SUMMARY
The immune response to a chronic viral infection is uniquely tailored to balance viral control and immunopathology. The role of myeloid cells in shaping the response to chronic viral infection, however, is poorly understood. We perform single-cell RNA sequencing of myeloid cells during acute and chronic lymphocytic choriomeningitis virus (LCMV) infection to address this question. Our analysis identifies a cluster of suppressive neutrophils that is enriched in chronic infection. Furthermore, suppressive neutrophils highly express the gene encoding Proviral integration site for Moloney murine leukemia virus-1 (PIM1), a kinase known to promote mitochondrial fitness and cell survival. Pharmacological inhibition of PIM1 selectively diminishes suppressive neutrophil-mediated immunosuppression without affecting the function of monocytic myeloid-derived suppressor cells (M-MDSCs). Decreased accumulation of suppressive neutrophils leads to increased CD8 T cell function and viral control. Mechanistically, PIM kinase activity is required for maintaining fused mitochondrial networks in suppressive neutrophils, but not in M-MDSCs, and loss of PIM kinase function causes increased suppressive neutrophil apoptosis.
In brief
Volberding et al. use scRNA-seq to identify and characterize a population of suppressive neutrophils that are present during chronic viral infection. Moreover, PIM1 is vital for the metabolic fitness and survival of suppressive neutrophils. Inhibition of PIM kinase reduces suppressive neutrophils and increases CD8 T cell function and viral control.
Graphical Abstract
INTRODUCTION
An immune response to a persistent viral infection requires functional adaptations that are tailored toward controlling viral replication while limiting destruction of vital tissues (Brooks et al., 2005; Kim and Ahmed, 2010; Pauken and Wherry, 2015). Among these are cells that have an anti-inflammatory function needed to attenuate the response to pathogens (Dorhoi and Du Plessis, 2018; Veiga-Parga et al., 2013). Recently, the tremendous plasticity of myeloid cells has become apparent; identification of myeloid-derived suppressor cells (MDSCs) demonstrates that myeloid cells exhibit inflammatory and immunosuppressive phenotypes (Dilek et al., 2012; Veglia et al., 2018). Supporting the notion that myeloid cell function is complex and keenly adapted to suit different immune challenges, suppressive myeloid cells are also present during chronic viral infections; however, the role of these cells during chronic infection is not completely understood (Dorhoi and Du Plessis, 2018; Medina and Hartl, 2018; Peñaloza et al., 2019).
MDSCs are a heterogenous population of immature myeloid cells that possess the ability to suppress T cell function (Almand et al., 2001; Dilek et al., 2012; Raber et al., 2014). This broad designation encompasses monocytic MDSCs (M-MDSCs), which have the surface marker phenotype CD11b+ F4/80− Ly6C+ Ly6G− in mice, and suppressive neutrophils, which are CD11b+ F4/80− Ly6C+ Ly6G+ in mice (Veglia et al., 2018; Zhou et al., 2018). It is challenging to study these individual cell types because they share surface markers with conventional monocytes and neutrophils. Furthermore, there is no consensus on how to separate conventional neutrophils from suppressive neutrophils. In humans, LOX1 is a potential marker that separates conventional neutrophils from suppressive neutrophils (Condamine et al., 2016); however, markers have yet to be identified that separate these cells in mice. Moreover, phenotypic designation of all CD11b+ F4/80− Ly6C+ Ly6G+ cells as polymorphonuclear leukocyte (PMN)-MDSCs supposes that there is no heterogeneity in the neutrophil compartment and that all neutrophils have a suppressive phenotype. For these reasons, in contrast to conventional inflammatory neutrophils, we refer to CD11b+ F4/80− Ly6C+ Ly6G+ cells with immunosuppressive activity as suppressive neutrophils. In addition to these surface markers, enhanced expression of STAT3 and its downstream target genes is considered to be indicative of a suppressive neutrophil-like signature (Gabrilovich and Nagaraj, 2009; Kortylewski et al., 2005; Nefedova et al., 2004, 2005). Understanding the mechanisms that regulate MDSC differentiation and function may enhance development of treatments for chronic viral infections.
Suppressive neutrophils can suppress T cells by producing prostaglandin E2 (PGE2), expressing programmed death ligand 1 (PD-L1), and reactive oxygen and nitrogen species (ROS and RNS, respectively) (Veglia et al., 2018). Of these, ROS production appears to be uniquely required for suppressive neutrophils to reduce T cell activation and proliferation in certain contexts (Youn et al., 2008). It is well understood that ROS have the ability to damage proteins, lipids, and nucleic acids (Redza-Dutordoir and Averill-Bates, 2016). In addition, sustained ROS and RNS production is detrimental to cellular metabolism through destruction of critical prosthetic groups, such as Rieske iron sulfur clusters present on enzymes in the electron transport chain (ETC) (Bulteau et al., 2003; Flint et al., 1993; Rose and O’Connell, 1967). Therefore, damage to mitochondria by ROS has the potential to induce apoptosis. Nevertheless, how suppressive neutrophils intrinsically mitigate the damaging effects of producing ROS is not understood.
STAT3 signaling is necessary for the differentiation and function of MDSCs, but the targets of this transcription factor that mediate the pathological activation and function of these cells have not been completely explored (Condamine et al., 2015; Gabrilovich and Nagaraj, 2009; Kortylewski et al., 2005). The PIM kinase family includes three isoforms, PIM1, PIM2, and PIM3, all of which are serine/threonine kinases that are expressed in response to Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling and are known to promote the survival of normal and transformed cells (Jinesh et al., 2016; Warfel and Kraft, 2015). Indeed, the viability of prostate cancer cells treated with a pharmacological inhibitor of PIM kinases is decreased in response to ROS (Warfel et al., 2016). Furthermore, PIM1 appears to be vitally important for mitochondrial fitness in cardiomyocytes by preventing mitochondrial fragmentation (Din et al., 2013, 2014; Samse et al., 2015). There is a strong correlation between mitochondrial morphology and function, where fused mitochondrial networks are generally more oxidative with increased mitochondrial membrane potential (Legros et al., 2002). In contrast, mitochondrial fragmentation is a key initial step of mitophagy and apoptosis (Cribbs and Strack, 2007).
Functional adaptation of T cell differentiation during chronic viral infection has been well studied (Fuller et al., 2004; Matloubian et al., 1994; Wherry et al., 2003; Zajac et al., 1998). However, how myeloid cells negatively regulate T cell function and differentiation in this context is not completely understood. Notably, reducing MDSC numbers or function may increase T cell effector function and improve viral control, indicating that MDSCs may play an important but largely unexplored role in the immune response to persistent infection (Dorhoi and Du Plessis, 2018; Dorhoi et al., 2019; Norris et al., 2013). Furthermore, it is unclear whether there are multiple transcriptionally and functionally distinct MDSC populations during chronic viral infection.
To address the role of myeloid cells in regulation of immune responses to chronic viral infection, we performed single-cell RNA sequencing (scRNA-seq) on myeloid cells isolated from mice with acute or persistent lymphocytic choriomeningitis virus (LCMV) infection. We identified two distinct subsets of neutrophils during both acute and persistent LCMV infection. These two subsets include a mature conventional neutrophil population and a second subset of suppressive neutrophils/polymorphonuclear leukocyte (PMN)-MDSCs. The suppressive neutrophils exhibited high expression of Pim1, a gene whose expression is dependent on STAT3 signaling. In support of this observation, treatment of chronic LCMV clone 13-infected mice with a PIM kinase inhibitor selectively reduced suppressive neutrophil-mediated immunosuppression, which led to increased CD8 T cell function and viral control. We showed that, mechanistically, suppressive neutrophils treated with PIM kinase inhibitor have increased fragmentation of mitochondria, which correlates with increased death of suppressive neutrophils.
