Abstract
The continuing pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, which causes coronavirus disease 2019 (COVID-19), has spread globally and its reliable diagnosis is one of the foremost priorities for protecting public health. Herein a rapid (<1 h), easy-to-implement, and accurate CRISPR-based evanescent wave fluorescence biosensing platform for detection of SARS-CoV-2 is reported. The collateral effect of Cas13a is combined with a universal autonomous enzyme-free hybridization chain reaction (HCR) by designing a cleavage hairpin reporter, which is cleaved upon target recognition, and hence releasing the initiator sequence to trigger the downstream HCR circuits. Detection of HCR assemblies is accomplished by first adsorbing to the desthiobiotin-modified optical fiber, followed by fluorescence emission induced by an evanescent field. Three Cas13a crRNAs targeting the genes of S, N and Orf1ab of SARS-CoV-2 are programmed to specifically target SARS-CoV-2 or broadly detect related coronavirus strains, such as MERS-CoV and SARS-CoV. The HCR amplification coupled Cas13a-based biosensing platform is capable of rapid detection of SARS-CoV-2 with attomolar sensitivity. This method is further validated by adding target RNA of SARS-CoV-2 in negative oropharyngeal swabs. The good discrimination capability of this technique demonstrates its promising potential for point-of-care diagnosis of COVID-19.
Keywords: SARS-CoV-2, CRISPR-Cas13a, Hybridization chain reaction, Evanescent wave, Biosensor
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, which causes coronavirus disease 2019 (COVID-19), has spread globally since its discovery in Wuhan city, China in December 2019 (Zhou et al., 2020). Up to January 3, 2021, the disease has spread to at least 219 countries/area, infected at least 83 million people, and has resulted in at least 1.8 million deaths globally. The apparent ease with it spreading from human to human presents an imminent threat to the global public health (Li et al., 2020; Wrapp et al., 2020). Simple, low-cost and yet accurate, sensitive, and quantitative detection of this virus has been one of the foremost priorities for facilitating public health interventions.
The SRAS-CoV-2 virus is enveloped and generally spherical with diameter ranging from 60 nm to 140 nm, and its solar corona-like morphology is consistent with the family Coronaviridae (Zhu et al., 2020). SARS-CoV-2 has a single-stranded positive-sense RNA genome that is ~30,000 nucleotides in length. Its first complete genome was discovered by using a combination of Sanger, Illumina, and nanopore sequencing, providing important data for researchers to design primers and to probe sequences for other nucleic acid tests (CDC, 2020; Tan et al., 2020). Furthermore, full-genome comparisons revealed that the SARS-CoV-2 has a similar genetic sequence to the beta coronavirus B lineage, showing ~80%, ~50%, and ~96% similarity to the genome of the severe acute respiratory syndrome virus (SARS-CoV), Middle East respiratory syndrome virus (MERS-CoV), and bat coronavirus RaTG13, respectively (Lu et al., 2020; Zhou et al., 2020). Although the high-throughput sequencing in diagnosis and identification of respiratory pathogens especially for patients with unknown origin infections is becoming more and more popular, it is not routinely used to detect the specific strains of virus due to the disadvantages in simplicity, cost- and time-effectiveness (Zhang et al., 2020b). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) is the current primary method of detecting SRAS-CoV-2 because of its sensitivity, specificity and ease of quantification (Corman et al., 2020; Shen et al., 2020b). A number of RT-qPCR kits have been designed to detect SARS-CoV-2 genetically since the genic sequence of SARS-CoV-2 was publicly disclosed on 12 January 2020 (WHO, 2020). However, the requirement of thermocycling limits its non-laboratory and low-cost applications under limited resources (Zhou et al., 2018).
Microbial adaptive immune systems use CRISPRs (clustered regularly interspaced short palindromic repeats) and CRISPR-associated (Cas) proteins for RNA-guided nucleic acid cleavage (Horvath and Barrangou, 2010; Li et al., 2019). The contained programmable endonucleases can adapt to target any sequence (Koonin et al., 2017). While some Cas enzymes target DNA (Chen et al., 2018; Li et al. 2018a, 2018b; Zhou et al., 2018), the RNA-guided, RNA-targeting CRISPR effector Cas13a (previously known as C2c2) can be reprogrammed with CRISPR RNAs (crRNAs) to cleave specific RNA target (Abudayyeh et al., 2016; Shmakov et al., 2017). Cas13a exhibits both target cleavage and a “collateral effect” of promiscuous RNase activity upon target recognition (Abudayyeh et al., 2016; East-Seletsky et al., 2016; Gootenberg et al., 2017; Shmakov et al., 2015). Revealed by the X-ray crystal diffraction, Cas13a and crRNA undergo a significant conformational change upon highly sequence-specific target binding, and hence activate Cas13a to cleave any single stranded RNA molecule in a non-specific manner (East-Seletsky et al., 2016; Liu et al., 2017). However, the collateral activity of Cas13a exhibits the base preferences and the cleavage could be improved with buffer and crRNA design optimization (Gootenberg et al., 2018; Wu et al., 2021). A platform, termed specific high-sensitivity enzymatic reporter unlocking (SHERLOCK), allows multiplexed, portable, and ultra-sensitive detection of viral RNA or DNA based on Cas13a-mediated collateral cleavage of a rationally designed nucleic acid reporter (East-Seletsky et al., 2016; Gootenberg et al., 2017). The CRISPR-based diagnostic technique has been designed to detect SARS-CoV-2 so as to accelerate virus outbreak response (Arizti-Sanz et al., 2020; Guo et al., 2020; Patchsung et al., 2020). To achieve high sensitivity and in-field application, different enzyme-based isothermal amplification steps were explored to amplify the target nucleic acids, followed by Cas13a detection (Gootenberg et al. 2017, 2018; Shen et al., 2020a). These amplification strategies employ expensive enzymes, tedious reverse transcription, and the proper design of species-specific primers is tricky, suffering from trade-offs among sensitivity, specificity, simplicity, cost, and speed (Corman et al., 2020; Huang et al., 2020a; Park et al., 2020; Vogels et al., 2020).
