Abstract
Human dental caries is an intractable biofilm-associated disease caused by microbial interactions and dietary sugars on the host’s teeth. Commensal bacteria help control opportunistic pathogens via bioactive products such as hydrogen peroxide (H2O2). However, high-sugar consumption disrupts homeostasis and promotes pathogen accumulation in acidic biofilms that cause tooth-decay. Here, we exploit the pathological (sugar-rich/acidic) conditions using a nanohybrid system to increase intrinsic H2O2 production and trigger pH-dependent reactive oxygen species (ROS) generation for efficient biofilm virulence targeting. The nanohybrid contains glucose-oxidase that catalyzes glucose present in biofilms to increase intrinsic H2O2, which is converted by iron oxide nanoparticles with peroxidase-like activity into ROS in acidic pH. Notably, it selectively kills Streptococcus mutans (pathogen) without affecting Streptococcus oralis (commensal) via preferential pathogen-binding and in situ ROS generation. Furthermore, nanohybrid treatments potently reduced dental caries in a rodent model. Compared to chlorhexidine (positive-control), which disrupted oral microbiota diversity, the nanohybrid had significant higher efficacy without affecting soft-tissues and the oral-gastrointestinal microbiomes, while modulating dental health-associated microbial activity in vivo. The data reveal therapeutic precision of a bi-functional hybrid nanozyme against a biofilm-related disease in a controlled-manner activated by pathological conditions.
Keywords: Hybrid nanozyme, Glucose oxidase, Catalytic nanoparticles, Biofilm, Polymicrobial, Dental caries
1. Introduction
Biofilms consist of microorganisms embedded in a matrix of extracellular polymeric substances (EPS), forming a protected community bound onto a surface that has enhanced antimicrobial tolerance [1] and can cause intractable human infections [2]. Dental caries is a biofilm-induced disease driven by the diet and microbe interactions that causes acid-dissolution of the mineralized teeth, leading to systemic complications if left untreated [3]. Despite advances, the disease remains unresolved and a major public health problem that affects more than 2.3 billion people globally with costs exceeding $140 billion/year in the United States alone [4,5]. Current modalities are inefficient and based on broad-spectrum antimicrobials, posing challenges for targeting of caries-causing pathogens without disrupting commensal bacteria [6]. (see Fig. 4).
Fig. 4.

Therapeutic effects of Dex-IONP-GOx on a biofilm-associated oral disease in vivo. a-c, Caries scores recorded from tooth surface according to carious lesion severity according to Larson’s modification of Keyes’ scoring system (initial lesions: enamel affected; moderate lesions: dentin affected; severe lesions: full cavitation) (n = 12). d, The heatmap shows main bacterial genera found across all samples, distributed by treatment groups. e, Analysis of richness and diversity, respectively (red asterisk highlights CHX effects). f, Weighted Unifrac principal coordinate analysis (PCoA). g, Ridge plot of predicted pathways in Dex-IONP-GOx. h, Pathways related to sugar metabolism depleted in Dex-IONP-GOx. *P < 0.05, **P < 0.01.
The diet plays a critical role in disrupting the interactions between commensals and opportunistic pathogens on the tooth surface. Frequent exposure to dietary sugars promotes EPS production and the acidification of the biofilm microenvironment associated with dental caries [7,8]. Pathogens such as Streptococcus mutans thrive in sugar-rich conditions and promote cariogenic biofilms due to its ability: (i) to synthesize large amounts of EPS, (ii) to metabolize carbohydrates into organic acids (acidogenicity), and (iii) to grow in acidic conditions (aciduricity) [9]. These properties facilitate the creation of an acidic biofilm environment conducive for disease [3]. Conversely, commensals can interfere with the establishment of pathogens [3]. For example, Mitis group streptococci (such as S. oralis) can antagonize S. mutans via production of hydrogen peroxide (H2O2) that has antibacterial activity against the pathogen, favoring commensal growth in the biofilm and maintaining dental health [10–12]. However, the effects of intrinsic H2O2 production are locally confined and relatively low in concentration [13]. In addition, H2O2 production by these organisms is slow, and S. mutans can overcome H2O2-killing under sugar-rich conditions due to enhanced growth, metabolic activity and acidification. Interestingly, iron oxide nanoparticles with high peroxidase-like activity at acidic pH are capable of catalyzing exogenously introduced H2O2 to produce reactive oxygen species (ROS) to disrupt pathogenic biofilm [14,15]. However, exogenous H2O2 provides challenges for clinical translation of nanocatalysis for topical oral use as it would require a two-step approach.
Here, we designed a bi-functional nanohybrid system whereby glucose oxidase (GOx) was covalently conjugated to dextran coated iron oxide nanoparticle (Dex-IONP) (Fig. 1a) to exploit the pathological conditions associated with disease (i.e. high sugar availability, low pH and high EPS amounts) to boost H2O2 production and to trigger ROS more efficiently in a controlled, acidic pH-dependent manner. This strategy resulted in dual catalytic activity. GOx can catalyze glucose that is abundantly available in cariogenic biofilms into H2O2, and the Dex-IONP cores catalyze the reaction of H2O2 in acidic conditions to locally generate ROS that kills bacteria and degrades the EPS matrix. This synergistic reaction and antibiofilm activity are illustrated in Fig. 1b. We found that Dex-IONP-GOx preferentially bind to S. mutans and kills the pathogen more effectively than the commensal, resulting in targeted biofilm disruption. Importantly, the nanohybrid system reduced caries development in vivo more effectively than the current gold standard oral antimicrobial agent (chlorhexidine) without deleterious effects on the surrounding soft tissues and the gastrointestinal microbiomes. Altogether, we propose a concept whereby the pathological conditions induce self-generation of an intrinsic commensal defensive measure via nanohybrid dual-catalysis that counter cariogenic biofilm establishment, without the need of exogenous H2O2 addition. This approach can facilitate clinical translation as a one-step nanocatalytic formulation for caries prevention, and highlights the potential of targeting pathogens within mixed community without disturbing the commensals and the host microbiota, a major challenge of antimicrobial therapy against polymicrobial diseases.
Fig. 1.
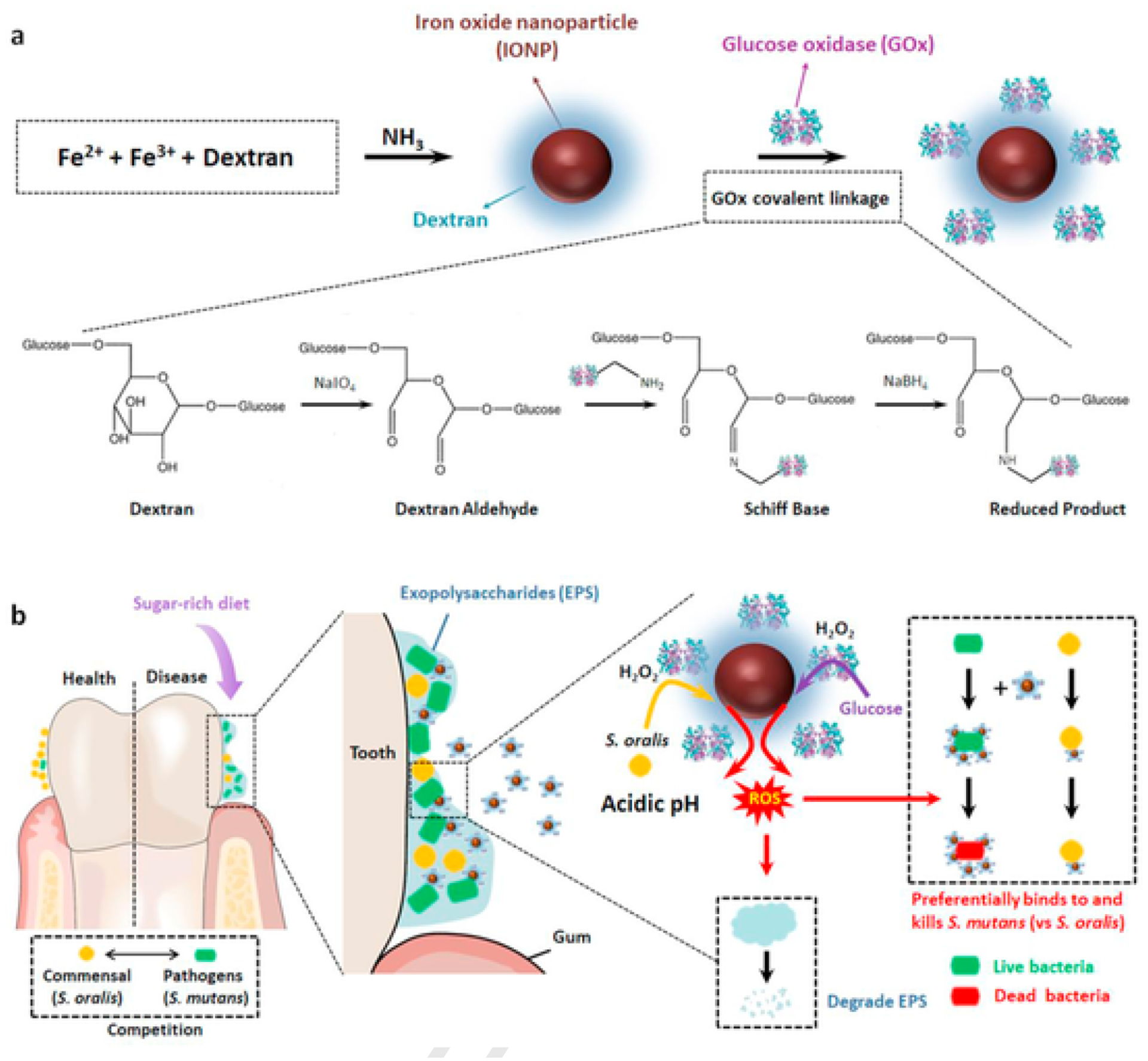
Proposed concept of a bi-functional hybrid nanozyme targeting pathogenic oral biofilm. a, Schematic depiction of the synthesis of the Dex-IONP-GOx nanohybrid. b, Schematic depiction of the selective catalytic–therapeutic mechanism of Dex-IONP-GOx for treatment of virulent acidogenic biofilms.
