mouse Alexa Fluor 488 conjugated (1:400, Cat. No. 16-308; Sigma-Aldrich) in NGS 5%, Triton X-100 0.1%, BSA 1% in 1 × PBS for 2–3 h at room temperature. After a final wash with 1 × PBS in the dark, sections were incubated with DAPI diluted in PBS (1:4,000) for 20 min and cover-slipped by using Fluromont-G (Electron Microscopy Sciences).
Images were acquired by using a Leica SP5 inverted and/or Leica SP8 upright confocal microscope. The ImageJ software (NIH, Bethesda, MD) was used to count stained cells. The expression of the test and control vectors was assessed with red and green fluorescence for mCherry and FLAG-tag, respectively. DAPI was used to stain nuclei, allowing identification of individual cells.41 For ATP1A3 transgene expression, we used FLAG-tag antibody staining as a marker. FLAG-tag, mCherry, and DAPI-positive cells were considered as cells expressing the transgene. The cells positive to DAPI and mCherry were considered as cells expressing reporter gene (lack of a transgene).
We assessed the degree of expression in each of five regions of the brain by using a semi-quantitative scale of 0 (no expression), +1 (1–25% of cells express transgene), +2 (25–50% of cells express the transgene), and +3 (50–100% of cells express the transgene). These regions were hippocampus, cortex, basal ganglia/thalamus, brainstem, and cerebellum. Specific brain regions were identified according to the mouse Allen Brain Atlas.
Behavioral testing
We performed the cold-water immersion test to induce hemiplegia and dystonia and a battery of tests, described next, that we previously demonstrated and that reflects the phenotype of AHC in mutant mice.15–17,42–44 In humans, hemiplegia and dystonia spells are two major causes of morbidity in AHC patients and these spells are often induced by stress, exposure to extreme temperatures, and exposure to cold water.13,14 Similarly, in our Mashl+/− D801N mutant mice, dystonia and hemiplegia spells occur spontaneously and with stressful stimuli.
The cold-water immersion test was described by Isaksen and associates to induce dystonia spells in mice carrying the Atp1a3 D801Y mutation.42 We have found that using a similar procedure in Mashl+/− mice induces spells consisting of a sequence of dystonia followed by hemiplegia in about half or more of our mice. Racine stages IV or V seizures also, often, follow. Thus, we compared the mutant control and mutant treatment groups as to the occurrence (yes/no) of hemiplegia and of dystonia spells and as to the occurrence (yes/no) of stage IV/V seizures induced by the test just cited and as previously performed by us. 17,42 For behavioral assessment, we performed the following tests as previously described by us and by others: balance beam (8 mm diameter round rod), grip strength (forelimb and hindlimb), T-Maze, gait analysis, rotarod, and open field tests.15–17,43,44 Behavioral tests were video-taped, and data were reviewed by a blinded observer.
Western blot
Mice were deeply anesthetized with urethane (1,000–2,000 mg/kg; IP injection) and decapitated. Hippocampus and cerebellum were dissected rapidly and quickly frozen in liquid nitrogen. The samples were homogenized by using a tissue homogenizer (47747-358; VWR) in RIPA buffer (10 μL/mg tissue) 150 mM NaCl, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 50 mM Tris, pH 8.0 containing ultra-tablet protease inhibitor cocktail (5892970001; Sigma-Aldrich). After incubating homogenate on ice for 10 min, the sample was centrifuged at 13,000 g for 15 min. The supernatant was collected and stored at −80°C. Protein concentration was measured by using the Pierce BCA protein assay kit. Ten micrograms (hippocampal) and 20 μg (cerebellar) of extracts were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, and probed with antibodies.
Primary antibodies used in this study include mouse monoclonal anti-FLAG M2 antibody (F3165; Sigma-Aldrich) at 1:400 dilution and rabbit anti-actin antibody (A2066; Sigma-Aldrich) at 1:10,000 dilutions. Secondary antibodies at 1:10,000 dilution used in this study were horseradish peroxidase-conjugated anti-mouse (115-036-003; Jackson ImmunoResearch Laboratories, West Grove, PA, USA) and horseradish peroxidase-conjugated anti-rabbit (Jackson ImmunoResearch Laboratories). For chemiluminencence the membranes were incubated in Amersham ECL™ western blotting detection kit (GERPN2232; Sigma-Aldrich) for ∼2 min in the dark. Amersham imager 600 was used to capture images. Quantification of protein bands was carried out by using ImageJ software version 1.52q (NIH).
Vector genome quantification
Mice were anesthetized deeply with urethane (1,000–2,000 mg/kg) and were transcardially perfused with cold PBS to remove blood. After that, the brain was extracted and selected brain regions were extracted and stored at −80°C. Genomic DNA (gDNA) was extracted by using a DNeasy Blood and Tissue Kit (Cat. No. 69506; QIAGEN, Valencia, CA) from the tissues harvested from AAV-injected animals at the study endpoint. The rAAV genomes were quantified similarly to previously described methods.45 Briefly, quantitative real-time PCR was carried out by using Power SYBR Green Master Mix and custom primers to the SV40 polyadenylation signal that follows the transgene complementary DNA (cDNA) in the rAAV genome (Forward: 5′gcagacatgataagatacattgatgagtt3′, Reverse: 5′agcaatagcatcacaaatttcacaa3′). Samples were run on a QuantStudio5 (Applied Biosystems) in triplicate and compared with a standard curve by using the rAAV genome plasmid. Analysis was completed with coordinating QuantStudio Design and Analysis Software.
Real-time quantitative polymerase chain reaction
To assess transgene expression, we performed real-time quantitative polymerase chain reaction (RT-qPCR) for FLAG. Mice were anesthetized deeply with urethane (1,000–2,000mg/kg) and were transcardially perfused with cold PBS to remove blood. The brain regions of interest were dissected from the perfused brain and stored at −80°C. RNA was extracted according to the manufacturer's directions by using an miRNeasy Kit (Cat. No. 217004; QIAGEN) from the tissues harvested from AAV-injected animals at the study endpoint. Messenger RNA (mRNA) was converted to cDNA according to the manufacturer's directions by using a High-Capacity cDNA Reverse Transcription Kit (Cat. No. 436814; Applied Biosystems). AAV-derived cDNA was quantified (Forward Primer: 5′-GAGGATCTGGCAGCAAATGATATCC-3′; Reverse Primer: 5′-ACGTCTCCTGCTTGCTTTAACAG-3′), and compared with endogenous levels of GAPDH (Cat. No. Mm99999915_g1; Applied Biossystems). Samples were run on a QuantStudio5 (Applied Biosystems). Analysis was completed with coordinating QuantStudio Design and Analysis Software.
Experiment I: effect of test vector on ATPase enzyme activity
In this experiment, we compared two groups of WT mice (n = 3/group). Group I mice were injected with 20 × 1010 vg of active vector (5 μL of 4 × 1013 vg/mL) in the right lateral ventricle at P10 and were sacrificed at P40. Group II mice were similarly injected with control vector at P10 and were sacrificed at P40. We, then, compared the enzyme activity of the ouabain-sensitive fraction of ATPase in the right hippocampus and in other ipsilateral brain regions (cortex, thalamus/basal ganglia, brain stem, and cerebellum) of both groups. We determined, as described earlier, the ouabain-sensitive fraction since ATP1A3-related ATPase activity is ouabain-sensitive.18,36
Experiment II: effects of test vector on transgene expression and dose optimization study
In this experiment, we injected WT P10 mice and tested mice at P40 for mCherry and FLAG-tag expression. We initially used unilateral intracerebroventricular (ICV) injection (four mice, dose 20 × 1010 vg of test vector, 5 μL of 4 × 1013 vg/mL) to assess for hippocampal expression. We then used ICM injection on a separate group of mice (four mutants, dose 15 × 1010 vg, 10 μL of 1.5 × 1013 vg/mL) to assess for cerebellar expression. Based on the observation that robust expression was achieved only in the areas contiguous to the injections sites, we performed the next experiment, using concurrent ICM and bilateral ICV injections in each animal. In this experiment, we studied three groups of WT mice (three mice/group), each receiving a different dose of the test vector. The doses were of 9 × 1010 vg (divided as 3 × 1010 vg in each site) for the first group, then 18 × 1010 vg (divided as 6 × 1010 vg in each site) for the second group, and finally 27 × 1010 vg (divided as 9 × 1010 vg in each site) for the last group (2, 4, and 6 μL per site of 1.5 × 1013 vg/mL, respectively). We then evaluated whether administration of an even higher ICV dose would increase reporter gene expression beyond the immediately contiguous areas. The two doses were: 7.5 × 1010 vg and 20 × 1010 vg (5 and 13.5 μL of 1.5 × 1013 vg/mL) applied ICV unilaterally. We then based the doses in experiment III on the results of this optimization experiment.