Owing to the high programmability and modularity, autonomous enzyme-free nucleic acid circuits are attracting ever-growing interest as signal amplifiers in developing ultrasensitive biosensing techniques. Nucleic acids themselves could be engineered as independent modules that carry out multiple functions (Memon et al., 2020; Seelig et al., 2006). In enzyme-free nucleic acid circuits, nucleic acids act as a set of “hardware” that executes an algorithm obeying Watson-Crick base pair rules and function as “circuits” in carbon/silicon-based computing. The nucleic acid circuits with different functions can be easily modularized and cascaded because of their high controllability and programmability, which is especially important for the in silico assisted precise design of complicated large-scale systems (Chirieleison et al., 2013; Wang et al., 2014). One of the most attractive functions of enzyme-free nucleic acid circuits is signal amplification. Based on the elementary toehold-mediated strand displacement (TSD) mechanism, several nucleic acid circuits-amplifiers have achieved impressive sensitivities in biosensing applications (Jung and Ellington, 2014; Zhuang et al., 2013). These enzyme-free amplification schemes can operate under conditions that might otherwise inhibit proteins (Li et al., 2011). In general, the execution of nucleic acid circuits requires an oligonucleotide as input to trigger downstream toehold-mediated strand displacement reactions, which has paved the way for ultra-sensitive nucleic acid detection since nucleic acid circuits could be precisely designed using specific target sequence as input (Li et al., 2011; Liu et al., 2013). The toehold-mediated strand displacement reactions can be cascaded with superior properties in cost, programmable, modularity, and universality (Zhang and Seelig, 2011). In particular, hybridization chain reaction (HCR) amplification, a type of toehold-mediated strand displacement reaction first developed by Dirks and Pierce (Dirks and Pierce, 2004), is widely explored because of its enzyme-free nature, isothermal conditions, simple protocols, structural flexibility, and excellent amplification efficiency (Bi et al., 2017; 2020a).
To combine the advantages of targeting RNA with CRISPR-Cas13a and enzyme-free nucleic acid amplification, we reported the development and validation of a HCR coupled CRISPR-Cas13a-based assay (Cas-HCR) for detection of SARS-CoV-2. The detection performance of Cas-HCR assay was demonstrated on a home-made optical-fiber evanescent wave fluorescence biosensor capable of collecting the fluorescence excited within an exponentially decaying evanescent field around the optical fiber, one of the earliest trials for reusable nucleic acid biosensing device (Liu et al., 2018; Wang et al. 2017, 2019).
2. Materials and methods
2.1. Material and apparatus
Detailed information on reagents, oligonucleotides, buffers and apparatus used in this work are provided in Supplementary Information S1. The optical-fiber evanescent wave fluorescence biosensor was described in our previous works (Liu et al., 2018; Wang et al., 2017), and its photograph was shown in Fig. S1. Briefly speaking, the 635 nm He–Ne laser was introduced into a biofunctionalized optical-fiber to generate the evanescent wave on the fiber surface. The interface-bound fluorescent dyes due to the presence of the targets were excited by the evanescent wave. The emitted fluorescence signals were then acquired by using the photodiode with a lock-in amplifier and recorded in a built-in industrial Tablet PC. In this biosensor system, a combination tapered optical-fiber was adopted as described in our previous work (Wang et al., 2015). Preparation of desthiobiotin (DTB) functionalized optical-fiber sensing surface are provided in Supplementary Information S2 and the surface modification schematic of optical-fiber with BSA-desthiobiotin (BSA-DTB) conjugate was shown in Fig. S2.
2.2. Nucleic acid preparation of CRISPR-Cas13a system
crRNAs and target RNAs of SARS-CoV-2, SARS-CoV and MERS-CoV were designed based on the reported guidelines (Freije et al., 2019; Patchsung et al., 2020). Specifically, a highly conserved 28 nt window was the one with ≤2 polymorphic positions (≤95% major allele frequency) and less than 50% data missing at all sites. The viral genomes were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) and then aligned using the mafft v7.31 (Katoh et al., 2019), and the consensus information was showed on Jalview v2.11 (Waterhouse et al., 2009). Next, LwaCas13a target sites on the SARS-CoV-2 were filtered by blasting against the GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Among the crRNA sequences, the ultimate 28 nt sequence was selected considering the complexity of secondary structure predicted by NUPACK web server (Zadeh et al., 2011).