RESULTS
scRNA-seq reveals heterogeneity in the myeloid compartment
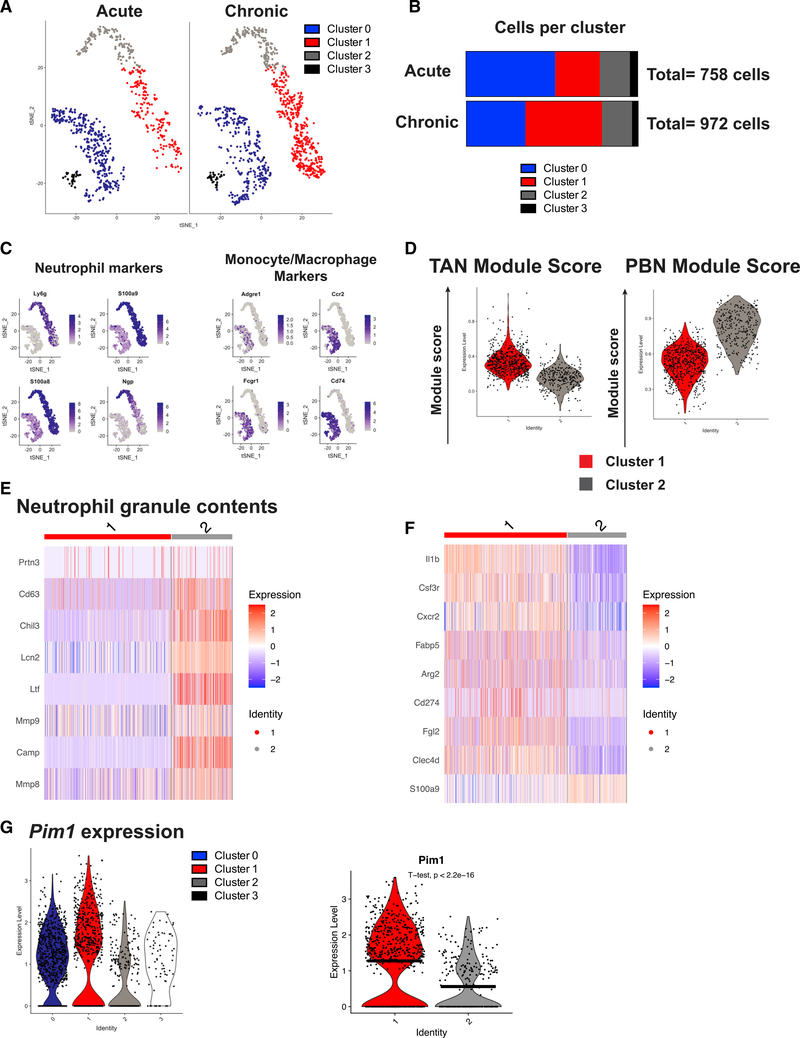
During chronic infection, viral control and immunopathology are balanced to promote host survival. Previous work has demonstrated that myeloid cells with the ability to suppress T cells are present during LCMV clone 13 infection but not during LCMV Armstrong infection (Norris et al., 2013). However, the heterogeneity of these cells and how they interact with the adaptive immune system to prevent immune pathology are not well understood. Because of slight inherent differences in tropism and infection route between the LCMV Armstrong and clone 13 models (Ahmed et al., 1984), we sought to directly compare cells from acute and chronic LCMV clone 13 infections. To this end, we performed scRNA-seq with cells isolated from the spleens of C57BL/6 mice infected with 2 × 102 plaque-forming units (PFUs) or 2 × 106 PFUs of LCMV clone 13, doses that cause an acute or persistent infection, respectively (Cornberg et al., 2013; Stamm et al., 2012). B220− CD3− NK1.1− CD11b+ cells were purified by fluorescence-activated cell sorting (FACS) from spleens 7 days post infection (d.p.i.), and single-cell transcriptomes were sequenced. We recovered transcripts from 2,470 cells from acute infection and 2,118 cells from chronic infection, with 2,088 and 2,041 median genes per cell for each sample, respectively. Analysis of the scRNA-seq library revealed cells readily identified as CD8 T cells based on expression of Cd8a, Cd3g, Pdcd1, and Gzma (Figure S1A). Therefore, these cells were excluded from the analysis. Based on transcriptional heterogeneity of the remaining cells, four distinct clusters were distinguished and visualized by t-distributed stochastic neighbor embedding (t-SNE) (Figure 1A). Cluster 0 was identified as macrophages/monocytes by expression of the gene encoding F4/80 (Adgre1), clusters 1 and 2 as neutrophils by expression of Ly6g, and cluster 3 as dendritic cells by expression of the genes for CD11c and MHCII (Itgax and H2aDMb2) (Figure S1B). Interestingly, although clusters 1 and 2 demonstrated high expression of Ly6g, cluster 1 was markedly enriched in chronically infected mice (Figure 1B). Further validating clusters 1 and 2 as neutrophils, cells in these clusters expressed the neutrophil-specific transcripts Ly6g, S100a9, S100a8, and Ngp (Figure 1C).
Figure 1. scRNA-seq reveals heterogeneity in the myeloid compartment.
(A) t-SNE projection of 1,730 myeloid cells from mice infected with 2 × 102 (acute) or 2 × 106 (chronic) PFUs LCMV clone 13 from the spleen.
(B) Cells from (A) separated by infection dose and quantified in a stacked bar graph.
(C) Feature plots showing expression of neutrophil-associated genes (Ly6g, S100a9, S100a8, and Ngp) and key markers of monocytes/macrophages (Adgre1, Ccr2, Fcgr1, and Cd74).
(D) TAN and PBN module scores were calculated using genes upregulated more than 1 log-fold change (logFC) in TANs versus PBNs or vice versa, respectively, from GSE118245.
(E) Heatmap depicting expression of genes encoding neutrophil granule components.
(F) Expression of selected known markers associated with suppressive neutrophils or transcripts known to regulate immune function.
(G) Violin plots showing expression of Pim1 in all four myeloid cell clusters from LCMV clone 13 infection (left) and violin plot comparing Pim1 expression between clusters 1 and 2 via a t test (right). Black horizontal lines denote mean expression.
To characterize the transcriptional phenotype of clusters 1 and 2, we analyzed the publicly available dataset GSE118245, a scRNA-seq library created from sorted CD45+ CD11b+ Ly6G+ cells from peripheral blood or tumors in a model of non-small cell lung cancer (NSCLC) in mice (Mollaoglu et al., 2018). We created module scores based on genes upregulated in tumor-associated neutrophils (TANs), which have potent immunosuppressive function, or peripheral blood neutrophils (PBNs). Interestingly, we found that the TANs were similar to cluster 1 from our dataset, and the PBNs were similar to cluster 2 (Figure 1D). Interestingly, neutrophils in cluster 2 expressed high levels of transcripts for proteins in immature neutrophils, such as Prtn3, Cd63, Chil3, Lcn2, Ltf, Mmp9, Mmp8, and Camp (Evrard et al., 2018; Figure 1E). Furthermore, several transcripts that were significantly upregulated in cluster 1 compared with cluster 2 encode proteins that have been shown to negatively regulate immune responses or are enriched in suppressive myeloid cells, including Il1b (Najjar et al., 2017), Csf3r (encodes the granulocyte colony-stimulating factor (G-CSF) receptor; Li et al., 2016; Trikha and Carson, 2014), Cxcr2 (Highfill et al., 2014; Katoh et al., 2013; Sun et al., 2019), Fabp5 (Bogdan et al., 2018; Veglia et al., 2019), Arg2 (encodes type II mitochondrial arginase; Vaccari et al., 2019), Fgl2 (encodes fibrinogen-like protein 2; Yan et al., 2019), Clec4d (encodes C-type lectin domain family 4 member d; Steichen et al., 2013), and Cd274 (encodes PD-L1) (Figure 1F). Of note, although S100a9 expression was detectable in clusters 1 and 2, cluster 2 had increased S100a9 expression compared with cluster 1 (Figure 1F). STAT3 plays a vital role in differentiation and survival of MDSCs, and we detected high expression in cluster 1 of the transcript for the G-CSF receptor (Csf3r), which signals through STAT3 (Gabrilovich and Nagaraj, 2009; Kortylewski et al., 2005; Nefedova et al., 2004, 2005). Interestingly, the gene Pim1, known to be regulated by STAT3 signaling (Jinesh et al., 2016; Warfel and Kraft, 2015), was also highly expressed in cluster 1 compared with cluster 2 (Figure 1G). PIM1 has documented roles in maintaining survival and mitochondrial fitness in cardiomyocytes (Borillo et al., 2010; Din et al., 2013, 2014), which prompted us to surmise that PIM1 may represent an important signaling node in suppressive neutrophils. However, the function of PIM kinase in suppressive neutrophils has not been studied. The single-cell transcriptomics indicated that cells in cluster 1 are suppressive neutrophils that may have an immunoregulatory role during chronic LCMV clone 13 infection. Overall, these results demonstrate that chronic LCMV clone 13 infection is characterized by enrichment of Pim1-expressing suppressive neutrophils that may be responsible for negatively regulating the response to persistent viral infection.
Chronic infection is associated with accumulation of M-MDSCs and suppressive neutrophils
To understand the kinetics of the myeloid response to acute and chronic LCMV infection, we performed flow cytometry analyses on circulating blood leukocytes during the first 14 days of infection and examined the cells in the spleen at 7 and 14 d.p.i. in acute and chronic infection. We observed a consistent increase in the frequency of monocytic cells (CD11b+ F4/80− Ly6C+ Ly6G−) in the blood and persistence of these cells after 7 d.p.i. in chronically infected animals (Figures S2A and S2B). This corresponds with persistent infection in chronically infected animals and resolution of infection and return to homeostasis in acutely infected animals. There was a rapid increase in the frequency of neutrophils (CD11b+ F4/80− Ly6C+ Ly6G+) during both infections; however, by 14 d.p.i., neutrophil frequency had contracted in acute infection, perhaps in response to clearance of virus. Indeed, we found that acutely infected mice had cleared their infection because viral titers were below the limit of detection in mice infected with 1 × 102 PFU LCMV clone 13 at 14 d.p.i. (Figure S2C). In contrast, neutrophil frequency remained more than 10-fold higher in the blood of chronically infected mice at 14 d.p.i. compared with the levels determined prior to infection. We also noted that there was a significant increase in the frequency of monocytic cells and neutrophils in the spleens of chronically infected mice at 7 and 14 d.p.i. compared with mice given acute infection (Figures S2D and S2E). Finally, consistent with our scRNA-seq data showing that suppressive neutrophils express S100a9 at intermediate rather than high levels, we found an increase of S100A9int suppressive neutrophils at 7 d.p.i. during chronic infection (Figure S2F). These data support our scRNA-seq findings showing that there are increased suppressive neutrophils in the spleen during chronic LCMV infection at 7 d.p.i. compared with acute infection.
Persistent antigen exposure, such as during chronic infection, leads to T cell exhaustion, a differentiation state that is marked by expression of inhibitory molecules such as PD-1 and a reduction in effector functions (Barber et al., 2006; Blackburn et al., 2009; Pauken and Wherry, 2015). As expected, analysis of CD8 T cells stimulated with the immunodominant LCMV epitopes GP-33, GP-276, and NP-396 in acutely infected mice demonstrated increased effector function, as measured by interferon γ (IFN-γ)+ CD107a+, and IFN-γ+ tumor necrosis factor alpha (TNF-α)+ double-positive cells (Figures S2G and S2H). In addition, we tested whether virus-specific CD8 T cells from acutely infected mice expressed distinguishable memory T cell markers compared with those from persistently infected mice. As expected, virus-specific CD8 T cells from acutely infected mice had a higher proportion of memory precursor CD127+ KLRG1− cells (Figure S2I; Kaech and Cui, 2012). Differentiation of CD8 T cells toward an exhausted phenotype is associated with an increase in PD-1 surface expression (Blackburn et al., 2009), which we also observed in the chronically infected mice (Figure S2I). These data demonstrate that suppressive neutrophils are enriched during chronic viral infection and are correlated with T cell exhaustion.