Experiment IIIA: effects of optimized test vector on phenotype during pubescence
In the first part, we compared five groups of animals at P40. After optimization of dose and route of injection as described in the earlier experiment II, we conducted this experiment III by using the following combined ICM and bilateral ICV injections dosing regimen (7.5 × 1010 vg per each site, 5 μL of 1.5 × 1013 vg/mL, into the ICM and bilateral ICV sites, 22.5 × 1010 vg/mouse). We injected mice at P10 and studied them as of P40. The five groups were the following, naive WT receiving no injections (n = 10), WT-control group receiving control vector (n = 10), WT-treatment group receiving test vector (n = 4), Het-control group (n = 10), and Het-treatment group (n = 9). We then performed the above battery of behavioral tests that reflect, as reported by us previously, the behavioral impairments in our mouse model corresponding to human AHC symptomatology.16,17 The different groups and respective numbers of animals are included in Table 1.
Table 1.
Summary of groups
| No. of Mice | Type of Injection at P10a | |
|---|---|---|
| Naïve WT | 10 | None |
| WT-control | 10 | Control vector |
| WT-treatment | 4 | Test vector |
| Het-Control | 10 | Control vector |
| Het-Treatment | 9 | Test vector |
For both control and test vectors, the doses injected were 7.5 × 1010 vg per each site, 5 μL of 1.5 × 10 13 vg/mL, into the ICM and bilateral ICV sites, 22.5 × 1010 vg/mouse.
ICM, intra-cisterna maga; ICV, intracerebroventricular; P10, postnatal day 10; WT, wild-type.
Experiment IIIB: effects of optimized test vector on phenotype during adulthood
After completing the behavioral studies, mice were allowed to mature into full adulthood and the behavioral tests were repeated on surviving mice as of P70 on the same groups. Survival among the groups was compared by using the Kaplan-Meier survival curves. Animals from each group were sacrificed after completion of the behavioral tests, and western blot experiments were performed by using FLAG-tag antibodies as a marker for ATP1A3 transgene expression.
Experiment IV
To perform vector genome quantification, we injected a parallel group of five mutant mice ICM and bilateral ICV with the same dosing regimen as was used in experiment III and sacrificed them at age P40. Brain regions were dissected as described earlier and internal organs, including heart, liver, lung, and diaphragm, were harvested for vector genome quantification by using the methodology described earlier.
Statistics
We performed the Student's t-test and analysis of variance (ANOVA) for normally distributed data or the Mann-Whitney and Kruskal-Wallis tests for data that failed the normality test. After completing ANOVA for multiple groups, we performed Student-Newman-Keuls test for post hoc pairwise comparisons. Kolmogorov-Smirnov test was used to determine normality. After Kruskal-Wallis test, post hoc pairwise comparisons were performed by using Dunn's method. For categorical variables, we performed the Fisher Exact test. For comparison of mortality data, we used the Kaplan-Meier survival analysis with the log-rank test. All statistics were performed by using the SigmaPlot 14.0 program (SYSTAT, Inc., Chicago, IL). Data are presented as mean ± standard error of the mean. Statistical significance was considered when p was <0.05.
Results
Experiment I: test vector increased ATPase activity
When we performed ATPase assays on P40 WT mice injected at P10 with 20 × 1010 vg of test vector (AAV9-hSyn-ATP1A3-Myc-FLAG-p2A-mCherry-SV40polyA), ICV unilaterally in the right ventricle, we found the test vector group's ouabain-sensitive ATPase activity to be 102% higher in the ipsilateral hippocampus than in hippocampus of the control vector group. Control vector: 57.24 ± 17 nmol Pi/mg/min, test vector: 115.9 ± 10 nmol Pi/mg/min (p = 0.046, n = 3/group). In other ipsilateral areas, the changes did not achieve statistical significance but, generally, the areas closer to the lateral ventricle showed higher activity; 37% higher in the cerebral cortex, 26% higher in the basal ganglia/thalamus, 16% higher in the brain stem, and none in the cerebellum (all comparisons p > 0.05).
Experiment II: optimized test vector dosing resulted in robust transgene expression
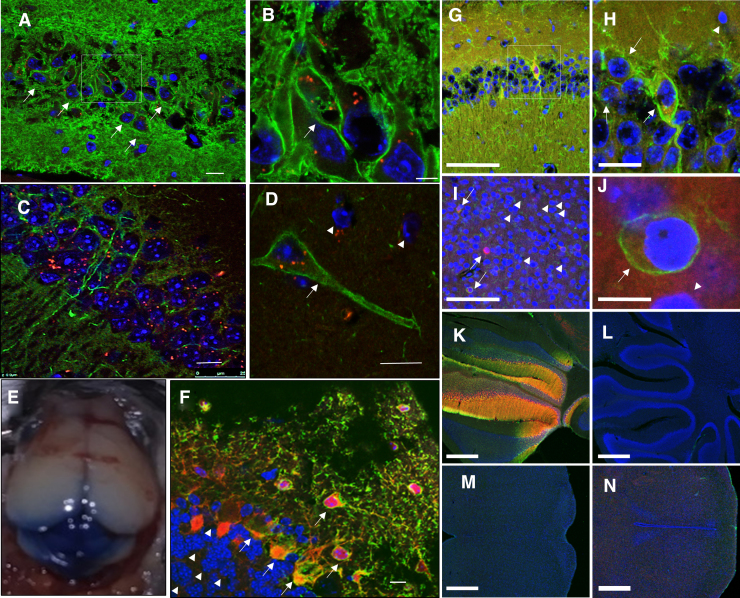
After unilateral ICV injection of 20 × 1010 vg of test vector, we found robust expression in the ipsi lateral hippocampus, +3 score for both mCherry and FLAG-Tag, but not in other brain areas (predominantly 0 scores) (Fig. 2A–D; n = 4). To establish ICM injection technique, trypan blue (0.4%) was injected in WT mice (n = 3). Two hours post-injection mice were sacrificed, and robust trypan blue staining was found in the cerebellum (Fig. 2E). After ICM injection of 15 × 1010 vg of test vector, we found, in all four mice, robust expression in cerebellar cortical areas contiguous to the injections site, +3 score, for both mCherry and FLAG, and only 0 to +1 expression in more distant areas (Fig. 2F). After combined injections (bilateral ICV+ICM), we found robust expression of both markers, particularly of FLAG, with the higher dose of 9 × 1010 vg per site (total 27 × 1010 vg per mouse) but not with the lower doses of 3 or 6 vg × 1010 of test vector per site (total 9 and 18 vg × 1010 of test vector per mouse).
Figure 2.
