Abstract
Lung cancer ranks first as the cause of cancer-associated deaths gobally. The American Cancer Society estimates for 228,820 new cases and 135,720 deaths from lung cancer in the United States for the year 2020. Targeted treatment options have rapidly emerged for non-small cell lung cancer (NSCLC) within the past decade. Screening for molecular aberrations is mainly done by tissue biopsy. However, in some cases a biopsy is not possible, or patients do not consent to it. Hence, liquid biopsy remains the only option. Relevant data about the topic of liquid biopsy, with a special focus on NSCLC, was obtained via a PubMed search. We included mainly literature published from 2010 onwards, omitting older studies whenever possible. With this review of the literature, we give an overview of different liquid biopsy approaches, as well as their respective advantages and disadvantages. We have reviewed the assessment of epidermal growth factor receptor (EGFR) mutation status in particular, and go into detail with current use of liquid biopsy in everyday clinical practice. Today, liquid biopsy is still infrequently used, depending on the treatment center, but popularity is steadily increasing. Various different approaches are already available, but costs and level of sensitivity significantly differ between techniques. By using liquid biopsy more widely in selected patients, complication rates can be reduced, and constant disease monitoring is made considerably easier.
Keywords: Liquid biopsy, non-small cell lung cancer (NSCLC), treatment, prognosis
Background
Targeted treatment modalities are quickly emerging in the clinical management of non-small cell lung cancer (NSCLC). Lung cancer is the leading cause of cancer-related deaths worldwide. For both men and women, lung cancer is the most frequently diagnosed cancer entity worldwide (11.6% of all cancers) and accounts for 18.4% of total cancer-related deaths, according to data from 2018 (1). The American Cancer Society estimates, that 228,820 new cases and 135,720 deaths from lung cancer are to be expected in the United States for the year 2020 (2). Increasing knowledge has been gained within the last decade about the molecular abnormalities in lung cancer, defining disease subsets based on molecular properties (3).
For several years, providing tailored treatment options, depending on certain molecular characteristics of the disease, has been the standard in everyday clinical practice. It is common, however, that over the course of the disease, resistance to targeted treatment is acquired. Thus, current research focuses on the establishment of next-generation therapeutics, potent enough to overpower mechanisms of drug resistance (4-11). Genetic information from tumors is used to predict the response to treatment with certain targeted therapeutics, e.g., epidermal growth factor tyrosine kinase inhibitors (EGFR TKIs).
The approval of first-generation EGFR TKIs consequently resulted in the development of second- and third-generation TKIs like osimertinib, having its specific point of impact against certain mutant forms of EGFR. Several advantages are provided by this novel group of treatment agents: common EGFR activating mutations are specifically and effectively addressed; inhibition of the EGFR protein harboring the T790M mutation is provided (this mutation is responsible for treatment failure of EGFR TKIs of the first or second generation); as well as the impact against wild-type (WT) EGFR is relatively low, which reduces treatment toxicity and adverse effects (12). Another example is the ever-expanding group of anaplastic lymphoma kinase (ALK) targeting drugs, which provide valid treatment modalities for each subject having developed resistance to crizotinib, a first-generation ALK TKI (12). These next generation ALK TKIs (alectinib, ceritinib, brigatinib, ensartinib and lorlatinib) have the potential to bind and inhibit mutant forms of ALK. Of note, all these novel ALK TKIs have different binding affinities, depending on specific resistance mutations, so as a method to find tailored treatment modalities for each individual, exact identification of these resistance mutations is mandatory (4). Since the evolution of the tumor microbiology over the course of the disease may lead to a change in mutations, allowing for additional therapeutic options, repeated tissue biopsies have been advocated. However, this approach comes with a considerable risk for the respective patient, and depending on performance status cannot be applied to each individual (4). For instance, computed tomography (CT)-guided lung biopsy has a 5% rate of major complications (13).
Until today, there is only one predictive tumor biomarker which is routinely tested to outline patients suitable for first line immunotherapy, i.e., programmed cell death 1 ligand 1 (PD-L1) as evaluated by means of immunohistochemistry from tissue sections (14). PD-L1 is thus routinely assessed in many pathology laboratories throughout the world, however the assessment can sometimes be challenging due to biologic or technical limitations (15). PD-L1 expression is found in malignant-, but also in immune-cells, rendering a careful assessment of the PD-L1 status even more complicated (16,17). Moreover, there is a considerable intratumoral heterogeneity regarding PD-L1 expression, and small biopsies may not be representative of the whole tumor (17).
Tumor mutational burden (TMB) in biopsies of cancer tissue has been outlined as a new biomarker, especially for outlining NSCLC patients for treatment with immune checkpoint inhibitors, like nivolumab and ipilimumab (18). However, until today cross laboratory technical standards and validation of TMB analysis are still lacking, making the implementation of TMB into everyday clinical practice difficult (19). The prediction of therapeutic response based on PD-L1 immunohistochemistry from tissue biopsies, or else, TMB assessment, is not always possible due to a very small amount of tumor tissue or a minor proportion of cancer cells in the biopsy specimen. Furthermore, in a few patients with a relatively high TMB and high PD-L1 expression, immunohistochemistry might still be false negative (17).
As an alternative, liquid biopsy is an emerging diagnostic tool, already used in clinical routine in lung cancer patients in some specialized treatment facilities. Originally, the term liquid biopsy defined circulating tumor cells (CTCs), but today also comprises circulating cell-free tumor DNA (cfDNA) as well as exosomes (3). Liquid biopsies are utilized either as a method for the diagnosis of lung cancer, or as a tool to monitor treatment response or for the detection of minimal residual disease after curative surgery (3).
A variety of liquid biopsy platforms have been established in order to outline mechanisms of drug resistance that have developed over time. The liquid biopsy approach has been recommended by the new College of American Pathologists (CAP)/International Association for the Study of Lung Cancer (IASLC)/Association for Molecular Pathology (AMP) guideline for the molecular testing of NSCLC patients (20). Of note, liquid biopsy cannot substitute for an initial diagnostic tissue biopsy. Only in rare cases where tissue cannot be obtained via tissue biopsy, liquid biopsy may serve as a tool to acquire the histologic diagnosis (21). In some cases, tissue biopsy material is small, which prevents the pathologist to carry out all the necessary molecular tests, so the acquisition of new tissue for conducting further analyses would be urgently required. Liquid biopsy is the alternative option in this scenario as well. Moreover, liquid biopsy is more cost-effective when compared to conventional tissue biopsy (22). It has also been found that molecular properties of CTCs provide a more accurate picture of the actual systemic tumor load, furthermore reflecting more accurately the heterogeneity within a given tumor specimen, as well as tumor biology of metastases, which cannot be covered with single-site biopsy only (23,24).
When making liquid biopsy techniques widely available in clinical routine, methods of sample collection, storage and shipping have to be optimized and standardized across treatment centers. In a recent study (25), the cell- and DNA-stabilizing properties of Streck Cell-Free DNA BCT blood collection tubes have been analyzed. These tubes allow for the shipping of whole blood at ambient temperature, while the integrity of cfDNA is still provided, preventing the dilution of cancer-derived DNA with WT DNA from the genome. According to this report, collection of whole blood from healthy individuals in cfDNA BCTs, followed by storing for a time period of five days or less, at room temperature, did not compromise DNA quality and mutation background levels. Mutant circulating tumor DNA (ctDNA) in the blood obtained from patients with colorectal cancer, and acquired using cfDNA BCTs, remained stable over a time period of three days of storage at room temperature. Still, as a consequence of storage at ≤10 °C and at 40 °C for a longer time period, levels of healthy DNA from the genome, and an unusually large cell plasma interface, along with reduced plasma volumes, were observed (25). Thus, correct handling and storing, a preferably short storage time and quick sample analysis are key factors to achieve the maximum diagnostic benefit from liquid biopsy.
