Abstract
Background
This study sought to investigate the association between the ERCC1/2 single-nucleotide polymorphisms (SNPs) and the efficacy of radiotherapy and prognosis in patients with non-small cell lung cancer (NSCLC).
Methods
We examined 6 SNPs in the ERCC1 and ERCC2 genes in 87 consecutive patients with NSCLC who were treated with definitive radiotherapy. The objective remission rates (ORR), overall survival (OS), and progressive-free survival (PFS) were assessed. A Cox regression analysis was conducted to analyze the independent factors related to death and recurrence.
Result
Patients with the G allele had better OS than patients with the A allele, and there was a statistical difference between the two groups (30.9 vs. 16.2 months; P=0.003). Patients with the AA genotype had significantly worse OS than patients with the AG or GG genotypes (6.8 vs. 19.8 vs. 30.9 months, respectively; P=0.000). The median PFS of the G allele was 18.9 months, which was significantly better than that of the A allele (P=0.040). The median PFS of patients with the GG genotype, the AG genotype, and the AA genotype was 18.9, 11.3, and 5.1 months, respectively; the difference among the three groups was statistically significant (P=0.019). Patients with the G allele also had better PFS than those with the A allele (18.9 vs. 11.3 months, P=0.040). The multivariate cox proportional hazard analysis showed that the ERCC1 gene rs11615 was an independent survival indicator [HR: 1.623, 95% confidence interval (CI): 1.018–2.591, P=0.042] but not an independent recurrence indicator (HR: 1.497, 95% CI: 0.932–2.404, P=0.095).
Conclusions
The ERCC1 rs11615 SNP may be a potential biomarker for predicting survival prognosis in Chinese NSCLC patients who have undergone definitive radiotherapy. Patients with the G allele had better OS than those with the A allele.
Keywords: Non-small cell lung cancer (NSCLC), single-nucleotide polymorphisms (SNPs), radiotherapy
Introduction
Ionizing radiation (IR) acts directly or indirectly to cause various forms of deoxyribonucleic acid (DNA) damage in cells. DNA damage repair is initiated after the cell is genetically damaged (1). Nucleotide-excision repair (NER) is one of the ways of DNA repair. ERCC1-xeroderma pigmentosum group F (XPF) is the complex expression of the ERCC1 and ERCC4 genes. It is a structure-specific endonuclease that cut DNA at single-stranded/double-stranded junctions with a specific polarity. The ERCC2 gene was found to complement the human cell xeroderma pigmentosum group D (XPD), which was identified as an adenosine triphosphate (ATP)-dependent helicase (5'-3' duplex DNA) during NER reaction (2) Single-nucleotide polymorphisms (SNPs) interrupt promoters, codons, or reading frames by altering single nucleotides, which affects the expression of protein amino acid sequences and thus reflects in the phenotype.
Research on the ERCC1/2 gene polymorphisms in non-small cell lung cancer (NSCLC) has mainly focused on disease susceptibility and the efficacy and prognosis of platinum-based chemotherapy (3-7). Numerous studies have shown that the low expression of ERCC1 protein in tumor tissue can improve the sensitivity and survival of platinum-based chemotherapy patients. The GG genotype of the ERCC1 gene rs11615 is the protective factor of NSCLC. The polymorphisms of the ERCC2 gene rs13181 A>C in exon 23 leads to the mutation of the codon 751 translation product, which increases the DNA binding force and decreases the DNA repair ability of cells. This genetic polymorphism is associated with the risk of radiation-induced esophagitis (8). To date, very few studies have investigated the association between the genetic polymorphisms of ERCC1/2 and the efficacy and prognosis of radiotherapy among Chinese NSCLC patients. This study sought to explore the correlation between ERCC1/2 SNPs and the radiotherapy efficacy and prognosis and the risk of radiation-induced lung injury (RILI) in NSCLC patients.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jtd-21-755).
Methods
Patient population
It’s a retrospective study. A total of 87 NSCLC patients who received definitive radiotherapy at the Cancer Hospital of University of Chinese Academy of Sciences between August 2014 and December 2017 were evaluable in this study. The eligibility criteria were: (I) have histologically or cytologically confirmed NSCLC; (II) have a lesion that could be measured by an imaging examination; (III) have no secondary primary malignant tumor; (IV) not have undergone any previous anti-cancer treatment; and (V) have a blood sample available. Patients signed informed consent forms before radiotherapy. Information on patient age, gender, smoking history, complications, tumor location, stage, pathology, chemotherapy and radiotherapy technology were collected. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by committee of Zhejiang Cancer Research Institute (Hangzhou, Zhejiang, China).
Radiotherapy
Patients with stage I NSCLC were treated with stereotactic body radiation therapy (SBRT) with a total dose of 50 to 60 Gy (the median biologically effective dose was 100 Gy). Patients with stages II–III NSCLC were treated with intensity modulated radiotherapy (IMRT) with a total dose of 60–70 Gy at 2 Gy/fraction. In relation to the SBRT, the volume of lung that received at least 20 Gy (V20) was kept to less than 10%, V10 was kept to less than 15%, V13.5 was kept less than 1,000 cc, and V12.5 was kept to less than 1,500 cc. Conversely, IMRT required a mean lung dose less than 13 Gy, and V20 was kept to less than 28%, and V30 was kept to less than 20%.
