ABSTRACT
COVID-19 pandemic has become a global public health priority. The rapid increase in infection numbers, along with a significant number of deaths, has made the virus a serious threat to human health. Rapid, reliable, and simple diagnostic methods are critical to controlling the disease. While Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) is the current diagnostic gold standard, Reverse Transcriptase Loop-Mediated Isothermal Amplification (RT-LAMP) appears to be a compelling alternative diagnostic test due to its greater simplicity, shorter time to obtain a result, and lower cost. This study examined RT-LAMP application for rapid identification of SARS-CoV-2 infection compared to the RT-PCR assay. A systematic review and meta-analysis was conducted over six scientific databases in accordance with PRISMA guidelines. Original studies published in English conducted on human clinical samples were included. Articles evaluating the sensitivity and specificity of RT-LAMP relative to RT-PCR were considered eligible. Quality assessment of bias and applicability was examined based on QUADAS-2. A total of 351 studies were found based on the keywords and search queries. Fourteen eligible case–control studies fit the mentioned criteria. Quality assessment using QUADAS-2 indicated alow risk of bias for all included studies. All case studies, containing 2,112 samples, had acumulative sensitivity of 95.5% (CI 97.5% = 90.8–97.9%) and cumulative specificity of 99.5% (CI 97.5% = 97.7–99.9%). The RT-LAMP assay could be areliable alternative COVID-19 diagnostic method due to its reduced cost and processing time compared to RT-PCR. RT-LAMP could potentially be utilized during critical high-throughput and high-demand situations.
KEYWORDS: RT-LAMP, RT-PCR, SARS-CoV-2, COVID-19, diagnostic test
INTRODUCTION
The recently emerged novel Coronavirus Disease (COVID-19), the disease caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection, has caused a pandemic and become a global public health emergency [1]. As of 3 January 2021, the COVID-19 pandemic has resulted in a total of 83,322,449 confirmed cases and 1,831,412 deaths, according to the WHO COVID-19 report [World Health 2]. SARS-CoV-2 is currently known to be transmitted among humans through respiratory droplets and aerosols produced from infected persons while sneezing, talking, and/or coughing [3]. At present, effective antiviral drugs and vaccines for SARS-CoV-2 are still undergoing research [4]. The absence of suitable therapeutic agents necessitates prompt diagnosis with simple, rapid, and reliable detection of SARS-CoV-2 infection [5].
Diagnosis of early infection is difficult because COVID-19 manifests with nonspecific clinical symptoms, such as fever, cough, or shortness of breath, which overlap (at least in mild and early-stage cases) with those found in the common cold and influenza [1,3]. In addition, patients in the early stages of SARS-CoV-2 infection can remain asymptomatic. Symptoms appear in as little as 2 days or as many as 2 weeks after exposure [4]. Thus, confirmation of SARS-CoV-2 infection relies on nucleic acid testing, which detects viral RNA [6]. Early, rapid, and accurate identification of SARS-CoV-2 infected patients, even prior to an immune response and for asymptomatic carriers, is crucial not only for providing appropriate and timely medical support for patients but also for mitigating further spread [1,7,8].
The current gold standard for COVID-19 molecular diagnosis is based on real-time reverse transcription-polymerase chain reaction (RT-PCR), which detects SARS-CoV-2 RNA [6]. RT-PCR produces outstanding analytical performance with highly sensitive and specific results [8]. Nevertheless, RT-PCR-based approaches for COVID-19 detection suffer from several limitations, as these methods require highly skilled personnel and sophisticated equipment with poor availability (restricted to public health laboratories) [4]. As a result, they are considered impractical, especially in remote areas and developing countries with limited resources [3]. Furthermore, the fact that PCR-based methods are relatively time-consuming and complicated limits their ability to meet the demand for testing in an ongoing pandemic [3].
Rapidly growing number of COVID-19 incidences urges substitution of RT-PCR testing with an equally reliable, rapid, and cost-effective molecular detection method to facilitate the detection of SARS-CoV-2 infection [1,3]. In seeking to meet this challenge, a test kit that offers shorter turnaround time and is simple to operate would be highly desirable. The test should be able to identify infected patients even at an early stage. Ideally, it would be mobile, reducing the need for complicated equipment, and its results would be interpretable by the naked eye, making it feasible for use at public facilities, particularly health centers in rural areas [W. E. 9].
Reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) is a rapid, one-step DNA amplification technique [7]. This method has been applied in the detection of numerous pathogens, such as viruses, bacteria, and malaria [W. E. 9]. RT-LAMP is regarded as a promising point-of-care test due to several features: high sensitivity and specificity, quick reaction time, and reduced dependence on sophisticated equipment [5]. The LAMP reaction takes place at a constant temperature of 60°C to 65°C in under an hour [8]. Since the DNA amplification process occurs at a constant temperature, RT-LAMP can be performed on simpler equipment and eliminates the need for the thermal cycle in PCR [6]. A single reaction in RT-LAMP significantly shortens the reaction time and bypasses the DNA purification step. Thus, RT-LAMP can be used to rapidly detect SARS-CoV-2 [W. E. 9]. RT-LAMP results can be detected by visual turbidity or fluorescence in real time [7]. As such, the results can be interpreted by the untrained naked eye with a short turnaround time [5]. Finally, the simplicity of the procedure means that it can easily be mastered by junior laboratory technicians or health-care workers with little training [8].