2.3. Cas13a collateral cleavage activity
To form annealed hairpins, crRNA and Reporter 1 beacon were individually denatured at 76 °C for 5 min, and then cooled at room temperature for over 1 h. Cas13a collateral cleavage activity was investigated by using 45 nM LwaCas13a, 22.5 nM crRNA, 125 nM Reporter 1 beacon, 1.25 μL RNase inhibitor, and varying amounts of nucleic acid target in nuclease assay buffer. The reactions were incubated at 37 °C for 1–2 h on a 384 well microplate reader with fluorescent measurements every 5 min.
2.4. Gel electrophoresis
The CRISPR-Cas13a was diluted by using 6 × loading buffer with a volume ratio of 5:1 for gel electrophoresis. A 15% urea-denaturing polyacrylamide gel was prepared with 10 × TBE buffer. After pre-running and warming the gel for 30 min at 50 V and then loading the samples, electrophoresis was run at 80 V for 100 min in 1 × TBE buffer. Gels were soaked in 1 × TBE for about 10 min and then stained with SYBR Green I Gel Stain and imaged under UV illumination using FLS-5100 film (Fuji Photo Film Co., Ltd., Tokyo, Japan).
To characterize the formation of HCR assemblies, a 15% fresh polyacrylamide gel was prepared with 10 × TBE buffer. Electrophoresis was run at 60 V for 100 min in 1 × TBE buffer. Gels were stained and imaged as the way in the denaturing polyacrylamide gel electrophoresis.
2.5. Detection of SARS-CoV-2 in buffer/human oropharyngeal swabs using the Cas-HCR biosensing technique
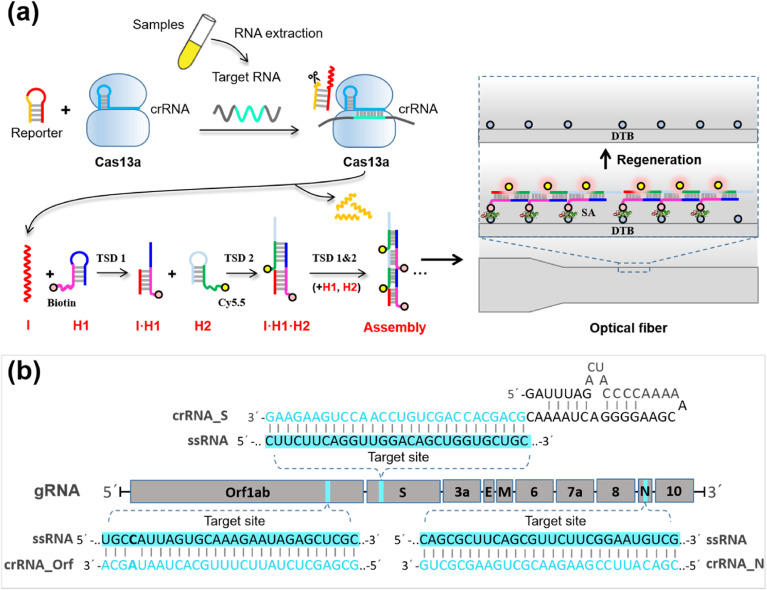
Under the optimized conditions, 2 μL LwaCas13a in storage buffer (63.3 μg ml−1 stock concentration), 2 μL crRNA (225 nM stock), 2 μL Reporter 2 (1.25 μM stock), 2 μL synthetic SARS-CoV-2 genes at varying concentrations, 0.5 μL RNase inhibitor and 11.5 μL nuclease assay buffer were mixed for Cas13a recognition. After incubation of the mixture for 20 min, 20 μL SPSC buffer, 5 μL annealed Bio-H1 (1 μM stock) and 5 μL annealed Cy5.5-H2 (1 μM stock) were added and then the mixtures further incubated at 37 °C for 30 min. The annealing process contained that 1 μM Bio-H1 and Cy5.5-H2 stock were denatured respectively at 95 °C for 5 min, and then cooled at room temperature for over 1 h to form annealed hairpins. Lastly, 500 μL SPSC buffer was added into the reaction tube that contained amplified HCR products to fit the volume requirement for evanescent wave fluorescent biosensing detection. The optical-fiber based detection proceeded as follows: 1) 20 nM streptavidin was first delivered into the flow cell to bind to DTB immobilized onto the sensing surface of fiber for 2 min; 2) SPSC buffer was pumped to elute the unbounded SA; 3) amplified HCR products with biotin labels were pumped to react with interface-bound streptavidin for 10 min; 4) washing buffer was pumped to elute interface-bound streptavidin and HCR products for 10 min; 5) equilibration buffer was pumped for 1 min to regenerate the sensing surface for next round of reaction cycle.