Follow-up examinations
Follow-up examinations were performed using patients’ clinical records every 3 months after patient discharge until mortality or withdrawal, or until the follow-up deadline (of 5 years). The objective response (OR) was determined according to the RECIST (version 1.1). RILI was determined according to the Common Terminology Criteria for Adverse Events (version 5.0).
Genotyping methods
Peripheral blood leukocytes were obtained from 2 mL of a whole blood sample and were used for DNA extraction. Genomic DNA was isolated using a Blood Genomic DNA Kit (Qiagen, Valencia, CA). We genotyped the SNPs of the ERCC1 gene (rs11615, rs3212961, and rs3212986) and ERCC2 gene (rs13181, rs238406, and rs1799793) using the polymerase chain reaction (PCR) restriction fragment-length polymorphism method. PCR-based assays were used to amplify the fragments that contained ERCC1/2 polymorphisms. The primers were designed and provided by the Hangzhou Molecular Detection Biotechnology Co. Ltd. (see Table S1 for further details). The SNPs were analyzed using the illuminaHiSeq2500 platform.
Statistical analysis
IBM SPSS 25.0 was used for the statistical analysis. A Hardy-Weinberg analysis was conducted to analyze the frequencies of the genotypes and alleles. The Chi-square test was used to analyze differences in the selected demographic variables, risk factors, alleles, and genotypes of the 6 SNPs ERCC1/2 between the OR group and non-OR groups, and any association between RILI and clinical or genetic variables in NSCLC patients who underwent radiotherapy. Kaplan-Meier curves were used to estimate the cumulative overall survival (OS) and progressive-free survival (PSF) between different genotype groups. A univariate Cox proportion hazards regression analysis was performed to analyze the risks of OS and PFS. A multivariate Cox hazards regression analysis was performed to adjust for other covariates. The univariate and multivariate logistic regression analyses were used to investigate the association between RILI and ERCC1/2 polymorphisms or clinicopathological factors. All of the tests were two-sided. A P value less than 0.05 was considered statistically significant.
Results
The distribution of the ERCC1/2 SNPs alleles and genotypes in the 87 NSCLC patients
Table 1 sets out details of the genotype frequency and the minor allele frequency (MAF) of each sequence variants of the ERCC1/2 gene in the 87 NSCLC patients. The MAFs of the 6 SNPs of the patients were similar to those of the general population in the National Center for Biotechnology Information (NCBI) dbSNP databases. The distributions of the ERCC1/2 SNPs in the 87 patients were within the parameters of the Hardy-Weinberg equilibrium (P>0.05).
Table 1. Primary information for the analyzed ERCC1/2 SNPs in our population.
| Genes | SNPs | Major allele (%) | Minor allele (%) | Genotypes | MAF | χ2 | P | ||
|---|---|---|---|---|---|---|---|---|---|
| Homozygous 1 | Heterozygote | Homozygous 2 | |||||||
| ERCC1 | rs11615 | G (79.3) | A (20.7) | GG [55] | GA [28] | AA [4] | 0.4117 | 0.03 | 0.98 |
| ERCC1 | rs3212961 | G (50.0) | T (50.0) | GG [22] | GT [43] | TT [22] | 0.1447 | 0.01 | 0.99 |
| ERCC1 | rs3212986 | C (70.7) | A (29.3) | CC [43] | CA [37] | AA [7] | 0.2568 | 0.06 | 0.97 |
| ERCC2 | rs13181 | T (93.1) | G (6.9) | TT [75] | TG [12] | GG [0] | 0.3689 | 0.48 | 0.79 |
| ERCC2 | rs238406 | G (51.1) | T (48.9) | GG [24] | GT [41] | TT [22] | 0.4205 | 0.28 | 0.87 |
| ERCC2 | rs1799793 | C (94.8) | T (5.2) | CC [78] | CT [9] | TT [0] | 0.3088 | 0.26 | 0.88 |
SNPs, single-nucleotide polymorphisms; MAF, minor allele frequency.
The correlation between the ERCC1/2 SNPs and the OR
Among the 87 NSCLC patients, 44 patients had an OR [including a complete response (CR) and a partial response (PR)] and 43 patients had a non-OR [including stable disease (SD) and progressive disease (PD)]. Table 2 sets out the correlations between the OR and the clinicopathological characteristics of the NSCLC patients who underwent definitive radiotherapy. There was no significant difference between the clinical, pathological, and therapeutic regimen factor and the OR (P>0.05). The correlations between the distribution of the ERCC1/2 gene SNPs and the OR are shown in Table S2. Neither the genotype nor the allele of ERCC1/2 SNPs was significantly correlated with the OR of NSCLC patients who received definitive radiotherapy (all P>0.05).
