Abstract
Purpose:
Transforming growth factor-βs (TGF-βs) are overexpressed in many advanced cancers and promote cancer progression through mechanisms that include suppression of immunosurveillance. Multiple strategies to antagonize the TGF-β pathway are in early phase oncology trials. However, TGF-βs also have tumor suppressive activities early in tumorigenesis, and the extent to which these might be retained in advanced disease has not been fully explored.
Experimental Design:
A panel of twelve immunocompetent mouse allograft models of metastatic breast cancer was tested for the effect of neutralizing anti-TGF-β antibodies on lung metastatic burden. Extensive correlative biology analyses were performed to assess potential predictive biomarkers and probe underlying mechanisms.
Results:
Heterogeneous responses to anti-TGF-β treatment were observed, with 5/12 models (42%) showing suppression of metastasis, 4/12 (33%) showing no response and 3/12 (25%) showing an undesirable stimulation (up to 9-fold) of metastasis. Inhibition of metastasis was immune-dependent, while stimulation of metastasis was immune-independent and targeted the tumor cell compartment, potentially affecting the cancer stem cell. Thus the integrated outcome of TGF-β antagonism depends on a complex balance between enhancing effective anti-tumor immunity and disrupting persistent tumor suppressive effects of TGF-β on the tumor cell. Applying transcriptomic signatures derived from treatment-naive mouse primary tumors to human breast cancer datasets suggested that breast cancer patients with high-grade, estrogen receptor-negative disease are most likely to benefit from anti-TGF-β therapy.
Conclusions:
Contrary to dogma, tumor suppressive responses to TGF-β are retained in some advanced metastatic tumors. Safe deployment of TGF-β antagonists in the clinic will require good predictive biomarkers.
Introduction
TGF-βs are overexpressed by many advanced human tumors, and high expression frequently correlates with poor prognosis, making the pathway a candidate for therapeutic targeting (1). Multiple biological activities of TGF-βs contribute to driving cancer progression. TGF-βs have been implicated in promoting invasion and migration of tumor cells, driving tumor cell plasticity and the epithelial to mesenchymal transition (EMT), expanding the cancer stem cell (CSC) compartment, and enhancing generation of cancer-associated fibroblasts (1–3). Importantly, TGF-βs have strong immunosuppressive activity, and extensive evidence suggests that elevated TGF-β expression by tumor or stromal cells may compromise anti-tumor immunity and limit the efficacy of immunotherapy (1,4–7). TGF-βs can also mediate resistance to chemotherapy (8) and radiation therapy (9). Compounding the problem, many therapeutic approaches themselves further increase TGF-β production, including radiation (10), chemotherapy (10), and immune checkpoint inhibition (11). Thus there is a compelling rationale to be made for attempting TGF-β pathway blockade. Based on encouraging pre-clinical data showing therapeutic benefit of targeting the TGF-β signaling axis (1,11–18), over 40 early phase clinical oncology trials are now ongoing, using various TGF-β pathway antagonists either as single agents, or in combination with other therapeutics, including immune checkpoint inhibitors (1)(https://clinicaltrials.gov). In general, TGF-β pathway blockade has been well-tolerated in the clinic, with some early signs of efficacy (19–21).
The situation is complicated by the highly pleiotropic nature of the biological processes that are regulated by TGF-β, as TGF-β has context-dependent tumor suppressor activity in addition to its pro-oncogenic properties (1–3). The prevailing dogma is that for most epithelial tumors, TGF-β functions as a tumor suppressor early in the carcinogenic process through homeostatic effects on cell proliferation, survival, genomic integrity and inflammatory cytokine production (1–3). However during tumor progression, genetic and epigenetic changes in the tumor cell, coupled with increased local levels of TGF-β and altered TGF-β responsiveness of the tumor cell, frequently lead to selective loss of these tumor suppressive responses. Pro-progression effects of TGF-β on tumor cells and stroma are progressively unmasked, and come to dominate in later stages of the disease. Given this complex dual role, there was initially reluctance to target the TGF-β pathway in cancer, until preclinical studies in the early 2000s suggested that it might be possible to target the pro-oncogenic TGF-β in advanced disease without disrupting effects on normal homeostasis and tumor suppression (12,13).
Preclinical studies in mouse models play an important role in supporting the clinical drug development process. However, there has been emerging awareness recently of limitations of the preclinical research endeavor in generating information that translates usefully into clinical practice (22,23). One significant issue is that much preclinical work is done using a relatively small number of well-studied models that fail to capture the heterogeneity of the human disease. Other limitations have included a heavy reliance on immunodeficient mice, and the failure to use metastatic burden as the clinically relevant endpoint. To overcome some of these issues, we recently assembled and characterized a panel of twelve immunocompetent allograft models of metastatic breast cancer that capture some of the heterogeneity of the human disease (24). Here we have tested the effect of anti-TGF-β neutralizing antibodies on the metastatic endpoint across all models. Using this expanded panel, we find that 3/12 models respond to anti-TGF-β therapy with an undesirable stimulation of metastasis, suggesting that tumor suppressive responses to TGF-β may be retained in advanced disease in some cases. We exploit the panel to gain insights into potential predictive biomarkers and underlying mechanisms.
Materials and Methods
Mouse cell line models of metastatic breast cancer
The twelve mouse metastatic mammary cell lines, 4T1, EMT6, TSAE1, MET1, R3T, HRM1, 6DT1, D2A1, E0771, F3II, M6 and MVT1 were obtained from the originating investigators as described in Yang et al. (24). Individual characteristics of the cell lines, including culture conditions, are given in Supplementary Table 1, with additional information about genomic characterization in (24).
Animal experiments
All animal experiments were conducted under protocol LC-070 approved by the Animal Care and Use Committee of the National Cancer Institute. For metastasis therapy studies, tumor cells were either orthotopically implanted into the mammary fat pad (4T1, EMT6, R3T, HRM1, 6DT1, D2A1, E0771, M6, MVT1), or delivered by tail-vein injection (TSAE1, F3II, MET1) into strain-matched mice. Unless otherwise indicated, primary tumors were resected when they reached 0.5–0.8 cm diameter. Mice were randomized to treatment groups of 10–15 mice/group, and treated with the neutralizing anti-TGF-β mouse monoclonal antibody 1D11 (Genzyme Corp, or BioXCell) or isotype control (13C4, Genzyme Corp. or MOPC21, BioXCell) at 5mg/Kg intraperitoneally 3x per week unless otherwise indicated. At resection or experimental endpoint, tumors were harvested for histology and molecular analyses. Metastatic burden in the lung was determined by counting histologically evident metastases in lung cross-sections. More details are available in Supplementary Methods.
Statistical analysis
Statistical significance in animal-based studies was determined by Mann-Whitney test, or Kruskal-Wallis test with Dunn’s multiple comparison correction, using Graphpad Prism 7 unless otherwise specified. Statistical significance in human datasets was determined by two-tailed Student t test for independent samples, or one-way ANOVA unless otherwise specified.
Supplementary methods
Additional methodologic details are available in the Supplementary Methods and Supplementary Tables.
Data availability
Gene expression microarray data is available from NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE96006. Copy number variant data and single nucleotide variant data are in GEO under accession # GSE69902.