Transgene expression of test vector. (A–F) Transgene expression after single ICV or ICM injections of test vector. Reporter gene expression as assessed by mCherry and ATP1A3 transgene expression assessed by using anti-FLAG-tag antibody. Injections were performed at P10, and sacrifice was done at P40 [confocal merged images in (A–D, F), FLAG, green; mCherry, red; DAPI, blue, n = 4 mice]. (A) Hippocampal CA3 region expression (unilateral ICV injection of 20 × 1010 vg). (B) Higher magnification image taken from selected region in (A). Note that mCherry is located in the cytoplasm, whereas FLAG-tag is located on the cell membrane. (C) Hippocampal CA1 region expression after injection of the ICV dose just cited. (D) Higher magnification image taken from the CA1 region after injection of the ICV dose just cited. (E) Trypan blue (0.4%) staining of cerebellum 2 h after ICM injection showing robust staining of the cerebellum (n = 3 mice). (F) Cerebellar reporter gene expression after ICM injection (15 × 1010 vg, n = 4 mice). Robust expression is seen in cerebellar Purkinje cells in cerebellar cortex but not in deeper areas. (G–N) Transgene expression after combined ICM and bilateral ICV injections. Reporter gene expression as assessed by FLAG-tag and mCherry after vector injections of test vector of 9 × 1010 in each (total 27 × 1010 vg/mouse). Injections were performed at P10, and sacrifice was at P40 (confocal merged images in all, FLAG, green; mCherry, red; DAPI, blue, n = 3 mice): FLAG staining is high in hippocampus (G, H) and in cerebellum close to ICM injection site (K). It is lower in cortex (I, J), and hardly any in cerebellum distant from ICM injection site (L), brainstem (M), and thalamus (N). In all figures, arrows show cells expressing transgene (FLAG-tag) whereas arrowheads show lack of transgene (positive only to reporter gene mCherry). Scale bar in (A, C, J) 10 μm, (B, D, H) 25 μm, (F) 20 μm, (G, I) 100 μm, and (K–N) 500 μm. ICM, intra-cisterna maga; ICV, intracerebroventricular; P10, postnatal day 10. Color images are available online.
Importantly, with the higher dose, the hippocampus showed FLAG expression score of +3 in CA3 and CA1 (Fig. 2G, H). Cortex and cerebellum showed overall expression scores of +1 but higher expression scores of +3 were in areas close to the injection sites in cerebral and cerebellar cortices (Fig. 2I–K) and lower expression scores of 0 to +1 in areas that are more distal (Fig. 2L, N). These low expression areas included basal ganglia/thalamus, and brainstem. In these experiments, we noted that mCherry was located in the cytoplasm since it is cleaved from the ATP1A3-FLAG, whereas ATP1A3-FLAG fusion protein was detected in the cell membrane. This indicated not only that the virally expressed ATP1A3 transgene is expressed but also that the ATP1A3 protein is transported to its functional locale, the cell membrane.
In our vector, since the FLAG coding DNA sequence is at the 3′ end of the ATP1A3 coding sequence, it is expected to be transcribed only after the ATP1A3 coding sequence is transcribed. Thus, the demonstration of a positive signal for FLAG implies the transcription and expression of exogenous ATP1A3 through the test vector. In addition, data from our western blot analysis, see next, based on the size of the band detected (110 kDa) demonstrated the expression of the exogenous ATP1A3-Flag Tag fusion protein. With the lower doses of 6 × 1010 vg per site and 3 × 1010 vg per site, expression levels were much lower, all scores in the range of 0 to +1 (data not shown), as compared with the expression levels observed and described after the just cited higher dose of 9 × 1010 vg per site. We then studied whether the administration of an even higher ICV dose would increase transgene expression beyond the immediately contiguous areas. We found no further increases in FLAG transgene expression with more than doubling of the just cited high dose of 9 × 1010 vg in the lateral ventricle (Supplementary Data S1).
Experiment IIIA: optimized test vector dosing resulted in reduction of hemiplegia spells and in improvement in balance beam performance at P40
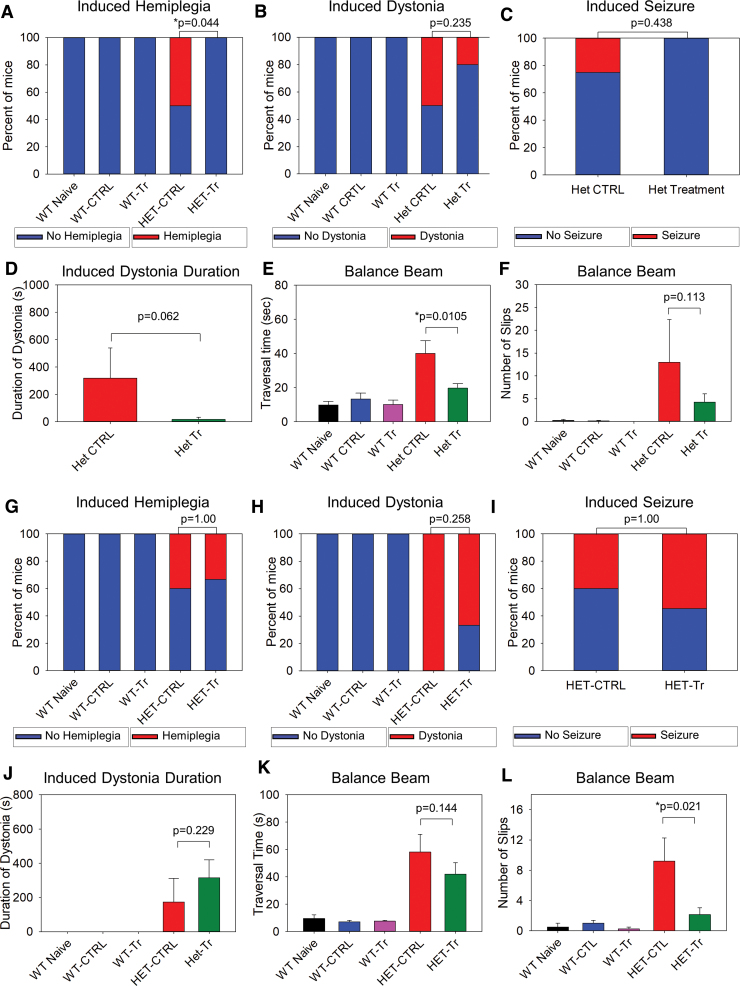
Here, we compared the just described five groups of mice (Table 1): naive WT receiving no injections, WT-control group receiving control vector, WT-treatment group receiving test vector, Het-control group, and Het-treatment group. We found that significantly decreased occurrence of cold water induced hemiplegic spells in the Het-treatment group as compared with the Het-control group (Fig. 3A). This indicated that exogenous delivery of WT ATP1A3 prevented cold-water induced hemiplegia: Hemiplegia occurred in 50% of Het-control animals and in none of the Het-treatment mice (p = 0.044, Fisher exact test). The mean duration of hemiplegia in the Het-control group was 70 ± 35 s.
Figure 3.