In this review article, we aim to elucidate the pros and cons of liquid biopsy, as well as current clinical relevance, use in everyday practice and limitations.
We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/tlcr-21-3).
Methods
Relevant data about the topic of liquid biopsy, with a special focus on NSCLC, was obtained via a PubMed search (26). We specifically searched for “liquid biopsy”, “non-small cell lung cancer”, and in this context for “circulating tumor DNA”, “circulating tumor cells”, “mutations”, “plasma micro RNAs”, “tumor-educated platelets”, “exosomes” and “clinical application”. Our focus was to include mainly literature published from 2010 onwards, omitting, if possible, older studies.
Results
Circulating tumor DNA
CtDNA counts among the group of cfDNA. In 1948 already, little quantities of cfDNA in blood plasma or -serum samples of human beings, have been detected. In 1977, cfDNA was outlined in blood samples of cancer patients, and after 17 further years it was genotyped as a tumor marker (27). ctDNA originates from tumor cell apoptosis, necrosis and extracellular vesicles which tumor cells secrete. Contrary to genomic DNA, ctDNA which is found in blood samples is at large part fragmented, ranging from 180 base pairs (bp) to 1,000 bp, if the origin is apoptosis, and 10,000 bp if the origin is necrosis (28). Extracellular vesicles are of crucial importance for intercellular communication, thus containing large fragments of double-stranded DNA (>10 kb) embodying KRAS-, p53- and EGFR-mutations (29,30). Of note, ctDNA only accounts for a very small fraction, namely <1% of total cfDNA, meaning that conventional sequencing approaches are not sensitive enough for the detection of EGFR mutations in cfDNA. Recently, novel approaches have been found to increase the sensitivity of assays for the detection of EGFR mutations (31,32). The amplification-refractory mutation system (ARMS)/Scorpion assay contains primers that use specific probes with enhanced allelic specificity, which are able to detect and differentiate WT, as opposed to mutant DNA, reducing the degree of background to a minimum. When primers match completely, a selective amplification of mutated gene sequences is realized. Peptide nucleic acids (PNAs), on the contrary, are utilized to selectively suppress the WT PCR manufacture. Next-generation sequencing (NGS) is a method for the detection of a subgroup of specific genes, analyzing the entire genome of a given tumor. Very differently to techniques which can only outline pre-known mutations, individual resistance mechanisms can be determined by NGS. NGS makes use of DNA polymerases, catalyzing the consolidation of fluorescence-labeled nucleotides into a DNA template, simultaneously with sequential DNA synthesis. This process is extended across millions of fragments contemporaneously (33).
Of note, also urine and saliva can be used for the detection of EGFR mutations, since DNA, mRNA, micro RNA (miRNA), proteins and metabolites are incorporated in saliva, which serve as potential predictive biomarkers for either cancer, or other systemic conditions (34). Short-length, tumor-derived DNA fragments can also be detected in the urine, which serves as a method, for example, to detect KRAS mutations in colon cancer patients (35). Recently, a mutation-enriched PCR coupled with NGS was used as a urine platform to detect T790M- and L858R mutations, and also exon 19 deletions in NSCLC (36). This urine platform proved good sensitivity with 93% for T790M mutations, 80% for L858R mutations and 83% for exon 19 deletions (with tissue-based results used as a reference) (36).
For ctDNA extraction and analysis, attention has to be paid to accurate blood sample collection, handling, and storage procedures, in order to enable an exact molecular analysis (4). Factors that influence the accuracy of ctDNA analysis are the way of storing and blood sample transport, but also how much time elapses between blood taking and the extraction of plasma. The two most prominent methods for the acquisition of ctDNA is the use of ethylenediaminetetra-acetic acid (EDTA) tubes for blood plasma acquisition, as well as utilizing preservative tubes, manufactured specifically for cell-free DNA extraction. The amount of blood that has to be drawn is not standardized yet, however, most institutions collect 20 mL (37). EDTA tubes are easily available and cost-efficient compared to other preservative tubes. However, the use of EDTA means that the blood sample has to be processed shortly after acquisition (38). Conversely, special preservative tubes for ctDNA, such as Streck (La Vista, Nevada) Cell-Free DNA BCT effectively preserves the quality of tiny fragments of DNA, lasting several days, upon storage at room temperature, without the requirement of on-site processing techniques (25).
CfDNA, on the contrary, reflects the sum of all ctDNA, irrespective of origin. CtDNA only describes a subset of DNA specifically originating from the tumor (4). CfDNA collection tubes contain preservative reagents stabilizing blood cells with nuclei, so their disintegration and the consequential leaking of genomic DNA into the tube is averted. Moreover, the way exonuclease normally works, is blocked in the conservant tube, so cfDNA degradation is inhibited (39). In case EDTA tubes are used instead of preservative tubes, samples have to be processed within 1 to 2 hours after collection (40). A major hazard to ctDNA analysis is impurity caused by genomic DNA, released from lysed leukocytes, which happens when blood samples are not immediately handled and processed. Notably, even upon optimal sample storing, shipping and processing, the proportion of tumor-derived mutant forms of an allele is very small, often less than 1% of the total DNA (mutant plus wild type) for a given gene sequence (41). Keeping this in mind, it becomes evident that the contamination of the cfDNA sample by DNA from the genome, caused by incautious sample processing, leads to a consequential dilution of mutant cfDNA fragments, even below detection level. It is also recommended to centrifuge the blood tube twice (the so-called double-spin technique), once in the original collection tube and then a second time in another tube after having transferred the plasma, ensuring a more efficient purification.
In clinical practice, more than one technique has been engineered to analyse the presence and quality of ctDNA. For example, the amplification of single gene loci, or else, a whole genome sequencing approach (42,43). In the first studies on this topic, polymerase chain reaction (PCR)-based amplification of certain mutations, specifically linked to cancer, was utilized (21). Allele-specific PCR makes the trustworthy amplification of certain and well-known cancer-associated mutations possible. Still, sensitivity especially in early-stage lung cancer is very low. Digital PCR poses an improvement, because samples are partitioned into multiple and smaller reactions, which considerably increases sensitivity (22). Conventional as well as digital PCR-based approaches test for a confined number of common and well-defined mutations. Hence, this approach is only applicable for patients with common driver mutations. NGS has expanded the applicability of ctDNA testing, utilizing a combination of multiplexed PCR assays which are designed to intensify only a minor number of hot-spot regions, screening for gene mutations and quantifying the fraction of mutated alleles (24,44). The limitation of this technique is, that only a small number of genes is interrogated and copy number variations or structural variants cannot be detected if the breakpoint sequence has not been previously characterized. There is also an approach combining hybrid capture followed by NGS, which maintains an extremely high level of sensitivity while it allows for a larger variety of gene aberrations to be quantified (45). Additionally, deep sequencing of the whole exome (46), or genome (47,48) can provide comprehensive ctDNA profiling. However, deep sequencing techniques are exclusively used in individuals with an advanced malignant condition, because the costs per sample are considerable, and the sensitivity is rather low.