Table 2. Comparison of 87 NSCLC patient characteristics between the OR (CR + PR) group and the non-OR (SD + PD) group.
| Factor | OR (N=44) | Non-OR (N=43) | P |
|---|---|---|---|
| Age (years) | |||
| >65 | 20 | 23 | 0.454 |
| ≤65 | 24 | 20 | |
| Gender | |||
| Female | 6 | 8 | 0.528 |
| Male | 38 | 35 | |
| Complication | |||
| Yes | 20 | 21 | 0.752 |
| No | 24 | 22 | |
| Smoking history (packs-year) | |||
| >30 | 28 | 24 | 0.457 |
| ≤30 | 16 | 19 | |
| Tumor location | |||
| Peripheral | 30 | 31 | 0.690 |
| Central | 14 | 12 | |
| Pathology | |||
| Squamous | 22 | 19 | 0.587 |
| Non-squamous | 22 | 24 | |
| T stage | |||
| 1 | 18 | 15 | 0.314 |
| 2 | 16 | 18 | |
| 3 | 2 | 6 | |
| 4 | 8 | 4 | |
| N stage | |||
| 0 | 21 | 17 | 0.597 |
| 1 | 3 | 1 | |
| 2 | 11 | 13 | |
| 3 | 9 | 12 | |
| Chemotherapy | |||
| Yes | 23 | 22 | 0.918 |
| No | 21 | 21 | |
| RT technology | |||
| SBRT | 15 | 12 | 0.533 |
| IMRT | 29 | 31 | |
| Concurrent CRT | |||
| Yes | 15 | 9 | 0.128 |
| No | 29 | 34 |
NSCLC, non-small cell lung cancer; OR, objective response; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; SBRT, stereotactic body radiation therapy; IMRT, intensity modulated radiotherapy; CRT, chemoradiotherapy.
The Association between the ERCC1/2 SNPs and OS and PFS
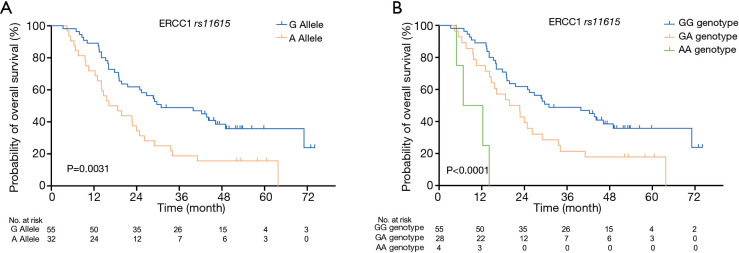
The median follow-up time was 24.9 months. The median survival time (MST) and PFS of the 87 patients was 24.9 and 16.1 months, respectively. The results of our study indicated that NSCLC patients with different ERCC1 rs11615 genotypes who received definitive radiotherapy had quite different survival curves (see Figure 1). Patients with the G allele [30.9 months, 95% confidence interval (CI): 12.832–48.968] had better OS than patients with the A allele (16.2 months, 95% CI: 9.102–23.238); the difference between the two groups was statistically significant (P=0.003). Patients with the AA genotype had significantly worse OS than that patients with the AG or GG genotypes (6.8 vs. 19.8 vs. 30.9 months, respectively, P=0.000). The results also indicated that patients with the ERCC2 rs3212986 variant A allele had better OS than patients with the C allele (MST: 30.9 vs. 21.5 months); however, the difference was not statistically significant (P=0.086).
Figure 1.
Kaplan-Meier curves of OS of the ERCC1 gene rs11615 SNP in 87 NSCLC patients who received definitive radiotherapy. (A) OS at allele gene level (P=0.003); (B) OS at genotype level (P=0.000). OS, overall survival; SNP, single-nucleotide polymorphism; NSCLC, non-small cell lung cancer.
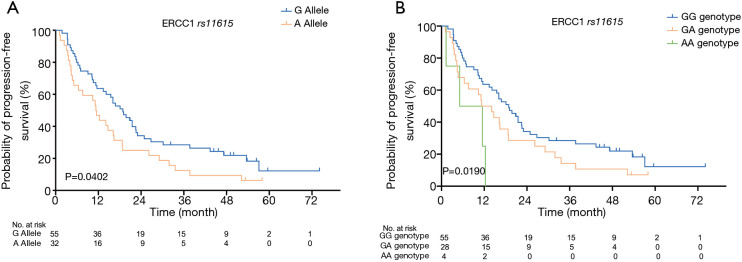
A similar association was found between PFS and ERCC1 rs11615 in our study (see Figure 2). The median PFS of patients with the G allele was 18.9 months (95% CI: 13.960–24.780), which was significantly higher than that of patients with the A allele (11.3 months, 95% CI: 8.655–14.005); the difference between the two groups was statistically significant (P=0.040). The median PFS of patients with the GG genotype, the AG genotype and the AA genotype was 18.9 months (95% CI: 12.556–19.584), 11.3 months (95% CI: 5.755–16.905), and 5.1 months (95% CI: 0–15.126), respectively; the difference among the three groups was statistically significant (P=0.019). Table 3 shows the association between the ERCC1/2 gene SNPs and prognosis (OS and PFS) for the NSCLC patients who received definitive radiotherapy.
Figure 2.
Kaplan-Meier curves of PFS of the ERCC1 gene rs11615 SNP in NSCLC 87 patients who received definitive radiotherapy. (A) PFS at allele gene level (P=0.040); (B) PFS at genotype level (P=0.019). PFS, progressive-free survival; SNP, single-nucleotide polymorphism; NSCLC, non-small cell lung cancer.