Behavioral effects of test vector on AHC phenotype of the same group of animals tested at P40 and again at P70. (A–F) During pubescence at P40 (Het-control vs. Het-treatment). (A) Hemiplegia induced by cold-water exposure (p = 0.044, Fisher exact test). (B) Dystonia induced by cold-water exposure (p = 0.235, Fisher exact test). (C) Stage IV/V seizures induced by cold-water exposure (p = 0.438, Fisher exact test). (D) Duration of cold-water induced dystonia (p = 0.062, Student's t-test). (E) Comparison of the time taken to complete the balance beam test. The Het-control group was different from each of the other groups (p ≤ 0.001 for differences among groups and p ≤ 0.0105 in all post hoc paired comparisons of the Het-control group, one way ANOVA). Importantly, with post hoc analysis, a statistically significant difference was present between the Het-control and Het-treatment groups (p = 0.0105). (F) Comparison of the number of slips during the balance beam test. There was a statistically significant difference among the groups (p < 0.001). With post hoc analysis with Mann-Whitney U test, there was no difference between the Het-control and Het-treatment groups (p = 0.113) and both groups were different from all the WT groups (p < 0.024 in all comparisons). WT naive (n = 6), WT CTRL (n = 10), WT treatment (n = 4), Het CTRL (n = 6), and Het treatment (n = 9) mice. (G–L) During adulthood at P70 (Het-control vs. Het-treatment). Only the number of beam slips showed significance whereas the other tests did not: The number of beam slips at P70 (L) was better in the Het-treatment group as compared with the Het-control group. (G) Hemiplegia induced by cold-water exposure (p = 1.00, Fisher exact test). (H) Dystonia induced by cold-water exposure (p = 0.258, Fisher exact test). (I) Stage IV/V seizures induced by cold-water exposure (p = 1.00, Fisher exact test). (J) Comparison of duration of cold-water induced dystonia spells (p = 0.229, Student's t-test). (K) Comparison of the time taken to complete the balance beam test. There was a difference among the groups (p ≤ 0.001), but the Het-control group was no longer different from the Het-treatment group (p = 0.144, Student's t-test). (L) Comparison in the number of slips during the balance beam test. There was a difference among the groups (p < 0.05). On post hoc analysis, using the Mann-Whitney U test, a statistically significant difference was present between the Het-control and the Het-treatment groups (p = 0.021). Both groups were also different from all three WT groups (p < 0.042 in all comparisons). WT naive (n = 6), WT-CTRL (n = 10), WT treatment (n = 4), Het CTRL (n = 5), and Het treatment (n = 9) mice. AHC, alternating hemiplegia of childhood; ANOVA, analysis of variance; WT, wild-type. Color images are available online.
The hemiplegia episodes were always preceded by dystonia in these Het-control mice. Only one mouse from Het-treatment (which received the transgene) group developed dystonia, but this animal did not develop subsequent hemiplegia. The occurrence of cold-water induced dystonic spells and of stage IV/V seizures was not significantly different between the Het-control and the Het-treatment groups (Fig. 3B, C, p = 0.235 and 0.438, respectively, Fisher exact test). There was a trend for a shorter duration of dystonia in the Het-treatment group than in the Het-control group (Fig. 3D, p = 0.062).
Importantly, we found a significant difference in the balance beam test time to traverse the beam. That time was shorter for the Het-treatment group as compared with the time taken by the Het-control group (Fig. 3E, p = 0.0105). The number of hindlimb slips that were exhibited by the Het-treatment group was lower than that exhibited by the Het-control group, but it did not achieve statistical significance (Fig. 3F, p = 0.113). We also performed additional behavioral tests (Table 2). The T-maze, open field, and grip strength tests showed significant differences between WT and mutants (p < 0.05 in all comparisons) but not between the Het-control and Het-treatment groups or the WT-control and the WT-treatment groups (p > 0.05 in all paired comparisons, one-way ANOVA). The rotarod test did not show any difference among any of the groups.
Table 2.
Results of the five behavioral tests at postnatal day 40 that showed no differences between mutant-control and mutant-treatment groups
| Test | WT Control | WT Treatment | Mutant Control | Mutant Treatment | p-Value, All Groups | p-Value, Mutants vs. WTs | p-Value, Mutant Control vs. Mutant Treatment |
|---|---|---|---|---|---|---|---|
| Rotarod (s) | 96.2 ± 18.5 | 144.8 ± 16.7 | 116 ± 17.3 | 117.1 ± 20.1 | 0.498 | 0.694 | 0.969 |
| Grip strength | |||||||
| Forelimb (N) | 1.1 ± 0.05 | 1.3 ± 0.07 | 0.7 ± 0.05 | 0.8 ± 0.08 | <0.001 | <0.001 | 0.391 |
| Hindlimb (N) | 0.7 ± 0.09 | 0.7 ± 0.1 | 0.5 ± 0.04 | 0.5 ± 0.05 | <0.001 | <0.001 | 0.781 |
| T-maze (%) | 65 ± 5.7 | 55 ± 6.5 | 35 ± 11.9 | 38 ± 8.6 | 0.035 | 0.002 | 0.84 |
| Gait analysis | |||||||
| HL stride (cm) | 6.3 ± 0.2 | 6.9 ± 0.07 | 4.4 ± 0.2 | 4.7 ± 0.2 | <0.001 | <0.001 | 0.375 |
| FL stride (cm) | 6.3 ± 0.2 | 6.8 ± 0.07 | 4.5 ± 0.3 | 4.4 ± 0.2 | <0.001 | <0.001 | 0.788 |
| HL BOS (cm) | 2.1 ± 0.07 | 2.2 ± 0.07 | 2.5 ± 0.2 | 2.6 ± 0.1 | 0.050 | 0.003 | 0.680 |
| FL BOS (cm) | 1.6 ± 0.05 | 1.9 ± 0.06 | 1.7 ± 0.2 | 1.5 ± 0.06 | 0.054 | 0.217 | 0.280 |
| Open field | |||||||
| Total crosses (normalizeda) | 1.3 ± 0.2 | 0.9 ± 0.1 | 1.8 ± 0.5 | 2.5 ± 0.2 | 0.003 | 0.001 | 0.068 |
| Time in center (normalizeda) | 0.6 ± 0.1 | 0.6 ± 0.2 | 1.3 ± 0.5 | 1.0 ± 0.2 | 0.128 | 0.02 | 0.953 |
Number of crosses and time in center was divided by the mean value of the WT naive mice group and then groups were compared to reduce variability due to non-group-related factors.
BOS, base of support; FL, forelimb; HL, hindlimb.
Since our prior studies had shown that there is a sex difference in behavioral testing in only gait analysis,16 we compared all parameters of gait testing, forelimb and hindlimb base of support (BOS), and stride length of males and females of the Het-treatment and Het-control group. We found a trend toward beneficial effect in the males' forelimb BOS (male Het-treatment 1.43 ± 0.088 cm, male Het-controls 1.77 ± 0.088 cm, n = 3 in each group, p = 0.0557, Student's t-test). All other comparisons of the two groups for males and females were not significant (p = 0.380–1.00 in all comparisons).
Experiment IIIB: optimized test vector dosing resulted in improvement in survival and in balance beam performance at P70
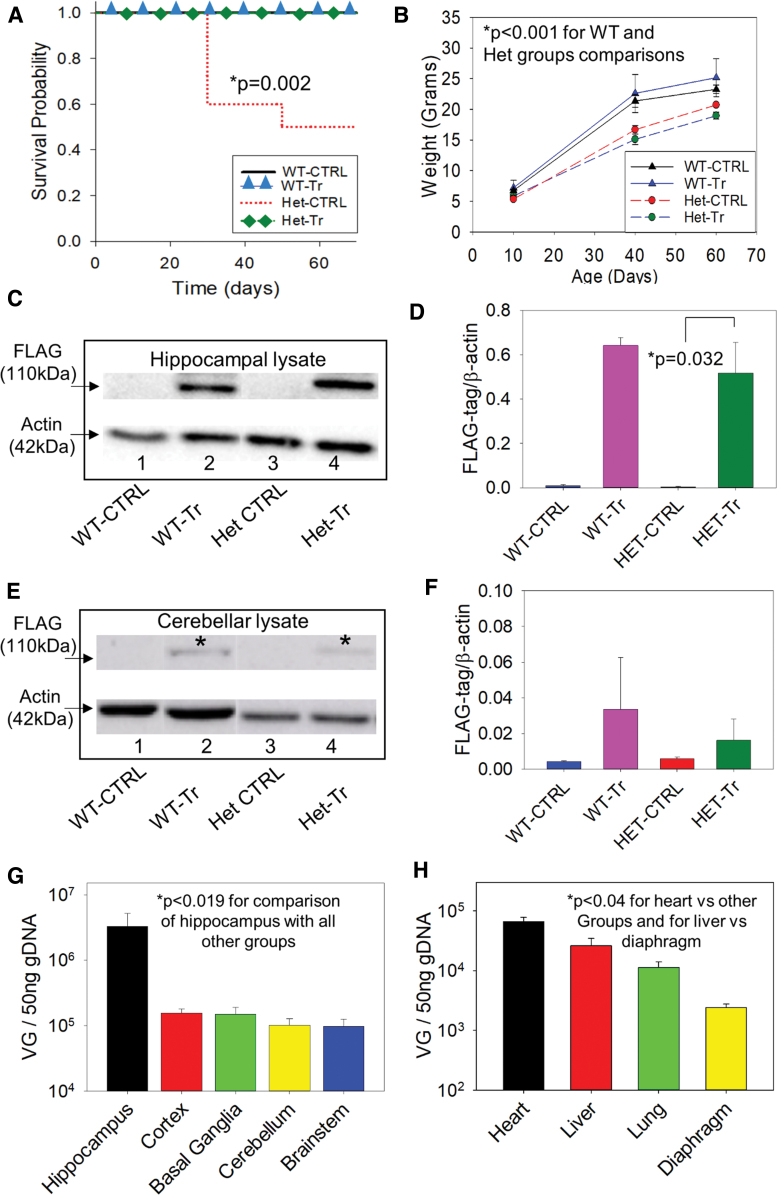
The results of the behavioral tests (Fig. 3G–L and Table 3) showed that the number of hindlimb slips on the balance beam test in the Het-treatment group was significantly lower than in the Het-control group (Fig. 3L, p = 0.021), consistent with a favorable effect of the test vector on the balance beam performance. None of the other behavioral tests, however, showed any difference. Importantly, however, none of these behavioral tests, nor the weight comparisons showed that the mutant-treatment group was worse than the mutant-control group. This argues against a deleterious effect of delivery of an extra copy of the gene. As seen in the Kaplan-Meier survival curve (Fig. 4A), there was a significant reduction in mortality of Hets that had received treatment compared with Het-controls (p = 0.002, the log-rank test).