CTCs
CTCs have to be enriched in blood samples for optimum detection, with can be carried out by label-dependent or label-independent strategies. The label-dependent method targets specific antigens on target cells, using complementary molecules, like antibodies. Conjugation of these complementary molecules is realized by magnetic beads or specific surfaces in a microfluidics platform. In terms of label-dependent approaches, assays based on immuno-magnetism, targeting EpCAM, are applied most widely (49). Label-independent CTC enrichment methods separate CTCs, making use of their physical rather than their biological properties (i.e., size, density, electrical properties, inertial effect of flow). Variations in size are utilized by filtering techniques, as well as the CTCs ability to deform, when compared to other blood cells (50). For example, the cell size in small cell lung cancer (SCLC) and NSCLC ranges from 7.2 to 15 µm in diameter, as it was measured in biopsies (51), however, CTCs can be considerably smaller.
CTCs as tumor markers usually provide a good-quality immunohistochemical staining. Thus, not only EGFR mutations can be outlined (as it is mainly done with ctDNA), but also ALK-, ROS1- and RET rearrangements, MET amplifications, as well as BRAF- and HER2 mutations (52). While it is rather difficult to detect ALK rearrangements in ctDNA (53), ALK status of CTCs can be assessed relatively easily by immunohistochemistry or by fluorescence in situ hybridization (FISH) (54). Still, the applications and studies that involve CTCs in genotyping of lung cancer remain few, as compared to the analysis techniques utilizing ctDNA. A considerable limitation would be the heterogeneous number of CTCs, varying widely among different cancer specimens, but also the fact that physical properties of CTCs change considerably during the disease course. In the majority of patients suffering from metastatic colorectal-, breast- or prostate cancer, CTCs can be outlined with good reliability. In lung cancer, however, CTC detection is still applicable only in selected cases, because only approximately 10% of patients suffering from NSCLC show ≥5 CTCs per 7.5 mL (55).
Testing for EGFR mutations
EGFR gene mutations have been found to occur in 43% of lung adenocarcinomas in subjects with a negative smoking history, and in 11% of lung adenocarcinomas of smokers (56). Worldwide, the highest rates of EGFR mutations have been reported in the Asian ethnicity (57). Multiple randomized trials have confirmed the benefit on progression-free survival (PFS) by treatment with the EGFR TKIs erlotinib, gefitinib and afatinib as compared to chemotherapy in the case of EGFR-mutant metastatic NSCLC (58-60). Thus, testing for EGFR mutations is one of the key factors in molecular analysis of NSCLC, necessary to provide optimum and tailored treatment for each patient. Tissue analysis remains the gold standard in screening for EGFR mutations. However, when tissue samples do not suffice for molecular testing, or the risk of biopsy is too high, liquid biopsy often becomes the only option.
The cobas EGFR Mutation Test v2 is a real-time PCR-based assay, originally applied on formalin-fixed paraffin-embedded tumor specimens (61). In the ENSURE clinical trial, comparing first line erlotinib administration with gemcitabine in combination with cisplatin, the application on plasma samples was validated (60).
The sensitivity of testing for EGFR mutations via ctDNA testing is dependent upon the total amount of ctDNA in the bloodstream, which can vary between patients, but also in one and the same patient at different timepoints. The half-life of ctDNA has been shown to be <2 hours (62). Tumor burden also strongly determines ctDNA concentration, and patients with more extensive disease tend to harbor higher levels of ctDNA (45). Previous data indicate, that extrathoracic disease (M1b) improves the likelihood of detecting EGFR mutations by liquid biopsy, as compared to intrathoracic disease (M1a or M0) (63). Of note, currently available ctDNA tests for EGFR prioritize specificity over sensitivity, so false negative results are far more common than false positives (64). Digital PCR- or NGS-based approaches screening for EGFR mutations may achieve lower detection limits compared to conventional PCR, thus providing a better sensitivity. It has to be kept in mind, that platforms testing for multiple common driver mutations in NSCLC are best suited to interpret negative results for EGFR mutations. For example, if a patient is tested negatively for EGFR mutations, but positive for KRAS mutation, the negative predictive value is much higher, as in a test for EGFR only which comes back negative (42).
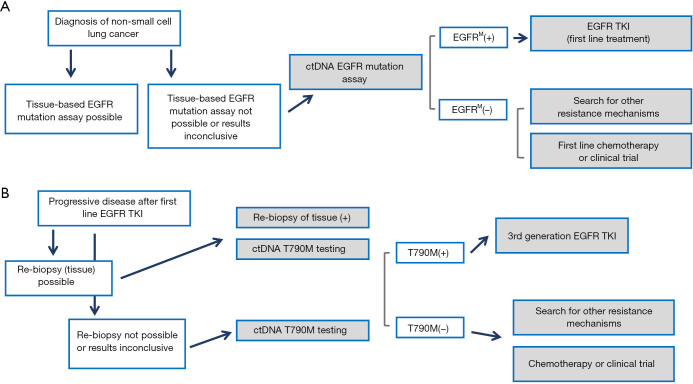
An algorithm showing the current recommendation for EGFR mutation status analysis is illustrated in Figure 1.
Figure 1.
Current paradigm for the use of plasma genotyping in clinical practice. (A) In patients diagnosed with NSCLC, when tissue samples cannot be used for mutation analysis, ctDNA genotyping by means of plasma analysis is recommended. EGFR tyrosine kinase inhibitor (TKI) treatment may be initiated based of plasma ctDNA testing. (B) When patients acquire resistance to first line EGFR TKI treatment, liquid biopsies can compensate tumor biopsies—given the fact that also tissues analysis is often not accurate in diagnosing the complex and heterogeneous pattern of acquired drug resistance. Figure adapted after Huang et al. (3).