The application ability of Cas-HCR biosensing technique was investigated by detecting the S genes of SARS-CoV-2 in negative oropharyngeal swabs samples that were acquired from healthy donors at the campus hospital with the approval of Tsinghua University. Total RNA in samples was extracted using Qiagen Viral RNA Mini kit following the manufacturer's instructions. 2 μL of the extracted sample RNA was added into 18 μL reaction mixtures contained Cas13a, crRNA, Reporter 2 and different amounts of target RNA as depicted in buffer, followed by the HCR amplification for fluorescence measurement. The relative signal intensity was calculated by the following equation.
| (1) |
where SMea means the measured signal intensity; SMin means the signal intensity corresponding to the control sample; SMax means the signal intensity corresponding to the maximum concentration.
2.6. Statistical analysis
Statistical analysis was performed with Origin 2018 software. Each experiment was conducted three times in parallel. Results were presented as measured mean value ± standard deviation. Error bars in all figures represent the standard deviations from independently triplicate measurements.
3. Results and discussion
3.1. Sensing mechanism of Cas-HCR biosensing technique
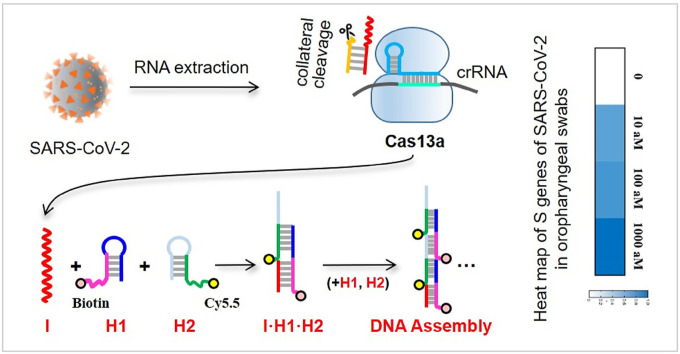
The Cas-HCR biosensing technique performed the recognition and cleavage of Cas13a towards its target RNA sequence as specified by the crRNA sequence, followed by the activation of the promiscuous RNase activity of Cas13a to cleave the rationally designed nucleic acid reporter and trigger the downstream HCR amplification. The biotinylated HCR products were captured by the streptavidin-functionalized optical fiber, after which the Cy5.5 fluorophores labeled on the HCR assemblies were excited by the evanescent wave, and collected to generate the signal by using the optical-fiber evanescent wave fluorescence biosensor platform to confirm the detection of the virus genes (Fig. 1 a). The specificity of the platform was not only derived from the base pairing but also from the high fidelity of Cas13a (Shan et al., 2019).
Fig. 1.
SARS-CoV-2 detection with enzyme-free amplification coupled CRISPR-Cas13a. (a) Schematic of SARS-CoV-2 detection workflow. Conventional RNA extraction can be used as an input to the Cas-HCR assay for detection of S gene, N gene and Orf1ab gene, respectively, which is detected by a home-made optical-fiber reusable evanescent wave fluorescence biosensor. (b) Visualization of targets and crRNAs on the SARS-CoV-2 genome.
We designed three Cas13a crRNAs with two specifically targeting the S (spiked) and N (nucleoprotein) genes of SARS-CoV-2 and one broadly targeting the Orf1ab gene of three highly pathogenic coronaviruses (SARS-CoV-2, MERS-CoV, SARS-CoV) (Fig. 1b) based on the previously reported CRISPR RNA design guidelines (Freije et al., 2019). The crRNAs consisting a 36 nt direct repeat region and a programmable 28 nt guide region (spacer) can assemble with Cas13a. Next, we designed the nucleic acid hairpin reporter containing RNA cleavage sites (UUUUUC) in yellow for the promiscuous RNase activity of Cas13a (Abudayyeh et al., 2016; Gootenberg et al., 2018). To demonstrate the specificity of CRISPR-Cas13a with different crRNA design, a dual-labeled nucleic acid reporter, Reporter 1 beacon with FAM-labeled 5′ end and a quencher molecule BHQ1-labled 3′ end and stem-loop hairpin constituting of 8 hybridized base pairs, was applied (Table S1). After annealing, the formation of hairpin reporter placed fluorophore in close proximity to its quencher, resulting in a low fluorescence intensity background; after the specific recognition, Reporter 1 beacon was cleaved and separated FAM from BHQ1, leading to an obvious fluorescence recovery.