Table 3.
Results of the five behavioral tests at postnatal day 70 that showed no differences between mutant-control and mutant-treatment groups
| Test | WT Control | WT Treatment | Mutant Control | Mutant Treatment | p-Value, All Groups | p-Value, Mutants vs. WTs | p-Value, Mutant Control vs. Mutant Treatment |
|---|---|---|---|---|---|---|---|
| Rotarod (s) | 107.6 ± 22.5 | 132.2 ± 53.8 | 89.2 ± 11.8 | 105.8 ± 20.8 | 0.504 | 0.291 | 0.500 |
| Grip strength | |||||||
| Forelimb (N) | 1.55 ± 0.03 | 1.55 ± 0.04 | 1.03 ± 0.10 | 0.83 ± 0.06 | <0.001 | <0.001 | 0.306 |
| Hindlimb (N) | 1.04 ± 0.16 | 1.06 ± 0.13 | 0.83 ± 0.07 | 0.67 ± 0.08 | 0.048 | 0.031 | 0.668 |
| T-maze (%) | 62.0 ± 4.9 | 62.5 ± 4.8 | 48.0 ± 8.6 | 30.0 ± 5.3 | 0.002 | <0.001 | 0.056 |
| Gait analysis | |||||||
| HL stride (cm) | 6.55 ± 0.05 | 6.967 ± 0.203 | 4.90 ± 0.339 | 4.886 ± 0.284 | <0.001 | <0.001 | 1.00 |
| FL stride (cm) | 6.55 ± 0.05 | 6.967 ± 0.260 | 4.150 ± 0.446 | 4.50 ± 0.231 | <0.001 | <0.001 | 0.904 |
| HL BOS (cm) | 2.00 ± 0.1 | 2.067 ± 0.208 | 2.825 ± 0.35 | 3.014 ± 0.249 | 0.028 | 0.0001 | 0.980 |
| FL BOS (cm) | 1.600 ± 0.2 | 1.833 ± 0.145 | 1.650 ± 0.132 | 1.614 ± 0.0857 | 0.741 | 0.905 | 0.999 |
| Open field | |||||||
| Total crosses | 62.9 ± 16.4 | 26.0 ± 25.0 | 91.8 ± 24.1 | 110.4 ± 27.7 | 0.283 | 0.052 | 0.960 |
| Time in center | 84.0 ± 20.6 | 147.4 ± 50.5 | 160.5 ± 48.5 | 216.5 ± 60.7 | 0.154 | 0.015 | 1.00 |
Figure 4.
Effect of test vector on survival, weight, vector genome quantification, and long-term transgene expression. (A) Kaplan-Meier Survival analysis revealed marked effect of therapy with worse survival for the Het-control group as compared with the other groups (p = 0.002, log-rank test). (B) Serial weights of surviving animals. There was no difference between the two mutant groups nor was there a difference between the two WT groups (p > 0.05 in both comparisons). Each of the WT groups did differ from each of the mutant groups (p < 0.001 in all comparisons). Note that the curves represent the means of surviving mice and, thus, are likely an over-representation of the weights of the Het-control group since animals that died in that group, and who could not be included in this analysis, had not thrived and very possibly had lower weights than those mutants who survived. (C) Illustration of western blot of hippocampal and cerebellar (E) samples from test and control vector treated mice. Transgene expression was detected by using FLAG-tag antibody. Note that FLAG band intensity was much stronger in hippocampal lysate than in the cerebellar lysates. This difference supports the continued presence of ATP1A3 protein expressed from DNA delivered through the test vector at P10, more so in the hippocampus than in the cerebellum. (D, F) Bar diagrams showing comparison of densitometry results of the treatment and control groups in hippocampal and cerebellar lysates, respectively (n = 3–6 mice/group). (G) Test vector genome quantification in different brain areas of a parallel group (n = 5) mutant mice injected at P10 with the same dosing regimen used for the behavioral experiments IIIA and IIIB and sacrificed at P40. (H) Test vector genome quantification in internal organs of the same parallel group of animals in 4G (n = 5). *Indicates the FLAG immunoreactive bands in cerebellar lysate. Color images are available online.
Analysis of serial weights (Fig. 4B) recorded at P10, P40, and P60 of surviving mice (Fig. 4B) revealed the following. There was an effect of mutation status (p < 0.001) and of age (p < 0.001) but not of treatment (p > 0.05). Specifically, weights at P10 of all groups were not different (p > 0.05 in all comparisons). Weights at P40 were not different between the WT-control and WT-treatment (p > 0.05) nor between the Het-control group and the Het-treatment group (p > 0.05). The WT-control group and the Het-control group were different (p < 0.001) as were the WT-treatment and Het-treatment groups (p < 0.001), indicating neither beneficial nor detrimental effects of the test vector on weight in the surviving animals (the animals that died were not able to be included in this analysis). Weights at P60 showed results similar to those at P40; WT-control and WT-treatment groups did not differ (p > 0.05) nor did the Het-control and the Het-treatment groups (p > 0.05). The WT-control and the Het-control groups were different (p < 0.001) as were the WT-treatment and Het-treatment groups (p < 0.001).
Transgene expression (Fig. 4C–F) was assessed by western blot after completion of the behavioral tests. Western blot of the hippocampus (Fig. 4C, D) and cerebellum (Fig. 4E, F) using FLAG-tag antibody showed that adult mice injected at P10 manifested FLAG-tag positivity, indicating expression and translation of FLAG. In the hippocampus, all six out of six tested Het-treatment mice had a positive FLAG band and all three out three tested WT-treatment mice had such a FLAG band. However, in the cerebellar samples, only one out of six tested Het-treatment mice had a positive FLAG band and only one out of three tested WT-treatment mice had such a FLAG band. As expected, none of the mice that had received control vector showed FLAG bands either in the cerebellar or in the hippocampal lysates (p = 0.032, Fig. 4C–F, 3–6/group). In addition, the intensity of FLAG bands was much stronger in the hippocampal than in the cerebellar lysates. This is consistent with continued expression of the transgene into adulthood, although more so in the hippocampus than in the cerebellum.