Given these facts, a review was conducted in order to examine the diagnostic performance of RT-LAMP compared to RT-PCR, the current gold standard for COVID-19 diagnosis. As of the time that the review was performed, the authors had not found other similar studies analyzing the use of RT-LAMP for COVID-19. The review results were expected to provide credible evidence for the use of the proposed diagnostic tool, RT-LAMP, as a potential alternative to address the current issue.
MATERIALS & METHODS
This systematic review and meta-analysis followed the Preferred Reporting Items for Systematic Review and Meta-Analyses (PRISMA) guidelines [10] to properly identify relevant studies and literature used in this review.
Search Strategy
Two independent investigators performed thorough literature searches in a blinded fashion. Discrepancies were resolved through discussion between the investigators. The literature search was done through six scientific databases – PubMed, ScienceDirect, Scopus, ProQuest, Wiley, and EBSCohost – for studies published through 12 October 2020. Literature search strategies were developed using medical subject headings (MeSH) in scientific databases with text words related to COVID-19 diagnosis, RT-PCR, and RT-LAMP. The search was performed using several keywords, consisting of ‘reverse transcriptase polymerase chain reaction,’ ‘RT-PCR,’ ‘reverse transcriptase loop mediated isothermal amplification,’ ‘RT-LAMP,’ ‘SARS-CoV-2,’ and ‘COVID-19’ in different combinations. We also produced a manual, hand-searching reference list from the included studies. The retrieved results were subsequently deduplicated and screened against the pre-specified eligibility criteria.
Study Eligibility Criteria
Inclusion criteria were set to filter for primary studies investigating the diagnostic study of RT-LAMP in COVID-19 diagnosis using PICO (Patient, Intervention, Control, Outcome) criteria in which patients were suspected or confirmed COVID-19 patients; intervention or index test using RT-LAMP kit, control or reference test using RT-PCR, and objective or outcome of sensitivity and specificity (diagnostic value). We included published studies consisting of prospective and retrospective, cross-sectional, and cohort studies from human clinical samples of COVID-19-suspected patients that evaluated the sensitivity and specificity of RT-LAMP for COVID-19 diagnosis in comparison to RT-PCR. Meanwhile, studies were excluded if any of the following criteria were met: (1) review articles, case series, or letters to editor; (2) in-vitro studies without clinical samples; (3) irretrievable full-text articles; and (4) non-English articles.
Study Selection
After the previous search in the proposed databases was completed, search results were stored in a Microsoft Excel spreadsheet (Microsoft Corp, Redmond, WA, USA). Authors manually checked and removed duplicate articles, and the spreadsheet was also utilized to manage the data. The authors independently reviewed the literature search. A screening of titles and abstracts of the selected articles was conducted to exclude studies in line with the exclusion criteria. The reviewers noted the reason for exclusion of items in the spreadsheet. If there was any uncertainty regarding a particular study, it was included for the following step. Next, the authors read the full text in order to exclude studies that did not meet the inclusion criteria. The selected studies were then validated by conducting a meeting among the reviewers to select which articles were considered eligible for review. A data extraction form was built to compile data from the included studies.
Data Extraction and Quality Assessment
Two reviewers (ADS, LW) independently collected the data. Data were justified and discussed by both authors to ensure completeness and plausibility before being synthesized. The variables reported included: authors, year published, location of study, study design, sample size, study sample/population, index test (RT-LAMP), reference test (RT-PCR), clinical setting (inpatient vs outpatient), and level of evidence. The main outcomes for this review were the diagnostic values of RT-LAMP, consisting of the true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) of all studies, providing their sensitivity and specificity values.
The included studies were then assessed for their methodological quality to reduce systematic biases and inferential errors from the data extracted. Two reviewers (ADS, LW) independently assessed the risk of bias of the included studies using Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) [11]. QUADAS-2 assesses four areas of bias [11]: patient selection, index test, reference standards, and flow/timing. All studies were subsequently judged to have low, unclear, or high risks of bias. The authors also assessed the concerns about applicability in terms of three areas: patient selection, index test, and reference standards, which were then judged to generate low, unclear, or high concern regarding applicability. A summary of the risk of bias is extrapolated into graphs that were generated in a Microsoft Excel spreadsheet.
Statistical Analysis
Quantitative analysis was performed using the outcomes of the included studies. Values of test accuracy were compared with RT-PCR methods, including sensitivity, specificity, true positives (TP), false negatives (FN), false positives (FP), and true negatives (TN) using data extracted from sources or calculated from the available data. RT-PCR was considered a reference test and comparator to the index test, RT-LAMP. Results of individual studies were graphically presented in a forest plot in addition to diagnostic value and random effects curve. Meta-analysis was conducted using MetaDTA software (University of Leicester, Leicester, England).
RESULTS
Study Selection and Characteristics
The literature search process is summarized in Figure 1 using the PRISMA guideline [10] flowchart. The initial search yielded 364 records, of which 16 were deduplicated and 332 were excluded following the title and abstract screening. A total of 16 articles were then properly assessed through full-text assessments, resulting in two articles being excluded due to inappropriate study design.