PIM1 is required for suppressive neutrophils to suppress CD8 T cell function
STAT3 signaling is required for suppressive neutrophil function (Gabrilovich and Nagaraj, 2009; Kortylewski et al., 2005; Nefedova et al., 2004, 2005), and suppressive neutrophils in our scRNA-seq data highly expressed Pim1, a gene that encodes a protein kinase and is known to be expressed downstream of JAK/STAT signaling. Therefore, we hypothesized that PIM1 might be a critical signaling node downstream of STAT3 for suppressive neutrophil function. To test this, we used a cell culture model to investigate whether PIM1 was required for suppressive neutrophils. In particular, we wanted to find out whether PIM1 was vital for production of ROS, a key mechanism by which suppressive neutrophils mediate immune suppression. ROS production by suppressive neutrophils is critical to suppress activation and proliferation of T cells (Veglia et al., 2018). Peroxynitrite (ONOO−), the product of the diffusion-controlled reaction between nitric oxide (NO) and superoxide, can nitrate and nitrosate proteins on T cells and transformed/infected cells and is thought to inhibit T cell-major histocompatibility complex (MHC) interactions to suppress T cell function (Nagaraj et al., 2007; Raber et al., 2014). However, to our knowledge, direct measurements of ONOO− production by suppressive neutrophils have not been performed. ROS production was measured using coumarin boronic acid (CBA), which reacts with ONOO− and hydrogen peroxide to produce a fluorescent product (Sikora et al., 2009; Zielonka et al., 2010). Total MDSC cultures (containing suppressive neutrophils and M-MDSCs) produced significant quantities of ROS when stimulated with phorbol 12-myristate 13-acetate (PMA) (Figure S3A). Surprisingly, ROS production was attenuated in the presence of catalase, an enzyme that converts peroxide to water and molecular oxygen, suggesting that hydrogen peroxide, but not ONOO−, is the primary form of ROS generated by suppressive myeloid cells (Figure S3A).
The potential role of PIM kinases in production of ROS by MDSCs was evaluated using the pan-PIM kinase inhibitor AZD1208. The concentration-dependent actions of this inhibitor identified an effective concentration of 1 μM in vitro (Figure S3B), consistent with concentrations used in a preclinical model of leukemia (Brasó-Maristany et al., 2016; Keeton et al., 2014). Indeed, we found that 3-day treatment with this PIM kinase inhibitor significantly reduced production of hydrogen peroxide by MDSCs (Figure 2A). Surprisingly, when we investigated the phenotype of the cells by flow cytometry, we found that genetic deletion of PIM1 and PIM2 (PIM1/2 double knockout (DKO)) or administration of the PIM kinase inhibitor caused an ~50% decrease in the proportion of suppressive neutrophils, whereas M-MDSC frequencies were largely unchanged (Figure 2B). Our scRNA-seq data suggested that suppressive neutrophils expressed an intermediate level of S100a9 (Figure 1F), and higher frequencies of S100A9int neutrophils were present in the spleens of chronically infected mice compared with acutely infected mice, as seen by flow cytometry analysis (Figure S2F). We therefore tested whether the proportion of these cells could be affected by PIM kinase inhibitor treatment in vitro. Indeed, the frequency of S100A9int neutrophils was decreased slightly and approached significance after PIM kinase inhibitor treatment in vitro (Figure S3C).
Figure 2. PIM1 is required for suppressive neutrophils to suppress CD8 T cell function.
(A) Cultured MDSCs were incubated with vehicle (DMSO) or the PIM inhibitor for 3 days. ROS production was measured by fluorescence by adding coumarin boronic acid (CBA) to cells in the presence (stimulated) or absence (unstimulated) of PMA. Measurements were conducted using a plate reader. Raw fluorescence data are summarized in graphical form.
(B) Representative flow cytometry plot and quantification of cultured MDSCs with vehicle or PIM inhibitor for 3 days. Combined data from 2 independent experiments. WT MDSCs, n = 3; DKO MDSCs, n = 2. *p < 0.05 by one-way ANOVA.
(C) In-vitro-cultured MDSCs were flow sorted using the markers CD11b+, Ly6C+, and Ly6G− for monocytes and CD11b+, Ly6C+, and Ly6G+ for neutrophils. Cells were cultured for 4 h and incubated with PMA or vehicle, and ROS production was measured by CBA fluorescence.
(D) Cultured suppressive neutrophils were incubated in the presence of vehicle or PIM inhibitor for 4 h, stimulated with PMA, and then CBA fluorescence was measured.
(E) CD8 T cells isolated from a C57BL/6 mouse spleen were labeled with Cell Trace Violet (CTV) dye and activated with plate-bound anti-CD3 and soluble anti-C28 antibody for 3 days in the presence of the indicated ratio of cultured MDSCs from vehicle control- or PIM inhibitor-treated cultures. Data are shown as mean ± SEM. *p < 0.05 by t test.
(A) is representative of five independent experiments. (B), (D), and (E) are representative of three independent experiments.
Using FACS-purified populations of monocytic and granulocytic myeloid cells (Figure S3D), we identified suppressive neutrophils as the primary source of ROS production (Figure 2C) and showed that administration of the PIM kinase inhibitor (4-h treatment) had no effect on ROS production by this population of cells (Figure 2D). We next examined whether PIM1 was required for MDSC-mediated regulation of CD8 T cell proliferation by pre-treating MDCSs for 3 days with the PIM inhibitor, followed by co-culture with CD8 T cells. Pre-treatment with the PIM kinase inhibitor reduced the ability of total cultured MDSCs to control expansion of CD8 T cells in vitro (Figure 2E). These data demonstrate that PIM kinase supports suppressive neutrophil function and that loss of PIM kinase reduces the frequency of hydrogen peroxide-producing suppressive neutrophils and the overall suppressive function of these cells.
Inhibition of PIM kinase selectively targets suppressive neutrophils during chronic viral infection
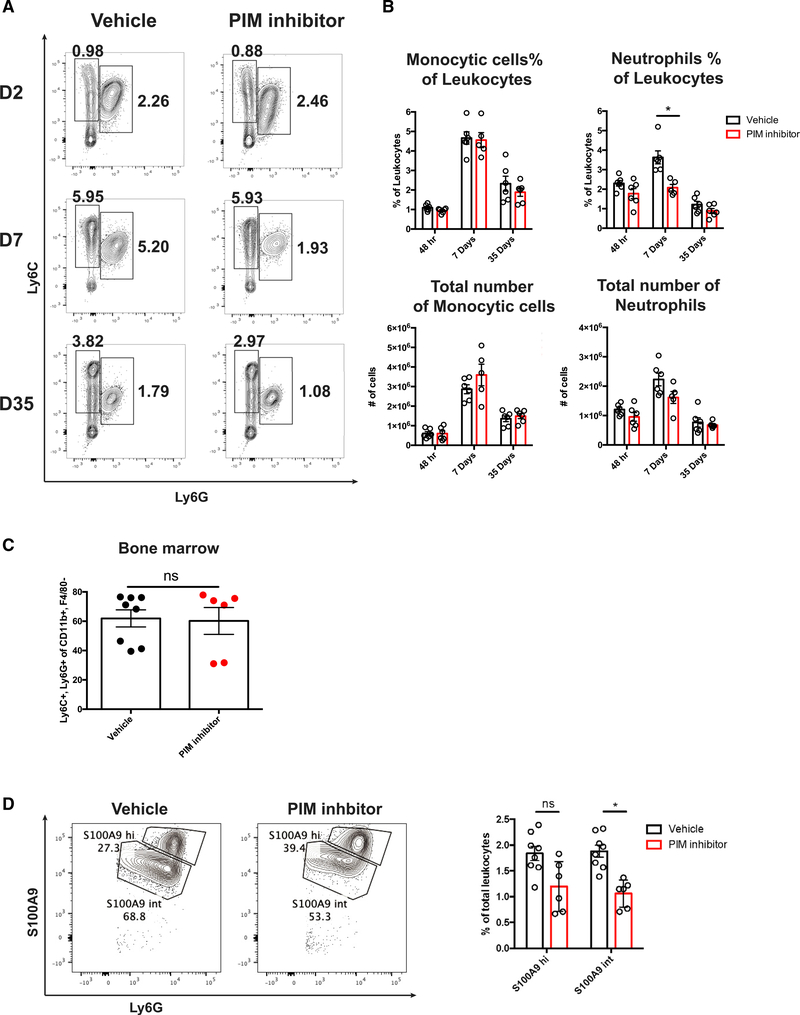
To determine whether PIM kinase function is required for suppressive neutrophil generation during chronic viral infection, LCMV clone 13-infected mice were treated with the PIM kinase inhibitor or vehicle by oral gavage, and the frequency and number of myeloid cells in the spleen at 2, 7, and 35 d.p.i. were determined. There was no significant change in the frequency and number of monocytic cells in the spleen at all time points examined. In stark contrast, the frequency of neutrophils was decreased as early as 2 d.p.i. in mice treated with the PIM inhibitor. This decrease achieved statistical significance by 7 d.p.i. and appeared to have resolved by 35 d.p.i. because the neutrophil numbers and frequencies were similar to those found in vehicle control-treated animals (Figures 3A and 3B). These findings indicate that PIM kinase inhibition has a transient rather than permanent effect on suppressive neutrophil frequency. Using our scRNA-seq data, we determined that the transcript for S100A9 was increased significantly in conventional compared with suppressive neutrophils. Therefore, we hypothesized that we could separate the two clusters of neutrophils present in the spleen during LCMV clone 13 infection using this intracellular marker. We found that PIM kinase inhibitor treatment significantly reduced the frequency of S100A9int but not S100A9hi neutrophils (Figure 3D). Importantly, these data support our in vitro findings that loss of PIM kinase reduces the frequency of suppressive neutrophils. Furthermore, we found that there was no detectable difference between the frequency of splenic neutrophils in naive WT and PIM1 knockout animals (Figure S4A). In addition, we examined the bone marrow from vehicle- and inhibitor-treated animals at 7 d.p.i. and found no significant differences in the frequency of total neutrophils (Figure 3C). These data support the hypothesis that S100A9int suppressive neutrophils are most susceptible to PIM kinase inhibitor treatment in vivo.