Results
Heterogeneous responses to TGF-β antagonism in a panel of immunocompetent metastatic breast cancer models
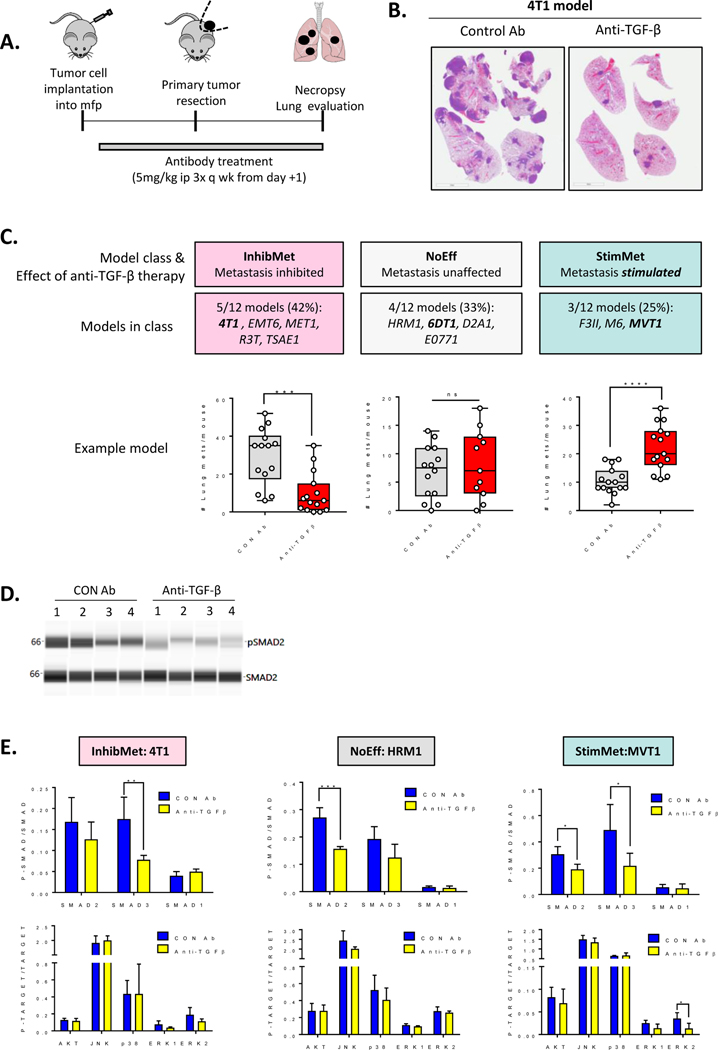
We have previously assembled and characterized a panel of twelve immunocompetent mouse allograft models of metastatic breast cancer (24). Here we assessed the effect of treatment with a pan-TGF-β neutralizing antibody on the metastatic endpoint across the panel. The antibody used was the mouse monoclonal, 1D11, which has similar properties to the fully human anti-TGF-β antibody Fresolimumab (25). Where possible (n=6 models), the models were run in the clinically relevant format of orthotopic tumor implantation with subsequent surgical resection. In some cases, adequate metastatic efficiency was only achieved if primary tumors were left unresected (n=3 models), or if tumor cells were injected via the tailvein (n=3 models). For all models, lung metastatic burden was quantitated in histologic cross-sections (Fig. 1A,B). The results are presented in Table 1, with representative data for each response class in Fig 1C, and full datasets in Suppl Fig S1. The effect of the antibody on primary tumor burden varied between models, but normalization of metastatic burden to primary tumor weight confirmed that effects on metastasis of 4T1, EMT6 and MVT1 models were independent of effects on the primary tumor (Suppl Fig S1). Five models (42%) showed inhibition of metastasis in response to anti-TGF-β antibody treatment (“InhibMet” class). Four models (33%) showed no effect of anti-TGF-β antibodies on metastasis (“NoEff” class), while three (25%) showed an undesirable stimulation of metastasis (“StimMet” class). The metastasis-stimulating effects of anti-TGF-β treatment were confirmed in at least one independent experiment for each StimMet model. Although antibody treatment was generally initiated on day+1 after tumor cell implantation for most experiments, similar results were obtained if antibody treatment was delayed until tumors were well-established (Suppl. Fig S2A–D).
Figure 1. Heterogeneous effects of anti-TGF-β antibody therapy on metastasis in breast cancer models.
A. Schematic for experimental format used for models in (C). B. Representative H&E stained sections of lungs with metastases (4T1 model; median mouse/group). C. Effects of anti-TGF-β therapy on metastatic burden. One representative model is shown for each the three different response categories (InhibMet, 4T1; NoEff, 6DT1; StimMet, MVT1). Median +/− IQ range for n=11–15/group; Mann-Whitney U-test. D. Representative virtual Western blot for quantitative CNIA assay assessing effect of anti-TGF-β antibodies on SMAD2 activation in 4T1 primary tumors for four independent tumors (#1 through #4) from each experimental group. E. Effect of anti-TGF-β antibody treatment on activation state of SMAD signaling pathways (upper panel) or non-SMAD signaling pathways (lower panel) in primary tumors from one representative model for each response class, assessed by quantitative CNIA. Mean +/− SD, n=4 tumors/model/treatment; unpaired t-test for treated vs control comparisons.
Table 1: Effect of anti-TGF-β therapy on lung metastasis burden in metastatic breast cancer model panel.
Models were treated with anti-TGF-β or control antibodies using the indicated metastasis assay format. Marker, mutation data and transcriptomic classification of the models is from Yang et al. (24). Effect size is the fold suppression or promotion of the number of metastases/lung following anti-TGF-β treatment. N=10–15 mice/treatment group/model; Mann-Whitney p-value; ns, not significant; n/a not applicable. The p-value for the metastasis index is the p-value for the difference in metastatic burden following correction for effect of therapy on primary tumor size, where applicable. GEM, genetically engineered mouse; Mut, mutant; ER-pos, estrogen receptor positive; ER-neg, estrogen-receptor negative. (All models were immunohistochemically negative for progesterone receptor and genomically negative for HER2 amplification).
| Cell line | Tumor Origin | Mouse Strain | Metastasis assay format | ER status | Intrinsic transcriptomic subtype | Tp53 status | Ras status | Myc status | PI3K pathway status | Effect of anti-TGF-β on lung metastasis | Metastasis effect size (absolute fold change in metastasis number) | p-value for metastasis number | p-value for metastasis index |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4T1 | Spontaneous | BALB/c | Orthotopic with resection | ER-neg | Luminal A | Nulla | WT | WT | WT | Inhibits | 5.8 | p=0.0003 | p=0.02 |
| EMT6 | Spontaneous | BALB/c | Orthotopic with resection | ER-pos | Luminal A | WT | WT | WT | Pten null | Inhibits | >2 | p=0.0006 | p=0.003 |
| TSAE1 | Spontaneous | BALB/c | Tail vein injection | ER-pos | Luminal A | Mutant | mutKras | Amp | WT | Inhibits | 2.4 | p<0.0001 | n/a |
| MET1 | MMTV-PVT GEM | FVB/N | Tail vein injection | ER-neg | Luminal A | Mutant | WT | WT | WTb | Inhibits | 7.5 | p=0.002 | n/a |
| R3T | DMBA treated OPN−/− GEM | 129S1 | Orthotopic with resection | ER-neg | Luminal A | Mutant | mutKras | WT | WT | Inhibits | 3.8 | p=0.05 | ns |
| HRM1 | PIK3CA-Y1047R GEM | FVB/N | Orthotopic with resection | ER-pos | Luminal A | WT | WT | Amp | mutPik3ca | No Effect | n/a | ns | |
| 6DT1 | MMTV-Myc GEM | FVB/N | Orthotopic, no resection | ER-neg | Luminal A/B | WT | mutKras | Amp† | mutPik3ca | No Effect | n/a | ns | |
| D2A1 | Spontaneous | BALB/c | Orthotopic with resection | ER-neg | Luminal B | WT | WT | Amp | WT | No Effect | n/a | ns | |
| E0771 | Spontaneous | C57Bl/6 | Orthotopic, no resection | ER-neg | Luminal B | Mutant | mutKras | Amp | WT | No Effect | n/a | ns | |
| F311 | Spontaneous | BALB/c | Tail vein injection | ER-pos | Luminal A | Mutant | WT | WT | WT | Stimulates | 4 | p=0.0007 | n/a |
| M6 | C3(1)Tag GEM | FVB/N | Orthotopic with resection | ER-neg | Basal | Nulla | WT | WT | WT | Stimulates | 9 | p=0.01 | ns |
| MVT1 | MMTV-Myc/VEGF GEM | FVB/N | Orthotopic, no resection | ER-pos | Luminal A/B | WT | mutKras | Amp† | mutPik3ca | Stimulates | 2 | p<0.0001 | p<0.0001 |
Genetically wildtype but functionally null
Genetically wildtype but functionally activated
Transgenically amplified.
The canonical TGF-β signaling pathway involves phosphorylation of the signaling transducers, SMAD2 and SMAD3, which associate with SMAD4 and regulate transcription (26). Non-canonical signaling can also occur through other SMADs (SMAD1,5,9), or non-SMAD pathways such as MAPK, JNK, p38 (MAPK14) and AKT (26). To confirm pharmacodynamic activity of the drug, quantitative capillary nano-immunoassays (CNIA; SimpleWestern™) for activation of TGF-β signaling pathways were performed on lysates from primary tumors treated with anti-TGF-β or control antibodies. The results showed that anti-TGF-β treatment could reduce phosphorylation of the canonical TGF-β signaling pathway components, SMAD2 and/or SMAD3, by 30–50% in tumors from all three response classes (Fig. 1D,E, Suppl. Fig S3A,B), indicating that lack of response in the NoEff class was not due to the drug failing to engage the target. In contrast, no effect of treatment was seen on non-canonical signaling through the AKT, JNK, p38 or MAPK/ERK pathways in any response class (Fig 1E). In summary, with this expanded panel of breast cancer models, we find that some models robustly respond to TGF-β antagonism with an undesirable increase in metastatic burden, raising the possibility that this phenomenon might also occur in human breast cancer patients.