Figure 1.
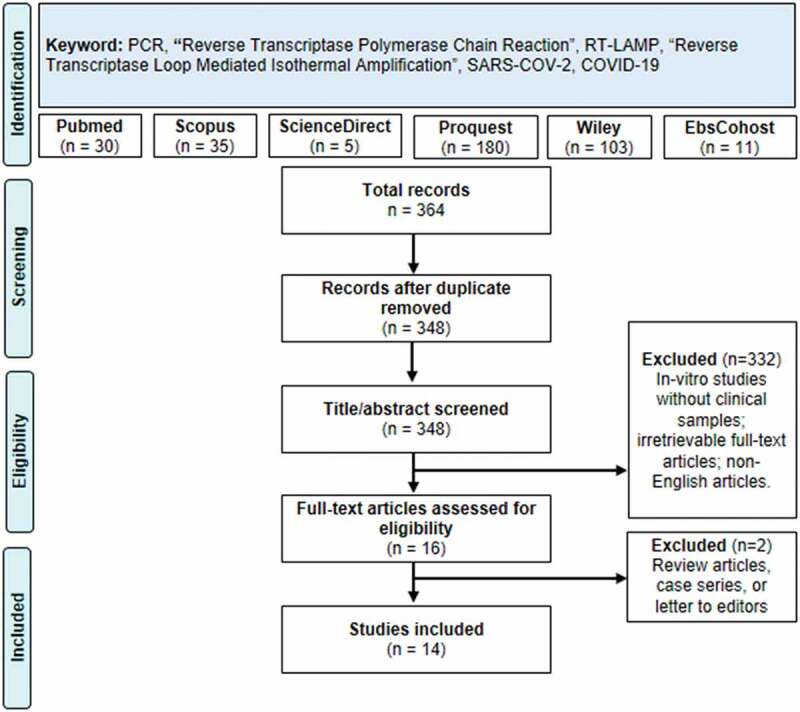
PRISMA Flow-chart describing the literature search and screening strategy of the review
The 14 articles included contained a total of 2,112 patients or samples, consisting of either suspected or confirmed COVID-19 patients. Patients’ characteristics were not mentioned in most studies; thus, the authors of this study did not extract characteristics. The included studies consist of a wide variety of samples: five studies from China; two studies from Germany [12,13]; and one study each from Hong Kong [8], China [6], Japan [14], Malaysia [15], South Korea [7], Canada [16], Australia [17], and the U.S. [4].
Regarding the index test, six studies did not report the RT-LAMP kit used [1, 7, 8, W. E. 4, 9, 15, 18], while other studies used the Warmstart RT-LAMP Assay Kit (n = 3) [5,7,16], the Variplex RT-LAMP Assay Kit (n = 1) [13], the Loopamp RNA Amplification Kit (n = 2) [6,14], RT-LAMP Mastermix kit (n = 1) [17], or the specific colorimetric and fluorescence RT-LAMP Kit (n = 1) [12]. As for the reference test, the majority of studies used the qRT-PCR Kit, with eight studies specifying the kit used (LightMix RT-PCR [13]; ABI COVID-19 qRT-PCR Kit [1]; NPMA qRT-PCR [18]; BGI qRT-PCR Kit [6]; Bio-rad iTaq Universal Probes One-Step Kit qRT-PCR [7]; Quantabio qScript XLT One-Step RT-qPCR ToughMix [17]; LightCycler® Multiplex RNAVirus Master kit [12]; Invitrogen Superscript III one step RT-PCR system [15]; Shanghai ZJ Bio-Tech 2019-nCoV RT-PCR kit [W. E. 9]). The remaining five studies did not specify the reference RT-PCR kit used [4,5,8,14,16]. The RT-PCR kits utilized in the studies were approved as COVID-19 diagnosis reference methods by the WHO (n = 4) [1, W. E. 9, 13, 15], CDC [190] (n = 1) [17]; and FDA (n = 2) [7,12]. Most studies used a cross-sectional study design (n = 13), with the one exception using prospective cohorts in its study [1].
Primers for RT-LAMP assays were designed to target several sequences of the SARS-CoV-2 gene, such as those encoding the envelope (E) (n = 3) [8, W. E. 9, 15]; nucleocapsid (N) (n = 6) [7, W. E. 5, 9, 12, 17, 18]; spike protein (S) (n = 3) [1,6,16]; membrane protein (M) (n = 1) [13]; open reading frame 1ab (ORF1ab) (n = 2) [W. E. 6, 9]; ORF3a (n = 1) [8]; RNA-dependent RNA polymerase (RdRP) (n = 1) [15]; and non-structural protein 3 (NSP 3) (n = 1) [4]. One study did not specify the targeted gene [14].
Meanwhile, RT-PCR, the reference test for validating the RT-LAMP test results, targeted the N gene (n = 6) [1, W. E. 4, 9, 14, 16, 18]; E gene (n = 5) [12,13,15–17]; S gene (n = 2) [W. E. 6, 9]; ORF1ab gene (n = 4) [1, W. E. 6, 9, 18]; and RdRP gene (n = 1) [15]. The included studies’ characteristics are summarized in Table 1.
Table 1.