Experiment IV: robust test vector viral genome expression
Vector genome quantification in different brain areas (Fig. 4G) showed higher test vector expression in the hippocampus than the cortex (p = 0.0181), basal ganglia (p = 0.0179), cerebellum (p = 0.0164), and brain stem (p = 0.0163). Paired comparisons of all other brain regions were not significant, p > 0.9613 in all pairwise comparisons. Internal organ vector genome quantification (Fig. 4H) showed that heart had higher transduction than liver, p = 0.0013, lung, p < 0.0001, and diaphragm, p < 0.001. In addition, the liver had higher transduction than the diaphragm, p = 0.0329. Other paired comparisons were not significant, p > 0.1631 in both comparisons. These results indicated efficient vector distribution and transduction into brain areas and the expected concurrent, although, lower transduction into studied organs.
Discussion
In this article, we demonstrated, as a proof of concept, that delivering a normal copy of the ATP1A3 gene can ameliorate some of the manifestations of AHC in the mouse model carrying the most common AHC-causing mutation. These favorable effects include improvement in the occurrence of hemiplegia spells at P40, improvement in balance beam performance at P40 and P70, and, most importantly, improved survival through P70. The potential underlying mechanism of improvement in survival and in hemiplegia spells may be related to amelioration in spreading depolarization, which has been linked to premature death and to hemiplegia spells in AHC, as discussed later.15–17 A potential mechanism of improvement of balance is an amelioration of abnormal cerebellar Purkinje cell function through the effects of transgene expression.
It is interesting that some of the beneficial effects noted at P40 were not observed in adulthood. There are several potential explanations. The first is that, by P70, the most severely affected mice in the Het-control group had died, and the less severely affected survivors of that group were the ones compared with the Het-treatment group. The second possible explanation is potential loss of effect of the test vector with age. It has been shown that ATP1A3 dysfunction can result in neuronal injury and death in multiple disease models.25–28,46–48 It is, therefore, conceivable that there is, in AHC, ongoing ATP1A3-related neurodegeneration and that this neurodegeneration failed to be completely reversed by the test vector, resulting in loss of beneficial effects over time. In support of this hypothesis is another study we recently completed (Uchitel et al., submitted). We have shown that Mashl+/− mice older than 3 months perform less well on behavioral tests than P30–P40 mice, indicating worsening function with age. In addition, because there is continued neurogenesis in the cerebellum into adulthood,48,49 newly formed cerebellar neurons would not have the transgene because AAV9 does not result in propagation into dividing cells and this could have contributed to the lack of observable difference in the time to traverse the balance at P70.
To investigate which of the two explanations cited above is more likely, we performed a statistical comparison of the balance beam performance results of the P40 mice studied in our experiment IIIA but with the elimination of any mice that died later after P40. We reasoned that if the first explanation were correct, then performing that analysis would result in loss of the difference, at P40, between the control and test treatment groups. On the other hand, if the difference was still found, with the elimination of any mice that died between P40 and P70, then this would strongly argue against that hypothesis. When we performed that analysis, we found that the difference was still present (mean beam transversal time 42.38 ± 19.7 s and 8.75 ± 2.58 s, p = 0.0084). This strongly supports that there is a decrease of active vector effect over time.
To investigate whether there is decreased transgene expression at the later age of P70, as compared with P40, in the cerebellum that could potentially explain the loss of effect in the balance beam performance over time we conducted the experiment described in Supplementary Data S2. Briefly, RT-qPCR for FLAG was performed on the cerebellar samples of three groups of mice (n = 5–6/group), all injected with the same doses of active vector as in experiment IIIA and sacrificed at P20, 40, and 70, respectively. The results showed an approximately fourfold higher transgene mRNA expression at P40 as compared with P70, suggesting that a reduction in transgene expression may be contributing to the observed loss of effect over time (Supplementary Data S2, Supplementary Figure S1).
The strengths of our study include our choice of AAV9, because it has good central nervous system transduction after both ICV and intravenous injections, improving the translational potential of our studies.29,30,50 We also chose the hSyn promoter, because it is neuron specific, as demonstrated in the brain and spinal cord of rodents and primates,51,52 and because the ATP1A3 gene is expressed in neurons rather than in glia.34,35 Another strength of our study is the use of our AHC mouse model that carries the disease's most common mutation and faithfully reproduces the manifestations of AHC. This approach will likely enhance the chances of success of future translation of AHC gene therapy into human application.
A novel aspect of our article is that prior studies of gene therapy in neurogenetic disorders have used viral vectors to correct abnormalities of proteins that are either one of the following: enzymes: structural, ion channel, or signal transduction proteins.24,53,54 ATP1A3-related Na+, K+-ATPase is at the same time an ATPase enzyme, an important cellular ion pump, and a signal transduction factor.55,56 Thus, our study introduces gene therapy as a method to correct abnormalities resulting from a mutation of an ATPase, which represents a new class of target molecules with multiple combined functions. This is the first study, to our knowledge, addressing a molecule that affects all of the functions cited earlier.
Our study points to future directions for the research of AHC. One of those directions is the investigation of the effect of transgene expression on the physiology of the hippocampus. We have previously shown that the hippocampus in mutant mice is predisposed to spreading depolarization.16 Spreading depolarization has been implicated as an underlying mechanism of hemiplegia and of sudden unexpected death in epilepsy and in AHC.2,16,57 The improvements we observed in decrease in hemiplegia and in improved survival may, thus, be secondary, at least in part, to the amelioration of the predisposition to spreading depolarization. This is a hypothesis that could be the focus of future investigations.
An additional area for future research raised by our findings is investigating the discordance in response between performance related to cerebellar function (balance beam, improvement) and performance related to one aspect of hippocampal function (T-Maze, no improvement). This discordance occurred despite higher transgene expression in the hippocampus than in the cerebellum. One possibility is that this discordance may be related to an underlying corresponding difference in the role of ATP1A3-related ATPase activity in the function of the hippocampus and cerebellum during the pubescent period. In support of this possibility is that in adolescent humans ATP1A3 expression in the cerebellum is high whereas it is moderate to low in the hippocampus.58 Increased expression of ATP1A3 likely implies more important contributions to normal function of the specific region at that stage of development. In addition, lack of detection of an effect on memory deficits on the T-maze test may be related to hippocampal injury, resulting from the effects of mutation that may have occurred before the test vector was injected. This potential explanation is supported by the observation in humans that in infancy at around the age of 4 months there is high expression of ATP1A3.58 Currently, there are no mouse data on the ontogeny of expression of Atp1a3 in hippocampus and cerebellum, similar to what is available on humans, so this should be an area of interest for future investigations.
Finally, another area to investigate is the effect of gene therapy on cellular biology and physiology of the mutant mice, including on misfolding of ATP1A3 protein and on electrophysiological properties of pyramidal cells and interneurons.17,59 Despite the encouraging results, not all impairments, which are manifestations of AHC, improved in our model. This is possibly related to the limited extent of transgene expression in multiple areas of the brain, despite the robust active vector spread demonstrated by our viral genome quantification results. This limited expression may be related to the size of the ATP1A3 gene, which is close to the limit of the capacity of AAV9. Thus, future studies need to concentrate on improving the expression by either modifying the vector or using alternative vectors. Another possibility is that there may be a dominant negative effect since such an effect has been hypothesized to explain in vitro findings in Xenopus oocytes expressing mutated ATP1A3.56 However, such an effect is likely not existent in vivo since mutant mice carrying disease-causing mutation have only a < 50% reduction in oubain-sensitive ATPase activity.
Other or even more long-term effects and potential toxicities of the therapy also need to be determined. For example, if modification of the test vector results in better expression of the transgene in areas distant from the injection sites, this may result in induction of gain-of-function disease. We believe that there is a need to perform further studies aimed at achieving more extensive transgene expression through either the use of multiple injection sites, of repeated doses, or of a different vector.