No robust correlation between TGF-β expression or pathway activation and response to anti-TGF-β therapy
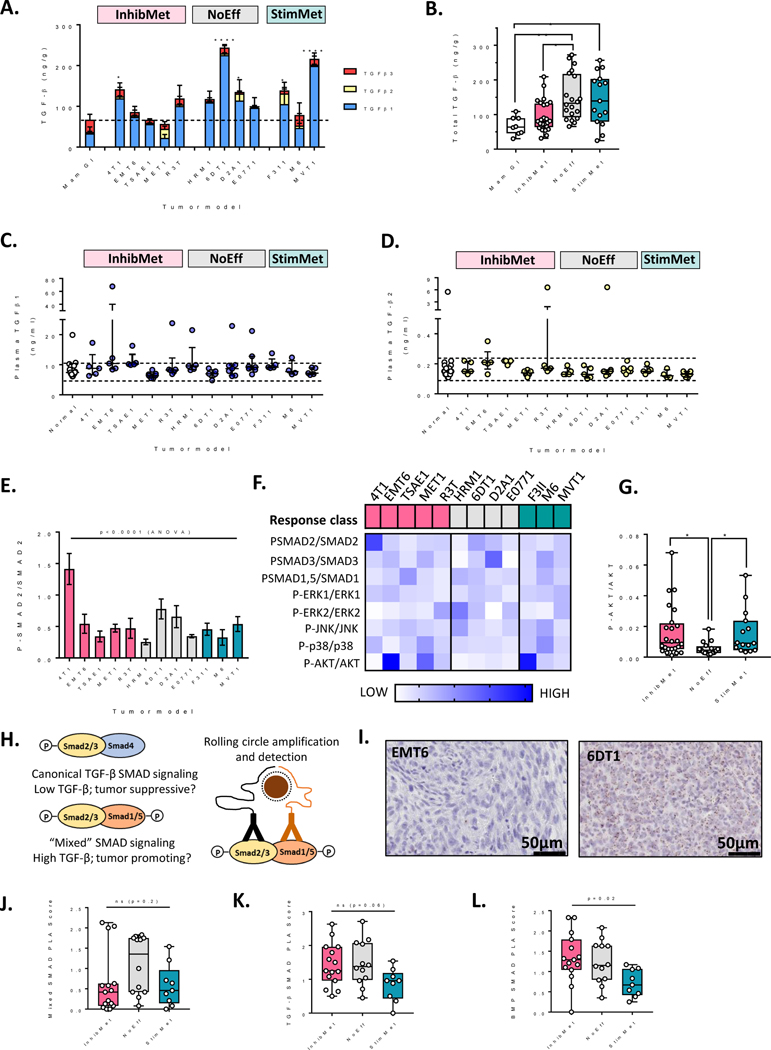
Our observation of undesirable stimulatory responses to TGF-β antagonism in 25% of models tested makes the development of good predictive biomarkers critical. We first assessed whether response-to-therapy correlated with any parameters of TGF-β production or signaling in the models. Protein levels for all three TGF-β isoforms were quantitated in treatment-naïve primary tumors, and in plasma from mice bearing large primary tumors (0.5–1cm diameter). Treatment-naïve primary tumors were used as this material is the most likely to be available in a clinical setting. Total TGF-β in the primary tumors varied over a four-fold range across the models (Fig 2A), and was unexpectedly lowest in the InhibMet class, though the classes were not well separated (Fig 2B). Analyzing by isoform, the NoEff response class tended towards higher TGF-β1 and lower TGF-β3 (Suppl. Fig S4A,B), with a similar, though less pronounced trend at the RNA level (Suppl. Fig S4C,D), but again, differences were not great. With few exceptions, circulating plasma TGF-β1 and TGF-β2 protein levels were in the normal range for all tumor-bearing animals (Fig 2C,D) and TGF-β3 was undetectable (<0.12 ng/ml). Thus there is no strong correlation between circulating or local TGF-β ligand levels in the treatment-naïve tumor models and therapeutic response.
Figure 2. TGF-β expression and signaling in primary tumors from models in the different therapeutic response classes.
A. Expression of TGF-β protein isoforms in acid-ethanol extracts of primary mammary tumors compared with normal mammary gland (Mam Gl). Results are mean +/− SD for n= 5–8 mice/group. ANOVA with Dunnett’s multiple comparisons test for total TGF-β in tumors from individual models vs normal mammary gland. Dotted line shows mean for normal mammary gland. B Total TGF-β (all three isoforms) for 5 tumors/model in each therapeutic response class. Results are median +/− IQ range; Kruskal-Wallis test. C,D. TGF-β1 (C) or TGF-β2 (D) in plasma from normal or tumor-bearing mice. Dotted lines represent +/− 2 SD from mean in normal mice. Data are median +/− IQ range for n=5–7 tumor-bearing mice and 21 normal mice. Response class for the tumor models is indicated. E. PhosphoSMAD2 activation status in untreated primary tumors assessed by quantitative CNIA. Mean +/−SD, n=3–5 tumors/model. F. Heatmap summarizing relative activation state of SMAD signaling and non-canonical TGF-β signaling pathways in tumors from the model panel. Each data point represents the mean phospho-target/target ratio for the model (n=3–5 tumors/model), after median centering of the data across the model panel for each target. G. AKT phosphorylation status in untreated individual tumors in the different response classes. Results are median +/− IQ range; Kruskal-Wallis test. H. Schematic of canonical and mixed SMAD complexes and their detection by brightfield proximity ligation assay (PLA). I. Representative images for proximity ligation assay staining for mixed SMAD complexes in representative primary tumors showing low (EMT6) and high (6DT1) mixed SMAD formation. Brown dots represent complex formation. Scale bar, 50μm. J,K,L. Semi-quantative PLA scores for non-canonical mixed SMAD complexes (J), canonical TGF-β SMAD complexes (K) and canonical BMP SMAD complexes (L) in the three therapeutic response classes. Results are median +/− IQ range within the response class, n=3 tumors/model; Kruskal-Wallis test.
To address whether TGF-β signaling in the tumor cells might differ between the different response classes, we first showed by exome sequencing that the core signaling components Tgfbr1, Tgfbr2, Tgfbr3, Smad2, Smad3 and Smad4 were not mutated or deleted in any of the models (data from (24)). Furthermore, TGF-β could induce phosphorylation of SMAD2 and/or SMAD3 in all the tumor cell lines in vitro (Suppl Fig S5A), so the canonical TGF-β signaling pathway was intact in all 12 tumor models. We then used quantitative SimpleWestern CNIA to assess the basal activation state of TGF-β signaling through SMAD and non-SMAD pathways in untreated primary tumors from all models. The endogenous activation state of the individual pathways in the tumors varied significantly across the model panel (shown for SMAD2 in Fig 2E). However, there was no strong correlation with response-to-therapy (Fig 2F and Suppl. Fig S5B). The NoEff class showed significantly lower AKT activation but the groups were not well-distinguished (Suppl Fig S2B).
Recent work has suggested that certain pro-oncogenic effects seen at high TGF-β levels may be mediated through activation of non-canonical “mixed SMAD” signaling complexes comprising activated TGF-β SMADs (SMAD2 or SMAD3) together with activated BMP SMADs (SMAD1, SMAD5 or SMAD9) (27,28). We hypothesized that the presence of these complexes might indicate that the pro-oncogenic arm of TGF-β signaling was dominant and hence identify the InhibMet class. Using a brightfield proximity ligation assay (PLA) that we developed (29), we detected mixed SMAD, canonical TGF-β SMAD and BMP SMAD complexes semi-quantitatively in treatment-naive tumor samples (Fig 2H,I and Suppl Fig S6A–C). Contrary to our hypothesis, no significant difference was seen for the mixed SMADs (Fig. 2J), while canonical TGF-β SMAD signaling (Fig. 2K) and canonical BMP signaling (Fig. 2L) trended to lower in the StimMet class. Thus overall, we were unable to identify any parameter of TGF-β production or signaling that could robustly distinguish the therapeutic response classes.
Tumor transcriptomics identify gene signatures associated with the different response classes.