Included studies characteristics
| Studies, year | Studies characteristics | ||||||||
| Location | Design | Sample size | Population/Sample | Index Test |
Index Test Gene Target |
Reference Test | Ref. Test Gene Target | Clinical settings | |
| 16 | Calgary, Canada | Cross- sectional |
124 | COVID-19 patients/Archived nasopharyngeal swab | Warmstart RT-LAMP assay | S and RdRP gene | RT-PCR | E gene and N2 gene | N/A |
| 13 | Penzberg, Germany | Cross-sectional | 137 | COVID-19 patients/Archived nasopharyngeal swab | Variplex RT-LAMP assay | [M] gene | LightMix E-Gene RT-PCR* | E gene | N/A |
| 8 | Hongkong | Cross-sectional | 362 | COVID-19 and other viral infection/Nasopharyngeal swab | COVID-19 RT-LAMP assay | orf3a and E gene | qRT-PCR | N/A | Inpatient |
| 14 | Saitama, Japan | Cross-sectional | 76 | Suspected COVID-19 patient/N/A | Loopamp SARS-COV-2 Detection Kit | N/A | qRT-PCR | N gene | Outpatient |
| 1 | Guangdong, China | Cohort | 481 | Suspected COVID-19 patient/N/A | RT-LAMP Kit | S gene | ABI COVID-19 qRT-PCR Kit* | ORF1ab geneN genes | Inpatient |
| 18 | Wenzhou, China | Cross-sectional | 260 | COVID-19 and other viral infection/Nasopharyngeal swab | RT-LAMP Kit | N gene | NPMA qRT-PCR | orf1ab gene, N gene | N/A |
| 6 | Beijing, China | Cross-sectional | 130 | COVID-19 and pneumonia patients/|Nasopharyngeal swab | Loopamp RNA amplification kit [RT-LAMP] | orf1ab gene; S gene | BGI RT-PCR Kit* | orf1ab and S genes | N/A |
| 4 | Belmont, USA | Cross-sectional | 20 | N/A/Serum, urine, oropharyngeal,nasopharyngeal swab | RT-LAMP Kit | NSP 3 gene; ORF1Ab gene | qRT-PCR | N1 and N2 gene | Outpatient |
| 5 | Nantong, China | Cross-sectional | 56 | Suspected COVID-19 patients/Nasopharyngeal swab | Warmstart RT-LAMP Assay | N gene | RT-qPCR kit | N/A | Inpatient |
| 7 | Seoul, South Korea | Cross-sectional | 154 | COVID-19 and other viral infection/Nasopharyngeal swab | Warmstart RT-LAMP Assay | N gene | Bio-rad iTaq Universal Probes One-Step Kit qRT-PCR*** | N/A | N/A |
| 17 | Melbourne, Australia | Cross-sectional | 157 | N/A/Nasopharyngeal swabs | RT-LAMP Mastermix | N gene | Quantabio qScript XLT One-Step RT-qPCR ToughMix** | E gene | N/A |
| 12 | Heidelberg, Germany | Cross-sectional | 77 | Random eligible sample/Nasopharyngeal swab | Colorimetric and Fluorescence RT-LAMP | N gene | LightCycler® Multiplex RNAVirus Master kit*** | E gene | N/A |
| 15 | Sungai Buloh, Malaysia | Cross-sectional | 89 | COVID-19 patients/Archived nasopharyngeal swab | RT-LAMP Kit | RdRP and E genes | Invitrogen Superscript III one step RT-PCR system* | RdRP and E genes | N/A |
| 9 | Ningbo, China | Cross-sectional | 16 | COVID-19 and other viral infection/Nasopharyngeal swab | RT-LAMP Kit | orf1ab, N, and E gene | Shanghai ZJ Bio- Tech 2019-nCoV RT-PCR kit* | orf1ab, S, and N gene | Outpatient |
Legend: E (envelope]; N (nucleocapsid), S (spike protein); M (membrane protein); ORF1ab (Open Reading Frame 1ab); RdRP (RNA dependent RNA polymerase gene); NSP3 (nonstructural protein 3)
Notes:
*Approved reference method by WHO
**Approved reference method by CDC*** Approved reference method by FDA
Risk of Bias Assessment
The risk of bias assessment using QUADAS-2 [11] revealed that most studies carried a low or unclear risk of bias. On the patient selection aspects, 10 out of 14 studies had unclear risk of bias, with 4 out of 14 showing a low risk of bias. This result can be explained by the fact that most studies did not explicitly declare their study design. Meanwhile, in terms of the reference test, 7 out of 14 studies had an unclear risk of bias, with another seven studies carrying a low risk of bias. Unclear risk of bias was assigned for studies that did not clearly state the blinded nature of the data analysis of reference tests without knowing index test results. In the index test assessment, 5 out of 14 studies had an unclear risk of bias, four because they did not mention the blinded fashion of data extraction and one because it had no pre-specified threshold on the index test. On the flow and timing aspects, 3 out of 14 studies demonstrated an unclear risk of bias due to unclear statements regarding the interval time between the reference test and the index test. All studies not mentioned above yielded a low risk of bias.