2. Materials and methods
2.1. Synthesis of Dex-IONP
Dex-IONP was prepared according to a previously reported protocol [15,16]. In brief, 12.5 g of dextran (10 kDa, Pharmacosmos, Holbaek, Denmark) was dissolved in deionized (DI) water under a nitrogen gas atmosphere. Then, 0.366 g of ferrous chloride tetrahydrate and 0.985 g of ferric chloride hexahydrate (Sigma-Aldrich, St. Louis, MO) were separately added to the above solution, and kept stirring for 45 min. Subsequently, 15 ml of ammonium hydroxide (28–30%, Sigma-Aldrich) was injected into the reaction mixture at different rates using a syringe pump. Afterwards, the reaction mixture was stirred overnight at room temperature after heating at 90 °C for 1 h. Finally, the nanoparticle suspension was purified by centrifugation using ultrafiltration tubes (100 kDa, Sartorius Stedim Biotech, Germany), followed by diafiltration columns (100 kDa, Spectrum Labs, CA).
2.2. Conjugation of Dex-IONP and GOx
GOx was conjugated to the Dex-IONP using a modified reported technique [17–19]. 48 mg of NaIO4 (Sigma-Aldrich) was added into a solution of dextran coated iron oxide nanoparticles (1 ml of 5 mg Fe/ml) and reacted overnight at room temperature in the dark. Afterwards, the Dex-IONP was collected and washed with distilled water using ultrafiltration tubes (100 kDa). The resulting modified dextran coated iron oxide nanoparticles were suspended in 0.1 M NaHCO3, pH 9.6, and incubated with 16 mg glucose oxidase (Sigma-Aldrich) at 4 °C overnight. At the end of this period, NaBH4 (Sigma-Aldrich) was added to a final concentration of 1 mg/ml to reduce the Schiff bases formed between the dextran and glucose oxidase. After 30 min, the resulting Dex-IONP-GOx were washed with distilled water using ultrafiltration tubes (300 kDa) and stored at 4 °C for further work.
2.3. Characterization of Dex-IONP-GOx
TEM was performed using a Tecnai F-20 and the hydrodynamic diameter and zeta potential were measured using a Nano-ZS 90 (Malvern Instrument, Malvern, UK). Inductively coupled plasma optical emission spectroscopy (ICP-OES) (Spectro Genesis ICP) was used to determine iron concentrations. The UV-VIS spectra were recorded using a UV-VIS spectrometer (Beckman Coulter, DU800). Pierce BCA Protein Assay Kit (Thermo Scientific) was used to determine concentrations of the GOx enzyme. Powder X-ray Diffraction (XRD) was recorded using a Rigaku GiegerFlex D/Max-B X-ray diffractometer operated at 45 kV and 30 mA with a monochromatized Cu Kα radiation wavelength of 1.5406 Å and a scan rate of 2° per minute. Energy-Dispersive X-ray (EDX) was performed using a Quanta 600 field emission gun scanning electron microscope (FEI) operated at 15 kV and equipped with EDX detectors (EDAX, Inc.).
2.4. Sequential catalytic activity of Dex-IONP-GOx
The sequential catalytic activities of the Dex-IONP-GOx were assessed via a colorimetric assay of 3,3′,5,5′-tetramethylbenzidine (TMB, Sigma-Aldrich). In brief, 10 μl of 2 mg/ml of Dex-IONP-GOx, 10 μl of 10 mg/ml TMB were added into 980 μl of NaAc (0.1 M) with or without 1% of glucose at pHs of 4.5, 5.5 and 6.5. The catalytic activities were measured at an absorbance of 652 nm. Each experiment was repeated three times and the data is presented as mean ± SD.
2.5. Iron ions release from Dex-IONP-GOx
The release of iron ions from Dex-IONP-GOx was investigated in 0.1 M NaAc buffer (pH 4.5). 1 ml of 0.5 mg/ml of Dex-IONP-GOx was incubated for 10, 30, 60, and 120 min (n = 3 per time point) at 37 °C. Next, the free iron ions and intact nanoparticles were separated under centrifugation using ultrafiltration tubes (10 kDa). ICP-OES was used to measure the iron content from the separated free iron ions and nanoparticles. The catalytic activity of the free iron and intact nanoparticle from each incubation time point was analyzed using the abovementioned TMB assay.
2.6. Michaelis–Menten kinetics measurement
Glucose oxidase kinetics was determined with 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) as substrate and a HRP-coupled colorimetric assay using a UV–VIS spectrophotometer based on a previous report [20], with minor modifications. Briefly, 10 μl of 2 mg/ml of Dex-IONP-GOx and 10 μl of 2 μM of HRP were added into the 970 μl of NaAc buffer (0.1 M, pH 4.5) containing 2 mM ABTS. Next, 10 μl of different concentrations of glucose (i.e., 10, 25, 50, 75, 100 mM) were added and mixed thoroughly by pipetting. The absorbance at 415 nm was recorded and the Michaelis–Menten constant (Km) was calculated through the Lineweaver-Burk plotting.
Dex-IONP peroxidase-like kinetics was studied using a method based on previous reports [15]. In brief, 10 μl of IONP-GOx (2 mg/ml) and 10 μl of TMB (10 mg/ml) were added to 970 μl of NaAc buffer (0.1 M, pH 4.5). Then, 10 μl of H2O2 with different concentrations (i.e., 2.5, 5, 10, 25, 50, 100 mM) was added to above solution to initiate the reaction. The absorbance at 652 nm was recorded immediately after addition of H2O2 and the resulting progress curves were linearly fitted to determine the Km value.
2.7. Assessment of Dex-IONP-GOx binding to bacterial cells
Streptococcus mutans UA159 (ATCC 700610), a virulent cariogenic pathogen and well-characterized biofilm-forming strain, or Streptococcus oralis J22, a commensal oral streptococcus generating hydrogen peroxide (H2O2)[21], was grown in ultra-filtered (10-kDa cutoff; Millipore) tryptone-yeast extract broth (UFTYE, pH 7.0) at 37 °C and 5% CO2 to mid-exponential phase. Subsequently, 1 ml of 109 CFU/ml of S. mutans or S. oralis was centrifuged and resuspended in 1 ml 0.1 M NaAc with 1 mg/ml Dex-IONP or Dex–IONO–GOx at pH 4.5, followed by incubation for 30 min with rocking at 37 °C. After incubation, the bacteria pellets were washed three times to remove unbound nanoparticles. The pellets were then dissolved with 1 ml of 70% HNO3 for iron quantification using ICP-OES. In addition, we also tested HRP binding to S. mutans or S. oralis using 0.1 mg/ml HRP following the same procedure as described above. Then, the bacteria pellets were collected for HRP quantification using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific).
2.8. Bacterial killing and EPS degradation by Dex-IONP-GOx
Time-lapse high-resolution confocal imaging was performed to assess the dynamics of bacterial killing and glucan degradation by the Dex-IONP-GOx in the presence of glucose. S. mutans or S. oralis was grown in UFTYE broth at 37 °C and 5% CO2 to mid-exponential phase. Dex-IONP-GOx was added to actively growing S. mutans (109 CFU/ml) or S. oralis (109 CFU/ml) at a concentration of 1 mg/ml in the presence of 1% glucose in 0.1 M NaAc buffer at pH 4.5. SYTO 9 (485/498 nm; Molecular Probes) and propidium iodide (PI) (535/617 nm; Molecular Probes) were used for live and dead cell labeling, respectively. Confocal images were acquired in the same field of view at different time intervals using Zeiss LSM 800 upright single photon laser scanning microscope with a 40 × (numerical aperture = 1.2) water immersion objective. Bacteria cells were also collected at 0, 1, 2, 3 h for colony forming units (CFU) counting. In another set of experiments, S. mutans or S. oralis was grown as described above, and then incubated with 0.1 mg/ml HRP in the presence of 1% H2O2 solution. The bacteria cells were collected and plated for CFU counting. For EPS degradation, insoluble glucans were produced by S. mutans-derived exoenzyme glucosyltransferase B (GtfB) immobilized on poly-l-lysine coated MatTek dish, and labelled with 1 μM Alexa Fluor 647-dextran conjugate (647/668 nm; Molecular Probes) as previously described [22]. The preformed fluorescently labelled insolubale glucans were then incubated with 1 mg/ml of Dex-IONP-GOx with 1% glucose in 0.1 M NaAc buffer at pH 4.5, and time-lapsed confocal imaging was performed using a 20 × (numerical aperture = 1.0) water immersion objective. Images were analyzed by ImageJ.
2.9. In vitro biofilm model
The mixed-species biofilm method was designed according to the “ecological-plaque” concept [23] as reported previously [24]. Briefly, hydroxyapatite discs (surface area of 2.7 ± 0.2 cm2; Clarkson Chromatography Inc., South Williamsport, PA) were coated with filter-sterilized, clarified human whole saliva for 1 h at 37 °C. The saliva-coated hydroxyapatite (sHA) discs were vertically suspended in 24-well plates using a custom-made disc holder to mimic the smooth surfaces of the pellicle-coated tooth. S. mutans and S. oralis were grown in UFTYE (pH 7.0) containing 1% glucose at 37 °C and 5% CO2 until mid-exponential growth phase. Each of the bacterial suspensions were then mixed to provide an inoculum with 106 CFU/ml of S. mutans and 102 CFU/ml of S. oralis. The mixed population was inoculated in 2.8 ml of UFTYE containing 1% (w/v) sucrose and the culture medium was changed twice daily at 19 and 29 h (8 a.m. and 6 p.m.) until the end of the experimental period (43 h). The biofilms were collected and analyzed for Dex-IONP-GOx binding and bioactivity as described below.