3.2. CRISPR-Cas13a system design and verification
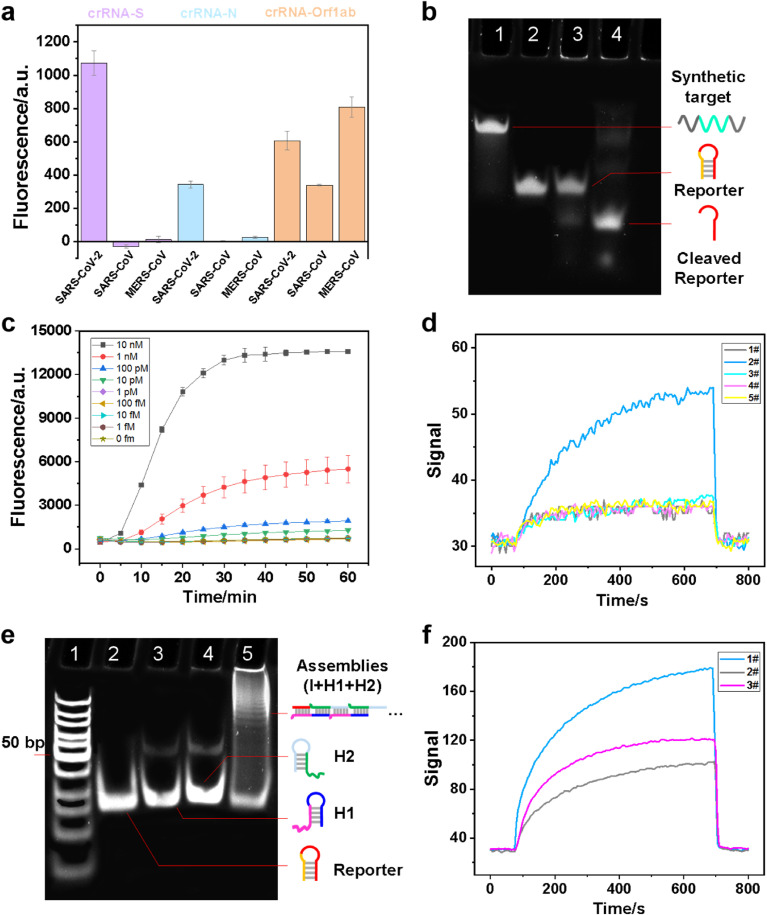
Cas13a crRNAs were programmed to specifically target SARS-CoV-2 or broadly detect related coronavirus strains. As shown in Fig. S3, the N gene and S gene of SARS-CoV and MERS-CoV were significantly different from that of SARS-CoV-2. While, there were only two-base difference between Orf1ab gene of three viral sequences. Therefore, theoretically, the N gene and S gene crRNAs used in the assay were specific for SARS-CoV-2, however failed to detect SARS-CoV and MERS-CoV, whereas the Orf1ab gene crRNA was capable to detect three highly pathogenic coronavirus strains. As revealed by the net fluorescence subtracting that of the blank sample in Fig. 2 a, we demonstrated that the CRISPR-Cas13a system was able to distinguish SARS-CoV-2 with no cross-reactivity for other two highly pathogenic coronaviruses using the crRNAs targeting N gene and S gene, however, with expected cross-reactivity for the related coronavirus strains using the crRNA targeting Orf1ab gene. The CRISPR-Cas13a system showed higher sensitivity when targeting S gene compared with that targeting N gene.
Fig. 2.
Design and validation of Cas-HCR assay for SARS-CoV-2 detection. (a) crRNA specificity by using dual-labeled Reporter 1 beacon. (b) Electrophoretic analysis of the cleavage ability of Cas13a/crRNA in the presence of target RNA.1: Synthetic target (1 μM); 2: Reporter 1 beacon (1.25 μM); 3: Cas13a (0.45 μM), crRNA_S (0.225 μM), Reporter 1 beacon (1.25 μM); 4: Cas13a (0.45 μM), crRNA_S (0.225 μM), Reporter 1 beacon (1.25 μM), Synthetic target (1 μM). (c) Real-time kinetic measurement of Cas13a reactions initiated by target RNA towards Reporter 1 beacon. (d) The signal generated by H1, H2 with/without reporters, referring to the fluorescent value measured by the evanescent wave biosensor. 1#: without reporter; 2#: Reporter 1; 3#: Reporter 2; 4#: Reporter 3; 5#: Reporter 4. (e) Electrophoretic analysis of the Cas-HCR products. 1: DNA ladder; 2: Reporter 2 (1.25 μM); 3: H1 (1 μM); 4: H2 (1 μM); 5: Cas13a (0.45 μM), crRNA_S (0.225 μM), Reporter 2 (1.25 μM), Synthetic target (1 μM), followed by adding H1 (1 μL 10 μM), H2 (1 μL 10 μM), SPSC buffer (13 μL). (f) Performance analysis of one-pot and two-step Cas-HCR assay on the evanescent wave biosensor. 1#: 30 min CRISPR followed with 1 h HCR; 2#: one-pot CRISPR and HCR reaction for 1 h; 3#: one-pot CRISPR and HCR reaction for 1.5 h.