Our review question did not focus on any particular patient demographics, and none of the included studies excluded patients based on demographic characteristics. Therefore, there was no concern regarding applicability for patient selection. The index test, RT-LAMP, was not specifically mentioned in our review question. Thus, all RT-LAMP kits are applicable for our review. The reference standard tests in nearly all of the included studies used qRT-PCR, the current gold standard test for COVID-19 diagnosis. As a result, these studies were determined to be of low concern regarding their applicability in terms of both index tests and standard tests. Therefore, all included studies generated only low concern as to applicability in all aspects. A summary of the QUADAS-2 assessment can be found in Supplementary (Figure S1).
Outcome of Studies
A summary of the outcome of each included study is listed in Table 2. Study outcomes, consisting of TP, FN, FP, and TN values, are listed. The sensitivity and specificity values for each study were then calculated. The sensitivity of RT-LAMP is found to range from 75% to 100%, while its specificity is found to range from 90% to 100%. All outcomes of each study were extracted to generate the cumulative outcome if the mentioned study used several subgroups for their analysis. Out of all 14 studies, 10 studies showed a sensitivity value of more than 90%, with 4 of them having a sensitivity value of 100% [W. E. 6, 9, 12, 15]. Three studies showed sensitivity values of more than 80% [1,7,17], and only one study had a sensitivity value below 80% [13]. In terms of specificity values, all studies showed a high degree of specificity, with nine studies showing 100% specificity while the other five studies showed a specificity value of more than 98% [1,4,5,7,18]. Both sensitivity value and specificity value were then displayed in a forest plot, pictured in Figure 2, to represent the outcomes of all studies. As shown in the plot, each of the studies’ sensitivity and specificity values is shown with a confidence interval (CI) of 95%. Data on limit of detection (LOD) were also extracted from each article and ranged from 1 to 304 RNA copies per reaction. However, one study used another unit of measurement with a result of 500 RNA copies/mL [18].