Table 3. Association between the ERCC1/2 gene polymorphisms and prognosis in 87 NSCLC patients who received definitive radiotherapy.
| Gene | MST (months) | 95% CI | P | Median PFS (months) | 95% CI | P |
|---|---|---|---|---|---|---|
| rs11615 | ||||||
| AA | 6.77 | 0–14.05 | 0.000 | 5.13 | 0–15.13 | 0.019 |
| AG | 19.77 | 11.14–28.40 | 11.33 | 5.76–16.91 | ||
| GG | 30.90 | 12.83–48.97 | 18.87 | 12.56–19.58 | ||
| A allele | 16.17 | 9.10–23.24 | 0.024 | 11.33 | 8.66–14.01 | 0.005 |
| G allele | 30.90 | 12.83–48.97 | 18.87 | 13.96–23.78 | ||
| rs3212961 | ||||||
| GG | 24.77 | 4.75–44.79 | 0.735 | 11.53 | 6.41–16.66 | 0.910 |
| GT | 23.97 | 15.75–32.19 | 18.07 | 12.67–23.47 | ||
| TT | 24.93 | 9.37–40.49 | 11.60 | 5.28–19.92 | ||
| G allele | 24.93 | 18.95–30.91 | 0.441 | 16.17 | 11.84–20.50 | 0.897 |
| T allele | 24.77 | 4.75–44.79 | 11.53 | 6.41–16.66 | ||
| rs3212986 | ||||||
| AA | 71.13 | 0–160.416 | 0.092 | 15.53 | 0–38.01 | 0.718 |
| AC | 29.10 | 18.92–39.28 | 18.70 | 11.91–25.49 | ||
| CC | 21.53 | 12.97–30.09 | 14.67 | 8.849–20.49 | ||
| A allele | 30.90 | 15.68–46.12 | 0.086 | 18.01 | 8.85–20.49 | 0.423 |
| C allele | 21.53 | 12.97–30.09 | 14.67 | 11.89–24.25 | ||
| rs13181 | ||||||
| TT | 24.93 | 12.67–36.87 | 0.692 | 19.07 | 9.96–28.19 | 0.921 |
| TG | 24.77 | 16.19–33.67 | 16.47 | 10.17–19.17 | ||
| rs238406 | ||||||
| TT | 16.17 | 0–36.26 | 0686 | 11.33 | 8.92–13.74 | 0.143 |
| TG | 28.67 | 22.72–34.62 | 21.50 | 16.86–26.14 | ||
| GG | 19.80 | 13.76–25.84 | 11.30 | 6.67–15.93 | ||
| T allele | 26.83 | 21.44–32.22 | 0.562 | 16.83 | 12.16–21.50 | 0.097 |
| G allele | 19.80 | 13.76–25.84 | 11.30 | 6.67–15.93 | ||
| rs1799793 | ||||||
| CC | 25.40 | 18.74–32.05 | 0.222 | 15.53 | 11.79–19.27 | 0.411 |
| CT | 19.07 | 10.48–27.66 | 16.13 | 2.89–29.37 |
NSCLC, non-small cell lung cancer; CI, confidence interval; MST, median overall survival time; PFS, progression-free survival.
Prognostic factors for NSCLC patients who received definitive radiotherapy
The analysis results of the associations between the prognosis, clinicopathology, and genetic factors in NSCLC patients are set out Table 4. A univariate Cox proportional hazards model was used to analyze the association between the survival or recurrent time and several factors, including gender, age, complication, smoking history, T stage, N stage, chemotherapy, radiation technology, and ERCC1/2 gene polymorphism in NSCLC patients treated with radiotherapy. The multivariate Cox proportional hazard analysis showed that T stage, ERCC1 rs11615, and ERCC2 rs3212986 were independent survival factors for NSCLC patients who received definitive radiotherapy (see Table 4). T3–4 stage (hazard ratio, HR): 3.215, 95% CI: 1.792–5.747, P=0.000), ERCC1 rs11615 A allele (HR: 2.451, 95% CI: 1.435–4.184, P=0.001) than G allele, and ERCC2 rs3212986 A allele (HR: 2.538, 95% CI: 1.149–5.618, P=0.021) had higher risks of OS than C allele. The univariate and multivariate Cox proportional hazard analyses showed that T stage was also an independent recurrence-related factor in NSCLC patients who received definitive radiotherapy (T3–4 stage, HR: 2.252, 95% CI: 1.300–3.921, P=0.004). ERCC1 rs11615 was an independent recurrence-related factor in the univariate Cox proportional hazard analysis (HR: 1.623, 95% CI: 1.018–2.591, P=0.042), but there was no significant difference in the multivariate Cox proportional hazard analysis (HR: 1.497, 95% CI: 0.932–2.404, P=0.095).