Plasma miRNAs, exosomes and tumor-educated platelets (TEPs)
Liquid biopsy not only comprises the analysis of CTCs or ctDNA, but also the analysis of miRNAs, exosomes, tumor-associated antigens and TEPs (65). Small non-coding RNAs, including miRNA, are stabilized by processing proteins in the circulation—contrary to cell-free RNA which is rapidly degraded in the bloodstream. miRNA can be quantified using quantitative reverse transcription PCR (RT-PCR) (66). However, in the previously published studies about the clinical application of miRNA screening as a liquid biopsy technique, different sets of markers and thresholds for positivity have been used and thus, circulating miRNA-based techniques are at present not applicable for everyday clinical routine. In essence, miRNAs are small regulatory RNA molecules (about 22 nt in size), modulating the activity of specific mRNA targets and playing an important role in a vast variety of biochemical processes (67). In a carefully designed study on prostate cancer by Mitchell and colleagues, the role of miRNA as cancer detection biomarkers was analyzed (66). MiRNAs have been shown to be dysregulated in cancers (68), their expression patterns in human cancers are tissue-specific (69), and they are usually highly preserved in formalin-fixed tissue samples (70). The authors of this study assumed that miRNAs exhibit good stability in plasma and serum as well. First, small RNAs with a size range of 18–24 nt were isolated from plasma samples using radioactive labeling (66). A fraction of RNAs with a size of ≈22 nt, characteristic of miRNAs, was isolated from the pool of blood-based RNA. RT-PCR analysis was performed in order to clarify whether these RNAs feature markers characteristic of miRNAs. Indeed, 93% of sequences comprised characteristic miRNA markers, serving as a proof that miRNAs are present in human plasma (66). Three miRNAs (miR-15b, miR-16 and miR-24) were specifically quantified using TaqMan quantitative RT-PCR analysis, and were all readily detectable in the plasma of each analyzed individual. Interestingly, when testing the stability of miRNAs in plasma, incubation of plasma at room temperature for up to 24 hours, or subjecting them to eight cycles of freeze-thawing had but a minimal effect on miRNA levels. Next, the presence of tumor-derived miRNA in plasma was proven in a mouse xenograft model of prostate cancer (71). Two miRNAs, miR-629 and miR-660 were both found to be expressed in prostate cancer cells. Levels of these miRNAs were basically not detectable in healthy control mice, but in each of the tumor-bearing mice, they were readily detected (66). Moreover, levels of these miRNAs were also moderately correlated with tumor mass, indicating that the abundance of plasma miRNAs reflects overall tumor burden. Noteworthy, in this xenograft experiment, miRNAs were found not to be associated with CTCs, as demonstrated by two centrifugation processes, which should pellet any intact cells. However, in the pellet material no miRNAs were found, but still, miRNAs were present in the supernatant remaining after the final 12,000 ×g centrifugation. As a final step, Mitchell et al. sought to outline miRNAs as disease biomarkers in patients suffering from prostate cancer. Candidate miRNAs were outlined based on previously published data on miRNA expression profiles in prostate cancer—and additionally, healthy subjects were screened for these miRNAs in their plasma, and miRNAs that were found in the healthy donors were excluded (66). As a result, miR-100, miR-125b, miR-141, miR-143, miR-205, and miR-296 remained as candidate prostate cancer biomarkers. Next, these candidate miRNAs were analyzed in 25 individuals suffering from metastatic prostate cancer, and in 25 healthy male control subjects. MiR-141 showed the most striking differential expression, being 46-fold overexpressed in the serum of prostate cancer patients, when compared with the healthy controls (66). Notably, serum levels of miR-141 could detect individuals with prostate cancer with a sensitivity of 60% and a specificity of 100%. Moreover, miR-141 and prostate-specific antigen (PSA) values moderately correlated, as shown by Pearson and Spearman correlation analysis.
Also for NSCLC, several studies have focused on the specific impact of circulating miRNAs, when used as biomarkers, applying high-throughput technologies (72,73). An increased expression of miR-29, as well as the decrease of seven miRNAs (miR-146b, miR-221, let-7a, miR-155, miR-17-5, miR-27a and miR-106a) was seen in a RT-PCR analysis in serum samples of early-stage NSCLC patients (74). When a certain panel of miRNAs from the blood plasma (miR-21, miR-126, miR-210 and miR-486-5p) was used as a diagnostic tool for the detection of NSCLC, sensitivity was 86.2% and specificity was 96.6%. Interestingly, adenocarcinomas were more likely diagnosed using these miRNAs, as opposed to squamous cell carcinomas (75). A panel comprising the miRNAs miR-483, miR-193a-3p, miR-25, miR-214 and miR-7 showed a significantly stronger expression in NSCLC patients when compared to control subjects (76). When used as detection tools for early pro-tumorigenic changes in high-risk individuals, circulating miRNA signatures, composed of reciprocal ratios of 24 miRNAs with both diagnostic and prognostic value could be outlined (77). Consecutively, this miRNA Signature Classifier (MSC) has been approved in a cohort comprising 1,000 consecutive blood plasma specimens, stemming from 4,099 tumor patients, showing a good diagnostic performance with a sensitivity of 87% and a specificity of 81% (78). Still, the application of routinely using circulating miRNAs in everyday clinical practice has not been implemented. The main reason is the heterogeneity of existing studies on miRNAs as cancer detection biomarkers, their limited sample size, a lack of prospective analysis and lack of a large external and unbiased validation (79).
Another approach to liquid biopsy is through exosome analysis (80). Exosomes are defined as round, nano-sized vesicles, their diameter ranging between 40 and 100 nm, and their density being about 1.13–1.19 g/mL (81). Immune cells, stem cells, as well as malignant cells count among the cell species that routinely release exosomes (82). Exosomes are created via the endocytic pathway, and certain multivesicular bodies, stemming from early endosome maturation, release them into the extracellular space. The release of exosomes takes place under the endosomal sorting complex, which is required for their transport, and with the aid of related proteins (83). Multivesicular bodies can fuse with the plasma membrane, inducing the release of exosomes into the extracellular space (84). Because of their endocytic origin, the composition of exosomes reflects that of their parental cells. Moreover, the exosomes’ lipid bilayer is stable and relatively resistant to degradation, allowing for the identification of the original cells (85). Tumor-derived exosomes are related to tumor progression (86). Exosome function in tumor cells may considerably differ from normal cells, and exosomes are generally more abundant in tumor cells. Exosome levels were found to be upregulated in body fluids of lung cancer patients, which means that exosomes might play a crucial role in the development and disease progression of lung cancer (80). One study (87) assessed the association of circulating cancer exosome levels, exosomal small RNAs and exosomal miRNAs with the presence of lung adenocarcinoma, and their influence on prognosis. Plasma samples of 27 individuals were analyzed. Exosome levels were indeed higher in the adenocarcinoma- compared to the control group, with mean plasma values of 2.85 versus 0.77 mg/mL, respectively. Rodriguez and colleagues demonstrated recently that exosome levels in bronchoalveolar lavage samples from lung cancer patients were significantly elevated, when compared to that of non-tumor bearing subjects (88). In a study on lung cancer cells, the TP53 pathway was found to be involved in exosome secretion into the medium (89). In general, tumor-derived exosomes play an important role in tumor angiogenesis and invasion, and they have the potential to proliferate in receptor cells, thereby facilitating disease progression and metastatic spread (90). In recent years, research has put increasing focus on the investigation of certain molecules in exosomes, which form the exosomes’ pathophysiological properties. Thereby it was found that tumor plasma contained a higher level of exosome-related miRNA, meaning that altered molecular profiles of exosomes are possibly involved in tumor-associated biological processes (80). The exosomal cargo, which is usually significantly dysregulated in cancer tissue, may serve as a diagnostic, prognostic and predictive biomarker for lung cancer, as accumulating evidence suggests. Modification of receptor cells by tumor-derived exosomes is a likely event in carcinogenesis, as shown in a study by Wang et al., where treatment with tumor-derived exosomes led to a differential expression of long non-coding RNAs and of protein-coding mRNA as well (91). All relevant exosome-related biomarker studies published have focused primarily on exosome-derived miRNAs and proteins. For instance, Cazzoli et al. outlined 742 different miRNAs for an in-depth analysis, comparing 10 patients with lung adenocarcinoma, 10 with lung granulomas and 10 current smokers with no known lung disease (92). Based on their findings, the authors of this study developed a screening panel comprising four miRNAs (miR-378a, miR-379, miR-139-5p and miR-200b-5p) to distinguish potentially malignant lung nodules from non-nodules. The panel showed 87.5% sensitivity and 72% specificity (92). Additionally, a diagnostic panel was developed, comprising six miRNAs (miR151a-5p, miR-30a-3p, miR-200b-5p, miR-629, miR-100, and miR-154-3p), to discriminate between lung adenocarcinoma and -granuloma. With this screening panel, a sensitivity of 96% and a specificity of 60% was achieved. Another research group (93) established a screening panel which consists of six miRNAs (miR-19b-3p, miR-21-5p, miR-221-3p, miR-409-3p, miR-425-5p, and miR-584-5p), for detection in plasma samples for the diagnosis of lung adenocarcinoma. In particular, miR-19-3p, miR-21-5p, and miR-221-3p were strikingly upregulated in exosomes, which had been extracted from peripheral blood plasma from patients with lung adenocarcinoma (93).