Table 2.
Outcome of studies
| Studies, year | Outcome of Studies | |||||||
| True Positive (TP) | False Negative (FN) | False Positive (FP) | True Negative (TN) | Total Sample | Sensitivity (Sn; %) | Specificity (Sp; %) | Limit of Detection (copies per reaction) | |
| 16 | 65 | 1 | 0 | 58 | 124 | 98.5 | 100 | 25 |
| 13 | 72 | 24 | 0 | 41 | 137 | 75 | 100 | N/A |
| 8 | 219 | 4 | 0 | 143 | 366 | 98.2 | 100 | 42 |
| 14 | 30 | 2 | 0 | 44 | 76 | 93.8 | 100 | 10 |
| 1 | 72 | 9 | 4 | 396 | 481 | 88.9 | 99 | 4.23 |
| 18 | 43 | 4 | 1 | 212 | 260 | 91.5 | 99.5 | 500* |
| 6 | 58 | 0 | 0 | 72 | 130 | 100 | 100 | 20 |
| 4 | 19 | 1 | 2 | 18 | 40 | 95 | 90 | 304 |
| 5 | 34 | 2 | 2 | 18 | 56 | 94.4 | 90 | 118.6 |
| 7 | 14 | 2 | 2 | 138 | 156 | 87.5 | 98.6 | 100 |
| 17 | 93 | 14 | 0 | 50 | 157 | 86.9 | 100 | 50 |
| 12 | 17 | 0 | 0 | 17 | 34 | 100 | 100 | 10 |
| 15 | 47 | 0 | 0 | 42 | 89 | 100 | 100 | 1 |
| 9 | 8 | 0 | 0 | 8 | 16 | 100 | 100 | 80 |
Note:
* Used unit of measurement: RNA copies/mL
Figure 2.
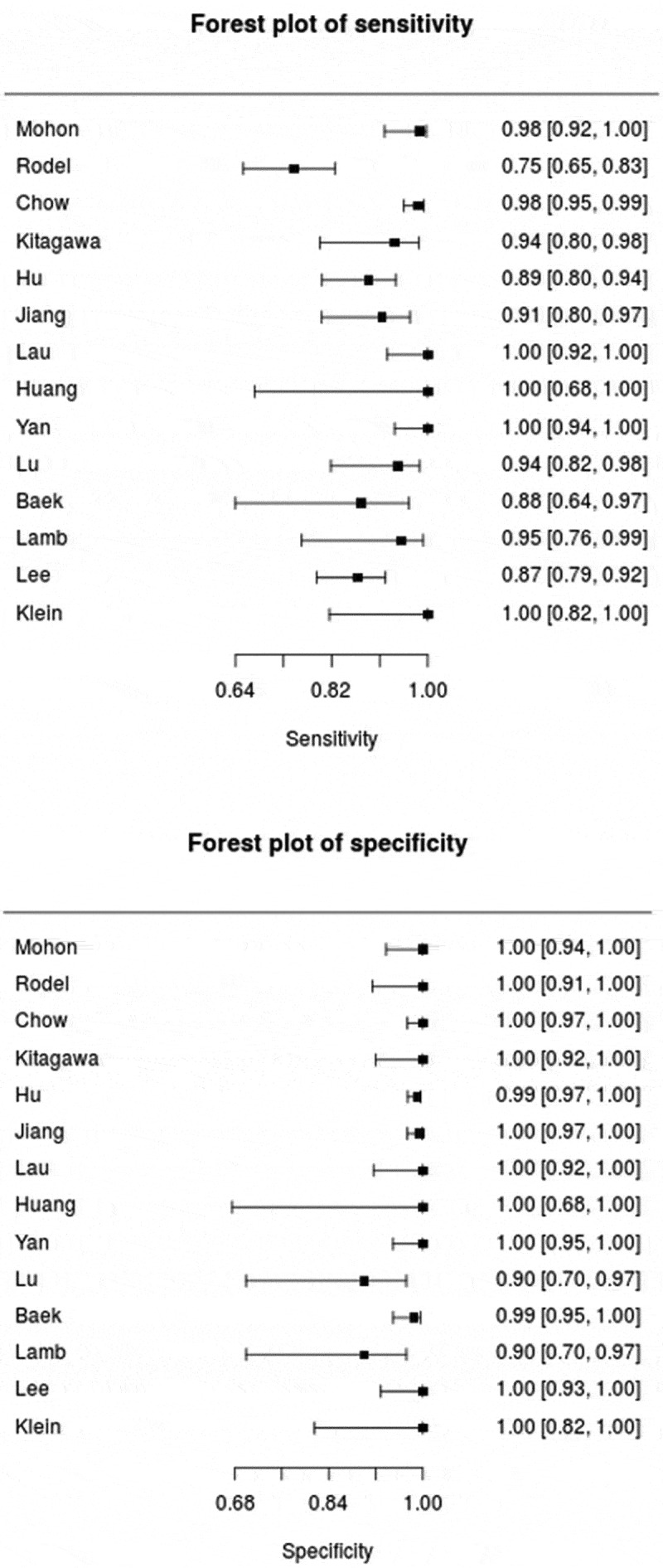
The forest plot of sensitivity and specificity of included studies on RT-LAMP diagnostic performance
Meta-Analysis of Sensitivity and Specificity
We performed a pool analysis on 14 included studies using RT-LAMP as the index test and RT-PCR as the reference test. In the pooled analysis, we estimate the pooled sensitivity and specificity for all studies and report the data in a SROC curve, as seen in Figure 3. The result of the pooled analysis is summarized in Table 3. Pooled sensitivity and specificity are found to be 99.5% (CI 97.5%: 90.8–97.9%) and 95.5% (CI 97.5%: 97.7–99.9%) respectively, indicating good overall performance by RT-LAMP as a diagnostic test thus far. The false positive rate is also considered low, at 0.5% (CI 97.5%: 0.1–0.23%).
Figure 3.
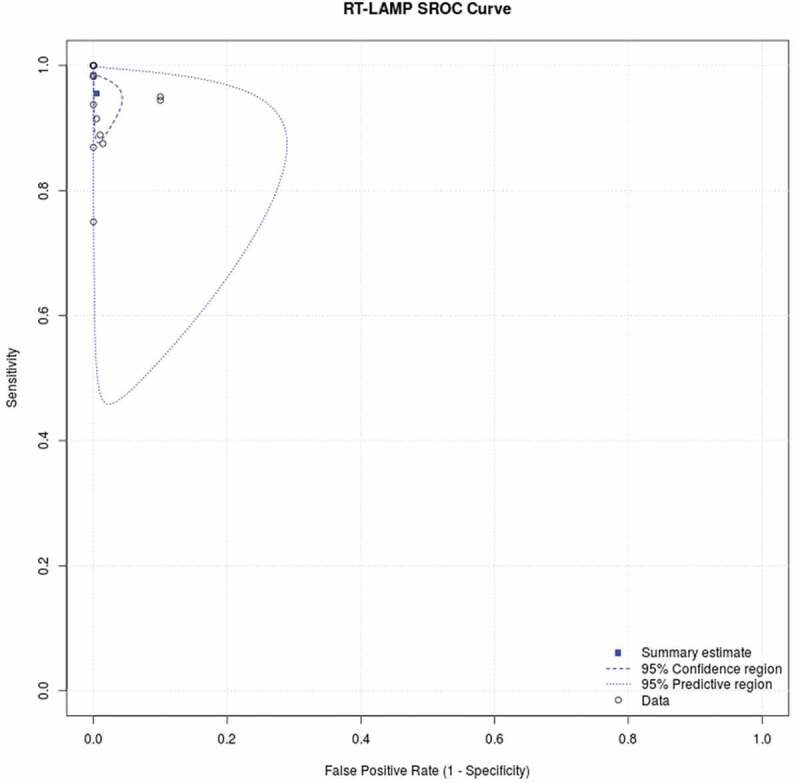
Summary Receiver Operating Characteristic (SROC) illustrates RT-LAMP diagnostic performance as compared to RT-PCR in COVID-19 diagnosis
Table 3.