Denaturing polyacrylamide gel electrophoresis (PAGE) assay revealed that the targets and nucleic acid reporters, Reporter 1 beacon, were cleaved into short RNA fragments due to the cleavage activity of CRISPR-Cas13a (Fig. 2b). However, a slight band of cleaved reporter fragments was observed even in the absence of the targets (lane 3), indicating the potential non-target-induced promiscuous RNase activity of Cas13a. The similar phenomenon was also found by other researchers (East-Seletsky et al., 2016; Huang et al., 2018). Reporter 1 beacon released the I sequence, which exhibited a much higher migration speed than other longer strands as shown in lane 4. The time-dependent fluorescent intensity changes of the reporter were measured in the presence of SRAS-CoV-2 RNA sequences at different concentrations (Fig. 2c). The fluorescence significantly increased in the first 20–30 min because of the promiscuous RNase activity of Cas13a, exhibiting the time-dependent self-amplification process (Bruch et al., 2019; Wu et al., 2021). For the sake of time saving, the optimal time for the CRISPR-Cas13a recognition was selected to be 20 min. The obvious increase of fluorescent intensity was observed with the increased RNA concentrations (Fig. S4), further confirming Reporter 1 beacon was cleaved under the promiscuous RNase activity of Cas13a and released the BHQ1-labeled I sequences. The limit of detection (LOD) of the CRISPR-Cas13a system for the S genes of SARS-CoV-2 without signal amplification was 25 pM, i.e. approximately 1.5 × 107 copies/μL, which was regarded as the minimum concentration in which the signal was significantly different from the cutoff one defined as the average signal of blank samples plus three times of the standard deviation. Doubtlessly, the CRISPR-Cas13a system not only exhibit the accurate nucleic acid target recognition capability, but also amplify the signal relying on the promiscuous RNase activity.
3.3. Nucleic acid hairpin reporter design
We designed the nucleic acid hairpin reporter containing the initiator (I) region in red to trigger the downstream HCR amplification. The hairpin reporter should release the I sequence properly relying upon the presence of target rival RNA sequence, thereby triggering the formation of Cy5.5-labeled assemblies. The stem-loop hairpins constituted of 8, 12, 14 and 16 hybridized base pairs, named Reporter 1, Reporter 2, Reporter 3 and Reporter 4, respectively, as shown in Table S1, were examined by using the home-made evanescent wave biosensor (Insert of Fig. 2d). After that, we designed the I sequence-triggered TSD of hairpins (Bio-H1 and H2-Cy5.5) to generate multiple linear duplex I·(H1·H2)n assemblies as signal amplification products. By using a home-made optical-fiber evanescent wave fluorescence biosensor as the signal collector, the spontaneous hybridization of H1 and H2 hairpins was negligible (baseline, line 1) because of the locked complementary domains in the absence of I (Dirks and Pierce, 2004). The robust cascade pathways can be achieved on the basis of computational tools (Wolfe et al., 2017) and practical design guidelines (Ang and Yung, 2016). The hairpin with a stem of 8 hybridized base pairs, i.e. Reporter 1, exhibited an obvious signal increase compared with baseline (line 2), most likely resulting from the instability of stem-loop structure, thereby triggering the HCR reactions independently. The other three hairpins remained stable, thereby showing the similar signal response as baseline (lines 3–5). The reporter with a stem of 12 hybridized base pairs, i.e. Reporter 2, was used in the following experiments. The formation of assembled products was also be demonstrated by native PAGE (Fig. 2e), where the formed HCR assemblies in lane 5 exhibited lower and variable migration speeds than other short strands. Annealing of sequences showed a slight improvement on the biosensor signal (Fig. S5a). For the sake of time and cost saving, the concentrations of H1 and H2, temperature and optimal reaction time for the viral recognition triggered HCR reactions were 5 μL, 37 °C, 0.5 h, respectively (Figs. S5b–d). We also compared the performance of adopting two individual steps (i.e. CRISPR-Cas13a recognition and HCR amplification) with that of merging them into one-pot. The biosensor signal exhibited only half of the original value after merging two steps together (Fig. 2f). Hence, one-pot reaction was not recommended even though prolonging the reaction time helped to improve the signal to some extent. More investigations to develop a common buffer that could accommodate both the CRISPR-mediated recognition and the HCR amplification are needed (Ladha et al., 2020).
3.4. Signal detection using evanescent wave-induced emission
To reduce the cost of single detection, we used DTB molecules with moderate binding affinity toward streptavidin to prepare a reusable optical-fiber surface (Wang et al., 2019). Biotinylated HCR assemblies were designed to be anchored onto DTB-functionalized fiber surface via streptavidin “glue”, providing approximately unified physical distances for labeled fluorophores. Signals emitted by these Cy5.5-labeled HCR assemblies under the evanescent wave excitation can be measured by the biosensing platform. Moreover, streptavidin-HCR complexes can be effectively washed off to regenerate the DTB modified fiber surface. The performances of DTB-functionalized optical fiber in governing the balance of HCR assembly capture and surface regeneration were evaluated. Specifically, BSA-DTB conjugates with optimal molar ratios of 50 X were synthesized and covalently immobilized on the fiber surface. The “glue” streptavidin layer was robust enough to resist buffer elution (Wang et al., 2019), which was applied before the HCR assembly injection to guarantee the minimal interferences from unbound SA, and could be efficiently washed off using washing buffer (0.5% SDS, pH 1.9) (Wang et al., 2019).