2.10. Biofilm treatment and quantitative analysis
The treatment solutions containing Dex-IONP and Dex-IONP-GOx were prepared in sodium acetate (NaAc) buffer (0.1 M, pH 4.5) at a final concentration of 1 mg/ml based on preliminary experiments and previous studies [14,15]. GOx was diluted to the equivalent of 2.7 mg/ml in NaAc (0.1 M, pH 4.5) to match amount of GOx conjugated in Dex-IONP-GOx. The biofilms were topically treated twice-daily by placing them in 2.8 ml of treatment agents in 0.1 M NaAc (pH 4.5) or vehicle control (buffer only) for 10 min at room temperature as described previously [14,15]. After each treatment, the biofilms were washed 3 times with sterile saline to remove unbound material, and transferred to the culture medium. At the end of the experiment period (43 h), the biofilms were removed and homogenized via water bath sonication followed by probe sonication (30 s pulse at an output of 7 W; Branson Sonifier 150, Branson Ultrasonics, Danbury, CT) as described previously [14, 25]; the sonication procedure does not kill bacterial cells, while providing optimum dispersal and maximum recoverable counts. The homogenized suspension was serially diluted and plated for colony forming units (CFU). The remaining suspension was washed and oven-dried for biomass (dry-weight) analysis. In a separate experiment, we tested whether the antibiofilm effect was caused by ROS generated from Dex-IONP-GOx using l-ascorbic acid as ROS scavenger. The Dex-IONP-GOx treatment was similar as described above and l-ascorbic acid at difference concentrations (i.e. 0, 6.25, 12.5, 25 μg/ml) were added in the culture medium.
2.11. Quantification of glucose generated from EPS degradation
The glucose is a byproduct of degradation of EPS by dextranase produced by S. mutans. To assess the amount of glucose generated by EPS degradation, 1 mg/ml glucans synthesized by streptococcal Gtfs, as previously reported, [25] was mixed with purified dextranase (α-(1 → 6) glucanase, EC3.2.1.11; Sigma-Aldrich) at a concentration of 10 U/ml in 0.1 M sodium acetate buffer, and incubated at 37 °C overnight. In a separate experiment, we also used the dextranase produced by S. mutans single species or S. mutans-S. oralis mixed-species biofilm to degrade glucans. In brief, the biofilms were grown to 19 h and homogenized as described above. Then, the homogenized suspension was centrifuged at 5500 rpm for 10 min, and 1 ml of the supernatant was mixed with 1 mg of glucans and incubated at 37 °C overnight. In both experiments, the amount of glucose released from glucan degradation was determined using a glucose assay kit (Sigma-Aldrich).
2.12. Fluorescence in situ hybridization (FISH) microscopy
To visualize the distribution of the different species in the mixed-species model, fluorescence in situ hybridization (FISH) was performed using confocal microscopy [26]. In brief, the treated biofilms on sHA discs were washed with PBS and fixed with 4% paraformaldehyde at 4 °C for 4 h. Next, the biofilms were transferred into 50% ethanol after washing with PBS and stored at −20 °C. The biofilm architecture was analyzed via FISH as detailed previously [14, 26]. Bacterial cells were labelled by species-specific FISH probes: MUT590, 5′-ACTCCA-GACTTTCCTGAC-3′ with Cy5 for S. mutans; MIT588, 5′-ACAGCCTT-TAACTTCAGACTTATCTAA-3′ with Cy3 for S. oralis. The samples were dissolved in the hybridization buffer including 0.9 M NaCl, 0.01% SDS, 25% formamide, 20 mM Tris-HCl, and incubated with the probes at 46 °C for 2 h. After incubation, the hybridized cells were washed with buffer (0.2 M NaCl, 0.01% SDS, 5 mM EDTA, 20 mM Tris-HCl (pH 7.5)), followed by incubation at 46 °C for 15 min. The biofilms were analyzed by sequentially scanning at 561 nm and 640 nm with lasers, and the fluorescence emitted was collected with hybrid detectors (550–653 nm for Cy3 and 644–752 nm for Cy5, respectively).
2.13. Confocal fluorescence microscopy
To visualize the spatial distribution of Dex-IONP-GOx, bacterial cells and the EPS within intact biofilm, high resolution confocal imaging was performed using LSM 800 upright microscope with a 20 × (numerical aperture = 1.0) water immersion objective [15]. Dex-IONP-GOx was conjugated with Alexa Fluor 488 Hydrozide (ThermoFisher Scientific) following the manufacturer’s instructions. UV–VIS Spectra (Beckman Coulter, DU800) was used to confirm the successful conjugation of Alexa-488 with Dex-IONP-GOx (Supplementary Fig. 16a), and the conjugation did not affect the catalytic activity Dex-IONP-GOx (Supplementary Fig. 16b). SYTO 82 (541/560 nm; Molecular Probes) was used for labeling bacteria, and Alexa Fluor 647-dextran conjugate (647/668 nm; Molecular Probes) was used for labeling insoluble EPS. Each component was illuminated sequentially to minimize cross-talk as follows: Alexa Fluor 488 (Dex-IONP-GOx) was excited at 488 nm and collected using a 480/40 nm emission filter; SYTO 82 (bacterial cells) was excited at 560 nm, and collected using a 560/40 nm emission filter; Alexa Fluor 647 (EPS) was excited at 640 nm and collected using a 670/40 nm emission filter [15]. Images were visualized using Amira 5.4.1 software (Visage Imaging, San Diego, CA).
2.14. Detection of H2O2 and ROS
H2O2 produced by S. oralis was assessed using Prussian blue production in the presence of H2O2 on agar plates containing ferric chloride and potassium hexacyanoferrate (III) [27,28]. Briefly, 10 μl of actively growing S. oralis was diluted (105 CFU/ml) and spotted onto the agar plate with 0.2% or 2% glucose, and incubated at 37 °C overnight. Similarly, S. oralis (105 CFU/ml) mixed with Dex-IONP-GOx (0.1 μg Fe/ml) were also spotted onto the agar plate. H2O2 production was quantified (in millimeters) by measuring the blue halos between the border of the colony and the border of the halo formed by the Prussian blue on the indicator plates. In addition, pentafluorobenzenesulfonyl fluorescein (PBSF, 98+%, Cayman Chemical Company) was also used for localized detection of H2O2 production. 1 ml of actively growing S. mutans (109 CFU/ml) or S. oralis (109 CFU/ml) was incubated with PBSF (5 μM) in the presence of SYTO 60 (10 μM) for 4 h at 37 °C. Next, 20 μl of the bacterial suspension was placed onto glass slides for confocal microscopy visualization and quantification. Confocal images were obtained using LSM 800 upright single photon laser scanning microscope with a 40 × (numerical aperture = 1.2) water immersion objective. The H2O2 was detected at 488/505 nm and bacteria cells were visualized at 652/678 nm. ImageJ was employed to quantify the H2O2 production based on the fluorescent intensity of PBSF.
For observation of ROS generation, hydroxyphenyl fluorescein (HPF) was used as fluorescent probe to detect hydroxyl radicals. S. mutans or S. oralis was labelled by SYTO 60 (652/678 nm; Molecular Probes) and incubated for 30 min at 37 °C. Then, the bacterial cells were washed three times and resuspended in 1 ml 0.1 M NaAc (pH 4.5) containing 1 mg Fe/ml of Dex-IONP-GOx, 10 μM of HPF, 30 μM of propidium iodide (PI) in the presence of 1% of glucose. 20 μl of the mixture was immediately placed onto glass slides for visualization with confocal microscopy. Micrographs were acquired in the same field of view over time using LSM 800 upright single photon laser scanning microscope with a 40 × (numerical aperture = 1.2) water immersion objective.
For the ROS quantification in vitro, the biofilm was treated with Dex-IONP-GOx at 0 h and 6 h. After 19 h incubation, the culture medium was removed and replaced with 0.1 M NaAc (pH 4.5) containing 1% glucose in each of the 24-well plate. Subsequently, 28 μl of TMB (10 mg/ml) was added at the final concentration of 0.1 mg/ml, and the color change in the treated biofilm observed over time for up to 8 h. Following TMB reaction, biofilms were removed and homogenized for measurement of total ROS amounts as determined spectrophotometrically at the absorbance of 652 nm.
2.15. Human oral gingival cell viability
Primary human gingival epithelial cells were received from Dr. Manju Benakanakere (School of Dental Medicine, University of Pennsylvania), and were cultured in keratinocyte serum-free medium (Invitrogen, NY). Human fibroblast cells (from ATCC, Manassas, VA, USA) were cultured in DMEM and medium 199 in a 4 to 1 ratio and supplemented with 10% heat-inactivated FBS (Gibco, NY) and 0.01 mg/ml of hygromycin B (Sigma-Aldrich). To determine the cytotoxicity of Dex-IONP-GOx, the human primary oral gingival cells and fibroblast cells were seeded in 96-wells at a density of approximately 1 × 104 per well. Cells were allowed to incubate at 37 °C in a 5% CO2 atmosphere overnight and then cells were washed with sterile phosphate buffered saline (PBS) and the media was carefully changed with the fresh media containing nanoparticles with different concentrations for 10 min and 24 h (n = 6 wells per condition with three replicates). Finally, media was removed and cells were washed with PBS and then added with fresh media and subjected to a standard MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay. The results were expressed as the percentage of cell viability by comparing to the average absorbance at 490 nm of the cells treated with media only (n = 6 wells per plate used, with three replicates).