Pooled analysis outcome of meta-analysis on 14 studies, summarized in this table with sensitivity, specificity, false positive rate, and logit of sensitivity and specificity
| Pooled Analysis Outcome (n = 14) | |||
| Parameter | Estimate | 2.5% CI | 97.5% CI |
| Sensitivity | 0.955 | 0.908 | 0.979 |
| Specificity | 0.995 | 0.977 | 0.999 |
| False Positive Rate | 0.005 | 0.001 | 0.023 |
| logit(sensitivity) | 3.053 | 2.286 | 3.819 |
| logit(specificity) | 5.26 | 3.738 | 6.782 |
DISCUSSION
As of the authors’ latest search, this study is the first systematic review and meta-analysis examining the performance and diagnostic value of RT-LAMP as a test for COVID-19. Most included studies used RT-LAMP as the index test with RT-PCR as the reference test and found a sensitivity of at least 90%, with four studies reporting 100% sensitivity and specificity. While there are three studies with sensitivity values of <90%, with one study reporting 75% sensitivity, we can safely assume that all studies reported a high diagnostic value for RT-LAMP in the diagnosis of COVID-19. At the time of this writing, the only published meta-analysis focusing on using RT-LAMP for diagnosis was the recent article by [20], which primarily investigated the performance of isothermal nucleic acid tests for human coronaviruses. The authors used 81 articles for the systematic review and 26 articles for subgroup analysis in the meta-analysis. The study reported a pooled sensitivity of 0.94 (CI 95%: 0.90–0.96%) for purified RNA from samples and sensitivity at 0.78 (CI 95%: 0.65–0.87%) for crude samples. Our review obtains the same result regarding the performance of RT-LAMP, finding a high sensitivity value. However, our review does not analyze the difference between the samples used in our analysis.
For all the studies, the QUADAS-2 assessment returned a low or unclear rating for risk of bias and concerns about applicability. As most studies yielded a low risk of bias, all studies were considered suitable to be included in the review and pooled analysis, as there is no indication of any high risk of bias or inapplicable study results.
While the majority of the studies seemed to be predominantly similar, there are some notable studies that evaluate various aspects of the performance of RT-LAMP. Rodel et al. [13] performed a diagnostic test to assess RT-LAMP in various respiratory samples and compared with RT-PCR and a combination of RT-LAMP and RT-PCR. They reported a combination of RT-PCR and RT-LAMP having a sensitivity value of 92–100%. The other 13 studies compared the diagnostic value of RT-LAMP kit alone to that of RT-PCR. Most studies used nasopharyngeal swabs obtained previously, while 4,used various samples, from respiratory samples to urine. However, the authors chose nasopharyngeal samples for the data extraction to ensure similarity to other studies.
RT-LAMP is considered to be a novel approach in diagnostic tests due to its rapid and low-cost kit [21], as mentioned by [22]. It has been used for detecting both DNA and RNA viral pathogens, such as foot-and-mouth disease [23], human immunodeficiency virus (HIV), Japanese encephalitis virus, chikungunya virus, human papillomavirus, dengue virus, West Nile virus, and mumps virus [3]. Furthermore, RT-LAMP has been reported to be effective for detecting other coronaviruses, such as severe acute respiratory syndrome coronavirus (SARS-CoV) [24] and the Middle East respiratory syndrome coronavirus (MERS-CoV) [P. 25]. The most recent study on the potential for using RT-LAMP as an emerging diagnostic test for COVID-19 was a mini-review by Thompson et al. [26] and Kashir et al. [27]. The lack of facilities that accommodate RT-PCR has given birth to cheaper and affordable diagnostic kits for COVID-19. Rodel et al. [13] also mentioned a faster cycle of RT-LAMP, as there is no need for the thermal cycling process. However, all included studies have the same view on the potential of RT-LAMP.
Our study identified all relevant peer-reviewed studies published to derive objective conclusions on the potential of RT-LAMP for the diagnosis of COVID-19. The authors have ensured that this study adheres to PRISMA guidelines to ensure objective review and a low risk of bias. The systematic review was conducted with an objective screening process using pre-determined keywords and guidelines to ensure the reproducibility of the authors’ method. The screening process, from article screening and selection to data extraction, was documented properly. Pooled analysis is conducted to report the objective cumulative outcome (sensitivity and specificity) of all included studies to properly highlight the pooled conclusion of the included articles. Nevertheless, this review has several limitations. The authors have noted that this review primarily uses RT-LAMP diagnostic value data on each study by extracting the cumulative data from all studies. As such, there was no extraction on subgroup analysis, which has been used in some studies. In our search strategy, the authors used several databases to find peer-reviewed articles relevant to the RT-LAMP diagnostic value for COVID-19. A systematic review and meta-analysis using preprints and ongoing clinical trials could be performed to gather more data for the systematic review while keeping in mind the need to properly assess study quality. Subgroup analysis, particularly on types of RT-LAMP used in trials, could also be used.
CONCLUSION
In conclusion, this systematic review and meta-analysis revealed the performance of RT-LAMP in diagnosing COVID-19 as compared with RT-PCR, the current gold-standard diagnostic tool. A systematic review of 14 studies showed a comparably high diagnostic value for RT-LAMP, as seen in its sensitivity and specificity values. The pooled analysis of all included studies has revealed a sensitivity value of 99.5% (CI 95%: 90.8–97.9%) and a specificity value of 95.5% (CI 95%: 97.7–99.9%), respectively. Thus, the authors conclude that RT-LAMP performs well and has high potential in the diagnosis of COVID-19. The authors have noted several limitations, including only using peer-reviewed studies and the lack of subgroup analysis. This leaves room for further improvement for further studies and reviews on the same topics.