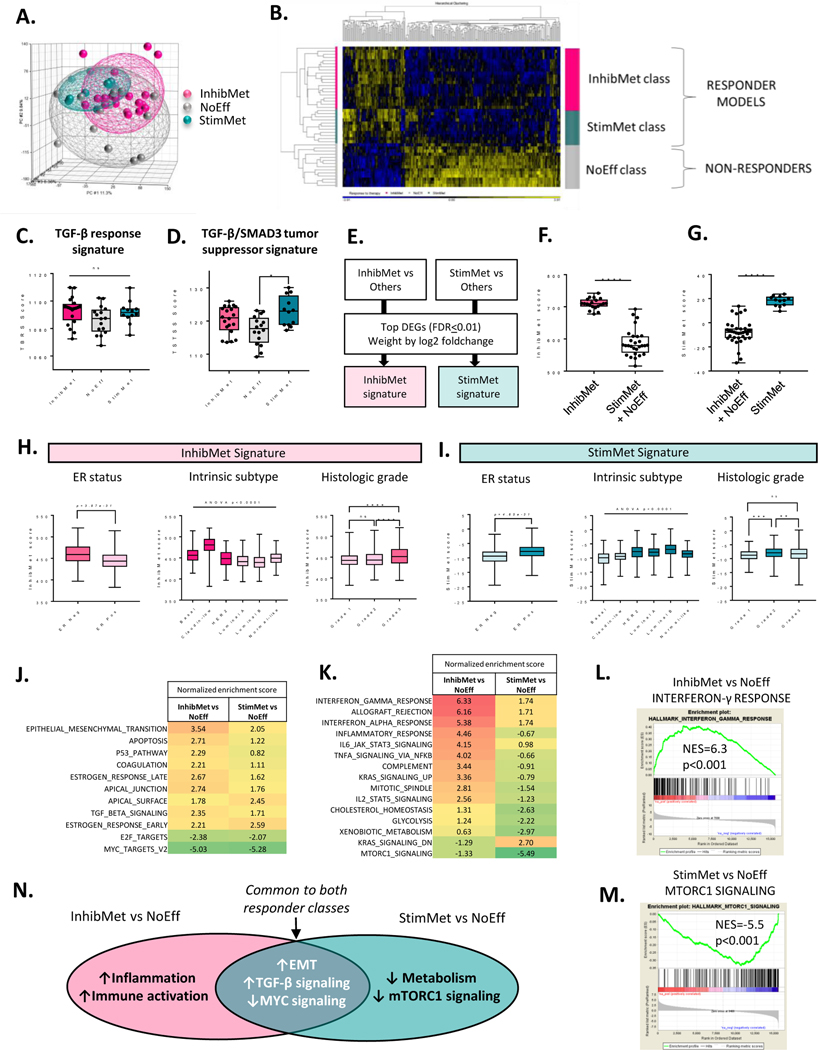
Since the candidate approach was unfruitful, we performed transcriptomic analysis on treatment-naive primary tumors from all models (n=4/model), as a discovery approach to biomarker identification, and to gain mechanistic insights. The tumor transcriptomes did not clearly segregate by therapeutic response class (Fig. 3A). Supervised hierarchical clustering showed that the two responder classes (InhibMet and StimMet) were transcriptomically more similar to each other than to the non-responder class (NoEff) (Fig. 3B). We started by testing whether existing TGF-β-related transcriptomic markers and/or signatures could discriminate the response classes. A number of prognostic TGF-β response signatures have been generated in preclinical model systems, of which the most widely used is the “TBRS” from the Massague laboratory (30). The TBRS which, like most TGF-β response signatures, reflects a mix of tumor promoting and tumor suppressive effects, showed similar scores in all three response classes (Fig. 3C). In contrast, we previously generated a signature (TSTSS) that specifically reflects just the tumor cell-autonomous suppressive effects of TGF-β, and associates with good outcome in human breast cancer datasets (31). Here we found that tumors from the StimMet response class show significantly higher expression of the TSTSS signature (Fig. 3D), suggesting that tumor cells in the StimMet models may retain tumor suppressive responses to TGF-β despite being high grade and aggressively metastatic. However, the TSTSS still only weakly discriminated the StimMet class. We also tested several genes (Pspc1, Klf5, Ywhaz (14–3-3ζ), Six1, Peak1, Rassf1, Dab2) that have been previously proposed to function as molecular “switch” determinants of whether TGF-β acts predominantly as a tumor suppressor or a tumor promoter (32). Of these, only Six1 and Ywhaz showed a significant association with response-to-therapy (Suppl. Fig S7A–C), with the direction of the Ywhaz association being opposite to that predicted from the literature (33). Similarly mutant p53, which has been proposed to cause TGF-β to switch toward oncogenic signaling (34), was not associated with response-to-therapy (Table 1).
Figure 3. Transcriptomes of untreated primary tumors distinguish the three therapeutic response classes.
A. Principal component analysis of primary tumor transcriptomes (n=4/model). B. Heatmap of top 257 most differentially expressed genes between the three response classes (ANOVA p<0.0001) Yellow, upregulated; blue, repressed. C. TGF-β response signature score (TBRS) in each response class (median+/−IQ range, n=4 tumors/model; Kruskal-Wallis test with Dunn’s multiple comparison correction) D. TGF-β/SMAD3 tumor suppressor signature score (TSTSS) in primary tumors. Statistics as for C. E. Schematic for generation of response class-specific gene signatures. F,G. InhibMet (F) or StimMet (G) signature score in the different response classes. Mann-Whitney test. H,I Scores for InhibMet (H) and StimMet (I) signatures in breast cancer patients from the METABRIC dataset (n=2000 patients), stratified by hormone receptor status, intrinsic subtype or histopathologic grade. ER, estrogen receptor. ER status, t test; others, ANOVA with Tukey’s multiple comparison test. ER-neg, n=439; ER-pos, n=1498; Basal, n=209; Claudin-low, n=218, HER2, n-224; Luminal A, n=700; luminal B, n=475; normal-like, n=148; grade1, n=169; grade2, n=771; grade3, n=952 J. Geneset enrichment analysis (GSEA) showing Hallmark genesets that are similarly enriched in both responder groups. NES, normalized enrichment score. Results shown for genesets with absolute NES>2 in at least one group and difference in NES between groups <1.5 K. GSEA for Hallmark genesets that are differentially enriched (difference in NES>=3) in the two responder groups. L,M. Representative enrichment plots for the top most affected Hallmark geneset in each responder group. N. Summary of predicted biological differences between response classes.
In an alternative approach to identify strategies for patient stratification, we generated weighted gene signatures from the most differentially expressed genes (FDR=0.01 cutoff) in the StimMet class (StimMet signature) or InhibMet class (InhibMet signature) when each responder class was individually compared against the other two classes combined (Fig. 3E,F,G and Suppl. Table S2), and we used these signatures to query the METABRIC human breast cancer transcriptomic datasets (35). The InhibMet signature was more highly expressed in estrogen receptor (ER)-negative breast cancer, and in the basal and claudin-low transcriptomic subtypes, with expression increasing with increasing tumor grade (Fig. 3H). Conversely, the StimMet signature was more highly expressed in ER-positive breast cancer, and in luminal A and luminal B tumors, with highest expression in grade 2 tumors (Fig 3I). HER2 tumors showed relatively high expression of both signatures suggesting this class may be heterogeneous. Thus our data suggest that anti-TGF-β therapy in breast cancer might be most safely and effectively applied to patients with high grade, ER-negative disease of the claudin-low and basal subtypes.
Transcriptomic analyses identify biological features associated with the different response classes.
To gain insight into biological properties of tumors in the different response classes, we performed Hallmark geneset enrichment analysis (GSEA). Here we wished to highlight commonalities and differences between the two responder classes, so we compared the StimMet and InhibMet classes individually with the NoEff class. For commonalities, both responder classes exhibited transcriptomic evidence of increased TGF-β signaling, EMT, apoptosis, p53 pathway, and estrogen response, while having decreased MYC and E2F signaling (Fig. 3J). The enrichment for TGF-β signaling in both responder classes is as expected, given that TGF-β antagonism affects both classes, albeit in opposite ways. Transcriptomic approaches are clearly a more sensitive readout of TGF-β signaling than the single target biochemical approaches that we employed earlier. The low relative expression of MYC target genes in both responder classes may reflect the enrichment (p<0.05, chi-square test) for Myc amplification in the NoEff class (Table 1). KRAS signaling was elevated in both responder classes, although the InhibMet class was enriched for genes upregulated by KRAS and the StimMet class for genes downregulated by KRAS. However, these transcriptomic differences did not correlate with the prevalence of mutant Kras, which was similar across all three response classes (Table 1), suggesting that Kras mutation status alone will not be predictive of response.
Despite these commonalities, the two responder classes also showed important differences from each other (Fig 3K,L,M, and summary in Fig 3N). The InhibMet class was strongly enriched for genesets associated with immunity and inflammation, suggesting that tumors in this class are immunologically active prior to treatment. In contrast, the StimMet class was distinguished by a geneset enrichment profile that reflected strongly suppressed mTORC1 signaling and a relatively inactive metabolic state. The predicted biological differences between the two responder groups give insights into possible differences in mechanism of action of the TGF-β antagonists in these groups (see below), which may be further exploitable for future biomarker development.