Apart from miRNAs, the peripheral blood of cancer patients is also a pool of so-called TEPs (44). Healthy cells present in the tumor microenvironment are continuously released into the blood stream of cancer patients, and TEPs count among them (94). There is a well-known interaction between blood platelets and tumor cells, affecting tumor growth and dissemination (95). This interaction affects the RNA profile of blood platelets (94). In a study from 2015, mRNA sequencing of TEPs from 283 platelet samples was performed, and sequencing profiles of 228 patients with localized and metastatic disease were compared to profiles of 55 healthy controls. TEP sequencing could distinguish tumor patients from the healthy individuals with 96% accuracy (94). Across six different tumor types (among them also NSCLC), the location of the primary tumor could be determined with an accuracy of 71%. Tumor-specific educational programs in TEPs were primarily dependent on tumor type, less so by tumor stage or the presence of metastases. Still, previous research has shown an influence of blood platelets on tumor cell dissemination (96), as well as metastatic spread (95). Although the above-mentioned study on TEPs suggests a possible future application as another liquid biopsy approach, more research on larger patient cohorts is still warranted until TEP-based liquid biopsy techniques are ready for implementation into clinical routine.
Liquid biopsy in today’s everyday clinical practice
Currently, applications for liquid biopsy testing in NSCLC are increasingly emerging. CfDNA testing is mainly performed in patients with early-stage and curable disease (97). The presence of circulating tumor cfDNA after treatment with a curative intention, strongly correlates with minimal or molecular residual disease (45,98). Recurrence of disease is predicted by the presence of cfDNA, signifying a worse outcome. A recent study evaluated the benefit of consolidation immunotherapy in patients with locally advanced NSCLC who underwent chemo-radiotherapy with a curative intention (42). Consolidation therapy improved outcomes in patients with cfDNA minimal residual disease-positive status, whereas patients who had no evidence of minimal residual disease in cfDNA assessment, no benefit of consolidation immunotherapy was seen. Hence, cfDNA status as an indicator of minimal residual disease could guide decisions regarding adjuvant therapy after curative-intent treatment regimens (97). According to recent investigations, about 40–50% of early-stage and curable NSCLC may be detected using NGS-based cfDNA screening techniques (45).
In a study by Heeke et al., an assessment of routine clinical practice of liquid biopsy for testing of EGFR status in NSCLC was conducted in a single-center trial (99). The authors analyzed 345 blood plasma samples by means of the US Food and Drug Administration (FDA) approved Cobas EGFR mutation test V2, and 103 samples by means of the Therascreen EGFR Plasma RSQ PCR Kit. The trial was performed over the time period of three years (comprising in total 395 plasma samples of 324 patients). Eleven plasma samples were independently validated using the Cobas Test, and 130 samples were further analyzed using Stilla digital PCR (99). According to this study, the median time from blood puncture to the validated clinical report of the liquid biopsy was 4 working days. In 119 (30%) of the specimens, an activating EGFR mutation was found. The authors suggest, on the basis of their findings, that the Cobas test kit is more robust and produced more reproducible assays for the detection of EGFR mutations in plasma samples as compared to the Therascreen kit. Thus, it was primarily implemented as the principal testing device (99). The additional use of digital polymerase chain reaction (dPCR) testing improved the detection of T790M mutations in the plasma samples, but not the detection of primary EGFR mutations. In this analysis, it was evident that liquid biopsies were mostly recommended for patients receiving EGFR TKI treatment. PCR-based assays were specifically requested for 183 (46%) of cases to search for T790M mutations – mostly because of progressive disease upon EGFR-TKI treatment. In 144 (36%) of cases, PCR-based assays were requested due to the impossibility of acquiring a conventional tissue sample (99). The authors conclude that PCR-based assays are effective, especially when used supplementary to the Cobas EGFR mutation test kit. Turnaround time from blood taking to clinical report is short in liquid biopsy, and acceptance in patients is high because of the limited invasiveness and small risk of the procedure. The most effective use, according to the authors, is the search for resistance mutations in patients receiving EGFR-targeted treatment. Another promising opportunity for the use of liquid biopsy is NGS analysis of cfDNA, for the follow-up of the genomic profile of the tumor over the disease course (99).
In 2018, the IASLC released a consensus paper with their current recommendations on performing liquid biopsy in lung cancer (37). According to the IASLC, liquid biopsy comprises CTCs, circulating exosomes, platelet RNA and circulating tumor RNA (ctRNA) as well (4,100). CtDNA still remains the most widely investigated method in liquid biopsy for lung cancer patients. It is crucial to point out, that the accessibility to different techniques, platforms, test reimbursements and drugs varies considerably between different countries and treatment facilities. Thus, the current standard of care and the clinicians’ and pathologists’ experience in a given treatment center have to be taken into account when giving recommendations on liquid biopsy as well.
At present, the difference between serum- and plasma analysis for variant detection in ctDNA is under investigation (101,102). Randomized clinical phase III trials are currently ongoing for gefitinib (IPASS) and afatinib (LUX-Lung 3), which examine the EGFR mutation status in tumor DNA extracted from standard tissue biopsy specimens, comparing the results to EGFR analysis of ctDNA extracted from serum liquid biopsy specimens (5,103). Sensitivity levels of ctDNA EGFR mutation analysis using a real-time PCR (qPCR)-based assay, according to these studies, added up to 43.1%, considering tissue-based analysis as a reference. In the phase IV clinical trial for gefitinib (IFUM) and the phase III trials for afatinib (LUX-Lung 6 and 3), EGFR mutation status was determined in plasma samples (103-105). Sensitivity of plasma tests in these studies was 65.7% and 60.5%, respectively (the mutation detection rate using serum samples, however, was considerably lower with only 28.6%) (106).
In the ASSESS trial, which included 1,288 patients from Japan and Europe, the concordance among plasma samples and tissue-based analysis of EGFR mutations was 89%, showing a high specificity of 97%, yet a low sensitivity of only 46% (107). In the EURTAC phase III study assessing erlotinib, the feasibility of ctDNA testing from blood samples (serum and plasma for each patient), evaluation of the EGFR mutation status and consecutive correlation with outcome, was carried out (59). In ctDNA drawn from 97 blood samples at baseline, EGFR mutation status was evaluated by means of a peptide nucleic acid-mediated qPCR assay, and EGFR mutations were discovered in 78% of patients. The specificity, according to this study, reached 100%. In this analysis, both serum and plasma samples were used for analysis, however, the authors did not state whether there was a difference between the two options (59).
For everyday clinical practice, the IASLC recommends the following claims, based on the data mentioned above (4):
First, plasma analysis is preferred over serum for ctDNA extraction. Second, the maximum time limit from blood taking to the extraction of plasma must not exceed two hours in case of EDTA tubes, and three days in case of special ctDNA sampling tubes. Third, a double-spin plasma isolation technique is strongly suggested. Fourth, blood samples must not be frozen if the plasma has not yet been extracted, irrespective of the type of sampling tube. Fifth, EDTA tubes and cfDNA preservative tubes can both be used for ctDNA extraction – however, if using EDTA tubes, rapid sample processing is crucial. Sixth, two standard 10 mL tubes per patients should be drawn to enhance the accuracy of analysis. Seventh, DNA extraction has to be carried out with protocols or kits specifically designed for small fragmented DNA (4).