Importantly, is that in our current study none of the tests we performed demonstrated worsening of the Het-treatment group as compared with the Het-control group. This provides preliminary evidence against marked deleterious effects of delivery of an extra copy of the gene. In the behavioral experiments, we did not use higher doses of the AAV9 vector since results of our experiment showed that increasing the dose further did not increase reporter gene expression. Future studies will need to concentrate on optimizing vector constructs to enhance the translational potential AHC gene therapy, as has been done for other diseases.29–32,60 For example, we have already performed preliminary experiments using AAV8 and are planning to test other vectors also.
Conclusion
In conclusion, our study demonstrates the feasibility to achieve improvements when gene therapy is applied to a disease model resulting from a mutation in an ATP1A3 gene. This has potential implications in the therapy of ATPase-related diseases, not only in the brain but also in ATPase-related diseases of other organs. In addition, our study supports the performance of further studies that would optimize transgene expression in the therapy of the AHC mouse model to, hopefully, lead to translation to human applications. The AHC is a devastating disease in urgent need of an effective therapy.3–8,11 Because it is a severe orphan disease, it will likely qualify for the special facilitated FDA (U.S. Food and Drug Administration) programs for such diseases. In addition, therapies that may be effective in AHC could potentially benefit the growing number of syndromes under the umbrella of ATP1A3-Related Spectrum Disorders.1 ATP1A3-related ATPase dysfunction is a newly recognized mechanism for neuronal injury, not only in disorders manifesting energy crises such as status epilepticus or hypoxia46,47 but also, perhaps more importantly, in neurodegenerative disorders such as Parkinson's Disease and Alzheimer's Disease.25–28,47 Thus, therapies developed for AHC may have implications in other disorders with secondary ATP1A3-related ATPase dysfunction. Conceivably, similar gene therapy approaches can also potentially be used in the therapy of deficiency diseases of other ATPases, such as V-ATPases, which are important in multiple types of human disease.61
Supplementary Material
Acknowledgments
The authors thank all members of Dr. McNamara's laboratory for their technical expertise and support.
Author Disclosure
The authors M.A.M., A.S.H., B.K., D.D.K., A.A., and R.S.P. have a pending patent application for gene therapy of ATPase-related diseases. All other authors have no competing financial interests exist.
Funding Information
This work was supported by grants from Duke Clinical and Translational Science Institute (CTSI) as a part of NIH grant number UL1TR002553 awarded by the National Center for Advancing Translational Sciences, the Duke Institute for Brain Sciences (DIBS), and a donation from the Cure AHC Foundation.
Supplementary Material
References
- 1. Fernandes C, Mikati MA. The expanding spectrum of ATP1A3 related disease. Eur J Paediatr Neurol 2019;23:345–346 [DOI] [PubMed] [Google Scholar]
- 2. Masoud M, Prange L, Wuchich J, et al. Diagnosis and treatment of alternating hemiplegia of childhood. Curr Treat Options Neurol 2017;19:8. [DOI] [PubMed] [Google Scholar]
- 3. Jasien JM, Bonner M, D'alli R, et al. Cognitive, adaptive, and behavioral profiles and management of alternating hemiplegia of childhood. Dev Med Child Neurol 2019;61:547–554 [DOI] [PubMed] [Google Scholar]
- 4. Kansagra S, Ghusayni R, Kherallah B, et al. Polysomnography findings and sleep disorders in children with alternating hemiplegia of childhood. J Clin Sleep Med 2019;15:65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Masoud M, Gordon K, Hall A, et al. Motor function domains in alternating hemiplegia of childhood. Dev Med Child Neurol 2017;59:822–828 [DOI] [PubMed] [Google Scholar]
- 6. Panagiotakaki E, Gobbi G, Neville B, et al. Evidence of a non-progressive course of alternating hemiplegia of childhood: study of a large cohort of children and adults. Brain 2010;133:3598–3610 [DOI] [PubMed] [Google Scholar]
- 7. Uchitel J, Abdelnour E, Boggs A, et al. Social responsiveness in alternating hemiplegia if childhood. Dev Med Child Neurol 2020. 2020;62:820–826 [DOI] [PubMed] [Google Scholar]
- 8. Uchitel J, Helseth A, Prange L, et al. The epileptology of alternating hemiplegia of childhood. Neurology 2019;93:e1248–e1259 [DOI] [PubMed] [Google Scholar]
- 9. Heinzen EL, Arzimanoglou A, Brashear A, et al. ATP1A3 Working Group. Distinct neurological disorders with ATP1A3 mutations. Lancet Neurol 2014;13:503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heinzen EL, Swoboda KJ, Hitomi Y, et al. De novo mutations in ATP1A3 cause alternating hemiplegia of childhood. Nat Genet 2012;44:1030–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Panagiotakaki E, De Grandis E, Stagnaro M, et al. Clinical profile of patients with ATP1A3 mutations in Alternating Hemiplegia of Childhood-a study of 155 patients. Orphanet J Rare Dis 2015;10:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kansagra S, Mikati MA, Vigevano F. Alternating hemiplegia of childhood. Handb Clin Neurol 2013;112:821–826 [DOI] [PubMed] [Google Scholar]
- 13. Mikati MA, Kramer U, Zupanc ML, et al. Alternating hemiplegia of childhood: clinical manifestations and long-term outcome. Pediatr Neurol 2000;23:134–141 [DOI] [PubMed] [Google Scholar]
- 14. Mikati MA, Maguire H, Barlow CF, et al. A syndrome of autosomal dominant alternating hemiplegia: clinical presentation mimicking intractable epilepsy; chromosomal studies; and physiologic investigations. Neurology 1992;42:2251–2257 [DOI] [PubMed] [Google Scholar]
- 15. Helseth AR, Hunanyan AS, Adil S, et al. Novel E815K knock-in mouse model of alternating hemiplegia of childhood. Neurobiol Dis 2018;119:100–112 [DOI] [PubMed] [Google Scholar]
- 16. Hunanyan AS, Fainberg NA, Linabarger M, et al. Knock-in mouse model of alternating hemiplegia of childhood: behavioral and electrophysiologic characterization. Epilepsia 2015;56:82–93 [DOI] [PubMed] [Google Scholar]
- 17. Hunanyan AS, Helseth AR, Abdelnour E, et al. Mechanisms of increased hippocampal excitability in the Mashl+/− mouse model of Na+/K+-ATPase dysfunction. Epilepsia 2018;59:1455–1468 [DOI] [PubMed] [Google Scholar]
- 18. Clapcote SJ, Duffy S, Xie G, et al. Mutation I810N in the alpha3 isoform of Na+,K+-ATPase causes impairments in the sodium pump and hyperexcitability in the CNS. Proc Natl Acad Sci U S A 2009:106:14085–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirshenbaum GS, Clapcote SJ, Duffy S, et al. Mania-like behavior induced by genetic dysfunction of the neuron-specific Na+,K+-ATPase α3 sodium pump. Proc Natl Acad Sci U S A 2011;108:18144–18149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kirshenbaum GS, Dachtler J, Roder JC, et al. Transgenic rescue of phenotypic deficits in a mouse model of alternating hemiplegia of childhood. Neurogenetics 2016;17:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marshall MS, Issa Y, Jakubauskas B, et al. Long-term improvement of neurological signs and metabolic dysfunction in a mouse model of Krabbe's disease after global gene therapy. Mol Ther 2018;26:874–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mendell JR, Sahenk Z, Malik V, et al. A phase ½a follistatin gene therapy trial for Becker muscular dystrophy. Mol Ther 2015;23:192–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura S, Muramatsu SI, Takino N, et al. Gene therapy for Glut1-deficient mouse using an adeno-associated virus vector with the human intrinsic GLUT1 promoter. J Gene Med 2018;20:e3013. [DOI] [PubMed] [Google Scholar]
- 24. Uchitel J, Kantor B, Smith EC, et al. Viral-mediated gene replacement therapy in the developing central nervous system: current status and future directions. Pediatr Neurol. 2020;110:5–19 [DOI] [PubMed] [Google Scholar]