Table 4. Univariate and multivariate regression analyses of survival and recurrence in patients with NSCLC.
| Factor | Overall survival | Progression-free survival | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | ||||||||||||
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | ||||
| Gender (male vs. female) | 1.177 | 0.580–2.388 | 0.652 | 1.145 | 0.616–2.127 | 0.669 | |||||||||
| Age (>65 vs. ≤65 years) | 1.320 | 0.797–2.184 | 0.281 | 1.155 | 0.731–1.826 | 0.537 | |||||||||
| Complication (yea vs. no) | 1.188 | 0.724–1.946 | 0.497 | 1.093 | 0.692–1.754 | 0.705 | |||||||||
| Smoking (>30 vs. ≤30 pack-year) | 1.406 | 0.833–2.375 | 0.201 | 1.125 | 0.703–1.799 | 0.624 | |||||||||
| T stage (T3–4 vs. T1–2) | 2.976 | 1.689–5.263 | 0.000 | 3.215 | 1.792–5.747 | 0.000 | 2.392 | 1.389–4.115 | 0.002 | 2.252 | 1.300–3.921 | 0.004 | |||
| N stage (N2–3 vs. N0–1) | 1.721 | 1.034–2.865 | 0.037 | 1.616 | 1.016–2.564 | 0.042 | |||||||||
| Chemotherapy (no vs. yes) | 0.578 | 0.346–0.964 | 0.036 | 1.587 | 0.998–2.525 | 0.051 | |||||||||
| Concurrent chemoradiotherapy (no vs. yes) | 1.227 | 0.707–2.128 | 0.468 | 1.336 | 0.824–2.262 | 0.226 | |||||||||
| Radiotherapy technology (IMRT vs. SBRT) | 1.756 | 0.988–3.123 | 0.055 | 1.773 | 1.059–2.968 | 0.029 | |||||||||
| Target therapy (no vs. yes) | 1.047 | 0.475–2.304 | 0.910 | 1.114 | 0.532–2.326 | 0.776 | |||||||||
| ERCC1 rs11615 A vs. G | 2.105 | 1.271–3.484 | 0.004 | 2.451 | 1.435–4.184 | 0.001 | 1.623 | 1.018–2.591 | 0.042 | 1.497 | 0.932–2.404 | 0.095 | |||
| ERCC1 rs3212961 T vs. G | 1.276 | 0.686–2.370 | 0.442 | 1.035 | 0.610–1.756 | 0.897 | |||||||||
| ERCC1 rs3212986 A vs. C | 1.554 | 0.936–2.581 | 0.088 | 2.538 | 1.149–5.618 | 0.021 | 1.206 | 0.763–1.905 | 0.423 | ||||||
| ERCC2 rs13181 G vs. T | 1.153 | 0.568–2.342 | 0.692 | 1.033 | 0.542–1.969 | 0.921 | |||||||||
| ERCC2 rs238406 T vs. G | 1.177 | 0.678–2.043 | 0.562 | 1.516 | 0.924–2.488 | 0.100 | |||||||||
| ERCC2 rs1799793 T vs. C | 1.585 | 0.752–3.333 | 0.226 | 1.342 | 0.664–2.717 | 0.413 | |||||||||
NSCLC, non-small cell lung cancer. CI, confidence interval; IMRT, intensity modulated radiotherapy; SBRT, stereotactic body radiation therapy.
Association between ERCC1/2 polymorphisms and RILI in NSCLC patients who underwent definitive radiotherapy
The clinical characteristics of patients with grade ≥2 RILI and those with grade <2 RILI are set out in Table 5. There were no significant differences in terms of smoking history, complication, tumor location, pathological type, T stage, N stage, radiotherapy technology, and ERCC1/2 polymorphisms variates between the two groups (P>0.05). A logistics regression analysis was conducted to analyze the association between the RILI and ERCC1/2 gene polymorphism in NSCLC patients who underwent definitive radiotherapy. As Table 6 shows, no association was observed in relation to the polymorphisms and adverse effects due to radiotherapy.
Table 5. Comparison of 87 NSCLC patient characteristics between the grade ≥2 RILI group and the grade <2 RILI group.
| Factors | Grade ≥2 RILI (N=16) | Grade <2 RILI (N=71) | P value |
|---|---|---|---|
| Gender | 0.045 | ||
| Male | 16 | 57 | |
| Female | 0 | 14 | |
| Age (years) | 0.372 | ||
| >65 | 7 | 37 | |
| ≤65 | 9 | 34 | |
| Complication | 0.129 | ||
| Yes | 5 | 36 | |
| No | 11 | 35 | |
| Smoking history (packs-year) | 0.137 | ||
| >30 | 12 | 40 | |
| ≤30 | 4 | 31 | |
| Tumor location | 1.000 | ||
| Peripheral | 11 | 50 | |
| Central | 5 | 21 | |
| Pathology | 0.139 | ||
| Squamous | 6 | 40 | |
| Non-squamous | 10 | 31 | |
| T stage | 0.531 | ||
| T1–2 | 12 | 55 | |
| T3–4 | 4 | 16 | |
| N stage | 0.548 | ||
| N0–1 | 8 | 34 | |
| N2-3 | 8 | 37 | |
| Radiotherapy technology | 0.600 | ||
| SBRT | 5 | 22 | |
| IMRT | 11 | 49 | |
| Chemotherapy | 0.372 | ||
| Yes | 7 | 37 | |
| No | 9 | 34 | |
| Concurrent CRT | 0.533 | ||
| Yes | 4 | 20 | |
| No | 12 | 51 | |
| Target therapy | 0.523 | ||
| Yes | 2 | 7 | |
| No | 14 | 64 | |
| rs11615 | 0.581 | ||
| G allele | 10 | 45 | |
| A allele | 6 | 26 | |
| rs3212961 | 0.376 | ||
| G allele | 3 | 19 | |
| T allele | 13 | 52 | |
| rs3212986 | 0.372 | ||
| C allele | 9 | 34 | |
| A allele | 7 | 37 | |
| rs13181 | 0.616 | ||
| T allele | 14 | 61 | |
| G allele | 2 | 10 | |
| rs238406 | 0.533 | ||
| G allele | 4 | 20 | |
| T allele | 12 | 51 | |
| rs1799793 | 0.523 | ||
| C allele | 14 | 64 | |
| T allele | 2 | 7 |
NSCLC, non-small cell lung cancer; RILI, radiation-induced lung injury; CRT, chemoradiotherapy.