Figure 3. Inhibition of PIM kinase selectively targets suppressive neutrophils during chronic viral infection.
(A) Representative flow cytometry plots from 2, 7, and 35 d.p.i. LCMV clone 13-infected spleens treated with vehicle control or PIM inhibitor daily from days 0–21. Cells were gated on CD11b+ F4/80− single cells.
(B) Quantification of flow cytometry data from (A).
(C) Frequency of neutrophils in bone marrow after vehicle or PIM kinase inhibitor treatment.
(D) Representative flow cytometry plots showing S100A9hi and S100A9int neutrophils at 7 d.p.i. (left) and quantification of S100A9hi and S100A9int neutrophil frequency after PIM kinase inhibitor treatment (right). Data are shown as mean ± SEM. *p < 0.05 by t test.
(A) is pooled from 2 independent experiments. (C) and (D) are pooled from 3 independent experiments.
PIM kinase inhibition enhances virus-specific CD8 T cell response and control of chronic viral infection
To determine whether the reduction in suppressive neutrophils by PIM kinase inhibition correlated with increased CD8 T cell function, virus-specific CD8 T cell frequencies and numbers were evaluated using the MHC class I tetramer H2-Db GP-33. Total CD8 T cells were also re-stimulated using GP-33, GP-276, and NP-396 peptides ex vivo at 7 and 35 d.p.i. There was no significant difference in the frequencies or numbers of H2-Db GP-33+ virus-specific CD8 T cells at 7 d.p.i. between vehicle control- and PIM inhibitor-treated mice (Figure 4A). Functionally, CD8 T cells that recognized each of the three viral epitopes had a greater capacity to degranulate (CD107a cell surface staining, IFN-γ production) and produce inflammatory cytokines (IFN-γ and TNF-α) at 7 d.p.i. Interestingly, the frequency of GP-33-specific CD8 T cells was increased in PIM-inhibitor treated animals at 35 d.p.i. and the frequency of IFN-γ +TNFα+ CD8 T cells was increased significantly in response to stimulation with the GP-33 epitope (Figure 4B). To ensure that these changes in CD8 T cells were not mediated by the effects of PIM inhibitor on cells besides suppressive neutrophils, we also examined other major immune cells. We were unable to detect any changes in the frequencies of germinal center (GC) B cells, T follicular helper (TFH) cells, or TH1 cells (Figure S4B). Furthermore, there was no significant difference in neutralizing antibody titer at 14 d.p.i. in serum from vehicle- and inhibitor-treated animals (Figure S4C), suggesting that changes in CD8 T cell responses were not due to alterations in humoral immunity. Consistent with the increased frequency and function of virus-specific CD8 T cells, PIM inhibitor treatment led to a 2-fold reduction in serum viral titers at 7 d.p.i. (Figure S4D) and a more than 10-fold reduction in viral titer at 35 d.p.i. (Figure 4C). These findings suggest that PIM kinase inhibition can significantly improve antiviral CD8 T cell function and control over chronic viral infection.
Figure 4. PIM inhibition enhances virus-specific CD8 T cell response and chronic viral control.
(A and B) Representative flow cytometry plots of H2-Db GP-33 tetramer+ and GP-33 peptide-stimulated CD8+ T cells from spleens of C57/BL6 mice infected with 2 × 106 PFU LCMV clone 13 at 7 (A) and 35 (B) d.p.i. (left) and quantification of IFN-γ, CD107a+ and IFN-γ, TNF-α+ CD8 T cells stimulated with GP-33, GP-276, and NP-396 peptides (right).
(C) Serum viral titer was quantified by viral plaque assay using samples from mice treated with vehicle or PIM inhibitor at 35 d.p.i. Data are shown as mean ± SEM; *p < 0.05 by t test. Data were pooled from two independent experiments.
Suppressive neutrophils in LCMV clone 13 infection have an oxidative metabolic phenotype
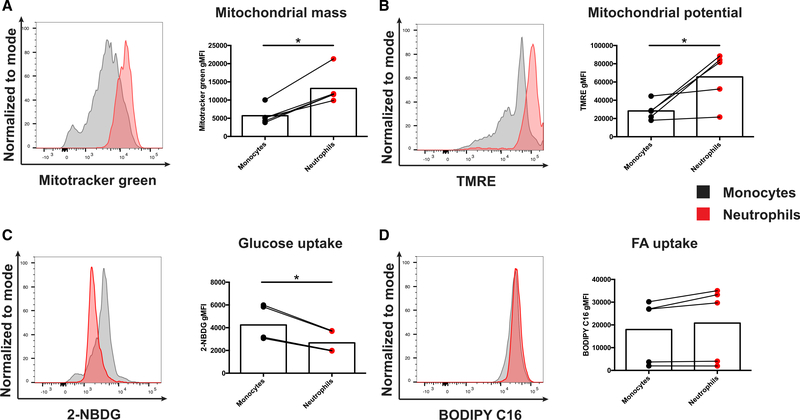
Although the metabolic state has been strongly linked to function in T cells as well as in macrophages (Buck et al., 2017; Orecchioni et al., 2019), how metabolic fitness regulates the function of suppressive neutrophils is poorly understood. PIM1 is known to regulate mitochondrial morphology and function (Din et al., 2013, 2014) and was highly expressed in suppressive neutrophils compared with conventional neutrophils (Figure 1G). Indeed, the suppressive neutrophils in cluster 1 had the highest expression of Pim1 of all myeloid cell clusters examined. Because treatment with the PIM inhibitor selectively reduced the frequency of suppressive neutrophils in vitro and in vivo but had little to no effect on monocytic cells, we next compared the metabolic state of neutrophils and monocytic cells directly ex vivo from spleens of C57BL/6 mice infected with persistent clone 13 infection at 7 d.p.i. Surprisingly, compared with monocytic cells in the spleen, neutrophils had increased mitochondrial mass, as measured by MitoTracker Green staining (Figure 5A), increased mitochondrial potential by tetramethyl rhodamine ethyl ester (TMRE) staining (Figure 5B), and decreased glucose uptake by staining with the fluorescent glucose analog 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino)-2-deoxyglucose (2-NBDG) (Figure 5C) but no noticeable change in fatty acid uptake, as measured by boron-dipyrromethene (BODIPY)-conjugated palmitate, a fluorescent fatty acid (FA) analog (Figure 5D). These data suggest that suppressive neutrophils in LCMV clone 13 infection have high mitochondrial content and potential compared with monocytic cells, suggesting that high mitochondrial content or potential may be required for supporting their suppressive function and/or survival. These data agree with similar characteristics of suppressive neutrophils in the setting of the tumor microenvironment (Rice et al., 2018).
Figure 5. Suppressive neutrophils in LCMV clone 13 infection have an oxidative metabolic phenotype.
(A–D) Representative flow cytometry plots and histograms depicting (A) MitoTracker Green, (B) tetramethylrhodamine ethyl ester (TMRE), (C) 2-NDBG, and (D) BODIPY C-16 staining in Ly6C+ Ly6G− monocytes and Ly6C+ Ly6G+ neutrophils from 14 d.p.i. with chronic LCMV clone 13. Cells were gated on CD11b+ F4/80− single cells. Data are shown as mean ± SEM. *p < 0.05 by paired t test. n = 5 for each condition. Data were collected from 2 independent experiments.
PIM1 kinase promotes mitochondrial fusion and fitness in suppressive neutrophils
Suppressive neutrophils had higher mitochondrial potential and mitochondrial mass than monocytic cells. In addition, suppressive neutrophils in cluster 1 (Figure 1) express high levels of Pim1 and are uniquely susceptible to PIM kinase inhibition (Figure 3). Furthermore, dynamin-related protein 1 (DRP1), a PIM1 substrate and key mediator of mitochondrial fission, can be inactivated by PIM1-mediated phosphorylation in cardiomyocytes (Borillo et al., 2010; Din et al., 2013; Samse et al., 2015). Previous studies have associated mitochondrial morphology and function, linking fused mitochondrial networks with enhanced mitochondrial oxidative metabolism (Buck et al., 2016; Rambold et al., 2011, 2015). This led us to hypothesize that PIM kinase increases fusion of mitochondria by inactivating DRP1, promoting survival of suppressive neutrophils. Treatment of suppressive neutrophils with the PIM kinase inhibitor decreased serine 637 phosphorylation of DRP1 (Figure 6A), and this was associated with an apparent increase in fragmentation of suppressive neutrophils’ mitochondria (Figure 6B). Strikingly, PIM kinase inhibition did not alter mitochondrial networks in M-MDSCs after 4 h of treatment (Figure 6B). Mitograph, which uses high-resolution, three-dimensional image stacks to create computational models of mitochondrial networks (Harwig et al., 2018; Rafelski et al., 2012; Viana et al., 2015), was used to quantify the morphology of mitochondrial networks in suppressive neutrophils and M-MDSCs in an unbiased manner. The Mitograph connectivity score (MCS), an index of mitochondrial morphology correlated with mitochondrial fusion, was decreased significantly in suppressive neutrophils treated with the PIM inhibitor, indicating that these cells have more fragmented mitochondrial networks (Figure 6C). Further, statistically significant increases in the number of nodes normalized to length and decreases in the PHI score (ratio of the largest connected component to overall mitochondrial size) were detected in PIM inhibitor-treated cells (Figure 6C). Increases in the number of nodes normalized to length and a decrease in PHI score have been associated with increased fragmentation of mitochondria (Harwig et al., 2018). These measurements suggest that PIM kinases are required to maintain fused mitochondrial morphology in suppressive neutrophils, which correlates with increased oxidative metabolism. Mitochondrial morphology is a tightly regulated process; mitochondria that are more fused have an increased ability to use alternative fuels, such as fatty acids (Rambold et al., 2015). This metabolic flexibility may aid cellular survival in times of metabolic stress, such as during an ongoing inflammatory response. To evaluate this hypothesis, we measured apoptosis in suppressive neutrophils in vitro. We found that the PIM kinase inhibitor correlated with increased Annexin V+ cells specifically in suppressive neutrophils but not in M-MDSCs (Figure 6D). Therefore, the enhanced mitochondrial function provided by PIM kinases may promote survival of suppressive neutrophils; conversely, the PIM inhibitor may be used to induce selective apoptosis of suppressive neutrophils to bolster the antiviral response.