Table 1 illustrates different liquid biopsy products currently tested in clinical studies, or already applied in everyday clinical routine.
Table 1. List of liquid biopsy companies (108).
| Company | Product/clinical trial | Mode of application | Reference |
|---|---|---|---|
| Grail | SUMMIT trial | Blood test aiming at the detection of various cancer types, including lung cancer | (109) |
| Guardant 360 | Lunar-2 | Detection of early-stage malignant disease in asymptomatic subjects at increased risk | (110) |
| Freenome | Screening test | An algorithm for the early detection of colorectal carcinoma and precancerous lesions of the colon, i.e., advanced colon adenomas | (111) |
| Biocept | Target SelectorTM ctDNA EGFR Kit | Detection of EGFR mutations | (112) |
| Inivata | InVisionSeqTM; InVisionFirstTM-Lung | A panel comprising about 40 biomarkers (mutations, CNVs, SNVs, fusions and indels) for the molecular profiling, monitoring and diagnosis of advanced cancers | (113) |
| Cynvenio | The LiquidBiopsy® Platform | NGS-based technique for the detection of mutations; very high sensitivity (only 1 target cell per mL/blood required) | (114) |
| CellMaxLife | FirstSightCRCTM | CTCs are used for colorectal cancer profiling, and for screening of colorectal adenomas and colorectal cancer | (115) |
| Exosomedx | Exosome-based biomarker tests | Used for the diagnosis of non-small cell lung cancer and prostate cancer | (116) |
| Biodesix | GeneStrat® test | Providing blood-based mutation results of EGFR, ALK, ROS1, RET, BRAF and KRAS for cancer diagnosis | (117) |
| Personal Genome Diagnostics | PlasmaSELECTTM-R 64 | Cancer diagnosis using NGS with a panel of 64 genes | (118) |
Table adapted after Chen and Zhao (108).
Discussion and conclusion
Although today still sporadically used in clinical routine, liquid biopsy gains increasing popularity. Various different approaches and techniques are already available, differing considerably with regards to cost and level of sensitivity.
Liquid biopsy may be used for the primary assessment of mutations status, with a special focus on EGFR mutations – or else, as a screening tool during the disease course to assess treatment response or the secondary development of resistance mutations.
With this review of the literature, we sought to shed light onto the complex landscape of liquid biopsy techniques, and current applicability. In future, it is likely that the development of evermore precise, as well as cost-efficient liquid biopsy approaches will allow it to become standard in everyday practice, probably replacing tissue biopsy at large part. Thus, complication rates for patients could be minimized and constant disease monitoring would become considerably easier.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-21-3
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-21-3
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-21-3). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.The American Cancer Society. Cancer Facts & Figures 2020. Atlanta: 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang WL, Chen YL, Yang SC, et al. Liquid biopsy genotyping in lung cancer: ready for clinical utility? Oncotarget 2017;8:18590-608. 10.18632/oncotarget.14613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. 10.1016/j.jtho.2018.05.030 [DOI] [PubMed] [Google Scholar]
- 5.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017;376:629-40. 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. 10.1200/JCO.2012.44.2806 [DOI] [PubMed] [Google Scholar]
- 8.Yang JC, Ahn MJ, Kim DW, et al. Osimertinib in pretreated T790M-positive advanced non–small-cell lung cancer: AURA study phase II extension component. J Clin Oncol 2017;35:1288-96. 10.1200/JCO.2016.70.3223 [DOI] [PubMed] [Google Scholar]
- 9.Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 10.Goss G, Tsai CM, Shepherd FA, et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol 2016;17:1643-52. 10.1016/S1470-2045(16)30508-3 [DOI] [PubMed] [Google Scholar]
- 11.Jia Y, Yun CH, Park E, et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 2016;534:129-32. 10.1038/nature17960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gainor JF, Dardaei L, Yoda S, et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov 2016;6:1118-33. 10.1158/2159-8290.CD-16-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heerink WJ, de Bock GH, de Jonge GJ, Groen HJ, Vliegenthart R, Oudkerk M. Complication rates of CT-guided transthoracic lung biopsy: meta-analysis. Eur Radiol 2017;27:138-48. 10.1007/s00330-016-4357-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prescribing Information - TECENTRIQ (atezolizumab). [Internet]. [cited 25 April 2019]. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/761034s019lbl.pdf
- 15.Hofman P. PD-L1 immunohistochemistry for non-small cell lung carcinoma: which strategy should be adopted? Expert Rev Mol Diagn 2017;17:1097-108. 10.1080/14737159.2017.1398083 [DOI] [PubMed] [Google Scholar]
- 16.Hofman P, Heeke S, Alix-Panabières C, et al. Liquid biopsy in the era of immuno-oncology: is it ready for prime-time use for cancer patients? Ann Oncol 2019;30:1448-59. 10.1093/annonc/mdz196 [DOI] [PubMed] [Google Scholar]
- 17.Ilie M, Long-Mira E, Bence C, et al. Comparative study of the PD-L1 status between surgically resected specimens and matched biopsies of NSCLC patients reveal major discordances: a potential issue for anti-PD-L1 therapeutic strategies. Ann Oncol 2016;27:147-53. 10.1093/annonc/mdv489 [DOI] [PubMed] [Google Scholar]
- 18.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093-104. 10.1056/NEJMoa1801946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heeke S, Hofman P. Tumor mutational burden assessment as a predictive biomarker for immunotherapy in lung cancer patients: getting ready for prime-time or not? Transl Lung Cancer Res 2018;7:631-8. 10.21037/tlcr.2018.08.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018;142:321-46. 10.5858/arpa.2017-0388-CP [DOI] [PubMed] [Google Scholar]
- 21.Chouaid C, Dujon C, Do P, et al. Feasibility and clinical impact of re-biopsy in advanced non small-cell lung cancer: a prospective multicenter study in a real-world setting (GFPC study 12-01). Lung Cancer 2014;86:170-3. 10.1016/j.lungcan.2014.08.016 [DOI] [PubMed] [Google Scholar]
- 22.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. 10.1001/jamaoncol.2016.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai W, Lin D, Wu C, et al. Intratumoral heterogeneity of ALK-rearranged and ALK/EGFR coaltered lung adenocarcinoma. J Clin Oncol 2015;33:3701-9. 10.1200/JCO.2014.58.8293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med 2017;376:2109-21. 10.1056/NEJMoa1616288 [DOI] [PubMed] [Google Scholar]
- 25.Medina Diaz I, Nocon A, Mehnert DH, et al. Performance of Streck cfDNA Blood Collection Tubes for Liquid Biopsy Testing. PLoS One 2016;11:e0166354. 10.1371/journal.pone.0166354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Library of Medicine [Internet].; 2020 [cited 10.12.2020]. Available online: https://pubmed.ncbi.nlm.nih.gov/
- 27.Yong E. Cancer biomarkers: Written in blood. Nature 2014;511:524-6. 10.1038/511524a [DOI] [PubMed] [Google Scholar]
- 28.Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. 10.1056/NEJMoa1408440 [DOI] [PubMed] [Google Scholar]