Table 6. Logistic regression analysis of the relationship between the ERCC1/2 polymorphisms and RILI.
| SNP | Allele | OR | 95% CI | P value |
|---|---|---|---|---|
| rs11615 | G allele | 1 | Ref. | 0.925 |
| A allele | 1.063 | 0.299–3.774 | ||
| rs3212961 | C allele | 1 | Ref. | 0.454 |
| A allele | 1.739 | 0.408–7.411 | ||
| rs3212986 | C allele | 1 | Ref. | 0.555 |
| A allele | 1.432 | 0.430–4.739 | ||
| rs13181 | A allele | 1 | Ref. | 0.755 |
| C allele | 1.331 | 0.220–8.065 | ||
| rs238406 | G allele | 1 | Ref. | 0.887 |
| T allele | 1.105 | 0.278–4.393 | ||
| rs1799793 | C allele | 1 | Ref. | 0.261 |
| T allele | 1.445 | 0.247–8.439 |
RILI, radiation-induced lung injury; SNP, single-nucleotide polymorphism.
Discussion
In this study, we assessed the association of prognosis with clinical factors and genetic factors among NSCLC patients who underwent definitive radiotherapy. Our results showed that a genetic polymorphic site (i.e., rs11615), which is a functional variant of the NER pathway that repairs the DNA gene ERCC1, may be an independent indicator of survival in NSCLC patients who undergo definitive radiotherapy. As the key enzymes in the NER pathway, the proteins expressed by the ERCC1 and ERCC2/XPD genes play an important role in this pathway. The protein expressed by ERCC1 is the rate-limiting enzyme of the NER pathway. By forming a tight heterodimer with specific 5' terminal endonuclease function by removing a complementary gene F (XPF) with the DNA repair enzyme, the damaged DNA can be identified and removed to ensure the fidelity of the gene (9). The protein expressed by ERCC2 is an ATP-dependent DNA helicase, which recognizes and opens the damaged DNA double strand and then assists the endonuclease to remove the damaged DNA (10).
The SNPs of ERCC1 and ERCC2 were associated with platinum-based chemosensitivity and prognosis in patients with NSCLC, but the results that followed were heterogeneous. The A allele of ERCC1 rs11615 was associated with poor OS. In relation to ERCC2, the T allele of rs1799793 was linked to an unfavorable OS, and the G allele of rs13181 had a worse OS and PFS in NSCLC patients who underwent platinum-based chemotherapy (10). Tan et al. (11) found that the ERCC1 rs11615 A allele was associated with poor OS in advanced NSCLC patients who received platinum chemotherapy. Conversely, Ren et al. (12) found that the presence of the rs11615 polymorphism (CT or TT) was related to better survival than CC. Additionally, some research has shown that the ERCC1 SNPs, especially rs11615, are associated with chemoradiotherapy rather than chemotherapy. A meta-analysis showed that ERCC1 had no significant correlation with platinum-based chemotherapy, but there was a significant correlation in platinum-based radiochemotherapy and prognosis (13); however, it should be noted that the meta-analysis focused on the Caucasian race. A study by Sullivan et al. (14) also showed that ERCC1 rs11615 was correlated with the efficacy of radiochemotherapy in NSCLC patients, but not in chemotherapy. Similarly, in esophageal cancer, rs11615 was found to be significantly associated with the short-term therapeutic efficacy (the CR rate) and survival time of ESCC patients who received docetaxel plus cisplatin regimen-based concurrent radiochemotherapy (15).
The results on the effects of SNPs of ERCC1/2 in relation to the efficacy of radiotherapy were conflicting. ERCC1/2 was identified and considered a potential biomarker in response to radiation (16). However, Jin et al. (17) developed an OS model and a methodology to identify a biomarker for radiosensitivity in ERCC1/2, and found the ERCC1 rs11615 GG genotype and the ERCC2 rs238406 CC genotype were correlated with radioresistance, and those patients could benefit from higher dose of radiation. However, it should be noted that the main subjects in Spring et al.’s study were Caucasians.