3.5. Performance evaluation of Cas-HCR biosensing technique
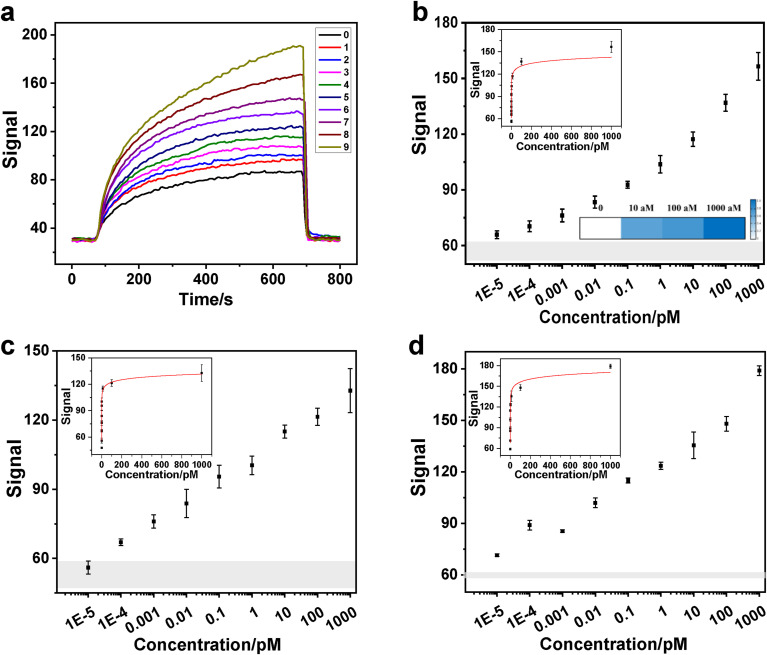
Based on the above investigation, the Cas-HCR assay can be run and visualized on a home-made optical-fiber evanescent wave fluorescence biosensor in approximately 1 h, which consists of an CRISPR-Cas13a recognition reaction at 37 °C for 20 min, HCR amplification reaction at 37 °C for 30 min and signal detection by using fiber at room temperature for approximately 10 min. Given the specific high fluorescence amplification toward biotinylated assemblies, the sensing performances using crRNA_S, crRNA_N and crRNA_Orf1ab towards the synthetic SARS-CoV-2 genes were compared. As expected, the original sensorgrams corresponding to the different concentrations of SARS-CoV-2 genes demonstrated that the Cas-HCR assay using crRNA_S operated reliably with biosensor signal deviations in the presence of 0.01 fM to 1 nM of SARS-CoV-2 target RNA sequence (Fig. 3 a). A rapid signal decrease after using washing buffer elution also indicated that the fiber surface regeneration could be efficiently achieved. By plotting the biosensor signals against the gene concentrations in Fig. 3b, a gradual signal increase was observed with the increased gene concentrations. The Cas-HCR biosensing technique could rapidly detect S genes of SARS-CoV-2 at concentrations as low as 10 aM, i.e. approximately 6 copies/μL, by obeying the similar rule that LOD was regarded as the minimum concentration in which the signal was significantly different from the cutoff one defined as the average signal of blank samples plus three times of the standard deviation. The experimental data points were further analyzed and fitted by a function of y = a-b × ln (x + c), where y was the signal and x was the gene concentration of SARS-CoV-2 (Left inset of Fig. 3b). The correlation coefficient (R2) reached 0.962 over the range of 0–1 nM. Using the similar approach, the performance of this Cas-HCR biosensing technique using crRNA_N towards the N genes of SARS-CoV-2 and crRNA_Orf1ab towards the Orf1ab of SARS-CoV-2 were investigated, respectively. Results showed that this technique was able to detect down to 100 aM, i.e. 60 copies/μL, SARS-CoV-2 N gene and 10 aM, i.e. 6 copies/μL, SARS-CoV-2 Orf1ab gene, respectively (Fig. 3c and d). The R2 of curve-fitting between the signal and the concentrations of N genes and Orf1ab genes were 0.997 and 0.966, respectively, over the range of 0–1 nM.
Fig. 3.
Performance evaluation of the Cas-HCR biosensing technique by using a home-made evanescent wave biosensor. (a) Representative real-time fluorescent signal of original sensorgrams corresponding to different concentrations of the S genes of SARS-CoV-2.0: no target; 1–9: ten-fold diluted RNA from 10 aM to 1 nM. (b) Sensitivities of this technique towards the S genes of SARS-CoV-2. Left inset: Standard curve towards S genes of SARS-CoV-2 (R2 = 0.962) fitted by y = a-b × ln (x + c). Right inset: Heat map of three concentration levels of S genes of SARS-CoV-2 spiked in RNA extracts of negative oropharyngeal swabs. The color bar ratio represents the relative signal intensity calculated by Eq. 1 (c-d) Sensitivities of this technique towards the N and Orf1ab genes of SARS-CoV-2, respectively. Each data point represents the average signal with standard deviation for independently triplicate experiments. Inset: Standard curves towards N genes (R2 = 0.997) and Orf1ab genes of SARS-CoV-2 (R2 = 0.966), respectively, fitted by y = a-b × ln (x + c). Gray shadow indicates three times of the standard deviation of blank samples, in which the top edge represents the cutoff signal defined as the average fluorescence of blank samples plus three times of the standard deviation. And original sensorgrams corresponding to c and d were shown in Figs. S6 and S7, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Besides, in light of the moderate binding affinity between DTB and streptavidin and the strategy of immobilizing DTB molecule on the solid surface (Wang et al., 2019), the optical-fiber allowed to be reused over 100 successive cycles with less than 2.5% signal decrease using washing buffer (Fig. S8a). In general, we used the fiber for testing if we continued the experiment within one month for no more than 150 successive cycles. For example, three optical fibers were used to get all data in Fig. 3. Moreover, we evaluated the carryover issue to treat the fiber with 4 nM Cy5.5-labeled streptavidin protein. By observing the sensorgram during the binding, washing buffer elution, and equilibration buffer regeneration as shown in Fig. S8b, the fluorescent signal went down to the baseline when eluting with washing buffer for no more than 10 min, and then equilibration buffer for 1 min, indicating the washing buffer was effective to break the binding interaction between the DTB and streptavidin, hence the potential carryover issue should be negligible to ensure the surface regeneration efficiently.