2.16. In vivo efficacy of IONP-GOx
The therapeutic efficacy of Dex-IONP-GOx was assessed using a well-established rodent model of dental caries disease, as reported previously [29,30]. In brief, 15 days-old specific pathogen free Sprague-Dawley rat pups were purchased with their dams from Harlan Laboratories (Madison, WI, USA). Upon arrival, animals were screened for S. mutans and were determined not to be infected with the pathogen by plating oral swabs on mitis salivarius agar plus bacitracin (MSB). Then, the animals were orally infected with actively growing (mid-logarithmic) culture of 1:1 mixture of S. mutans UA159 and S. oralis J22, and their infection was confirmed at 21 days through oral swabbing and culturing/PCR. To simulate clinical situation, we developed a topical treatment regimen with short time exposure (30 s) using a custom-made applicator. All infected pups were randomly placed into five treatment groups (12 rats/group), and their teeth were treated twice daily. The treatment groups included: (1) control (0.1 M NaAc buffer, pH 4.5), (2) Dex-IONP only (1 mg Fe/ml), (3) GOx only (2.7 mg/ml), (4) Dex-IONP-GOx (1 mg Fe/ml) and (5) chlorhexidine (1.2 mg/ml). The treatments were blinded by placing the test agents in color-coded vials. Each group was provided the National Institute of Health cariogenic diet 2000 and 5% sucrose water ad libitum. The experiment proceeded for 3 weeks; all animals were weighed weekly, and their physical appearance was recorded daily. At the end of the experiment period, oral swabs and fecal samples were collected for oral and gut microbiome analysis. After sacrificing, the jaws were surgically removed and aseptically dissected, followed by sonication to recover total oral microbiota for microbiological and microbiome analyses as reported previously [31]. All of the jaws were defleshed and the teeth were prepared for caries scoring based on Larson’s modification of Keyes’ system [29,30]. Determination of caries score of the jaws was performed by a calibrated examiner who was blind for the study by using codified samples. Moreover, both gingival and palatal tissues were collected and processed for H&E staining for histopathological analysis by an oral pathologist at Penn Oral Pathology. This research was reviewed and approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC #805529).
2.17. 16S RNA analysis
Oral swab collection was made using moisten swab tip in microbial DNA free water pretreated with UV cross-linker for 1 h before use to prevent DNA contamination (519C FLOQSwab, COPAN). Fecal samples were collected the same day than oral swab. One fecal pellet was collected by cage. Since the rats were in suspended cages, we could collect pellet from the clean surface on the bottom of the cage and placed in sterile 1.5 ml tube. Dental plaque samples were obtained after the animal were sacrificed as previously reported [14]. All samples were kept in −80 °C until DNA extraction. Contamination control samples were included during the animal experiments as blank oral swab and DNA-free water and reagent controls as negative controls. Also, mock DNA sample as positive controls were included following the reported method article. DNA was extracted from the pellets using the DNeasy PowerSoil kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions within a sterile class II laminar flow hood. PCR amplification of V1–V2 region of 16S rRNA gene following previously reported methods (REF). The library was sequenced to obtain 2 × 250 bp paired-end reads using the MiSeq Illumina. To analyze 16S RNA gene sequences, we used QI-IME2 v19.4. We obtained taxonomic assignments based on GreenGenes 16S rRNA gene database v. 13_8. We also obtained amplicon sequence variant (ASV) for analysis of shared and unique bacterial taxa in each body-site through DADA2. Principal coordinate analysis was performed using library ape for R programming language.
2.18. Shotgun metagenomics analysis
Shotgun metagenomic data were analyzed using Sunbeam, a user-extendable bioinformatics pipeline that we developed for this purpose [32]. Quality control steps were performed by the default workflows in Sunbeam, which are optimized to remove host-derived sequences and reads of low sequence complexity. The abundance of bacteria was estimated using Kraken [33]. Reads were mapped to the KEGG database [34] using Diamond [35] to estimate the abundance of bacterial gene orthologs, as well as to curated databases of genes involved in butyrate production, polysaccharide utilization, and secondary bile acid production. The abundance of genes and taxa were analyzed at a community level using pairwise distance between samples, and visualized with Principal Coordinates Analysis. Linear models were used to detect differences in logit-transformed gene and taxon abundance between sample groups. P-values from multiple testing procedures were corrected to control for a specified false discovery rate. The KEGG pathway heatmap was created using the pheatmap package in R based on per sample pathway abundances scaled to the mean and S.D. of each pathway. To analyze whole genome sequence data from isolated bacterial strains, we performed quality control to remove non-genomic sequence and low-quality reads. We assembled de novo S. oralis and S. mutans genomes using Spades. We assessed the quality of assembled genomes with CheckM and Quast. Reads from metagenomic samples were aligned to assemblies using bwa mem to determine counts of the strains.
2.19. Statistical analysis
All the results were conducted through at least three independent experiment and the data were shown as means ± s.d. Data were analyzed using the Student’s t-test, with the exception of the animal study, which was analyzed using the pairwise Wilcoxon rank sum tests. Statistically significant differences between groups were considered when P < 0.05.
3. Results and discussion
3.1. Synthesis of Dex-IONP-GOx conjugates and characterization
The sequential catalyst was generated via chemical conjugation of glucose oxidase (GOx) to dextran-coated iron oxide nanoparticles (Dex-IONP). Dex-IONP was prepared based on the Molday synthesis for iron oxide nanoparticles. The reaction of ferrous chloride with ferric chloride in the presence of dextran (10 kD) under alkaline conditions yielded a suspension of dextran coated Fe3O4 colloidal particles. Dextran not only can provide excellent water solubility, high biocompatibility, but can be also functionalized to allow binding with the amino groups of enzymes [36–37]. X-ray diffraction (XRD) measurement confirmed crystalline nature of the Dex-IONP (Supplementary Fig. 1a), energy-dispersive X-ray (EDX) was performed and indicated that the Fe elements were present in the nanohybrid system (Supplementary Fig. 1b). Afterwards, GOx was conjugated to Dex-IONP by oxidation of the dextran coating (Fig. 1a). Their average core diameter was found to be 7.3 ± 0.9 nm (Fig. 2a). After conjugation with GOx, no obvious changes in size and morphology were observed in transmission electron microscopy (TEM) (Fig. 2b). The conjugation between Dex-IONP and GOx was achieved via reaction of the amine groups of GOx with aldehyde groups that had been introduced into the dextran coating of the Dex-IONP. After conjugation, the samples were washed with distilled water using ultrafiltration tubes (300 kDa) to remove excess, unbound GOx, and then kept at 4 °C. The success of this conjugation reaction was established after purification based on the absorbance of GOx at 280 nm in the final product (Fig. 2c). To further confirm conjugation between Dex-IONP and GOx, we conducted the protein detection using a colorimetric Bicinchoninic Acid (BCA) Protein assay. The result shows protein detection of GOx and Dex-IONP-GOx (equivalent amounts of GOx) at 562 nm, whereas negligible absorbance was detected with Dex-IONP, indicating the presence of GOx conjugated onto Dex-IONP (Supplementary Fig. 2). The hydrodynamic diameters of Dex-IONP, Dex-IONP-GOx were 36.5 ± 1.1 nm, 48.5 ± 3.8 nm, and the zeta potentials were −10.8 ± 0.2 mV, −19.8 ± 0.4 mV, respectively (Supplementary Fig. 3). The diameter of Dex-IONP-GOx measured using dynamic light scattering (DLS) was larger than that of Dex-IONP, which is likely due to the immobilization of GOx onto the surface of Dex-IONP. Moreover, the amount of GOx conjugated to Dex-IONP was determined to be 2.7 ± 0.7 mg of GOx per mg of Fe (see Fig. 3).
Fig. 2.