The distribution of ERCC1 rs11615 genotype frequency is highly dependent upon race. Africans have the highest ERCC1 rs11615 GG genotype frequency (more than 90%), while the GG genotype frequency in Asians is 20–60%, which is higher than that in Europeans (7–20%) (17). In the present study, which was the first to focus on Chinese NSCLC patients who underwent radiotherapy, we found that the OS and PFS of patients with the ERCC1 rs11615 G allele were significantly better than those with the A allele (P<0.05). The univariate and multivariate Cox proportional hazard model analyses also showed that rs11615 was an independent factor related to survival, but not to PFS (P=0.095). Similar results were found for the subgroup of patients who underwent IMRT; thus, rs11615 may be a potential biomarker that predicts survival outcomes in NSCLC patients, and patients with A allele or the AA genotype will have worse OS than those with the G allele or the GG/GA genotype. The subgroup of patients who received SBRT had similar results to those who received IMRT. There was a trend towards poorer clinical outcomes for patients with the A allele; however, it should be noted that these results were not significant (this may be attribute to the study’s small sample size). In relation to ERCC2, only one study has shown that the rs13181 T allele significantly increased tumor recurrence risk in gastric cancer, and it should be noted that some studies have found opposite results (16). In relation to rs1799793, which is described as Asp312Asn in some studies, several pre-clinical studies reported that the Asn/Asn genotype had a sub-optimal DNA repair capacity relative to the Asp-containing genotypes. In the present study, no association with clinical outcomes was found for ERCC2 SNP.
The cell-death mechanism related to ionizing radiation is mainly lethal DNA damage, which leads to cell-cycle arrest and apoptosis. Theoretically, as they are involved in DNA damage repair, the ERCC1/2 SNPs genes could be associated with the sensitivity, efficacy, and prognosis of radiotherapy. The SNPs of ERCC1 could affect micro-ribonucleic acid (mRNA) expression, and have attracted increasing interest as potential predictors of cancer therapy outcomes and patient prognosis indicators for lung cancer patients. rs11615 A>G, which is described as T>C in some articles, is a synonymous mutation and aspartic acid in transcription and translation (the ‘AAT’ and ‘AAC’ codons, respectively). As the usage of ‘AAT’ codon increases, the expression and protein level of mRNA increases (by about 50%), which improves the DNA damage repair ability of tumor cells and improves tumor resistance to radiotherapy and platinum-based chemotherapy (1,14).
Some studies have shown an association between ERCC1/2 polymorphisms and radiation-induced adverse effects in malignant tumors. One study reported that patients bearing the rs11615 T allele (TT or CT genotypes) variant had a significantly reduced risk of developing RILI compared to those carrying the CC genotype in lung cancer (18). In addition, research has shown that ERCC2 rs13181 is related to acute radiation-induced esophageal toxicity in lung cancer patients (19). The results of the present study showed that the ERCC1/2 polymorphisms had no statistically significant effect on RILI risk. This supports the results of a previous study on rs13181, but does not support the results of a previous study on rs11615.
In addition to rs11615, we also found that rs3212986 was associated with OS in NSCLC patients who received radiotherapy, and that the A allele patients had better OS than the C allele patients; however, the difference was not statistically significant. In the Cox regression analysis, rs3212986 was found to be an independent predictor of prognosis. It has been hypothesized that this is related to reduced ERCC1 synthesis due to the mRNA instability of the A allele. This may lead to an accumulation of DNA damage, which may make tumors more immunogenic and more responsive to immunotherapy (20).
This study had a number of limitations. First, the sample size of the study was small, which may have caused selection bias and affected the results. Second, this was a retrospective study, and the radiotherapy technology received by patients was not consistent; thus, the correlation between the radio-physical parameters and ERCC1/2 SNP could not be systematically analyzed. Finally, this study only analyzed the ERCC1/2 SNP, but the DNA damage mediated by ionizing radiation is the result of multiple pathway mechanisms and multiple genes. It is difficult to predict efficacy by relying on a specific biomarker. In the future, we intend to investigate more potential biomarkers for different pathways, and develop prediction models by combining multiple biomarkers.
In conclusion, the study provided the first clinical evidence that the ERCC1 rs11615 SNP may be a potential biomarker for predicting the survival prognosis of Chinese NSCLC patients who have undergone definitive radiotherapy. Patients with the G allele had better OS than those with the A allele. Further investigations need to be conducted to confirm these findings.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This article was supported by the National Natural Science Foundation of China (81372438).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by committee of Zhejiang Cancer Research Institute (Hangzhou, Zhejiang, China) and informed consent was taken from all the patients.
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jtd-21-755
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-21-755
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-21-755). The authors have no conflicts of interest to declare.