Figure 6. PIM1 kinase promotes mitochondrial fusion and fitness in suppressive neutrophils.
(A) Western blot of isolated Ly6G+ in-vitro-cultured suppressive neutrophils after 0, 2, and 4 h of culture with 1 μM AZD1208 (PIM inhibitor).
(B) Representative images of flow-sorted granulocytes or monocytes cultured with vehicle (DMSO) or PIM inhibitor for 4 h, stained with MitoTracker CMX rosamine and DAPI, and imaged with a spinning-disk confocal microscope. MitoTracker staining is pseudo-colored green, and DAPI staining is pseudo-colored blue.
(C) Quantification of mitochondrial networks using the Mitograph program.
(D) Flow cytometry analysis of apoptosis and cell death. Cells were cultured with the PIM kinase inhibitor for the indicated time. All samples were collected and analyzed on day 6 of culture.
(A) and (B) are representative of 3 independent experiments, (C) contains pooled data from 3 independent experiments, and (D) contains data from 2 independent experiments; n = 3. Data are shown as mean ± SEM. *p < 0.05 by one-way ANOVA with multiple comparisons (C) or t test (D).
DISCUSSION
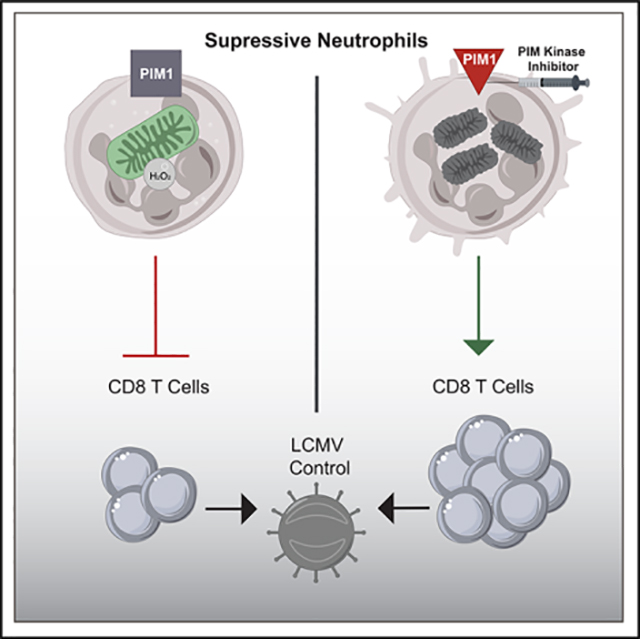
In this study, we identified a previously underappreciated population of suppressive neutrophils during chronic LCMV infection. Interestingly, this cluster of neutrophils bears similar transcriptional signatures as suppressive neutrophils found in cancer. We also demonstrated that suppressive neutrophils uniquely depend on PIM1 for their survival and function. Moreover, by selectively reducing suppressive neutrophils during chronic LCMV infection using a PIM kinase inhibitor, we were able to increase CD8 T cell function and improve viral control. Mechanistically, PIM kinase was required for maintaining fused mitochondrial networks, which correlated with increased survival of suppressive neutrophils. These findings underscore a previously unknown role of PIM1 in neutrophils that regulate the host immune response during chronic viral infection.
Although the cellular heterogeneity of myeloid cells in the tumor microenvironment and the bone marrow is increasingly well understood, little is known about how myeloid cells differentiate and function during chronic viral infection (Mollaoglu et al., 2018; Zhu et al., 2018). In a model of NSCLC in mice, 7 distinct clusters of Ly6G+ cells were found in the blood, and two additional clusters were found in the tumor microenvironment (Mollaoglu et al., 2018). In comparison, we found 2 neutrophil clusters in the spleen during chronic infection. Interestingly, the conventional neutrophils in cluster 2 express higher levels of transcripts for neutrophil granular components such as Ltf, Cd63, and Lcn compared with cells in cluster 1 (suppressive neutrophils). Expression of these transcripts has been associated with immature neutrophils (Evrard et al., 2018). It is generally thought that suppressive neutrophils exhibit a more “immature” phenotype, but it is unclear whether suppressive neutrophils in cluster 1 are more mature or whether they have an alternative activation or differentiation state. Indeed, it is possible that modification of neutrophil differentiation takes place, where neutrophil progenitors synthesize multiple signals that drive formation of suppressive neutrophils in addition to conventional neutrophils.
Suppressive neutrophils produce ROS to suppress CD8 T cell proliferation and function. Interestingly, a surrogate marker for ONOO− production, nitrosation of tyrosine residues, has been detected in MDSCs isolated from tumor samples (Raber et al., 2014; Youn et al., 2008). However, direct measurement of ONOO− production by MDSCs has not been performed previously. Surprisingly, we found that in-vitro-generated suppressive neutrophils produce hydrogen peroxide and not ONOO−. One possible explanation for the discrepancies between these observations is that contextual signals are required for production of ONOO− by suppressive neutrophils. However, further investigation of the ability of suppressive neutrophils to produce ONOO−, using direct measurement methods such as CBA, are warranted. Interestingly, conventional inflammatory neutrophils also produce ROS. However, a key feature that may lead to suppressive neutrophils’ ability to reduce CD8 T cell function may be their proximity to T cells and ability to transfer cellular constituents containing suppressive molecules directly to CD8 T cells (Baumann et al., 2020).
Mitochondrial dynamics and metabolic functions are interlinked. Fused mitochondrial networks have a higher oxidative capacity and can more efficiently use fatty acids as substrates (Buck et al., 2016; Rambold et al., 2015). Furthermore, mitochondrial fragmentation is a key first step in induction of apoptosis (Cribbs and Strack, 2007). We observed that short-term PIM kinase inhibition increased mitochondrial fragmentation in suppressive neutrophils but had no observable effect on the mitochondrial dynamics of M-MDSCs. PIM kinases promote fusion in mitochondrial networks through phosphorylation of DRP1 on S637 (Din et al., 2013, 2014). This post-translational modification inactivates DRP1 and, therefore, promotes mitochondrial fusion. However, it is not clear whether apoptosis was caused by the alterations of mitochondrial dynamics observed in suppressive neutrophils treated with the PIM kinase inhibitor. One potential explanation for the susceptibility of suppressive neutrophils to the PIM kinase inhibitor is their production of ROS because PIM inhibition may sensitize suppressive neutrophils to ROS-induced damage. Indeed, PIM kinases have been shown to activate Nrf2, a regulator of cellular resistance to oxidative damage (Warfel et al., 2016). Additionally, production of ROS from nicotinamide adenine dinucleotide phosphate (NADPH) by NOX2 is an energy-intensive process. Therefore, mitochondria may play a role in sustaining metabolic fitness and ATP production during ROS release through highly coupled oxidative respiration. The observation that loss of PIM kinase function compromises survival of suppressive neutrophils warrants further investigation.
In mice treated with the PIM inhibitor, we observed an increase in CD8 T cell function and enhanced viral control associated with a decrease in the population of suppressive neutrophils. Although the ensuing increase in T cell effector function was well tolerated by inhibitor-treated mice, PIM1/2 DKO animals infected with LCMV clone 13 die around 7 d.p.i. (P.J.V. and W.C., unpublished data). The reasons for the decreased viability are unknown; however, potential mechanisms to explain this response include immune pathology or a variety of factors, including reduced platelet function, that confound use of total PIM1/2 knockout mice as a model system (An et al., 2013). Further, it is not possible to use these animals as bone marrow donors to create mixed bone marrow chimeras because PIM1 controls the function of CXCR4, which is required for homing to the bone marrow niche and engraftment (Grundler et al., 2009). Future studies using conditional knockout systems would be beneficial; however, pharmacological inhibitors of PIM kinase have several advantages, including temporal control of PIM function and applicability to human diseases. Given that PIM kinases are also highly expressed in certain malignancies (Lu et al., 2013; Morishita et al., 2008), a pharmacological inhibitor of PIM kinases may reduce tumor growth and immune suppression by targeting tumor-associated suppressive neutrophils. An alternative treatment method besides small-molecule inhibitors is use of depleting antibodies. Interestingly, depletion of neutrophils and monocytes using an anti-Gr-1 antibody during LCMV clone 13 infection has been shown to lead to increased CD8 T cell antiviral function (Norris et al., 2013). However, this strategy did not lead to a significant increase in viral control. This may be due to expression of Ly6C and Ly6G among pro-inflammatory neutrophils and monocytic subsets as well as suppressive cells. In contrast, we target suppressive neutrophils specifically by exploiting a key kinase that is vital for their survival.