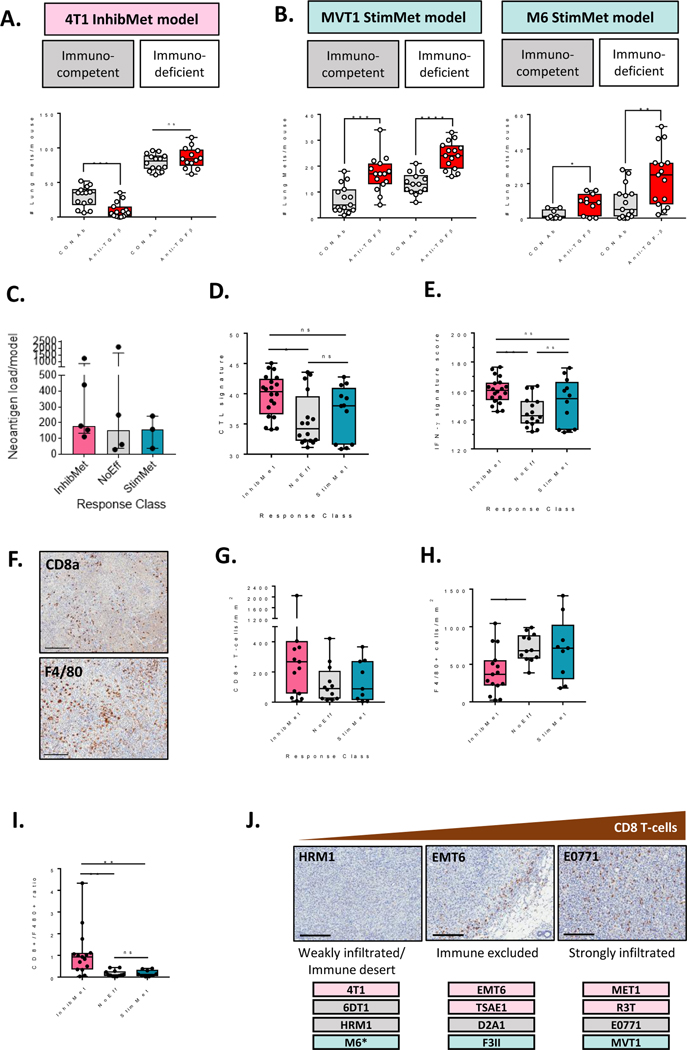
Metastasis-inhibiting effects of anti-TGF-β antibodies are immune-dependent while stimulating effects are immune-independent.
Transcriptomes of primary tumors from InhibMet models were characterized by a high degree of immune activation and inflammation, and we confirmed that the inhibitory effect of the anti-TGF-β antibodies in the 4T1 InhibMet model was lost in fully immunodeficient NSG mice (Fig. 4A), consistent with therapeutic efficacy being dependent on unmasking effective anti-tumor immunity. We and others have previously shown that the efficacy of TGF-β antagonists in responsive breast cancer models is dependent on both T-cells and NK cells (14,36). In contrast, the stimulatory effect of TGF-β antagonism was immune-independent in the two StimMet models tested (Fig. 4B). The therapeutic response was not correlated with tumor cell neoantigen load, which was similar across response classes (Fig. 4C). To assess whether we could find other markers reflecting the different immune dependence of the response classes, we scored the models for a presence of a transcriptomic cytotoxic T-cell signature (37), and a pan-tumor, T-cell inflamed, interferon-γ driven gene expression signature that predicted clinical response to PD1 blockade (38). The InhibMet models showed highest scores for both signatures in their untreated primary tumors (Fig. 4D,E), and immunohistochemical staining also showed a trend towards increased density of CD8+ T-cells in tumors from this response class (Fig. 4F,G). The results are consistent with the enrichment in InhibMet models of a T-cell inflamed microenvironment, similar to that shown to be necessary but not sufficient for clinical response to immune checkpoint blockade (38). The density of other immune cell markers was similar across all model classes (Suppl. Fig S8), with the exception of reduced F4/80+ cells in the InhibMet models (Fig 4H), leading to a significantly higher CD8+/F480+ ratio in InhibMet tumors (Fig. 4I). Recent studies have suggested that anti-TGF-β therapy might be particularly effective in tumors with an “immune-excluded” phenotype, since stromally-derived TGF-β can reduce immune cell infiltration into the tumor (4,5). Based on CD8+ cell distribution in the primary tumors, we classified the models as immune-excluded (EMT6, TSAE1, D2A1, F3II), strongly infiltrated (MET1, R3T, E0771, MVT1) and weakly infiltrated/immune desert (4T1, 6DT1, HRM1, M6). There was a very weak trend towards InhibMet models being more infiltrated with CD8+ T-cells (Fig 4J). In summary, InhibMet models show evidence of pre-existing immune activation, and the therapeutic efficacy of the anti-TGF-β antibodies is dependent on further unmasking of effective anti-tumor immunity. In the StimMet models, while there is transcriptomic evidence of some level of pre-existing immune activation when compared with the NoEff models, this appears not to be limited by TGF-β, and the stimulatory effect of anti-TGF-β therapy in the StimMet models is immune-independent.
Figure 4. Metastasis inhibition by anti-TGF-β antibodies is immune-dependent while stimulation is immune-independent and targets the tumor cell.
A, B. Effects of anti-TGF-β antibodies on metastasis endpoint in the 4T1 InhibMet model (A) or the MVT1 and M6 StimMet models (B) in immunocompetent or immunodeficient mice (NSG mice for 4T1 and M6; NOD/SCID for MVT1). Median +/− IQ range for 9–15 mice/group. Mann-Whitney test. C. Predicted neoantigen burden in each model (median +/− IQ range). D. Cytotoxic T-cell (CTL) signature score (median +/− IQ range, n=4 tumors/model within response class, Dunn’s multiple comparison test). E. Interferon-γ (IFN-γ) signature score (statistics as for D). F. Representative images for CD8a and F4/80 immunostaining in a MET1 primary tumor. Scale bar, 200μm G,H CD8a+ T-cell density (G) or F4/80+ macrophage density (H) in primary tumors determined by immunohistochemistry (median +/− IQ range, n=3 tumors/model, Dunn’s multiple comparison test). I. Ratio of CD8+ to F480+ cells in primary tumors assessed by immunohistochemistry. Stats as for G. J. Spatial distribution of CD8+ T-cells in primary tumors assessed by IHC. A representative tumor for each distribution pattern is shown, with models showing that pattern listed below. Scale bar, 200μm. * M6 was heterogeneous with 2/3 tumors having the immune desert phenotype and 1/3 being strongly infiltrated.
Metastasis-stimulating effects of anti-TGF-β antibodies target the tumor cell
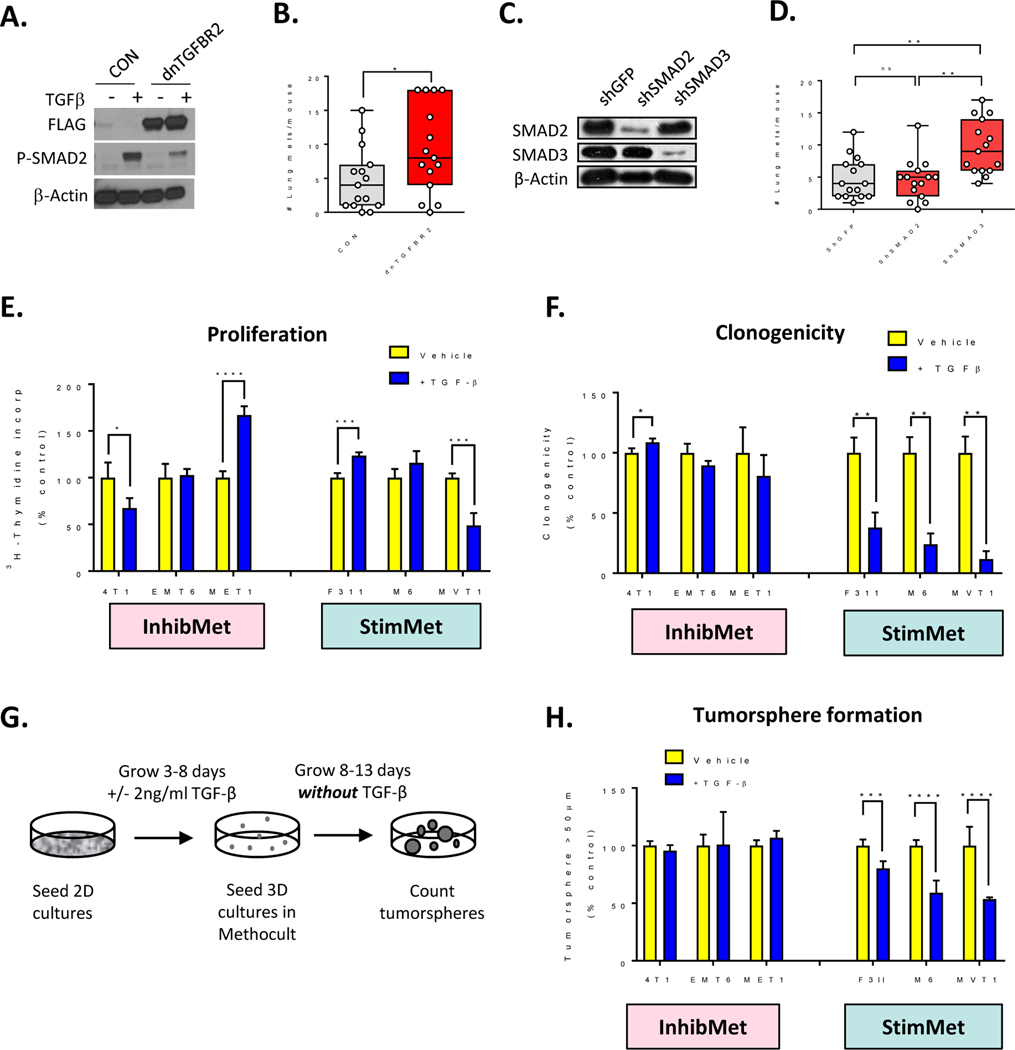
Since the metastasis-stimulating effects of TGF-β antagonism are immune-independent, we sought other cellular targets. TGF-βs are known to have direct tumor suppressive effects on the tumor cell in the early stages of tumorigenesis (1). To address whether the StimMet models retain these tumor cell-targeted tumor suppressive responses despite their advanced stage of progression, we knocked down TGF-β signaling in the MVT1 StimMet model by over-expression of a dominant negative type II TGF-β receptor (dnTGFBR2), or by silencing of SMAD2 or SMAD3 with shRNA. Blockade of the TGF-β receptor or knockdown of SMAD3, but not SMAD2, increased metastasis, suggesting that TGF-β signaling via the TGFBR2/SMAD3 axis has direct anti-metastatic effects on the tumor cell in the MVT1 StimMet model (Fig. 5A–D). Thus in the StimMet models, anti-TGF-β antibodies are likely promoting metastasis by interfering with metastasis-suppressive effects of TGF-β on the tumor parenchyma.