Acknowledgments
We would like to thank Jeremy Rafael Tandaju and Brenda Cristie Edina (Faculty of Medicine Universitas Indonesia, Jakarta, Indonesia) for their methodological and statistical advice.
Funding Statement
The authors declare that this paper was not supported by any funding agency or grant.
Disclosure of potential conflicts of interest
In accordance with the Taylor & Francis policy and my ethical obligation as a researcher, I am reporting that I have not received funding from a company that may be affected by the research reported in the enclosed paper. I have disclosed those interests fully to Taylor & Francis, and I have in place an approved plan for managing any potential conflicts arising from that involvement.
DECLARATION OF DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author, ADS, upon reasonable request.
References
- [1].Hu X, Deng Q, Li J, et al. Development and clinical application of a rapid and sensitive loop-mediated isothermal amplification test for SARS-CoV-2 infection. MSphere. 2020;5(4):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard.Geneva: World Health Organization. 2021. [Google Scholar]
- [3].Augustine R, Hasan A, Das S, et al. (2020). Loop-mediated isothermal amplification (LAMP): a rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. In Biology (Basel) (Vol. 9, Issue 8, pp. 1–17). MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lamb LE, Bartolone SN, Ward E, et al. Rapid detection of novel coronavirus/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS ONE. 2020;15(6):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lu R, Wu X, Wan Z, et al. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-COV-2. Int J Mol Sci. 2020;21(8):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yan C, Cui J, Huang L, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26(6):773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Baek YH, Um J, Antigua KJC, et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerging Microbes and Infections. 2020;9(1):998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chow FWN, Chan TTY, Tam AR, et al. A rapid, simple, inexpensive, and mobile colorimetric assay COVID-19-LAMP for mass on-site screening of COVID-19. Int J Mol Sci. 2020;21(15):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Huang WE, Lim B, Hsu C, et al. RT‐LAMP for rapid diagnosis of coronavirus SARS‐CoV‐2. Microb Biotechnol. 2020;13(4):950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Reitsma JB, Leeflang MMG, Sterne JAC, et al. Research and reporting methods accuracy studies. Ann Intern Med. 2011;155(4):529–536. [DOI] [PubMed] [Google Scholar]
- [12].Klein S, Müller TG, Khalid D, et al. SARS-CoV-2 RNA extraction using magnetic beads for rapid large-scale testing by RT-qPCR and RT-LAMP. Viruses. 2020;12(8):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rödel J, Egerer R, Suleyman A, et al. Use of the variplex™ SARS-CoV-2 RT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J Clin Virol. 2020;132:104616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kitagawa Y, Orihara Y, Kawamura R, et al. Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J Clin Virol. 2020;129:104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lau YL, Ismail I, Mustapa NI, et al. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of SARS-CoV-2. PeerJ. 2020;(2020(6):e9278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mohon AN, Oberding L, Hundt J, et al. Optimization and clinical validation of dual-target RT-LAMP for SARS-CoV-2. J Virol Methods. 2020;286:113972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lee JYH, Best N, McAuley J, et al. Validation of a single-step, single-tube reverse transcription loop-mediated isothermal amplification assay for rapid detection of SARS-CoV-2 RNA. J Med Microbiol. 2020;69(9):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jiang M, Pan W, Arasthfer A, et al. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. Front Cell Infect Microbiol. 2020;10:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Redfiield R. Centers for Disease Control and Prevention (CDC) 2019-Novel Coronavirus (2019-nCoV) Real-Time Reverse Transcriptase (RT)-PCR Diagnostic Panel Centers. Maryland: U.S. Food and Drug Administration. 2020:1–12. [Google Scholar]
- [20].Subsoontorn P, Lohitnavy M, Kongkaew C. The diagnostic accuracy of nucleic acid point-of-care tests for human coronavirus: a systematic review and meta-analysis. Sci Rep. 2020;10(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sahoo PR, Sethy K, Mohapatra S, et al. Loop mediated isothermal amplification: an innovative gene amplification technique for animal diseases. Vet World. 2016;9(5):465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ganguli A, Mostafa A, Berger J, et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(37):22727–22735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Farooq U, Latif A, Irshad H, et al. Loop-mediated isothermal amplification (RT-LAMP): a new approach for the detection of foot-and-mouth disease virus and its sero-types in Pakistan. Iranian Journal of Veterinary Research. 2015;16(4):331–334. Latif,;, Irshad,;, Ullah,;, Zahur,;, Naeem,;, Khan,;, Ahmed,;, Rodriguez,;, & Smoliga, G. [PMC free article] [PubMed] [Google Scholar]
- [24].Kim JH, Kang M, Park E, et al. A simple and multiplex loop-mediated isothermal amplification (LAMP) assay for rapid detection of SARS-CoV. BioChip J. 2019;13(4):341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huang P, Wang H, Cao Z, et al. A rapid and specific assay for the detection of MERS-CoV. Front Microbiol. 2018;9. (MAY). 10.3389/fmicb.2018.01101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Thompson D, Lei Y. Mini review: recent progress in RT-LAMP enabled COVID-19 detection. Sensors and Actuators Reports. 2020;2(1):100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kashir J, Yaqinuddin A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med Hypotheses. 2020;141:109786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, ADS, upon reasonable request.