Chronic LCMV infection of mice has been employed to discover key aspects of T cell differentiation in response to persistent antigen exposure (Ahmed et al., 1984; Barber et al., 2006; Pauken and Wherry, 2015; Wherry et al., 2004). In this study, we expanded use of this model system of host response to persistent infection to demonstrate an interplay between neutrophils and T cells. Suppressive neutrophils function to critically balance control of chronic virus infection with collateral damage of vital host tissues. We also provide a mechanism by which this balance can be altered in favor of a more vigorous immune response by specifically targeting a PIM-dependent subset of neutrophils. Our exciting findings support a potential therapeutic role of targeting and inhibiting PIM-dependent suppressive neutrophils as a novel and promising strategy for treatment of chronic viral infection and cancer.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Weiguo Cui (weiguo.cui@versiti.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Single-cell RNA sequencing data from this paper are available in the GEO database with accession number GEO: GSE167204. All other raw data and scripts are available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All mice were housed at the Medical College of Wisconsin Biological Resource Center (Milwaukee, WI). C57BL/6 mice were purchased from Charles River/NCI (Wilmington, MA) and maintained on a standard diet. LCMV Clone 13 infections were established by infecting 6- to 8-week-old female mice with 2×102 or 2×106 PFU intravenously for an acute and chronic infection, respectively. All mice were maintained in accordance with protocols approved by the Medical College of Wisconsin Animal Care and Use Committee. Viral titers were performed by assessing plaque formation on cultured Vero cells and analysis was performed blinded to the identity of the sample. Focus forming assay was performed by infection of Vero cells with serum at a dilution of 1:400 in DMEM with 10% serum. Cells were then fixed and stained with 2.53 mg/mL rat anti-LCMV nucleoprotein InVivo mAb clone VL-4 (BioXCell, NH) and 1 mg/mL goat anti-rat FITC-conjugated IgG2a (Bethyl Laboratories, TX). Foci were imaged and quantified using an Incucyte imager (Essen BioScience, MI).
METHOD DETAILS
Single-cell RNA sequencing
CD3ε− B220− NK1.1− CD11b+ cells were sorted by FACS and loaded onto a 10X Chromium controller (10X Genomics, CA). Single-cell libraries were prepared using the Chromium Single Cell 3′ Reagent kit (10X Genomics, CA) according to the manufacturer’s standard protocol. Sorted cells were loaded onto the controller with a target of 10,000 cells. Libraries were quantified using the Kapa library quantification kit (Roche, Switzerland). Libraries were sequenced with Illumina NextSeq with the NextSeq 500/550 High Output Kit v2 (150 cycles) (Illumina, CA) with the following conditions: 26 cycles for read 1, 98 cycles for read 2, and 8 cycles for i7 indexing. Sequencing data was downloaded with Python Run Downloader (Illumina, CA) and de-multiplexed using mkfastq and count functions in Cell Ranger V3.0 (10X Genomics, CA). Downstream analysis was performed using R version 3.6, and Seurat V3.0 (Butler et al., 2018). t-SNE was performed with a perplexity parameter of 30, and clustering was performed using a resolution of 0.1.
Flow cytometry
Mouse blood was collected in 4% citrate buffer and subjected to gradient centrifugation to remove red blood cells. Mouse splenocytes were collected by processing the organ through wire mesh, and lysing red blood cells using ACK lysing buffer as previously described (Xin et al., 2015). H2-Db GP-33, GP-276, and NP-396 PE conjugated tetramers were used to stain virus-specific CD8 T cells for 30 minutes on ice. Flow cytometry antibodies used in this study are listed in Table S1. Metabolic parameters were measured direct ex vivo on splenocytes. For glucose uptake, cells were stained with 100 μg/ml 2-NBDG (Cayman Chemical, MI) in glucose-free RPMI medium for 15 minutes at 37 degrees. Mitochondrial mass was measured by staining splenocytes with 40 nM Mitotracker green (Life Technologies, CA) for 15 minutes in RPMI containing 10% FBS at 37 degrees. Mitochondrial potential was assessed by staining splenocytes with 20 nM Tetramethyl rhodamine ethyl ester (TMRE) (Cayman Chemical, MI) in RPMI containing 10% FBS for 15 minutes at 37 degrees. Fatty acid uptake was assessed by incubating splenocytes in RPMI with 10% FBS containing 1 μg/ml BODIPY FL C16 (Life Technologies, CA) for 2 minutes at room temperature. Flow cytometry data was acquired on either a BD (BD Biosciences, CA) LSRII, or Celesta and analyzed using FlowJo (Treestar, OR).
Cell culture
Bone marrow derived MDSCs were generated by collecting bone marrow from C57BL/6 mice by flushing the bone cavities with RPMI (Lonza, Switzerland) with 1% fetal bovine serum, lysing red blood cells with ACK lysing buffer (Lonza, Switzerland), and culturing cells for 3 days in 10% RPMI (Lonza, Switzerland) with 40 ng/mL IL-6 and 40 ng/mL GM-CSF (Shenandoah Biotechnology, PA), then scraping, splitting the cells by a factor of two, and re-culturing cells in the same concentration IL-6 and GM-CSF for a further 3 days. AZD1208 was purchased from Selleckchem (Selleckchem, TX) and dissolved in DMSO and used at a final concentration of 1 μM. ROS production was quantified using Coumarin Boronic acid (CBA) (Cayman Chemical, MI) at 100 μM concentration, with absorbance taken using a BioTek SynergyMx fluorescent plate reader (BioTek Instruments, VT). Cells were stimulated with 200 ng/mL Phorbol 12-myristate 13-acetate (PMA) and measurements were taken 1 hour after stimulation according to previously published reports (Sikora et al., 2009). Polyethylene glycol catalase (PEG-Catalase) (Sigma, MO) was used at a concentration of 500 μM.
Microscopy and Mitograph analysis
Bone marrow derived MDSCs were separated into M-MDCSs and suppressive neutrophils by FACS, then plated in RPMI with 10% FBS in the presence of DMSO or AZD1208 for 4 hours on coverslips. Cells were stained with 40 nM Mitotracker CMX Rosamine (Thermo Fisher. MA) for 15 minutes, then fixed with 4% formaldehyde dissolved in PBS (Electron Microscopy Sciences, PA) for 20 minutes at room temperature, washed with PBS and stained with DAPI (4′,6-diamidino-2-phenylindole) (Thermo Fisher. MA). Coverslips were inverted and placed onto standard microscopy slides (Fisher Scientific, MA) with Pro-Long Diamond (Thermo Fisher. MA) as a mounting medium. Images were acquired on a 3I (Intelligent Imaging Innovations, CO) Vivo spinning disk confocal microscope using a 100X oil immersion lens. Images were cropped to isolate single cells using ImageJ program (Schneider et al., 2012). Single cells were analyzed with Mitograph (Viana et al., 2015) as previously described by Harwig et al., 2018.
Western blotting
Cells were lysed in RIPA buffer (Boston Bioproducts, MA) with Phosstop Phosphatase inhibitor (Roche, Switzerland) and COmplete protease inhibitor (Roche, Switzerland). Lysates were cleared and BCA assay was performed to normalize protein content between samples. SDS-PAGE gels were run according to standard procedure and transferred to PVDF membrane, blocked with 5% milk in TBST and imaged using a Super Signal West Femto chemiluminescence reagent (Fisher, MA) on a GE AI6800 Imager (GE, MA).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using Graphpad Prism version 6. All t tests were performed as unpaired two-tailed t tests with p-values considered significant if p < 0.05. ANOVA tests were calculated with corrections for multiple comparisons with p-values considered significant if p < 0.05. Statistical details of individual experiments can be found in the figures and legends.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Pacific Blue anti-mouse/human CD11b | BioLegend | Cat#101224; RRID:AB_755986 |
| FITC anti-mouse Ly6C | BioLegend | Cat#128006; RRID:AB_1186135 |
| APC-Cy7 anti-mouse Ly6G | BioLegend | Cat#127624; RRID:AB_10640819 |
| PE-Cy7 anti-mouse F4/80 | BioLegend | Cat#123113; RRID:AB_893490 |
| FITC anti-mouse CD8a | BioLegend | Cat#100706; RRID:AB_312745 |
| PE-Cy7 anti-mouse PD-1 | BioLegend | Cat#109110; RRID:AB_572017 |
| APC anti-mouse CD127 | BioLegend | Cat#135012; RRID:AB_1937216 |
| FITC anti-mouse/human KLRG1 | BioLegend | Cat#138410; RRID:AB_10643582 |
| PE anti-mouse TNFa | BioLegend | Cat#506306; RRID:AB_315427 |
| APC-Cy7 anti-mouse IFNg | BioLegend | Cat#505826; RRID:AB_2295770 |
| AlexaFluor 647 anti-mouse S100A9 | BD | Cat#565833; RRID:AB_2739373 |
| FITC anti-mouse CD107a | BioLegend | Cat#121606; RRID:AB_572007 |
| FITC anti-mouse CD4 | BioLegend | Cat#100406; RRID:AB_312691 |
| PE-Dazzle anti-mouse PD-1 | BioLegend | Cat#109115; RRID:AB_2566547 |
| APC anti-mouse CXCR5 | BioLegend | Cat#145506; RRID:AB_2561970 |
| PE-Cy7 anti-mouse CXCR6 | BioLegend | Cat#151119; RRID:AB_2721670 |
| PE anti-mouse FOXP3 | ThermoFisher | Cat#12-5773-82; RRID:AB_465936 |
| APC anti-mouse CD25 | BioLegend | Cat#102012; RRID:AB_312861 |
| APC-Cy7 anti-mouse/human CD44 | BioLegend | Cat#103028; RRID:AB_830785 |
| FITC anti-mouse/human GL7 | BioLegend | Cat#144603; RRID:AB_2561696 |
| PE-Cy7 anti-mouse/human B220 | BioLegend | Cat#103222; RRID:AB_313005 |
| APC-Cy7 anti-mouse CD19 | BioLegend | Cat#115529; RRID:AB_830706 |
| FITC goat anti-rat IgG2a | Bethyl Laboratories | Cat#A110-106F; RRID:AB_67282 |
| Rat anti-LCMV nucleoprotein | BioXCell | Cat#BE0106; RRID:AB_10949017 |
| Bacterial and virus strains | ||
| LCMV Clone 13 | Rafi Ahmed, PhD | Grown in house |
| Chemicals, peptides, and recombinant proteins | ||
| AZD1208 | Selleckchem | Cat#S7104 |
| LCMV GP33 tetramer | Made in house | N/A |
| LCMV GP276 tetramer | Made in house | N/A |
| LCMV NP396 tetramer | Made in house | N/A |
| Propidium iodide | BioLegend | Cat#421301; RRID:AB_2868885 |
| APC Annexin V | BD PharMingen | Cat#550474 |
| Catalase | Sigma | Cat#C4963 |
| BODIPY-C16 | Thermo Fisher | Cat#D3821 |
| 2-NBDG | Cayman Chemical | Cat#11046 |
| DAPI | Thermo Fisher | Cat#D1306; RRID:AB_2629482 |
| Mitotracker CMX Rosamine | Thermo Fisher | Cat#M7512 |
| Mitotracker Green | Thermo Fisher | Cat#M7514 |
| RIPA buffer | Boston Bioproducts | Cat#BP-115 |
| Phosstop | Roche/Sigma | Cat#4906845001 |
| COmplete protease inhibitor | Roche/Sigma | Cat#11697498001 |
| Super Signal West Femto | Thermo Fisher | Cat#34094 |
| Coumarin boronic acid | Cayman Chemical | Cat#14051 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma | Cat#P1585 |
| Critical commercial assays | ||
| Chromium Single Cell 30 Library & Gel Bead Kit v2 | 10x Genomics | Cat#PN-120267 |
| Chromium Single Cell A Chip Kit | 10x Genomics | Cat#PN-1000009 |
| Chromium i7 Multiplex Kit | 10x Genomics | Cat#PN-120262 |
| SPRIselect Reagent Kit | Beckman Coulter | Cat#B23318 |
| Kappa NGS quantification kit | KAPABiosystems | Cat#KK4824 |
| NextSeq 500/550 High Output Kit v2.5 (150 cycles) | Illumina | Cat#20024907 |
| Deposited data | ||
| scRNA-seq of myeloid cells 7 days post-infection with LCMV Armstrong or LCMV Cl13 | This paper | GEO: GSE167204 |
| Experimental models: Cell lines | ||
| Vero E6 cells | ATCC | Cat#CRL-1586; RRID:CVCL_0574 |
| Experimental models: Organisms/strains | ||
| C57BL/6 mice | Charles River | N/A |
| Software and algorithms | ||
| Cell Ranger 3.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| Seurat 3.0 | Butler et al., 2018 | https://satijalab.org/seurat/ |
| FlowJo 10.7.1 | Tree Star | N/A |
| Prism 6 | Graphpad Software | N/A |
| ImageJ 1.53 | NIH | https://imagej.nih.gov |
| MitoGraph 3.0 | Viana et al., 2015 | https://github.com/vianamp/MitoGraph |
Highlights.