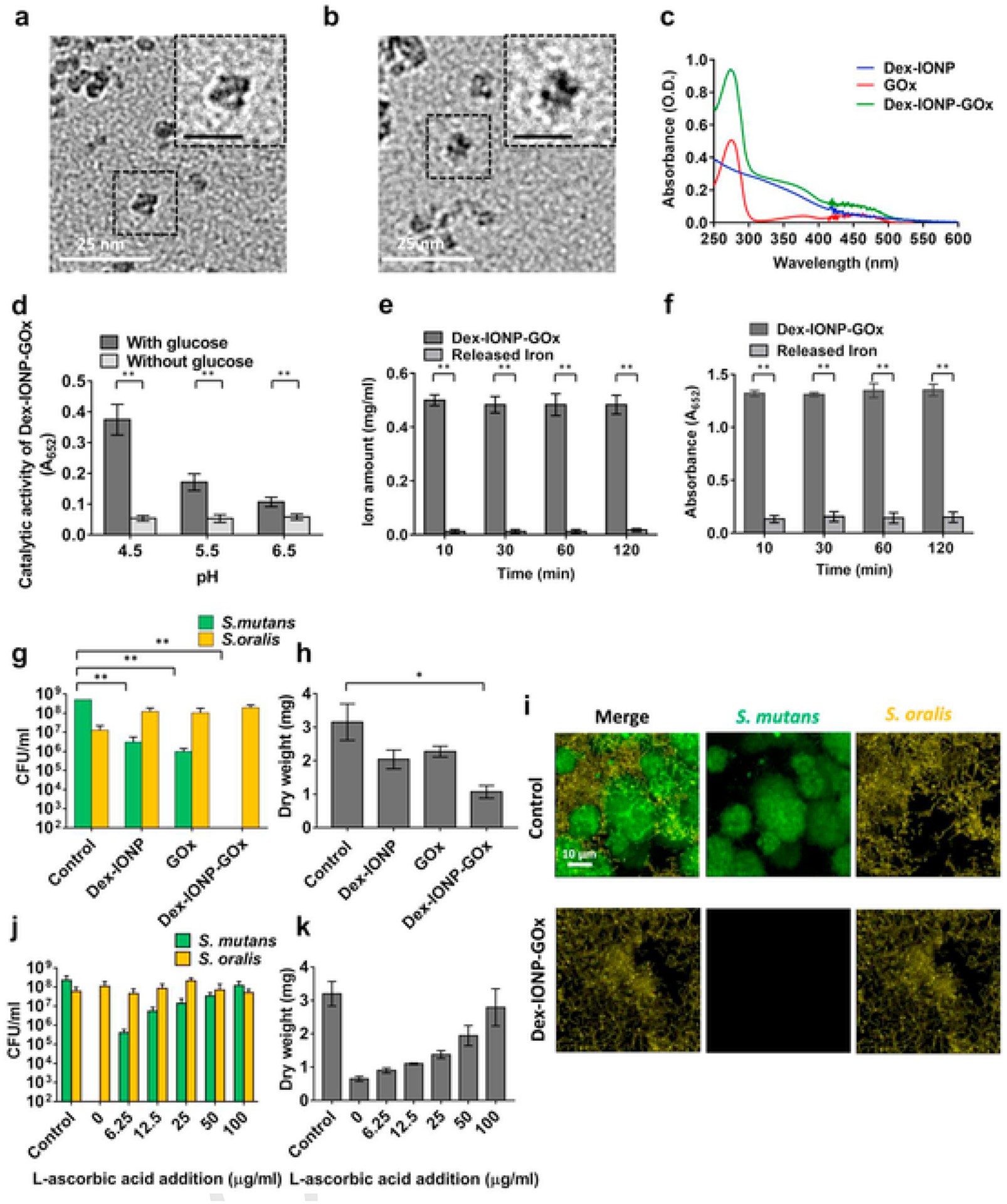
Catalytic activity and antibiofilm effects of hybrid nanozyme. a,b, TEM (Scale bar represents 10 nm in the insets) of Dex-IONP (a) and Dex-IONP-GOx (b). c, UV-VIS spectra of Dex-IONP, GOx and Dex-IONP-GOx. d, Catalytic activity of Dex-IONP-GOx at different pH values with or without glucose based on ROS production via sequential reaction (n = 3). e,f, Relative amounts of iron in Dex-IONP-GOx or released at pH 4.5 (e) and their respective catalytic activities (f) (n = 3). g,h, Effect of different formulations on bacterial viability (g) and the mass of biofilms (h) (n = 6). i, FISH microscopy of vehicle control-treated biofilm (top row) and biofilm treated with Dex-IONP-GOx (bottom row). S. mutans were stained with STR405-Cy5 (in green), S. oralis were stained with MIT588-Cy3 (in yellow). j,k, Effect of IONP-GOx on bacterial viability (j) and the mass of biofilm (k) after addition of different concentrations of l-ascorbic acid (n = 6). *P < 0.05, **P < 0.01.
Fig. 3.
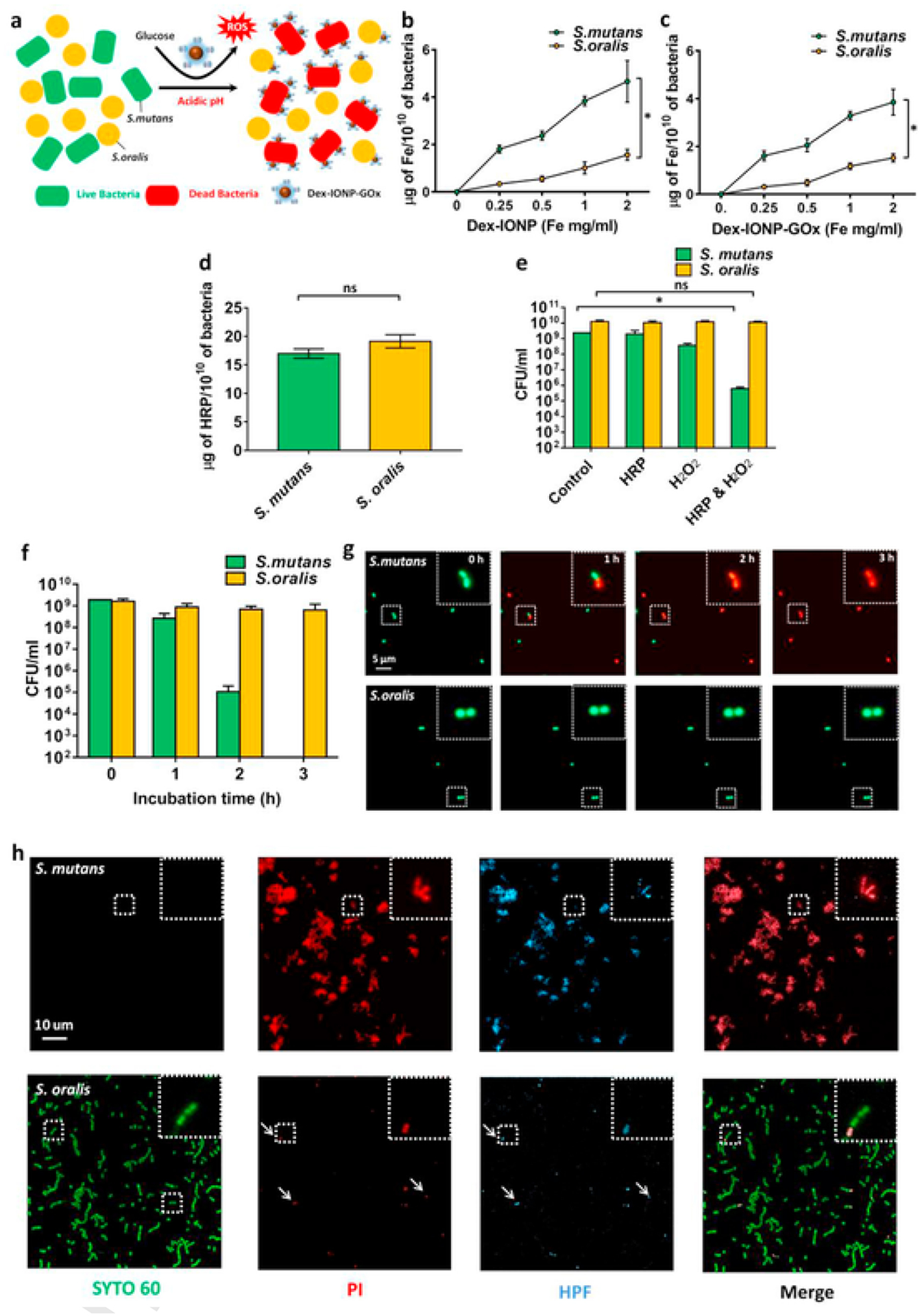
Mechanisms of bacterial killing by the hybrid nanozyme. a, Schematic depiction of the proposed Dex-IONP-GOx preferential binding and killing of S. mutans versus S. oralis. b,c, Binding of Dex-IONP (b) and Dex-IONP-GOx (c) to S. mutans or S. oralis (n = 3) d, Binding of HRP to S. mutans or S. oralis. e, Effect of HRP on bacteria viability after addition of H2O2 (n = 3). f, Effect of Dex-IONP-GOx on bacteria viability over time (n = 3). g, Micrographs of bacteria incubated with Dex-IONP-GOx at different time points. Live cells were labelled by SYTO 60 (green) and dead cells were stained with propidium iodide (PI) and displayed in red. h, Detection of ROS in S mutans or S. oralis after treatment with Dex-IONP-GOx. Live cells were stained with SYTO 60 (green), dead cells were stained with PI (red), and ROS was stained using HPF (light blue). *P < 0.05.
The proposed catalytic process of Dex-IONP-GOx is shown in Fig. 1b, i.e. GOx catalyzes the oxidation of glucose to generate H2O2, whose reduction is catalyzed by the Dex-IONP core. In order to investigate the catalytic activity of Dex-IONP-GOx based on sequential catalysis, 3,3′,5,5′-tetramethybenzidine (TMB) was used as a chromogenic substrate. After incubation of Dex-IONP-GOx with or without glucose under different pH conditions, it was observed that Dex-IONP-GOx exerted greater catalytic activity under acidic conditions (pH 4.5) than at other pH conditions in the presence of glucose (Fig. 2d). Conversely, there was no catalytic activity observed at different pH conditions without addition of glucose. This result demonstrates that sequential catalysis by Dex-IONP-GOx was favorable in the presence of glucose under acidic conditions. We further evaluated the catalytic activity of Dex-IONP-GOx under varied glucose concentrations, and found that catalysis is concentration dependent (Supplementary Fig. 4). In addition, we also examined whether the catalytic activity of Dex-IONP was generated from nanoparticles themselves rather than iron ions via the Fenton reaction. Fig. 2e demonstrates that low proportions (3.3 ± 1.0%) of the iron ions were released over 120 min, and the catalytic activity which associated with released iron was marginal (Fig. 2f), suggesting that the observed activity is primarily derived from the Dex-IONP itself.
We further probed the enzyme kinetics of Dex-IONP-GOx via applying Michaelis–Menten steady-state kinetics. The initial catalytic activity of GOx with variable concentration of glucose was measured according to the horseradish peroxidase (HRP)-coupled colorimetric assay [20]. The Km of GOx was determined as 1.68 mM (Supplementary Fig. 5), while the Km of IONP was found to be 41.4 μM (Supplementary Fig.6).