Figure 5. StimMet models retain tumor cell-autonomous tumor suppressive responses to TGF-β.
A. Western blot showing expression of FLAG-tagged dominant negative TGF-β receptor (dnTGFBR2) and suppression of SMAD signaling in MVT1 cells. B. Effect on lung metastasis of TGF-β pathway blockade in MVT1 cells with a dominant negative TGF-β receptor (dnTGFBR2). (median +/− IQ range, n=15 mice/group, Mann-Whitney test. C. Western blot showing shRNA knockdown of SMAD2 and SMAD3 in MVT1 cells. D. Effect on lung metastasis of TGF-β pathway blockade in MVT1 cells with shSMAD2 or shSMAD3. Median +/− IQ range, n=15 mice/group, Dunn’s multiple comparison test. E. Effect of TGF-β on cell proliferation in vitro in 3 representative models in the InhibMet and StimMet classes. Mean +/− SD, n=4, t-test F. Effect of TGF-β on clonogenicity in vitro. Mean +/− SD, n=3, t-test. G. Schematic for tumorsphere formation assay. H. Effect of TGF-β on tumorsphere formation in vitro. Mean +/− SD, n=6.
To address mechanisms underlying the metastasis suppressive effects of TGF-β on the tumor cell, we took three models in each of the InhibMet and StimMet classes and assessed their biological responses to TGF-β in vitro. There was no correlation between the response to anti-TGF-β antibodies in vivo and effects of TGF-β on tumor cells in vitro with respect to growth inhibition (Fig 5E), cell survival, migration or invasion (Suppl Fig S9A–C). In contrast, TGF-β consistently inhibited clonogenicity and tumorsphere formation by the StimMet models in vitro while having little or no effect on the InhibMet models (Fig 5F,G,H). Note that for the tumorsphere assay, tumor cells were pre-treated with TGF-β but tumorsphere formation was assessed in the absence of TGF-β, so as to disambiguate effects of TGF-β specifically on the cancer stem cell from more generalized effects on proliferation of all tumor cells. Since tumorsphere formation is a surrogate assay for cancer stem cell activity, the data suggested that TGF-β may have inhibitory effects on the cancer stem cell (CSC) population in the StimMet models, and that TGF-β antibodies release the brakes on CSC expansion. This lead will be pursued in future studies.
Discussion
TGF-βs have many tumor promoting activities, including immunosuppressive effects on multiple immune cell targets (1). Recent clinical successes with immune checkpoint inhibitors targeting CTLA4, PD1 or PD-L1 (39) have fueled a renewed surge of interest in targeting the TGF-β pathway, as an alternative or complementary approach to reactivating effective anti-tumor immunity. TGF-β pathway antagonists of various types, including neutralizing antibodies (Fresolimumab, Sanofi-Aventis; NIS793, Novartis), TGF-β traps (AVID200, Forbius), small molecule receptor kinase inhibitors (Galunisertib, Eli Lilly) and a bifunctional anti-PDL1/TGF-β trap (M7824, GlaxoSmithKline/Merck KGaA) are in early phase clinical oncology trials. They have been generally well-tolerated, with some early signs of efficacy (19–21). In breast cancer, patients with metastatic disease who received the highest dose of Fresolimumab in combination with focal radiotherapy had a favorable systemic immune response and longer median overall survival compared with the low dose arm (40). Five additional trials in breast cancer with TGF-β antagonists are ongoing (clinicaltrials.gov).
However, despite these encouraging results, the TGF-β pathway remains an unusually complex signaling axis to target clinically. Not only is TGF-β known to have tumor suppressive as well as pro-oncogenic effects, but nearly every cell type in the tumor ecosystem responds to TGF-β in highly context-dependent ways, making the integrated outcome of these effects in vivo is difficult to predict (1). In this study we have used a large panel of mouse models of metastatic breast cancer to explore whether preclinical therapy studies that capture more disease heterogeneity can give new insights to better support and inform clinical trials that target this highly contextual regulatory network. Our test agent was a pan-TGF-β neutralizing antibody, but we anticipate that our results may be more broadly applicable to other pathway antagonists.
Previous preclinical work on a smaller scale has shown anti-tumor efficacy of TGF-β antagonists in multiple tumor types (1,11–14,16,17,41). Consistent with these results, a plurality (5/12 or 44%) of models in our panel responded to TGF-β antagonism with the desired reduction in metastatic burden. Of these models, four had been independently shown to respond to TGF-β antagonism in other labs, either as allografts (4T1, EMT6, R3T (13–15)) or in the original GEM model (MMTV-PyVT for MET1 (13)). Crucially however, use of the expanded panel also revealed adverse metastasis-promoting responses to anti-TGF-β in 25% of the models. Although this effect had not previously been seen with pharmacologic TGF-β pathway inhibition in pre-clinical studies, tumor cell-specific genetic ablation of the TGF-β pathway enhanced metastasis in the MMTV-PyVT transgenic mouse model of metastatic breast cancer (42). Thus the mouse model data suggests that, contrary to dogma, tumor-suppressive responses to TGF-β may be retained and dominant in some instances of advanced metastatic breast cancer. We previously showed that high expression of a gene signature specifically reflecting tumor suppressive effects of TGF-β was associated with improved metastasis-free survival in clinical breast cancer cohorts (31), which supports the possibility that retention of TGF-β tumor suppressive responses may also be a feature of the human disease. In the context of ongoing clinical trials with TGF-β pathway antagonists, our data suggest that good predictive biomarkers will be critical for safe and effective use of these agents, not just to identify patients who will respond therapeutically, but more importantly to eliminate those patients who are at risk for adverse on-target responses.
To address this need, we used the mouse model panel as a platform for identification of potential predictive biomarkers. A summary of our results across multiple approaches is given in Suppl. Fig S10, with potential leads highlighted for further exploration. Broadly we found that at the larger scale of the present study, many individual biomarkers that had looked promising from small studies did not perform well. We were unable to find a robust correlation between response-to-therapy and any individual parameter relating to TGF-β ligand expression or downstream signaling events in the tumor. Similar observations have recently been made in skin cancer models treated with a TGF-β receptor kinase inhibitor (11). Furthermore, published transcriptomic signatures of TGF-β response, and the expression levels or mutation status of genes that were previously proposed to be involved in the switch of TGF-β from tumor suppressor to pro-metastatic factor (eg. Dab2, Klf5, Peak1, Pspc1, Rassf1, Six1, mutant Tp53), did not robustly correlate with response to anti-TGF-β therapy.
Moving beyond the candidate approach, we looked at transcriptomes from treatment-naïve primary tumors for biomarker discovery, reasoning that targeted transcriptomic information might be feasible to acquire in a clinical setting. Interestingly, the tumor transcriptomes of models in the two responder classes (InhibMet and StimMet) were much more similar to each other than to the non-responder (NoEff) class, which poses challenges for discriminating between them. The association of Myc gene amplification and increased MYC pathway activation with lack of therapeutic response in the NoEff class may be worth further assessment. Interestingly, both responder classes were enriched for genesets relating to increased TGF-β signaling when compared to the non-responder class, although our biochemical assays of the signaling pathway had shown no clear differences between them. Thus in general, the transcriptomic approach, which integrates signal over multiple targets, may be more robust to inter-individual heterogeneity than assessment of single targets. Where practical, acquisition of pre-treatment tumor transcriptomics in the early phase clinical trials might provide the biggest return on investment for biomarker identification.