PIM1 is highly expressed in suppressive neutrophils during chronic viral infection
PIM1 promotes mitochondrial fitness and cell survival in suppressive neutrophils
PIM kinase inhibition diminishes suppressive neutrophil-mediated immunosuppression
ACKNOWLEDGMENTS
This work is supported by NIH grants AI125741 and AI148403 (to W.C.) and DK127526 (to M.Y.K.); by an American Cancer Society research scholar grant (to W.C.); and by an Advancing a Healthier Wisconsin Endowment (AHW) grant (to W.C). G.X. is supported by The Elizabeth Elser Doolittle Postdoctoral Fellowship. M.Y.K. and A.K.B. are members of the Medical Scientist Training Program at the Medical College of Wisconsin (MCW), which is partially supported by a training grant from NIGMS (T32-GM080202). This research was completed in part with computational resources and technical support provided by the Research Computing Center at MCW.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109160.
REFERENCES
- Ahmed R, Salmi A, Butler LD, Chiller JM, and Oldstone MB (1984). Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J. Exp. Med 160, 521–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almand B, Clark JI, Nikitina E, van Beynen J, English NR, Knight SC, Carbone DP, and Gabrilovich DI (2001). Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. J. Immunol 166, 678–689. [DOI] [PubMed] [Google Scholar]
- An N, Kraft AS, and Kang Y (2013). Abnormal hematopoietic phenotypes in Pim kinase triple knockout mice. J. Hematol. Oncol 6, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, and Ahmed R (2006). Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439, 682–687. [DOI] [PubMed] [Google Scholar]
- Baumann T, Dunkel A, Schmid C, Schmitt S, Hiltensperger M, Lohr K, Laketa V, Donakonda S, Ahting U, Lorenz-Depiereux B, et al. (2020). Regulatory myeloid cells paralyze T cells through cell-cell transfer of the metabolite methylglyoxal. Nat. Immunol 21, 555–566. [DOI] [PubMed] [Google Scholar]
- Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DAA, and Wherry EJ (2009). Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol 10, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan D, Falcone J, Kanjiya MP, Park SH, Carbonetti G, Studholme K, Gomez M, Lu Y, Elmes MW, Smietalo N, et al. (2018). Fatty acid-binding protein 5 controls microsomal prostaglandin E synthase 1 (mPGES-1) induction during inflammation. J. Biol. Chem 293, 5295–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borillo GA, Mason M, Quijada P, Völkers M, Cottage C, McGregor M, Din S, Fischer K, Gude N, Avitabile D, et al. (2010). Pim-1 kinase protects mitochondrial integrity in cardiomyocytes. Circ. Res 106, 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasó-Maristany F, Filosto S, Catchpole S, Marlow R, Quist J, Francesch-Domenech E, Plumb DA, Zakka L, Gazinska P, Liccardi G, et al. (2016). PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat. Med 22, 1303–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DG, Teyton L, Oldstone MBA, and McGavern DB (2005). Intrinsic functional dysregulation of CD4 T cells occurs rapidly following persistent viral infection. J. Virol 79, 10514–10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, O’Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJ, et al. (2016). Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell 166, 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, Sowell RT, Kaech SM, and Pearce EL (2017). Metabolic Instruction of Immunity. Cell 169, 570–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau AL, Ikeda-Saito M, and Szweda LI (2003). Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry 42, 14846–14855. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamine T, Mastio J, and Gabrilovich DI (2015). Transcriptional regulation of myeloid-derived suppressor cells. J. Leukoc. Biol 98, 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamine T, Dominguez GA, Youn J-I, Kossenkov AV, Mony S, Alicea-Torres K, Tcyganov E, Hashimoto A, Nefedova Y, Lin C, et al. (2016). Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci. Immunol 1, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornberg M, Kenney LL, Chen AT, Waggoner SN, Kim SK, Dienes HP, Welsh RM, and Selin LK (2013). Clonal exhaustion as a mechanism to protect against severe immunopathology and death from an overwhelming CD8 T cell response. Front. Immunol 4, 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs JT, and Strack S (2007). Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 8, 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilek N, Vuillefroy de Silly R, Blancho G, and Vanhove B (2012). Myeloid-derived suppressor cells: mechanisms of action and recent advances in their role in transplant tolerance. Front. Immunol 3, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din S, Mason M, Völkers M, Johnson B, Cottage CT, Wang Z, Joyo AY, Quijada P, Erhardt P, Magnuson NS, et al. (2013). Pim-1 preserves mitochondrial morphology by inhibiting dynamin-related protein 1 translocation. Proc. Natl. Acad. Sci. USA 110, 5969–5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din S, Konstandin MH, Johnson B, Emathinger J, Völkers M, Toko H, Collins B, Ormachea L, Samse K, Kubli DA, et al. (2014). Metabolic dysfunction consistent with premature aging results from deletion of Pim kinases. Circ. Res 115, 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorhoi A, and Du Plessis N (2018). Monocytic Myeloid-Derived Suppressor Cells in Chronic Infections. Front. Immunol. 8, 1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorhoi A, Glaría E, Garcia-Tellez T, Nieuwenhuizen NE, Zelinskyy G, Favier B, Singh A, Ehrchen J, Gujer C, Münz C, et al. (2019). MDSCs in infectious diseases: regulation, roles, and readjustment. Cancer Immunol. Immunother. 68, 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard M, Kwok IWH, Chong SZ, Teng KWW, Becht E, Chen J, Sieow JL, Penny HL, Ching GC, Devi S, et al. (2018). Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity 48, 364–379.e8. [DOI] [PubMed] [Google Scholar]
- Flint DH, Tuminello JF, and Emptage MH (1993). The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem 268, 22369–22376. [PubMed] [Google Scholar]
- Fuller MJ, Khanolkar A, Tebo AE, and Zajac AJ (2004). Maintenance, loss, and resurgence of T cell responses during acute, protracted, and chronic viral infections. J. Immunol 172, 4204–4214. [DOI] [PubMed] [Google Scholar]
- Gabrilovich DI, and Nagaraj S (2009). Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol 9, 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundler R, Brault L, Gasser C, Bullock AN, Dechow T, Woetzel S, Pogacic V, Villa A, Ehret S, Berridge G, et al. (2009). Dissection of PIM serine/threonine kinases in FLT3-ITD-induced leukemogenesis reveals PIM1 as regulator of CXCL12-CXCR4-mediated homing and migration. J. Exp. Med 206, 1957–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwig MC, Viana MP, Egner JM, Harwig JJ, Widlansky ME, Rafelski SM, and Hill RB (2018). Methods for imaging mammalian mitochondrial morphology: A prospective on MitoGraph. Anal. Biochem 552, 81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, Kaplan RN, and Mackall CL (2014). Disruption of CXCR2-Mediated MDSC Tumor Trafficking Enhances Anti-PD1 Efficacy. Sci. Transl. Med 6, 237ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinesh GG, Mokkapati S, Zhu K, and Morales EE (2016). Pim kinase isoforms: devils defending cancer cells from therapeutic and immune attacks. Apoptosis 21, 1203–1213. [DOI] [PubMed] [Google Scholar]
- Kaech SM, and Cui W (2012). Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol 12, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Wang D, Daikoku T, Sun H, Dey SK, and Dubois RN (2013). CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell 24, 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton EK, McEachern K, Dillman KS, Palakurthi S, Cao Y, Grondine MR, Kaur S, Wang S, Chen Y, Wu A, et al. (2014). AZD1208, a potent and selective pan-Pim kinase inhibitor, demonstrates efficacy in preclinical models of acute myeloid leukemia. Blood 123, 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PS, and Ahmed R (2010). Features of responding T cells in cancer and chronic infection. Curr. Opin. Immunol 22, 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mulé J, Kerr WG, et al. (2005). Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med 11, 1314–1321. [DOI] [PubMed] [Google Scholar]
- Legros F, Lombès A, Frachon P, and Rojo M (2002). Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol. Biol. Cell 13, 4343–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang X, Chen Y, Xie Y, Liu J, Feng Q, Wang Y, Yuan W, and Ma J (2016). G-CSF is a key modulator of MDSC and could be a potential therapeutic target in colitis-associated colorectal cancers. Protein Cell 7, 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zavorotinskaya T, Dai Y, Niu X-H, Castillo J, Sim J, Yu J, Wang Y, Langowski JL, Holash J, et al. (2013). Pim2 is required for maintaining multiple myeloma cell growth through modulating TSC2 phosphorylation. Blood 122, 1610–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matloubian M, Concepcion RJ, and Ahmed R (1994). CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J. Virol 68, 8056–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina E, and Hartl D (2018). Myeloid-Derived Suppressor Cells in Infection: A General Overview. J. Innate Immun. 10, 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollaoglu G, Jones A, Wait SJ, Mukhopadhyay A, Jeong S, Arya R, Camolotto SA, Mosbruger TL, Stubben CJ, Conley CJ, et al. (2018). The Lineage-Defining Transcription Factors SOX2 and NKX2–1 Determine Lung Cancer Cell Fate and Shape the Tumor Immune Microenvironment. Immunity 49, 764–779.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita D, Katayama R, Sekimizu K, Tsuruo T, and Fujita N (2008). Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 68, 5076–5085. [DOI] [PubMed] [Google Scholar]