(English Language Editor: L. Huleatt)
Reference
- 1.Avinash Tejasvi ML, Maragathavalli G, Putcha UK, et al. Impact of ERCC1 gene polymorphisms on response to cisplatin based therapy in oral squamous cell carcinoma (OSCC) patients. Indian J Pathol Microbiol 2020;63:538. 10.4103/IJPM.IJPM_964_19 [DOI] [PubMed] [Google Scholar]
- 2.Gillet LC, Schärer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev 2006;106:253-76. 10.1021/cr040483f [DOI] [PubMed] [Google Scholar]
- 3.Liao WY, Ho CC, Tsai TH, et al. Combined effect of ERCC1 and ERCC2 polymorphisms on overall survival in non-squamous non-small-cell lung cancer patients treated with first-line pemetrexed/platinum. Lung Cancer 2018;118:90-6. 10.1016/j.lungcan.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 4.Grenda A, Błach J, Szczyrek M, et al. Promoter polymorphisms of TOP2A and ERCC1 genes as predictive factors for chemotherapy in non‐small cell lung cancer patients. Cancer Med 2020;9:605-14. 10.1002/cam4.2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushra MU, Rivu SF, Sifat AE, et al. Genetic polymorphisms of GSTP1, XRCC1, XPC and ERCC1: prediction of clinical outcome of platinum-based chemotherapy in advanced non-small cell lung cancer patients of Bangladesh. Mol Biol Rep 2020;47:7073-82. 10.1007/s11033-020-05771-2 [DOI] [PubMed] [Google Scholar]
- 6.Anoushirvani AA, Aghabozorgi R, Ahmadi A, et al. The relationship between rs3212986C> a polymorphism and tumor stage in lung cancer patients. Cureus 2019;11:e4423. 10.7759/cureus.4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez-Ramírez C, Cañadas-Garre M, Alnatsha A, et al. Pharmacogenetics of platinum-based chemotherapy: impact of DNA repair and folate metabolism gene polymorphisms on prognosis of non-small cell lung cancer patients. Pharmacogenomics J 2019;19:164-77. 10.1038/s41397-018-0014-8 [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Yang M, Bi N, et al. Association of TGF-β1 and XPD polymorphisms with severe acute radiation-induced esophageal toxicity in locally advanced lung cancer patients treated with radiotherapy. Radiother Oncol 2010;97:19-25. 10.1016/j.radonc.2010.08.015 [DOI] [PubMed] [Google Scholar]
- 9.Gillet LC, Schärer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev 2006;106:253-76. 10.1021/cr040483f [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Xian L. The association between the ERCC1/2 polymorphisms and the clinical outcomes of the platinum-based chemotherapy in non-small cell lung cancer (NSCLC): a systematic review and meta-analysis. Tumour Biol 2014;35:2905-21. 10.1007/s13277-013-1493-5 [DOI] [PubMed] [Google Scholar]
- 11.Tan LM, Qiu CF, Zhu T, et al. Genetic Polymorphisms and Platinum-based Chemotherapy Treatment Outcomes in Patients with Non-Small Cell Lung Cancer: A Genetic Epidemiology Study Based Meta-analysis. Sci Rep 2017;7:5593. 10.1038/s41598-017-05642-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren S, Zhou S, Wu F, et al. Association between polymorphisms of DNA repair genes and survival of advanced NSCLC patients treated with platinum-based chemotherapy. Lung Cancer 2012;75:102-9. 10.1016/j.lungcan.2011.05.023 [DOI] [PubMed] [Google Scholar]
- 13.Li F, Xie X, Ren X, et al. A meta-analysis identifies ERCC1 gene polymorphism as a predictor of better patient response to treatment with radiochemotherapy. Cancer Chemother Pharmacol 2016;77:1183-91. 10.1007/s00280-016-3015-9 [DOI] [PubMed] [Google Scholar]
- 14.Sullivan I, Salazar J, Majem M, et al. Pharmacogenetics of the DNA repair pathways in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Lett 2014;353:160-6. 10.1016/j.canlet.2014.07.023 [DOI] [PubMed] [Google Scholar]
- 15.Huang X, Liu C, Cui Y, et al. Association between XRCC1 and ERCC1 single-nucleotide polymorphisms and the efficacy of concurrent radiochemotherapy in patients with esophageal squamous cell carcinoma. Oncol Lett 2017;13:704-14. 10.3892/ol.2016.5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao L, He X, Shang Y, et al. Identification of potential radiation-responsive biomarkers based on human orthologous genes with possible roles in DNA repair pathways by comparison between Arabidopsis thaliana and homo sapiens. Sci Total Environ 2020;702:135076. 10.1016/j.scitotenv.2019.135076 [DOI] [PubMed] [Google Scholar]
- 17.Jin JY, Wang W, Ten Haken RK, et al. Use a survival model to correlate single-nucleotide polymorphisms of DNA repair genes with radiation dose-response in patients with non-small cell lung cancer. Radiother Oncol 2015;117:77-82. 10.1016/j.radonc.2015.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du L, Yu W, Dai X, et al. Association of DNA repair gene polymorphisms with the risk of radiation pneumonitis in lung cancer patients. Oncotarget 2017;9:958. 10.18632/oncotarget.22982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Yang M, Bi N, et al. ATM polymorphisms are associated with risk of radiation-induced pneumonitis. Int J Radiat Oncol Biol Phys 2010;77:1360-8. 10.1016/j.ijrobp.2009.07.1675 [DOI] [PubMed] [Google Scholar]
- 20.Aiello MM, Solinas C, Santoni M, et al. Excision Repair Cross Complementation Group 1 Single Nucleotide Polymorphisms and Nivolumab in Advanced Non-Small Cell Lung Cancer. Front Oncol 2020;10:1167. 10.3389/fonc.2020.01167 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as