- 29.Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014;289:3869-75. 10.1074/jbc.C113.532267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766-9. 10.1038/cr.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol 2012;7:115-21. 10.1097/JTO.0b013e3182307f98 [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Lu Y, Zhu G, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol 2013;66:1065-9. 10.1136/jclinpath-2013-201728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinde I, Wu J, Papadopoulos N, et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:9530-5. 10.1073/pnas.1105422108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Sun J, Lin CC, et al. The emerging landscape of salivary diagnostics. Periodontol 2000 2016;70:38-52. 10.1111/prd.12099 [DOI] [PubMed] [Google Scholar]
- 35.Su YH, Wang M, Brenner DE, et al. Detection of mutated K-ras DNA in urine, plasma, and serum of patients with colorectal carcinoma or adenomatous polyps. Ann N Y Acad Sci 2008;1137:197-206. 10.1196/annals.1448.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reckamp KL, Melnikova VO, Karlovich C, et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J Thorac Oncol 2016;11:1690-700. 10.1016/j.jtho.2016.05.035 [DOI] [PubMed] [Google Scholar]
- 37.Nikolaev S, Lemmens L, Koessler T, et al. Circulating tumoral DNA: Preanalytical validation and quality control in a diagnostic laboratory. Anal Biochem 2018;542:34-9. 10.1016/j.ab.2017.11.004 [DOI] [PubMed] [Google Scholar]
- 38.Lam NY, Rainer TH, Chiu RW, et al. EDTA is a better anticoagulant than heparin or citrate for delayed blood processing for plasma DNA analysis. Clin Chem 2004;50:256-7. 10.1373/clinchem.2003.026013 [DOI] [PubMed] [Google Scholar]
- 39.Kang Q, Henry NL, Paoletti C, et al. Comparative analysis of circulating tumor DNA stability In K(3)EDTA, Streck, and CellSave blood collection tubes. Clin Biochem 2016;49:1354-60. 10.1016/j.clinbiochem.2016.03.012 [DOI] [PubMed] [Google Scholar]
- 40.Sherwood JL, Corcoran C, Brown H, et al. Optimised Pre-Analytical Methods Improve KRAS Mutation Detection in Circulating Tumour DNA (ctDNA) from Patients with Non-Small Cell Lung Cancer (NSCLC). PLoS One 2016;11:e0150197. 10.1371/journal.pone.0150197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mack P, Banks KC, Riess JW, et al. WCLC 2016 — Clinical utility of circulating tumor DNA (ctDNA) analysis by digital next generation sequencing of over 5,000 advanced NSCLC patients. J Thorac Oncol 2017;12:263-4. 10.1016/j.jtho.2016.11.256 [DOI] [Google Scholar]
- 42.Moding EJ, Diehn M, Wakelee HA. Circulating tumor DNA testing in advanced non-small cell lung cancer. Lung Cancer 2018;119:42-7. 10.1016/j.lungcan.2018.02.019 [DOI] [PubMed] [Google Scholar]
- 43.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 44.Joosse SA, Pantel K. Tumor-Educated Platelets as Liquid Biopsy in Cancer Patients. Cancer Cell 2015;28:552-4. 10.1016/j.ccell.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 45.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. 10.1038/nature12065 [DOI] [PubMed] [Google Scholar]
- 47.Leary RJ, Sausen M, Kinde I, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 2012;4:162ra154. 10.1126/scitranslmed.3004742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heitzer E, Ulz P, Belic J, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med 2013;5:30. 10.1186/gm434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623-31. 10.1038/nrc3820 [DOI] [PubMed] [Google Scholar]
- 50.Zheng S, Lin HK, Lu B, et al. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices 2011;13:203-13. 10.1007/s10544-010-9485-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vollmer RT. The effect of cell size on the pathologic diagnosis of small and large cell carcinomas of the lung. Cancer 1982;50:1380-3. [DOI] [PubMed] [Google Scholar]
- 52.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol 2014;11:473-81. 10.1038/nrclinonc.2014.104 [DOI] [PubMed] [Google Scholar]
- 53.Ilie M, Hofman V, Long E, et al. Current challenges for detection of circulating tumor cells and cell-free circulating nucleic acids, and their characterization in non-small cell lung carcinoma patients. What is the best blood substrate for personalized medicine? Ann Transl Med 2014;2:107,5839.2014.08.11. [DOI] [PMC free article] [PubMed]
- 54.Pailler E, Adam J, Barthélémy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol 2013;31:2273-81. 10.1200/JCO.2012.44.5932 [DOI] [PubMed] [Google Scholar]
- 55.Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 2012;7:306-15. 10.1097/JTO.0b013e31823c5c16 [DOI] [PubMed] [Google Scholar]
- 56.Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 2012;18:6169-77. 10.1158/1078-0432.CCR-11-3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res 2015;5:2892-911. [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. 10.1016/S1470-2045(11)70184-X [DOI] [PubMed] [Google Scholar]
- 59.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 60.Wu YL, Zhou C, Liam CK, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol 2015;26:1883-9. 10.1093/annonc/mdv270 [DOI] [PubMed] [Google Scholar]
- 61.O'Donnell P, Ferguson J, Shyu R, et al. Analytic performance studies and clinical reproducibility of a real-time PCR assay for the detection of epidermal growth factor receptor gene mutations in formalin-fixed paraffin-embedded tissue specimens of non-small cell lung cancer. BMC Cancer 2013;13:210. 10.1186/1471-2407-13-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. 10.1038/nm.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR Mutation Status in Matched Plasma and Tumor Tissue of NSCLC Patients from a Phase I Study of Rociletinib (CO-1686). Clin Cancer Res 2016;22:2386-95. 10.1158/1078-0432.CCR-15-1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Normanno N, Denis MG, Thress KS, et al. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget 2017;8:12501-16. 10.18632/oncotarget.13915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liam CK, Mallawathantri S, Fong KM. Is tissue still the issue in detecting molecular alterations in lung cancer? Respirology 2020;25:933-43. 10.1111/resp.13823 [DOI] [PubMed] [Google Scholar]
- 66.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell 2006;11:441-50. 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 68.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69. 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- 69.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 70.Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 2007;13:1668-74. 10.1261/rna.642907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Attardi BJ, Burgenson J, Hild SA, et al. Steroid hormonal regulation of growth, prostate specific antigen secretion, and transcription mediated by the mutated androgen receptor in CWR22Rv1 human prostate carcinoma cells. Mol Cell Endocrinol 2004;222:121-32. 10.1016/j.mce.2004.04.013 [DOI] [PubMed] [Google Scholar]
- 72.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- 73.Foss KM, Sima C, Ugolini D, et al. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol 2011;6:482-8. 10.1097/JTO.0b013e318208c785 [DOI] [PubMed] [Google Scholar]
- 74.Heegaard NH, Schetter AJ, Welsh JA, et al. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer 2012;130:1378-86. 10.1002/ijc.26153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen J, Todd NW, Zhang H, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest 2011;91:579-87. 10.1038/labinvest.2010.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang P, Yang D, Zhang H, et al. Early Detection of Lung Cancer in Serum by a Panel of MicroRNA Biomarkers. Clin Lung Cancer 2015;16:313,9.e1. [DOI] [PubMed]