Although treatment-naïve primary tumors from the two responder classes were transcriptomically similar, we were able to generate gene signatures that individually distinguished the InhibMet or StimMet response classes. Applying those signatures to the human breast cancer datasets, we showed that the InhibMet signature is significantly enriched in ER-negative breast cancer, particularly in the claudin-low and basal intrinsic subtypes, and in tumors of higher grade. Conversely, the StimMet signature was significantly enriched in ER-positive tumors, with highest expression in luminal A, luminal B and normal-like tumors. The HER2 intrinsic subtype showed high expression of both signatures, possibly reflecting heterogeneity within this subtype. Thus, based on this analysis, we would predict that breast cancer patients most likely to benefit from anti-TGF-β therapy would be those with high grade, ER-negative tumors, particularly of the claudin-low or basal subtypes. As data come available from ongoing clinical trials with TGF-β antagonists in breast cancer, it will be important to validate these predictions.
A number of limitations of this study need to be acknowledged. The panel has no models of HER2+ disease, and in general the mapping of the mouse models onto human breast cancer subtypes is complex (24). Nevertheless, the mouse model studies allowed us to identify transcriptomic features of the treatment-naïve primary tumors that are associated with stimulatory or inhibitory responses to TGF-β antagonism, and to show that these features are enriched in specific human breast cancer subtypes. The paucity of human ER+ breast cancer cell lines that metastasize efficiently makes confirmation of the StimMet responses using human cell lines challenging currently, but may become possible with the development of metastatic patient-derived xenografts. A second major caveat is that we used anti-TGF-β antibodies as monotherapy, whereas they will likely be used in combination therapies in the clinic. However, we still see the undesirable metastasis-stimulating response to anti-TGF-β treatment in the MVT1 StimMet model when used in combination with otherwise efficacious doses of cyclophosphamide (Suppl Fig S11), so our results are likely also relevant in a combination therapy setting. Finally, in three of the models (TSAE1, MET1 and F3II), metastasis was established by tail vein injection rather than orthotopic implantation of tumor cells. We have previously shown in the 4T1 model that TGF-β antagonism has identical activity in both assay formats (14), suggesting that TGF-β antagonism primarily affects metastatic colonization rather than dissemination from the primary tumor, so this limitation may not be a major concern.
One place where mouse models can make a unique contribution is to understanding therapeutic mechanisms. We confirmed earlier findings by ourselves and others (1,14–16,43), that the therapeutic effect of TGF-β antagonists in the 4T1 InhibMet model is dependent on an intact immune system. The untreated primary tumors from the InhibMet models were highly enriched for genesets and signatures relating to inflammation and immune activation, suggesting that the desired inhibitory effect TGF-β antagonism as monotherapy was seen most strongly in the context of some level of pre-existing immune response. In contrast, StimMet tumors showed a weaker enrichment of genesets for inflammation and immunity, and we showed that the undesirable metastasis-stimulating effect of TGF-β antagonism in StimMet models was fully immune-independent. Notably, the StimMet tumor transcriptomes showed underexpression of pathways relating to metabolism and mTORC1 pathway activation. Since the mTOR pathway is a metabolite and nutrient sensor, and mTORC1 inhibitors such as rapamycin are immunosuppressive, in part by inhibiting expansion of effector T-cells (44), it is conceivable that StimMet tumors create a depleted metabolic environment that represses mTORC1 signaling, and hence prevents T-cell expansion and effective anti-tumor immunity by mechanisms that are independent of TGF-β. Furthermore, direct interactions between TGF-β and mTOR signaling pathways have been observed in other settings and may be worth pursuing (45). In the StimMet models, we found that metastasis-stimulating effects of the anti-TGF-β antibodies were due to interference with tumor suppressive effects of TGF-β on the tumor cell, with our in vitro data suggesting that the cancer stem cell subpopulation may be a key target of these effects. While most studies suggest a stimulatory effect of TGF-β on breast cancer stem cells (reviewed in (46)), we and others have previously shown that TGF-β can inhibit the cancer stem cell population in select breast cancer models (47,48), Further study will be necessary to determine the detailed underlying mechanisms.
In summary, our preclinical study has shown that the assumptions that tumor suppressive effects of TGF-β are either lost in advanced breast cancer, or are not susceptible to TGF-β antagonism, are not valid in a significant minority of cases. This raises the specter that some patients on anti-TGF-β therapy will have their disease course accelerated. While we do not know whether this phenomenon will also be observed for cancers other than breast, or for other classes of TGF-β pathway antagonists, we suspect that it will be the case. A complex balance between tumor suppressive effects of TGF-β on the tumor cell and tumor promoting effects of TGF-β on the immune stroma determines the outcome of anti-TGF-β therapy. Larger scale preclinical studies, such as the one we performed here for breast cancer, may help inform the safe use of TGF-β antagonists in other cancer histologies. While the TGF-β field was alert to the possibility of adverse outcomes with TGF-β antagonism, it is important to note that undesired stimulatory effects are also evident with other targeted therapies. Disease hyperprogression in response to treatment with immune checkpoint inhibitors (ICI) has been seen in several clinical studies, with incidence rates ranging from 9–29% of patients enrolled (49). Interestingly, gene expression profiling of pre-treatment tumors in one clinical study revealed an underexpression of pathways associated with cell metabolism in the hyperprogressors (50). Since we saw underexpression of these same pathways in the StimMet class of tumors, there may be mechanistic commonalities between hyperprogression on ICIs and on anti-TGF-β therapy that could be worth pursuing. Furthermore, given that we saw higher macrophage infiltration in StimMet and NoEff tumors, it is also intriguing that tumor-associated macrophages have been implicated in hyperprogression on, and resistance to, immune checkpoint inhibitors (50). The bottom line is that good predictive biomarkers will be crucial to the safe and effective deployment of TGF-β pathway antagonists, and that preclinical studies designed to capture more of the disease heterogeneity can provide useful information to complement and guide the clinical trials.
Supplementary Material
Translational Relevance.
Most novel cancer therapies are taken into clinical trials based on preclinical data from a limited number of models that capture little of the variability of the human disease. Antagonists of the TGF-β pathway are currently in early phase clinical oncology trials. However, this pathway is a particularly complex therapeutic target, as TGF-βs can have both stimulatory and suppressive effects on multiple cell types in the tumor ecosystem. Here we tested a neutralizing anti-TGF-β antibody in a large panel of twelve immunocompetent mouse models of metastatic breast cancer. While anti-TGF-β therapy suppressed metastasis in many of the models, 25% of models showed an undesirable increase in metastasis following TGF-β blockade. Good predictive biomarkers will be crucial for optimal deployment of TGF-β antagonists in the clinic. Our data suggest that breast cancer patients with high grade, hormone-receptor negative disease may be the most likely to benefit.
Acknowledgements
We acknowledge the expert technical assistance of Anthony Vieira, Elena Kuznetsova, Maria Figueroa, and Geneti Gaga (Laboratory Animal Sciences Program, National Cancer Institute) with the animal experiments, and of Dr. Xiaolin Wu (Laboratory of Molecular Technology, Frederick National Laboratory for Cancer Research) with the microarray analysis. This work was supported by funding from the Intramural Research Program of the National Cancer Institute Center for Cancer Research, NIH project ZIA BC 010881 (to LMW).
Abbreviations:
- BMP
bone morphogenetic protein
- CNIA
capillary nano-immunoassay
- CSC
cancer stem cell
- EMT
epithelial-to-mesenchymal transition
- ER
estrogen receptor
- GSEA
geneset enrichment analysis
- ICI
immune checkpoint inhibitor
- IHC
immunohistochemistry
- NES
normalized enrichment score
- NSG
NOD scid gamma
- PLA
proximity ligation assay
- TBRS
TGF-β response signature
- TGF-β
transforming growth factor-β
- TSTSS
TGF-β/SMAD3 tumor suppressor signature
Footnotes
Conflict of Interest Statement: The authors declare no potential conflicts of interest.