3.2. Antibiofilm activity of Dex-IONP-GOx in vitro
Next, we assessed the functional properties of the Dex-IONP-GOx system using a mixed-species biofilm model that mimics commensal-pathogen interactions under cariogenic conditions. In the in vitro model, S. mutans (pathogen) and S. oralis (commensal) were inoculated on saliva-coated hydroxyapatite (sHA) [24,38] discs and allowed to form a biofilm in the presence of sucrose. The data revealed that treatment with Dex-IONP-GOx potently inhibited S. mutans, but notably with minimal effects against the commensal S. oralis (Fig.2g). Moreover, Dex-IONP-GOx were significantly more effective than Dex-IONP or GOx alone, suggesting that the selective pathogen killing was caused by dual catalytic function. Consistent with bacterial viability data, the biomass of biofilms treated with Dex-IONP-GOx was significantly reduced when compared to control and more effective than Dex-IONP or GOx alone (Fig. 2h). Fluorescence in situ hybridization (FISH) microscopy further confirmed specificity towards S. mutans killing rather than S. oralis after treatment with Dex-IONP-GOx (vs control), whereby minimal S. mutans cells were detected post-treatment (Fig. 2i). We also found that Dex-IONP-GOx was capable of disrupting EPS as determined using time-lapsed confocal microscopy (Supplementary Fig. 7). High-resolution confocal images (Supplementary Fig. 8) showed that Dex-IONP-GOx treatment substantially reduced EPS matrix, impairing S. mutans accumulation and altering the biofilm 3D architecture (vs. control).
It is noteworthy that Dex-IONP alone was only moderately effective against S. mutans despite availability of H2O2 produced by S. oralis that could be catalyzed to ROS by Dex-IONP, indicating that the amount of H2O2 produced by S. oralis may not be enough to fully control S. mutans. Conversely, the inclusion of GOx in Dex-IONP-GOx could increase H2O2 production and maximize effects against S. mutans. To determine H2O2 production in our system, we employed confocal microscopy and the Prussian blue agar colorimertric method. Pentafluorobenzenesulfonyl fluorescein (PBSF) was used as a fluorescent probe to detect the production of H2O2 [39,40] by the bacterial cells. After incubation with different bacteria, fluorescence was observed in S. oralis rather than S. mutans (Supplementary Fig. 9a), which reveals the ability of H2O2 production by S. oralis. Next, S. oralis, Dex-IONP-GOx and S. oralis & Dex-IONP-GOx were spotted onto Prussian blue agar (Supplementary Fig. 9b). The amount of H2O2 produced by S. oralis & Dex-IONP-GOx in the presence of 2% of glucose was noticeably increased, compared to other conditions, as evidenced by the larger blue area. Quantification of the halo size confirmed the total amount of H2O2 was increased by S. oralis & Dex-IONP-GOx, which was dependent on the amount of glucose (Supplementary Fig. 9c). In addition, glucose is naturally present in cariogenic biofilms as a byproduct of sucrose metabolism and degradation of glucans, a major EPS in the matrix [41–43] by dextranase from S. mutans. We confirmed glucose generation by the action of dextranase on glucans extracted from biofilms, as well as detection of free glucose by dextranase activity from biofilms (Supplementary Fig. 10a, 10b). Thus, availability of glucose under cariogenic conditions can boost H2O2 production.
Given the enhanced production of H2O2, we further investigated whether the specific inhibition against S. mutans was related to ROS generated from Dex-IONP-GOx by adding l-ascorbic acid, an anti-oxidant agent known to suppress ROS production. We observed a dose-dependent effect whereby the killing of S. mutans and biomass reduction was nearly abrogated with increasing amounts of l-ascorbic acid (Fig. 2j, k), suggesting that antibiofilm activity results from the oxidative damage caused by ROS generation.
3.3. Preferential pathogen targeting by Dex-IONP-GOx
Notably, the treatment of Dex-IONP-GOx specifically targeted S. mutans while leaving S. oralis alive within the biofilm. We hypothesized that (1) Dex-IONP-GOx can preferentially bind to S. mutans compared to S. oralis, and thereby produce ROS in close proximity to more effectively kill S. mutans, and (2) S. oralis is more tolerant of ROS than S. mutans (Fig. 3a). To probe these questions, Dex-IONP or Dex-IONP-GOx at different concentrations were incubated with planktonic cells of S. mutans or S. oralis, as shown in Fig. 3b and c. The amount of Dex-IONP and Dex-IONP-GOx bound to both S. mutans and S. oralis increased in a dose-dependent pattern. However, Dex-IONP and Dex-IONP-GOx both bind preferentially to S. mutans compared to S. oralis (P < 0.05). The Dex-IONP surface coating with dextran could provide specific binding sites to S. mutans as this organism expresses several cell membrane associated glucan-binding proteins [44,45]. At least four glucan-binding proteins, in addition to surface-bound glucosyltransferase B (with a glucan binding domain), are uniquely expressed in high levels in S. mutans (rather than S. oralis), which can directly mediate binding interactions with dextran or dextran-coated surfaces [45–47]. Thus, the nanoparticles coated with dextran could provide preferential binding to S. mutans compared to S. oralis and other streptococcal species. This finding can partially explain the enhanced pathogen killing by Dex-IONP-GOx since its greater binding to S. mutans would result in localized ROS production, especially given that ROS: (1) has a brief life-span [48,49], and (2) does not diffuse over long distances [50]. In addition, to further investigate the sensitivity of S. mutans or S. oralis towards ROS, horseradish peroxidase (HRP) was incubated with S. mutans or S. oralis. In contrast to Dex-IONP-GOx, Fig. 3d shows the same amount of HRP binds to S. mutans and S. oralis, respectively. Cell viability was assessed with the addition of H2O2 after incubation of HRP. As seen in Fig. 3e, HRP was capable of reducing S. mutans viability after addition of H2O2 (>103 reduction in cell number) (P < 0.05), while S. oralis was not substantially affected. The data suggest that S. mutans is less tolerant to ROS than S. oralis.
To further examine susceptibility to ROS, Dex-IONP-GOx was incubated with planktonic cells of S. mutans or S. oralis in acidic conditions in the presence of glucose at 0, 1, 2 and 3 h, and the cell viability was assessed over time. As shown in Fig. 3f, Dex-IONP-GOx effectively killed S. mutans (>107 reduction), with little effect on S. oralis viability (less than 1-log killing after treatment within 3 h). This result was confirmed with high-resolution, time-lapsed fluorescence microscopy to detect live and dead cells (Fig. 3g). To visualize the ROS generation, we used a fluorescent probe, hydroxyphenyl fluorescein (HPF), capable of detecting hydroxyl radicals [51,52]. Planktonic cells (S. mutans or S. oralis) were incubated with Dex-IONP-GOx in the presence of glucose under acidic conditions, then observed using confocal microscopy. Fig. 3h shows micrographs of S. mutans and S. oralis after treatment with Dex-IONP-GOx. Most S. mutans cells were killed after treatment of Dex-IONP-GOx, which was accompanied by large amounts of ROS generation in situ. In sharp contrast, only a small proportion of S. oralis were damaged by treatment with Dex-IONP-GOx, with much less ROS observed. Interestingly, the position of the dead cells matched with that of ROS generation, implying that bacterial killing mainly originated from the oxidative damage. These findings provide additional evidence that S. mutans has more Dex-IONP-GOx binding and thereby more ROS generation in situ, leading to more effective killing compared to S. oralis. In addition, we also quantified the ROS generation within the biofilms using the TMB. Following TMB reaction, the intact biofilms were imaged for visual detection of ROS generated in situ, and then homogenized for measurement of total ROS amounts as determined spectrophotometrically at the absorbance of 652 nm (Supplementary Fig. 11).
3.4. Cytocompatibility of Dex-IONP-GOx in vitro
The effect of Dex-IONP-GOx on the viability of the human primary oral gingival cells, and fibroblast cells was tested via the MTS assay. The cells were incubated by Dex-IONP-GOx nanoparticles with concentrations of 0.25, 0.5, and 1.0 mg/ml of Fe for 10 min and 24 h (Supplementary Fig. 12). We found that Dex-IONP-GOx had no adverse effect on the cell viability, suggesting biocompatibility for in vivo experiments.
3.5. Dex-IONP-GOx suppress dental caries in vivo
We next sought to assess whether topical applications of Dex-IONP-GOx could inhibit the development of tooth decay in vivo using a rodent model that mimics the characteristics of human severe caries [29]. Rat pups were infected with the human oral pathogen (S. mutans) and commensal (S. oralis) bacteria and fed a sugar-rich diet. Afterwards, we applied the test solutions topically twice daily with a brief, 1 min exposure time to mimic the clinical use of a mouthwash. After 3 weeks’ treatment, the incidence and severity of caries lesions on teeth of rat pups was evaluated. Quantitative caries scoring analyses demonstrated that the treatment of Dex-IONP-GOx greatly attenuated both the initiation and severity of the lesions in smooth surfaces when compared to control, Dex-IONP or GOx alone (P < 0.01) (Fig. 4a–c). In sharp contrast, treatments with Dex-IONP or GOx were without significant effect when compared to control, which is consistent with in vitro biofilm results. It is also noteworthy that the efficacy of Dex-IONP-GOx treatment was significantly greater than the current standard oral antimicrobial agent known as chlorhexidine (CHX) (P < 0.05), showing potential antibiofilm activity to control tooth decay. Another important feature of the nanohybrid system is the ability to exert therapeutic effect without adding exogenous H2O2. This has many implications for clinical translation as two-component system (nanocatalyst + exogenous H2O2) would require specialized dual-chamber containers.