- 25. Komura H, Kakio S, Sasahara T, et al. Alzheimer Aβ assemblies accumulate in excitatory neurons upon proteasome inhibition and kill nearby NAKα3 neurons by secretion. iScience 2019;13:452–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohnishi T, Yanazawa M, Sasahara T, et al. Na, K-ATPase α3 is a death target of Alzheimer patient amyloid-β assembly. Proc Natl Acad Sci U S A 2015;112:E4465–E4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shrivastava AN, Redeker V, Fritz N, et al. α-Synuclein assemblies sequester neuronal α3-Na+/K+-ATPase and impair Na+ gradient. EMBO J 2015;34:2408–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yin, Q, Liu, JJ, Tian MT, et al. Protein fibrillation in neurodegenerative diseases and its chiral interaction with interfaces. Acta Polymer Sin 2019;50:575–587 [Google Scholar]
- 29. Bravo-Hernandez M, Tadokoro T, Navarro MR, et al. Spinal subpial delivery of AAV9 enables widespread gene silencing and blocks motoneuron degeneration in ALS. Nat Med 2020;26:118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hammond SL, Leek AN, Richman EH, et al. Cellular selectivity of AAV serotypes for gene delivery in neurons and astrocytes by neonatal intracerebroventricular injection. PloS One 2017;12:e0188830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang H, Yang B, Mu X, et al. Several rAAV vectors efficiently cross the blood–brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system Mol Ther 2011;19:1440–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zincarelli C, Soltys S, Rengo G, et al. Analysis of AAV serotypes 1–9 medicated gene expression and tropism in mice after systemic injection. Mol Ther 2008;16:1073–1080 [DOI] [PubMed] [Google Scholar]
- 33. Pattali R, Mou Y, Li XJ. AAV9 vector: a novel modality in gene therapy for spinal muscular atrophy. Gene Ther 2019;26:287–295 [DOI] [PubMed] [Google Scholar]
- 34. Bøttger P, Tracz Z, Heuck A, et al. Distribution of Na/K-ATPase alpha 3 isoform, a sodium-potassium P-type pump associated with rapid-onset of dystonia parkinsonism (RDP) in the adult mouse brain. J Comp Neurol 2011;519:376–404 [DOI] [PubMed] [Google Scholar]
- 35. Murata K, Kinoshita T, Ishikawa T, et al. Region- and neuronal-subtype-specific expression of Na,K-ATPase alpha and beta subunit isoforms in the mouse brain. J Comp Neurol 2020;528:2654–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Larsen BR, Stoica A, MacAulay N. Managing brain extracellular K(+) during neuronal activity: the physiological role of the Na(+)/K(+)-ATPase subunit isoforms. Front Physiol 2016;7:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Monahan PE, Lothrop CD, Sun J, et al. Proteasome inhibitors enhance gene delivery by AAV virus vectors expressing large genomes in hemophilia mouse and dog models: a strategy for broad clinical application. Mol Ther 2010;18:1907–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robbins KL, Glascock JJ, Osman EY, et al. Defining the therapeutic window in a severe animal model of spinal muscular atrophy. Hum Mol Genet 2014;23:4559–4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oliveira MS, Furian AF, Rambo LM, et al. Prostaglandin E2 modulates Na,K-ATPase activity in rat hippocampus: implications for neurological diseases. J Neurochem 2009;109:416–426 [DOI] [PubMed] [Google Scholar]
- 40. Zeinieh MP, Talhouk RS, El-Sabban ME, et al. Differential expression of hippocampal connexins after acute hypoxia in the developing brain. Brain Dev 2010;32:810–817 [DOI] [PubMed] [Google Scholar]
- 41. Roosing S, Hofree M, Kim S, et al. Functional genome-wide siRNA screen identifies KIAA0586 as mutated in Joubert syndrome. Elife 2015;4:e06602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Isaksen TJ, Kros L, Vedovato N, et al. Hypothermia-induced dystonia and abnormal cerebellar activityin a mouse model with a single disease-mutation in the sodium-potassium pump. PloS Genet 2017;13:e1006763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mikati MA, Holmes GL, Chronopoulos A, et al. Phenobarbital modifies seizure-related brain injury in the developing brain. Ann Neurol 1994;36:425–433 [DOI] [PubMed] [Google Scholar]
- 44. Pick CG, Yanai J. Long-term reduction in spontaneous alternations after early exposure to phenobarbital. Int J Dev Neurosci 1984;2:223–228 [DOI] [PubMed] [Google Scholar]
- 45. Keeler AM, Zieger M, Todeasa SH, et al. Systemic delivery of AAVB1-GAA clears glycogen and prolongs survival in a mouse model of Pompe disease. Hum Gene Ther 2019;30:57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Funck VR, Ribeiro LR, Pereira LM, et al. Long-term decrease in Na+,K+-ATPase activity after pilocarpine-induced status epilepticus is associated with nitration of its alpha subunit. Epilepsy Res 2014;108:1705–1710 [DOI] [PubMed] [Google Scholar]
- 47. Thornton C, Leaw B, Mallard C, et al. Cell death in the developing brain after hypoxia-ischemia. Front Cell Neurosci 2017;11:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shrivastava AN, Triller A, Melki R. Cell biology and dynamics of Neuronal Na+/K+-ATPase in health and diseases. Neuropharmacology 2020;169:107461. [DOI] [PubMed] [Google Scholar]
- 49. Ahlfeld J, Filser S, Schmidt F, et al. Neurogenesis from Sox2 expressing cells in the adult cerebellar cortex. Sci Rep 2017;7:6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rusanescu G, Mao J. Peripheral nerve injury induces adult brain neurogenesis and remodelling. J Cell Mol Med 2017;21:299–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Colella P, Sellier P, Costa Verdera H, et al. AAV gene transfer with tandem promoter design prevents anti-transgene immunity and provides persistent efficacy in neonate pompe mice. Mol Ther Methods Clin Dev 2018;12:85–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. De Camilli P, Cameron R, Greengard P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. I. Its general distribution in synapses of the central and peripheral nervous system demonstrated by immunofluorescence in frozen and plastic sections. J Cell Biol 1983;96:1337–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kantor B, Tagliafierro L, Gu J, et al. Downregulation of SNCA expression by targeted editing of DNA methylation: a potential strategy for precision therapy in PD. Mol Ther 2018;26:2638–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lykken EA, Shyng C, Edwards RJ, et al. Recent progress and considerations for AAV gene therapies targeting the central nervous system. J Neurodev Disord 2018;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reinhard L, Tidow H, Clausen MJ, et al. Na(+),K (+)-ATPase as a docking station: protein-protein complexes of the Na(+),K (+)-ATPase. Cell Mol Life Sci 2013;70:205–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li M, Jazayeri D, Corry B, et al. A functional correlate of severity in alternating hemiplegia of childhood. Neurobiol Dis 2015;77:88–93 [DOI] [PubMed] [Google Scholar]
- 57. Holt RL, Arehart E, Hunanyan A, et al. Pediatric sudden unexpected death in epilepsy: what have we learned from animal and human studies, and can we prevent it? Semin Pediatr Neurol 2016;23:127–133 [DOI] [PubMed] [Google Scholar]
- 58. BrainSpan Atlas of the Developing Human Brain. Developmental transcriptome, ATP1A3. www.brainspan.org/rnaseq/searches?exact_match=true&search_term=ATP1A3&search_type=gene&page_num=0 (accessed June16, 2020)
- 59. Arystarkhova E, Haq IU, Luebbert T, et al. Factors in the disease severity of ATP1A3 mutations: impairment, misfolding, and allele competition. Neurobiol Dis 2019;132:104577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Passini MA, Bu J, Richards AM, et al. Translational fidelity of intrathecal delivery of self-complementary AAV9-survival motor neuron 1 for spinal muscular atrophy. Hum Gene Ther 2014;25:619–630 [DOI] [PubMed] [Google Scholar]
- 61. Hinton A, Bond S, Forgac M. V-ATPase functions in normal and disease processes. Pflugers Arch 2009;457:589–598 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.