References
- 1.Akhurst RJ, Hata A. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov 2012;11(10):790–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massague J. TGFβ in Cancer. Cell 2008;134(2):215–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seoane J, Gomis RR. TGF-β Family Signaling in Tumor Suppression and Cancer Progression. Cold Spring Harb Perspect Biol 2017;9(12) doi 10.1101/cshperspect.a022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554(7693):544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554(7693):538–43. [DOI] [PubMed] [Google Scholar]
- 6.Batlle E, Massague J. Transforming Growth Factor-β Signaling in Immunity and Cancer. Immunity 2019;50(4):924–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miao Y, Yang H, Levorse J, Yuan S, Polak L, Sribour M, et al. Adaptive Immune Resistance Emerges from Tumor-Initiating Stem Cells. Cell 2019;177(5):1172–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JA, Yonekubo Y, Hanson N, Sastre-Perona A, Basin A, Rytlewski JA, et al. TGF-β-Induced Quiescence Mediates Chemoresistance of Tumor-Propagating Cells in Squamous Cell Carcinoma. Cell Stem Cell 2017;21(5):650–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardee ME, Marciscano AE, Medina-Ramirez CM, Zagzag D, Narayana A, Lonning SM, et al. Resistance of glioblastoma-initiating cells to radiation mediated by the tumor microenvironment can be abolished by inhibiting transforming growth factor-β. Cancer Res 2012;72(16):4119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas S, Guix M, Rinehart C, Dugger TC, Chytil A, Moses HL, et al. Inhibition of TGF-β with neutralizing antibodies prevents radiation-induced acceleration of metastatic cancer progression. J Clin Invest 2007;117(5):1305–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodagatta-Marri E, Meyer DS, Reeves MQ, Paniagua R, To MD, Binnewies M, et al. alpha-PD-1 therapy elevates Treg/Th balance and increases tumor cell pSMAD3 that are both targeted by anti-TGFβ antibody to promote durable rejection and immunity in squamous cell carcinomas. J Immunother Cancer 2019;7(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang YA, Dukhanina O, Tang B, Mamura M, Letterio JJ, MacGregor J, et al. Lifetime exposure to a soluble TGF-β antagonist protects mice against metastasis without adverse side effects. J Clin Invest 2002;109(12):1607–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muraoka RS, Dumont N, Ritter CA, Dugger TC, Brantley DM, Chen J, et al. Blockade of TGF-β inhibits mammary tumor cell viability, migration, and metastases. J Clin Invest 2002;109(12):1551–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam JS, Terabe M, Mamura M, Kang MJ, Chae H, Stuelten C, et al. An anti-transforming growth factor β antibody suppresses metastasis via cooperative effects on multiple cell compartments. Cancer Res 2008;68(10):3835–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge R, Rajeev V, Ray P, Lattime E, Rittling S, Medicherla S, et al. Inhibition of growth and metastasis of mouse mammary carcinoma by selective inhibitor of transforming growth factor-β type I receptor kinase in vivo. Clin Cancer Res 2006;12(14 Pt 1):4315–30. [DOI] [PubMed] [Google Scholar]
- 16.Yingling JM, McMillen WT, Yan L, Huang H, Sawyer JS, Graff J, et al. Preclinical assessment of galunisertib (LY2157299 monohydrate), a first-in-class transforming growth factor-β receptor type I inhibitor. Oncotarget 2018;9(6):6659–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostapoff KT, Cenik BK, Wang M, Ye R, Xu X, Nugent D, et al. Neutralizing murine TGFβR2 promotes a differentiated tumor cell phenotype and inhibits pancreatic cancer metastasis. Cancer Res 2014;74(18):4996–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanpouille-Box C, Diamond JM, Pilones KA, Zavadil J, Babb JS, Formenti SC, et al. TGFβ Is a Master Regulator of Radiation Therapy-Induced Antitumor Immunity. Cancer Res 2015;75(11):2232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strauss J, Heery CR, Schlom J, Madan RA, Cao L, Kang Z, et al. Phase I Trial of M7824 (MSB0011359C), a Bifunctional Fusion Protein Targeting PD-L1 and TGFβ, in Advanced Solid Tumors. Clin Cancer Res 2018;24(6):1287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris JC, Tan AR, Olencki TE, Shapiro GI, Dezube BJ, Reiss M, et al. Phase I study of GC1008 (fresolimumab): a human anti-transforming growth factor-β (TGFβ) monoclonal antibody in patients with advanced malignant melanoma or renal cell carcinoma. PLoS One 2014;9(3):e90353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda M, Takahashi H, Kondo S, Lahn MMF, Ogasawara K, Benhadji KA, et al. Phase 1b study of galunisertib in combination with gemcitabine in Japanese patients with metastatic or locally advanced pancreatic cancer. Cancer Chemother Pharmacol 2017;79(6):1169–77. [DOI] [PubMed] [Google Scholar]
- 22.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature 2012;483(7391):531–3. [DOI] [PubMed] [Google Scholar]
- 23.Ioannidis JPA, Kim BYS, Trounson A. How to design preclinical studies in nanomedicine and cell therapy to maximize the prospects of clinical translation. Nat Biomed Eng 2018;2(11):797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Yang HH, Hu Y, Watson PH, Liu H, Geiger TR, et al. Immunocompetent mouse allograft models for development of therapies to target breast cancer metastasis. Oncotarget 2017;8(19):30621–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grutter C, Wilkinson T, Turner R, Podichetty S, Finch D, McCourt M, et al. A cytokine-neutralizing antibody as a structural mimetic of 2 receptor interactions. Proc Natl Acad Sci U S A 2008;105(51):20251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derynck R, Budi EH. Specificity, versatility, and control of TGF-β family signaling. Sci Signal 2019;12(570) doi 10.1126/scisignal.aav5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gronroos E, Kingston IJ, Ramachandran A, Randall RA, Vizan P, Hill CS. Transforming growth factor β inhibits bone morphogenetic protein-induced transcription through novel phosphorylated SMAD1/5-SMAD3 complexes. Mol Cell Biol 2012;32(14):2904–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daly AC, Randall RA, Hill CS. Transforming growth factor β-induced SMAD1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth. Mol Cell Biol 2008;28(22):6889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flanders KC, Heger CD, Conway C, Tang B, Sato M, Dengler SL, et al. Brightfield proximity ligation assay reveals both canonical and mixed transforming growth factor-β/bone morphogenetic protein SMAD signaling complexes in tissue sections. J Histochem Cytochem 2014;62(12):846–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, et al. TGFβ primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 2008;133(1):66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato M, Kadota M, Tang B, Yang HH, Yang YA, Shan M, et al. An integrated genomic approach identifies persistent tumor suppressive effects of transforming growth factor-β in human breast cancer. Breast Cancer Res 2014;16(3):R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeh HW, Lee SS, Chang CY, Lang YD, Jou YS. A New Switch for TGFβ in Cancer. Cancer Res 2019;79(15):3797–805. [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Acharya S, Sahin O, Zhang Q, Saito Y, Yao J, et al. 14–3-3zeta turns TGF-β’s function from tumor suppressor to metastasis promoter in breast cancer by contextual changes of SMAD partners from p53 to Gli2. Cancer Cell 2015;27(2):177–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/SMAD complex opposes p63 to empower TGFβ-induced metastasis. Cell 2009;137(1):87–98. [DOI] [PubMed] [Google Scholar]
- 35.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486(7403):346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong Z, Carroll KD, Policarpio D, Osborn C, Gregory M, Bassi R, et al. Anti-transforming growth factor β receptor II antibody has therapeutic efficacy against primary tumor growth and metastasis through multieffects on cancer, stroma, and immune cells. Clin Cancer Res 2010;16(4):1191–205. [DOI] [PubMed] [Google Scholar]
- 37.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018;24(10):1550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127(8):2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359(6382):1350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Formenti SC, Lee P, Adams S, Goldberg JD, Li X, Xie MW, et al. Focal Irradiation and Systemic TGFβ Blockade in Metastatic Breast Cancer. Clin Cancer Res 2018;24(11):2493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganapathy V, Ge R, Grazioli A, Xie W, Banach-Petrosky W, Kang Y, et al. Targeting the Transforming Growth Factor-β pathway inhibits human basal-like breast cancer metastasis. Mol Cancer 2010;9:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forrester E, Chytil A, Bierie B, Aakre M, Gorska AE, Sharif-Afshar AR, et al. Effect of conditional knockout of the type II TGF-β receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer Res 2005;65(6):2296–302. [DOI] [PubMed] [Google Scholar]
- 43.Akhurst RJ. Targeting TGF-β Signaling for Therapeutic Gain. Cold Spring Harb Perspect Biol 2017;9(10) doi 10.1101/cshperspect.a022301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev 2012;249(1):43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asrani K, Sood A, Torres A, Georgess D, Phatak P, Kaur H, et al. mTORC1 loss impairs epidermal adhesion via TGF-β/Rho kinase activation. J Clin Invest 2017;127(11):4001–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellomo C, Caja L, Moustakas A. Transforming growth factor β as regulator of cancer stemness and metastasis. Br J Cancer 2016;115(7):761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang B, Yoo N, Vu M, Mamura M, Nam JS, Ooshima A, et al. Transforming growth factor-β can suppress tumorigenesis through effects on the putative cancer stem or early progenitor cell and committed progeny in a breast cancer xenograft model. Cancer Res 2007;67(18):8643–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruna A, Greenwood W, Le QJ, Teschendorff A, Miranda-Saavedra D, Rueda OM, et al. TGFβ induces the formation of tumour-initiating cells in claudin-low breast cancer. Nat Commun 2012;3:1055. [DOI] [PubMed] [Google Scholar]
- 49.Knorr DA, Ravetch JV. Immunotherapy and Hyperprogression: Unwanted Outcomes, Unclear Mechanism. Clin Cancer Res 2019;25(3):904–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo Russo G, Moro M, Sommariva M, Cancila V, Boeri M, Centonze G, et al. Antibody-Fc/FcR Interaction on Macrophages as a Mechanism for Hyperprogressive Disease in Non-small Cell Lung Cancer Subsequent to PD-1/PD-L1 Blockade. Clin Cancer Res 2019;25(3):989–99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression microarray data is available from NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE96006. Copy number variant data and single nucleotide variant data are in GEO under accession # GSE69902.