- Nagaraj S, Gupta K, Pisarev V, Kinarsky L, Sherman S, Kang L, Herber DL, Schneck J, and Gabrilovich DI (2007). Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat. Med 13, 828–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar YG, Rayman P, Jia X, Pavicic PG, Rini BI, Tannenbaum C, Ko J, Haywood S, Cohen P, Hamilton T, et al. (2017). Myeloid-Derived Suppressor Cell Subset Accumulation in Renal Cell Carcinoma Parenchyma Is Associated with Intratumoral Expression of IL1β, IL8, CXCL5, and Mip-1α. Clin. Cancer Res. 23, 2346–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefedova Y, Huang M, Kusmartsev S, Bhattacharya R, Cheng P, Salup R, Jove R, and Gabrilovich D (2004). Hyperactivation of STAT3 is involved in abnormal differentiation of dendritic cells in cancer. J. Immunol 172, 464–474. [DOI] [PubMed] [Google Scholar]
- Nefedova Y, Nagaraj S, Rosenbauer A, Muro-Cacho C, Sebti SM, and Gabrilovich DI (2005). Regulation of dendritic cell differentiation and antitumor immune response in cancer by pharmacologic-selective inhibition of the janus-activated kinase 2/signal transducers and activators of transcription 3 pathway. Cancer Res. 65, 9525–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris BA, Uebelhoer LS, Nakaya HI, Price AA, Grakoui A, and Pulendran B (2013). Chronic but not acute virus infection induces sustained expansion of myeloid suppressor cell numbers that inhibit viral-specific T cell immunity. Immunity 38, 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orecchioni M, Ghosheh Y, Pramod AB, and Ley K (2019). Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS−) vs. Alternatively Activated Macrophages. Front. Immunol 10, 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauken KE, and Wherry EJ (2015). Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 36, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñaloza HF, Alvarez D, Muñoz-Durango N, Schultz BM, González PA, Kalergis AM, and Bueno SM (2019). The role of myeloid-derived suppressor cells in chronic infectious diseases and the current methodology available for their study. J. Leukoc. Biol 105, 857–872. [DOI] [PubMed] [Google Scholar]
- Raber PL, Thevenot P, Sierra R, Wyczechowska D, Halle D, Ramirez ME, Ochoa AC, Fletcher M, Velasco C, Wilk A, et al. (2014). Subpopulations of myeloid-derived suppressor cells impair T cell responses through independent nitric oxide-related pathways. Int. J. Cancer 134, 2853–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafelski SM, Viana MP, Zhang Y, Chan Y-HM, Thorn KS, Yam P, Fung JC, Li H, Costa L.d.F., and Marshall WF (2012). Mitochondrial Network Size Scaling in Budding Yeast. Science 338, 822–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, and Lippincott-Schwartz J (2011). Fuse or die: Shaping mitochondrial fate during starvation. Commun. Integr. Biol 4, 752–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Cohen S, and Lippincott-Schwartz J (2015). Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redza-Dutordoir M, and Averill-Bates DA (2016). Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta 1863, 2977–2992. [DOI] [PubMed] [Google Scholar]
- Rice CM, Davies LC, Subleski JJ, Maio N, Gonzalez-Cotto M, Andrews C, Patel NL, Palmieri EM, Weiss JM, Lee JM, et al. (2018). Tumour-elicited neutrophils engage mitochondrial metabolism to circumvent nutrient limitations and maintain immune suppression. Nat. Commun 9, 5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose IA, and O’Connell EL (1967). Mechanism of aconitase action. I. The hydrogen transfer reaction. J. Biol. Chem 242, 1870–1879. [PubMed] [Google Scholar]
- Samse K, Emathinger J, Hariharan N, Quijada P, Ilves K, Völkers M, Ormachea L, De La Torre A, Orogo AM, Alvarez R, et al. (2015). Functional Effect of Pim1 Depends upon Intracellular Localization in Human Cardiac Progenitor Cells. J. Biol. Chem 290, 13935–13947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora A, Zielonka J, Lopez M, Joseph J, and Kalyanaraman B (2009). Direct oxidation of boronates by peroxynitrite: mechanism and implications in fluorescence imaging of peroxynitrite. Free Radic. Biol. Med 47, 1401–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm A, Valentine L, Potts R, and Premenko-Lanier M (2012). An intermediate dose of LCMV clone 13 causes prolonged morbidity that is maintained by CD4+ T cells. Virology 425, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen AL, Binstock BJ, Mishra BB, and Sharma J (2013). C-type lectin receptor Clec4d plays a protective role in resolution of Gram-negative pneumonia. J. Leukoc. Biol 94, 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Clavijo PE, Robbins Y, Patel P, Friedman J, Greene S, Das R, Silvin C, Van Waes C, Horn LA, et al. (2019). Inhibiting myeloid-derived suppressor cell trafficking enhances T cell immunotherapy. JCI Insight 4, e126853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trikha P, and Carson WE 3rd. (2014). Signaling pathways involved in MDSC regulation. Biochim. Biophys. Acta 1846, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari M, Fourati S, Brown DR, Silva de Castro I, Bissa M, Schifanella L, Doster MN, Foulds KE, Roederer M, Koup RA, et al. (2019). Myeloid Cell Crosstalk Regulates the Efficacy of the DNA/ALVAC/gp120 HIV Vaccine Candidate. Front. Immunol 10, 1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veglia F, Perego M, and Gabrilovich D (2018). Myeloid-derived suppressor cells coming of age. Nat. Immunol 19, 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veglia F, Tyurin VA, Blasi M, De Leo A, Kossenkov AV, Donthireddy L, To TKJ, Schug Z, Basu S, Wang F, et al. (2019). Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature 569, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga-Parga T, Sehrawat S, and Rouse BT (2013). Role of regulatory T cells during virus infection. Immunol. Rev 255, 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana MP, Lim S, and Rafelski SM (2015). Quantifying mitochondrial content in living cells. Methods Cell Biol. 125, 77–93. [DOI] [PubMed] [Google Scholar]
- Warfel NA, and Kraft AS (2015). PIM kinase (and Akt) biology and signaling in tumors. Pharmacol. Ther 151, 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfel NA, Sainz AG, Song JH, and Kraft AS (2016). PIM Kinase Inhibitors Kill Hypoxic Tumor Cells by Reducing Nrf2 Signaling and Increasing Reactive Oxygen Species. Mol. Cancer Ther. 15, 1637–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, and Ahmed R (2003). Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol 77, 4911–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Barber DL, Kaech SM, Blattman JN, and Ahmed R (2004). Antigen-independent memory CD8 T cells do not develop during chronic viral infection. Proc. Natl. Acad. Sci. USA 101, 16004–16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin G, Schauder DM, Lainez B, Weinstein JS, Dai Z, Chen Y, Esplugues E, Wen R, Wang D, Parish IA, et al. (2015). A Critical Role of IL-21-Induced BATF in Sustaining CD8-T-Cell-Mediated Chronic Viral Control. Cell Rep. 13, 1118–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhao Q, Gabrusiewicz K, Kong L-Y, Xia X, Wang J, Ott M, Xu J, Davis RE, Huo L, et al. (2019). FGL2 promotes tumor progression in the CNS by suppressing CD103+ dendritic cell differentiation. Nat. Commun 10, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J-I, Nagaraj S, Collazo M, and Gabrilovich DI (2008). Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J. Immunol 181, 5791–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, and Ahmed R (1998). Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med 188, 2205–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Nefedova Y, Lei A, and Gabrilovich D (2018). Neutrophils and PMN-MDSC: Their biological role and interaction with stromal cells. Semin. Immunol 35, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YP, Padgett L, Dinh HQ, Marcovecchio P, Blatchley A, Wu R, Ehinger E, Kim C, Mikulski Z, Seumois G, et al. (2018). Identification of an Early Unipotent Neutrophil Progenitor with Pro-tumoral Activity in Mouse and Human Bone Marrow. Cell Rep. 24, 2329–2341.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka J, Sikora A, Joseph J, and Kalyanaraman B (2010). Peroxynitrite is the major species formed from different flux ratios of co-generated nitric oxide and superoxide: direct reaction with boronate-based fluorescent probe. J. Biol. Chem 285, 14210–14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA sequencing data from this paper are available in the GEO database with accession number GEO: GSE167204. All other raw data and scripts are available from the lead contact upon request.