- 77.Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A 2011;108:3713-8. 10.1073/pnas.1100048108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sozzi G, Boeri M, Rossi M, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol 2014;32:768-73. 10.1200/JCO.2013.50.4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fortunato O, Gasparini P, Boeri M, et al. Exo-miRNAs as a New Tool for Liquid Biopsy in Lung Cancer. Cancers (Basel) 2019;11:888. 10.3390/cancers11060888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cui S, Cheng Z, Qin W, et al. Exosomes as a liquid biopsy for lung cancer. Lung Cancer 2018;116:46-54. 10.1016/j.lungcan.2017.12.012 [DOI] [PubMed] [Google Scholar]
- 81.Taverna S, Giallombardo M, Gil-Bazo I, et al. Exosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: critical analysis of evidence and potential role in clinical practice. Oncotarget 2016;7:28748-60. 10.18632/oncotarget.7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ventimiglia LN, Alonso MA. Biogenesis and function of t cell-Derived exosomes. Front Cell Dev Biol 2016;4:84. 10.3389/fcell.2016.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 2014;29:116-25. 10.1016/j.ceb.2014.05.004 [DOI] [PubMed] [Google Scholar]
- 84.Sahu R, Kaushik S, Clement CC, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell 2011;20:131-9. 10.1016/j.devcel.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Molina-Vila MA, Mayo-de-Las-Casas C, Giménez-Capitán A, et al. Liquid Biopsy in Non-Small Cell Lung Cancer. Front Med (Lausanne) 2016;3:69. 10.3389/fmed.2016.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200:373-83. 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rabinowits G, Gerçel-Taylor C, Day JM, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009;10:42-6. 10.3816/CLC.2009.n.006 [DOI] [PubMed] [Google Scholar]
- 88.Rodríguez M, Silva J, López-Alfonso A, et al. Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes Chromosomes Cancer 2014;53:713-24. 10.1002/gcc.22181 [DOI] [PubMed] [Google Scholar]
- 89.Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res 2006;66:4795-801. 10.1158/0008-5472.CAN-05-4579 [DOI] [PubMed] [Google Scholar]
- 90.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev 2013;32:623-42. 10.1007/s10555-013-9441-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang S, Li X, Zhu R, et al. Lung cancer exosomes initiate global long non-coding RNA changes in mesenchymal stem cells. Int J Oncol 2016;48:681-9. 10.3892/ijo.2015.3272 [DOI] [PubMed] [Google Scholar]
- 92.Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol 2013;8:1156-62. 10.1097/JTO.0b013e318299ac32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou X, Wen W, Shan X, et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget 2017;8:6513-25. 10.18632/oncotarget.14311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Best MG, Sol N, Kooi I, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015;28:666-76. 10.1016/j.ccell.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuznetsov HS, Marsh T, Markens BA, et al. Identification of luminal breast cancers that establish a tumor-supportive macroenvironment defined by proangiogenic platelets and bone marrow-derived cells. Cancer Discov 2012;2:1150-65. 10.1158/2159-8290.CD-12-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell 2011;20:576-90. 10.1016/j.ccr.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang SR, Schultheis AM, Yu H, et al. Precision medicine in non-small cell lung cancer: Current applications and future directions. Semin Cancer Biol 2020. [Epub ahead of print]. doi: . 10.1016/j.semcancer.2020.07.009 [DOI] [PubMed] [Google Scholar]
- 98.Chae YK, Oh MS. Detection of Minimal Residual Disease Using ctDNA in Lung Cancer: Current Evidence and Future Directions. J Thorac Oncol 2019;14:16-24. 10.1016/j.jtho.2018.09.022 [DOI] [PubMed] [Google Scholar]
- 99.Heeke S, Benzaquen J, Hofman V, et al. Critical Assessment in Routine Clinical Practice of Liquid Biopsy for EGFR Status Testing in Non-Small-Cell Lung Cancer: A Single-Laboratory Experience (LPCE, Nice, France). Clin Lung Cancer 2020;21:56,65.e8. [DOI] [PubMed]
- 100.Raez LE, Manca P, Rolfo C, et al. ROS-1 Rearrangements in Circulating Tumor Cells. J Thorac Oncol 2018;13:e71-2. 10.1016/j.jtho.2017.11.127 [DOI] [PubMed] [Google Scholar]
- 101.Lee TH, Montalvo L, Chrebtow V, Busch MP. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion 2001;41:276-82. 10.1046/j.1537-2995.2001.41020276.x [DOI] [PubMed] [Google Scholar]
- 102.Page K, Powles T, Slade MJ, et al. The importance of careful blood processing in isolation of cell-free DNA. Ann N Y Acad Sci 2006;1075:313-7. 10.1196/annals.1368.042 [DOI] [PubMed] [Google Scholar]
- 103.Wu YL, Sequist LV, Tan EH, et al. Afatinib as First-line Treatment of Older Patients With EGFR Mutation-Positive Non-Small-Cell Lung Cancer: Subgroup Analyses of the LUX-Lung 3, LUX-Lung 6, and LUX-Lung 7 Trials. Clin Lung Cancer 2018;19:e465-79. 10.1016/j.cllc.2018.03.009 [DOI] [PubMed] [Google Scholar]
- 104.Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. 10.1016/S1470-2045(13)70604-1 [DOI] [PubMed] [Google Scholar]
- 105.Douillard JY, Ostoros G, Cobo M, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer 2014;110:55-62. 10.1038/bjc.2013.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Malapelle U, Sirera R, Jantus-Lewintre E, et al. Profile of the Roche cobas® EGFR mutation test v2 for non-small cell lung cancer. Expert Rev Mol Diagn 2017;17:209-15. 10.1080/14737159.2017.1288568 [DOI] [PubMed] [Google Scholar]
- 107.Reck M, Popat S, Reinmuth N, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25 Suppl 3:iii27-39. 10.1093/annonc/mdu199 [DOI] [PubMed] [Google Scholar]
- 108.Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics 2019;13:34-8. 10.1186/s40246-019-0220-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Inc. Grail. ESUMMIT Study. [Internet].; 2018 [cited 21 Feb 2019]. Available online: https://grail.com/clinical-studies/summitstudy/
- 110.Inc. Guardant Health. Early detection LUNAR-2. [Internet].; 2018 [cited 21 Feb 2019]. Available online: https://guardanthealth. com/solutions/#lunar-2
- 111.Freenome. Clinical studies. [Internet].; 2019 [cited 21 Feb 2019]. Available online: https://www.freenome.com/clinicalstudies
- 112.Inc. Biocept. Target Selector™ ctDNA EGFR Kit*. [Internet].; 2019 [cited 21 Feb 2019]. Available online: https://biocept.com/ egfr-kit/
- 113.Inivata. Our products. [Internet].; 2019 [cited 21 Feb 2019]. Available online: https://www.inivata.com/our-products/
- 114.Inc. Cynvenio Biosystems. The LiquidBiopsy® Platform. [Internet]. 2019 [cited 21 Feb 2019]. Available online: https://www. cynvenio.com/instrumentation
- 115.CellMax Life. FirstSightCRC. [Internet].; 2019 [cited 21 Feb 2019]. Available online: https://cellmaxlife.com/test-firstsight-crc/
- 116.Exosome Diagnostics. Our diagnostics: for patients. [Internet].; 2019 [cited 21 Feb 2019]. Available online: http://www. exosomedx.com/patients
- 117.Inc. Biodesix. Genomic blood test. [Internet].; 2019 [cited 21 Feb 2019]. Available online: https://www.biodesix.com/ products/biodesix-lung-reflex/genestrat-2/
- 118.Inc. Personal Genome Diagnostics. Liquid biopsy. [Internet].; 2019 [cited 21 Feb 2019]. Available online: https://www. personalgenome.com/cap-clia.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as