In addition, we observed no deleterious effects on rats that received topical applications of Dex-IONP-GOx for 3 weeks. Histopathological analysis of gingival tissues as well as liver and kidney revealed no visible signs of harmful effects, such as proliferative changes, vascularization issues, necrosis or acute inflammatory responses, after treatment (Supplementary Fig. 13 and 14), suggesting biocompatibility of Dex-IONP-GOx treatment. The effects on oral and gut microbiome were also evaluated based on oral swabs and fecal samples, and all treatment groups showed similar microbiota composition (Fig. 4d). We also determined whether microbial diversity was affected by the treatments. Alpha diversity, which represents the taxonomical differences between groups, was assessed by richness index and Shannon index (Fig. 4e). When comparisons were performed among control, Dex-IONP, GOx and Dex-IONP-GOx, no significant differences were observed neither for richness index (indicating the treatments did not affect the number of bacterial species) nor for Shannon index (distribution of bacteria species was not affected by the treatments). However, CHX treatment resulted in significantly lower Shannon index when compared with other groups (P<0.05), indicating disruption of the ecological balance of the microbiota, whereby a few species of bacteria become dominant. This finding is consistent with the broad-antimicrobial spectrum of CHX and clinical recommendations for limited use due to potential of causing oral microbiota disturbances [53,54]. Hence, Dex-IONP-GOx was significantly more effective than CHX in reducing dental caries, but without the deleterious effects on the oral microbiota ecology. Furthermore, we also assessed the gut microbiota following the treatments. In our model, the animals are housed in wired cages to avoid cropophagia and reduce cross-contamination between oral and gut microbiota. Weighted UniFrac distance was employed to compare the composition of specie’s samples to each other (Fig. 4f). We found clear separation between the oral and gut microbiome consistent with the pattern found in these body sites in humans. In addition, topical oral treatments did not cause disruption of the gut microbiota composition or diversity.
Moreover, to investigate whether specific microbial pathways were affected, KEGG pathway enrichment was performed using the data set generated from metagenomics analysis. The top 50 KEGG enriched pathways included sugar metabolism, glycolysis/gluconeogenesis, oxidative phosphorylation among others (Supplementary Fig. 15). The ridge plot of predicted pathways with the treatment of Dex-IONP-GOx was further analyzed and found that several biosynthesis pathways were enriched, whereas sugar metabolism pathways were depleted compared to control group (Fig. 4g, h). Specifically, the increased biosynthesis of arginine (associated with commensal bacteria and healthy microbiota) and decreased sugar metabolism (associated with caries) suggest that treatments with Dex-IONP-GOx resulted in a microbiota activity associated with dental health while dampening biofilm pathogenicity, consistent with effective reduction of caries development.
4. Conclusions
In brief, we introduce a concept of exploiting the natural settings found in pathological conditions to trigger self-generating chemical weapons used by commensals against pathogenic bacteria to deter cariogenic biofilm establishment. We developed a dual-catalytic nanohybrid approach by integrating glucose oxidase (GOx) and dextran coated iron oxide nanoparticles (Dex-IONP) with peroxidase-like activity. GOx can self-fuel the hybrid system, in a controlled manner, by boosting intrinsic H2O2 production using readily available glucose that can be converted to ROS by the Dex-IONP catalytic cores with high catalysis at acidic pH 4.5 (but low at pH 6.5). It effectively kills S. mutans and degrades the EPS matrix while sparing the commensal S. oralis in pathogenic environment, resulting in localized biofilm disruption without the need of exogenous H2O2. Mechanistic studies revealed that: (i) enhanced H2O2 production was due to glucose availability that includes the sugar moiety released from EPS glucans found in high amounts within the cariogenic biofilm; (ii) Dex-IONP-GOx binds preferentially to S. mutans cells, producing more ROS in close proximity that result in enhanced pathogen killing; (iii) S. oralis is more tolerant of free-radicals, which contributed to its increased survival when treated with Dex-IONP-GOx. Then, we employed an animal disease model with simultaneous oral-gut microbiome and metagenome analyses that show remarkable efficacy of nanohybrid treatment to control caries (superior than chlorhexidine) without deleterious effect on the gastrointestinal microbiota and host tissues, while promoting microbial activity associated with dental health.
One of the challenges for clinical translation of nanocatalysis technology for topical oral use [14,15] has been the need to apply two-components, either in two steps (catalytic nanoparticle followed by H2O2 adminstration) or via use of containers with separate chambers housing each of the components separately. The latter, which would allow mixing of the two ingredients prior to the treatment, is feasible based on our preliminary testing (unpublished) but requires custom packaging. Thus, a one-step nanocatalytic formulation that relies on intrinsic H2O2 generation activated under pathological conditions might be a more practical and targeted strategy for oral disease prevention. Having shown the therapeutic precision and efficiency of the bi-functional hybrid nanozyme, further in vivo mechanistic and toxicity studies would lay the foundation for clinical applications for human caries prevention. This approach may be relevant in the post-microbiome era, whereby treatment of polymicrobial diseases requires specific targeting of opportunistic pathogens in mixed community without disturbing the commensals and the ecological diversity of the host microbiota.
Supplementary Material
Acknowledgements
This work was supported in part by the National Institutes of Health (R01DE025848). Additional support was provided by the University of Pennsylvania Research Foundation.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biomaterials.2020.120581.
References
- [1].Stewart PS, Mechanisms of antibiotic resistance in bacterial biofilms, Int. J. Med. Microbiol 292 (2002) 107–113, doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- [2].Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S, Biofilms: an emergent form of bacterial life, Nat. Rev. Microbiol 14 (2016) 563–575, doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- [3].Bowen WH, Burne RA, Wu H, Koo H, Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments, Trends Microbiol. 26 (2018) 229–242, doi: 10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Richards D, Oral diseases affect some 3.9 billion people, Evid. Base Dent 14 (2013) 35, doi: 10.1038/sj.ebd.6400925. [DOI] [PubMed] [Google Scholar]
- [5].Kassebaum NJ, Smith AGC, Bernabe E, Fleming TD, Reynolds AE, Vos T, Murray CJL, Marcenes W, Global, regional, and national prevalence, incidence, and disability-adjusted life years for oral conditions for 195 countries, 1990–2015: a systematic analysis for the global burden of diseases, injuries, and risk factors, J. Dent. Res 96 (2017) 380–387, doi: 10.1177/0022034517693566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L, Targeting microbial biofilms: current and prospective therapeutic strategies, Nat. Rev. Microbiol 15 (2017) 740–755, doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Duguid R, In-vitro acid production by the oral bacterium Sreptococcus mutans 10449 in various concentrations of glucose, fructose and sucrose, Arch. Oral Biol 30 (1985) 319–324, doi: 10.1016/0003-9969(85)90004-4. [DOI] [PubMed] [Google Scholar]
- [8].Newbrun E, Sugar and dental-caries - a review of human studies, Science 217 (1982) 418–423, doi: 10.1126/science.7046052. [DOI] [PubMed] [Google Scholar]
- [9].Lemos JA, Palmer SR, Zeng L, Wen ZT, Kajfasz JK, Freires IA, Abranches J, Brady LJ, The biology of Streptococcus mutans, Microbiol. Spectr 7 (2019), doi: 10.1128/microbiolspec.GPP3-0051-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Baldeck JD, Marquis RE, Targets for hydrogen-peroxide-induced damage to suspension and biofilm cells of Streptococcus mutans, Can. J. Microbiol 54 (2008) 868–875, doi: 10.1139/W08-078. [DOI] [PubMed] [Google Scholar]
- [11].Thurnheer T, Belibasakis GN, Streptococcus oralis maintains homeostasis in oral biofilms by antagonizing the cariogenic pathogen Streptococcus mutans, Mol. Oral Microbiol 33 (2018) 234–239, doi: 10.1111/omi.12216. [DOI] [PubMed] [Google Scholar]
- [12].Chen LL, Chakraborty B, Zou J, Burne RA, Zeng L, Amino sugars modify antagonistic interactions between commensal oral Streptococci and Streptococcus mutans, Appl. Environ. Microbiol 85 (2019), doi: 10.1128/AEM.00370-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhu L, Kreth J, The role of hydrogen peroxide in environmental adaptation of oral microbial communities, Oxid. Med. Cell. Longev (2012), doi: 10.1155/2012/717843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu Y, Naha PC, Hwang G, Kim D, Huang Y, Simon-Soro A, Jung HI, Ren Z, Li Y, Gubara S, Alawi F, Zero D, Hara AT, Cormode DP, Koo H, Topical ferumoxytol nanoparticles disrupt biofilms and prevent tooth decay in vivo via intrinsic catalytic activity, Nat. Commun 9 (2018) 2920, doi: 10.1038/s41467-018-05342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Naha PC, Liu Y, Hwang G, Huang Y, Gubara S, Jonnakuti V, Simon-Soro A, Kim D, Gao LZ, Koo H, Cormode DP, Dextran-coated iron oxide nanoparticles as biomimetic catalysts for localized and pH-activated biofilm disruption, ACS Nano 13 (2019) 4960–4971, doi: 10.1021/acsnano.8b08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Naha PC, Zaki AA, Hecht E, Chorny M, Chhour P, Blankemeyer E, Yates DM, Witschey WR, Litt HI, Tsourkas A, Cormode DP, Dextran coated bismuth-iron oxide nanohybrid contrast agents for computed tomography and magnetic resonance imaging, J. Mater. Chem. B 2 (2014) 8239–8248, doi: 10.1039/C4TB01159G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Altikatoglu M, Basaran Y, Arioz C, Ogan A, Kuzu H, Glucose oxidase-dextran conjugates with enhanced stabilities against temperature and pH, Appl. Biochem. Biotechnol 160 (2010) 2187–2197, doi: 10.1007/s12010-009-8812-8. [DOI] [PubMed] [Google Scholar]
- [18].Orrego AH, Ghobadi R, Moreno-Perez S, Mendoza AJ, Fernandez-Lorente G, Guisan JM, Rocha-Martin J, Stabilization of immobilized lipases by intense intramolecular cross-linking of their surfaces by using aldehyde-dextran polymers, Int. J. Mol. Sci 19 (2018), doi: 10.3390/ijms19020553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gao L, Zhuang J, Nie L, Zhang J, Zhang Y, Gu N, Wang T, Feng J, Yang D, Perrett S, Yan X, Intrinsic peroxidase-like activity of ferromagnetic nanoparticles, Nat. Nanotechnol 2 (2007), doi: 10.1038/nnano.2007.260. [DOI] [PubMed] [Google Scholar]
- [20].Zhang Y, Tsitkov S, Hess H, Proximity does not contribute to activity enhancement in the glucose oxidase-horseradish peroxidase cascade, Nat. Commun 7 (2016) 13982, doi: 10.1038/ncomms13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Redanz S, Cheng X, Giacaman RA, Pfeifer CS, Merritt J, Kreth J, Live and let die: hydrogen peroxide production by the commensal flora and its role in maintaining a symbiotic microbiome, Mol. Oral Microbiol 33 (2018) 337–352, doi: 10.1111/omi.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hwang G, Koltisko B, Jin XM, Koo H, Nonleachable imidazolium-incorporated composite for disruption of bacterial clustering, exopolysaccharide-matrix assembly, and enhanced biofilm removal, ACS Appl. Mater. Interfaces 9 (2017) 38270–38280, doi: 10.1021/acsami.7b11558. [DOI] [PubMed] [Google Scholar]
- [23].Marsh PD, Moter A, Devine DA, Dental plaque biofilms: communities, conflict and control, Periodontol 55 (2000) 16–35 2011, doi: 10.1111/j.1600-0757.2009.00339.x. [DOI] [PubMed] [Google Scholar]
- [24].Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR 3rd, Heydorn A, Koo H, The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm, PLoS Pathog. 8 (2012) e1002623, doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gao L, Liu Y, Kim D, Li Y, Hwang G, Naha PC, Cormode DP, Koo H, Nanocatalysts promote Streptococcus mutans biofilm matrix degradation and enhance bacterial killing to suppress dental caries in vivo, Biomaterials 101 (2016) 272–284, doi: 10.1016/j.biomaterials.2016.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xiao J, Hara AT, Kim D, Zero DT, Koo H, Hwang G, Biofilm three-dimensional architecture influences in situ pH distribution pattern on the human enamel surface, Int. J. Oral Sci 9 (2017) 74–79, doi: 10.1038/ijos.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Saito M, Seki M, Iida K, Nakayama H, Yoshida S, A novel agar medium to detect hydrogen peroxide-producing bacteria based on the prussian blue-forming reaction, Microbiol. Immunol 51 (2007) 889–892, doi: 10.1111/j.1348-0421.2007.tb03971.x. [DOI] [PubMed] [Google Scholar]
- [28].Sumioka R, Nakata M, Okahashi N, Li Y, Wada S, Yamaguchi M, Sumitomo T, Hayashi M, Kawabata S, Streptococcus sanguinis induces neutrophil cell death by production of hydrogen peroxide, PloS One 12 (2017) e0172223, doi: 10.1371/journal.pone.0172223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bowen WH, Rodent model in caries research, Odontology 101 (2013) 9–14, doi: 10.1007/s10266-012-0091-0. [DOI] [PubMed] [Google Scholar]
- [30].Horev B, Klein MI, Hwang G, Li Y, Kim D, Koo H, Benoit DS, pH-activated nanoparticles for controlled topical delivery of farnesol to disrupt oral biofilm virulence, ACS Nano 9 (2015) 2390–2404, doi: 10.1021/nn507170s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Klein MI, Scott-Anne KM, Gregoire S, Rosalen PL, Koo H, Molecular approaches for viable bacterial population and transcriptional analyses in a rodent model of dental caries, Mol. Oral Microbiol 27 (2012) 350–361, doi: 10.1111/j.2041-1014.2012.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Clarke EL, Taylor LJ, Zhao C, Connell A, Lee JJ, Fett B, Bushman FD, Bittinger K, Sunbeam: an extensible pipeline for analyzing metagenomic sequencing experiments, Microbiome 7 (2019) 46, doi: 10.1186/s40168-019-0658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wood DE, Salzberg Kraken SL, Ultrafast metagenomic sequence classification using exact alignments, Genome Biol. 15 (2014) R46, doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kanehisa M, Goto S, KEGG: kyoto encyclopedia of genes and genomes, Nucleic Acids Res. 28 (2000) 27–30, doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Buchfink B, Xie C, Huson DH, Fast and sensitive protein alignment using DIAMOND, Nat. Methods 12 (2015) 59–60, doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- [36].Molday RS, MacKenzie D, Immunospecific ferromagnetic iron-dextran reagents for the labeling and magnetic separation of cells, J. Immunol. Methods 52 (1982) 353–367, doi: 10.1016/0022-1759(82)90007-2. [DOI] [PubMed] [Google Scholar]
- [37].Thorek DL, Tsourkas A, Size, charge and concentration dependent uptake of iron oxide particles by non-phagocytic cells, Biomaterials 29 (2008) 3583–3590, doi: 10.1016/j.biomaterials.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, Koo H, Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo, Infect. Immun 82 (2014) 1968–1981, doi: 10.1128/IAI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rhee SG, Measuring H2O2 produced in response to cell surface receptor activation, Nat. Chem. Biol 3 (2007) 244–246, doi: 10.1038/nchembio0507-244. [DOI] [PubMed] [Google Scholar]
- [40].Leichnitz S, Heinrich J, Kulak N, A fluorescence assay for the detection of hydrogen peroxide and hydroxyl radicals generated by metallonucleases, Chem. Commun 54 (2018) 13411–13414, doi: 10.1039/C8CC06996D. [DOI] [PubMed] [Google Scholar]
- [41].Colby SM, Russell RR, Sugar metabolism by mutans Streptococci, Soc. Appl. Bacteriol. Symp. Ser 26 (1997) 80S–88S, doi: 10.1046/j.1365-2672.83.s1.9.x. [DOI] [PubMed] [Google Scholar]
- [42].Dewar MD, Walker GJ, Metabolism of the polysaccharides of human dental plaque. I. Dextranase activity of streptococci, and the extracellular polysaccharides synthesized from sucrose, Caries Res. 9 (1975) 21–35, doi: 10.1159/000260139. [DOI] [PubMed] [Google Scholar]
- [43].Germaine GR, Harlander SK, Leung WL, Schachtele CF, Streptococcus mutans dextransucrase: functioning of primer dextran and endogenous dextranase in water-soluble and water-insoluble glucan synthesis, Infect. Immun 16 (1977) 637–648, doi: 10.1128/IAI.16.2.637-648.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Matsumoto-Nakano M, Role of Streptococcus mutans surface proteins for biofilm formation, Jpn. Dent. Sci. Rev 54 (2018) 22–29, doi: 10.1016/j.jdsr.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Banas JA, Vickerman MM, Glucan-binding proteins of the oral streptococci, Crit. Rev. Oral Biol. Med 14 (2003) 89–99, doi: 10.1177/154411130301400203. [DOI] [PubMed] [Google Scholar]
- [46].Lynch DJ, Fountain TL, Mazurkiewicz JE, Banas JA, Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture, FEMS Microbiol. Lett 268 (2007) 158–165, doi: 10.1111/j.1574-6968.2006.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lynch DJ, Michalek SM, Zhu M, Drake D, Qian F, Banas JA, Cariogenicity of Streptococcus mutans glucan-binding protein deletion mutants, Oral Health Dent, OR Manag. 12 (2013) 191–199, doi: 10.4172/2247-2452.1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Koppenol WH, The reaction of ferrous EDTA with hydrogen peroxide: evidence against hydroxyl radical formation, J. Free Radic. Biol. Med 1 (1985) 281–285, doi: 10.1016/0748-5514(85)90132-1. [DOI] [PubMed] [Google Scholar]
- [49].Attri P, Kim YH, Park DH, Park JH, Hong YJ, Uhm HS, Kim KN, Fridman A, Choi EH, Generation mechanism of hydroxyl radical species and its lifetime prediction during the plasma-initiated ultraviolet (UV) photolysis, Sci. Rep 5 (2015) 9332, doi: 10.1038/srep09332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Guo Q, Yue Q, Zhao J, Wang L, Wang H, Wei X, Liu J, Jia J, How far can hydroxyl radicals travel? An electrochemical study based on a DNA mediated electron transfer process, Chem. Commun 47 (2011) 11906–11908, doi: 10.1039/C1CC14699H. [DOI] [PubMed] [Google Scholar]
- [51].Price M, Reiners JJ, Santiago AM, Kessel D, Monitoring singlet oxygen and hydroxyl radical formation with fluorescent probes during photodynamic therapy, Photochem. Photobiol 85 (2009) 1177–1181, doi: 10.1111/j.1751-1097.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Carlisi D, D’Anneo A, Martinez R, Emanuele S, Buttitta G, Di Fiore R, Vento R, Tesoriere G, Lauricella M, The oxygen radicals involved in the toxicity induced by parthenolide in MDA-MB-231 cells, Oncol. Rep 32 (2014) 167–172, doi: 10.3892/or.2014.3212. [DOI] [PubMed] [Google Scholar]
- [53].Burnham CA, Hogan PG, Wallace MA, Deych E, Shannon W, Warren DK, Fritz SA, Topical decolonization does not eradicate the skin microbiota of community-dwelling or hospitalized adults, Antimicrob. Agents Chemother 60 (2016) 7303–7312, doi: 10.1128/AAC.01289-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bescos R, Ashworth A, Cutler C, Brookes ZL, Belfield L, Rodiles A, Casas-Agustench P, Farnham G, Liddle L, Burleigh M, White D, Easton C, Hickson M, Effects of chlorhexidine mouthwash on the oral microbiome, Sci. Rep 10 (2020) 5254, doi: 10.1038/s41598-020-61912-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