Last, we investigated the application ability of this Cas-HCR biosensing technique by detecting S genes of SARS-CoV-2 spiked in RNA extracts of oropharyngeal swabs from three healthy donors. The SARS-CoV-2 RNA was confirmed negative by the RT-qPCR technique. Right inset of Fig. 3b shows the heat map of three concentration levels (10 aM, 100 aM and 1000 aM) of S genes of SARS-CoV-2 measure by using this technique. The color bar ratio represents the relative signal intensity calculated by Eq. 1, which showed significant difference from the control one and also good correlation between the target concentration and Cas-HCR signal. Notably, we observed the decreased signal against the same concentration in the oropharyngeal swabs compared with those obtained in buffer, reflecting the possible matrix interference (Figs. S9 and S10). Despite the issue, we still observed signal discrimination against different concentrations of target in all oropharyngeal swab samples. The comparable standard derivations both in oropharyngeal swabs and buffers confirmed the satisfactory sensitivity and applicability of the developed Cas-HCR biosensing platform for SARS-CoV-2 detection in clinical diagnostics.
Since the outbreak of COVID-19, many CRISPR-based technologies have been developed for the detection of the SARS-CoV-2 gene, which mainly rely on a pre-nucleic acid amplification step to achieve ultra-high sensitivities (Chen et al., 2020; Huang et al., 2020b; Tian et al., 2021; Wang et al., 2021; Xiong et al. 2020, 2021). The CRISPR biosensor without pre-amplification only showed LODs at nM or pM level (Li et al., 2019; Wang et al., 2020). Here, we presented a new strategy, which employed the specific product of the CRISPR reaction to trigger the enzyme-free HCR reaction for signal amplification. Notably, both LOD and turnaround time of this technique were comparable to those of reported CRISPR-based viral nucleic acid detections (Table S2). This attempt provides a valuable reference for the subsequent development of more post amplified CRISPR technologies.
4. Conclusion
In summary, a HCR coupled CRISPR-Cas13a-based biosensing technique was established to detect SARS-CoV-2 with high sensitivity and low testing cost on a home-made optical-fiber evanescent wave fluorescence biosensing platform. This technique combined the merits of not only the prominent viral gene-specific recognition capability of CRISPR-Cas13a system but also the cheap, enzyme-free and isothermal nucleic acid amplification by HCR without the need of temperature cycling operation to dramatically improve the detection sensitivity. Under the optimized conditions, this technique provided a LOD of attomolar level towards three specific genes (S genes, N genes and Orf1ab genes) of SARS-CoV-2. Analysis of SARS-CoV-2 in negative oropharyngeal swabs convincingly demonstrated the biosensing technique held a great potential for point-of-care diagnosis of COVID-19. This attempt provides a valuable reference for the subsequent development of more post amplified CRISPR technologies.
CRediT authorship contribution statement
Yihan Yang: Methodology, Validation, Formal analysis, Writing – original draft. Jinchuan Liu: Validation, Formal analysis, Writing – original draft. Xiaohong Zhou: Methodology, Writing – original draft, Supervision, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Tsinghua University Spring Breeze Fund (2020Z99CFY038) and National Nature Science Foundation of China (52091541).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113418.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., Severinov K., Regev A., Lander E.S., Koonin E.V., Zhang F. Science. 2016;353(6299) doi: 10.1126/science.aaf5573. aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang Y.S., Yung L.-Y.L. Chem. Commun. (Camb.) 2016;52(22):4219–4222. doi: 10.1039/c5cc08907g. [DOI] [PubMed] [Google Scholar]
- Arizti-Sanz J., Freije C.A., Stanton A.C., Petros B.A., Boehm C.K., Siddiqui S., Shaw B.M., Adams G., Kosoko-Thoroddsen T.F., Kemball M.E., Uwanibe J.N., Ajogbasile F.V., Eromon P.E., Gross R., Wronka L., Caviness K., Hensley L.E., Bergman N.H., MacInnis B.L., Happi C.T., Lemieux J.E., Sabeti P.C., Myhrvold C. Nat. Commun. 2020;11(1):5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi S., Yue S., Zhang S. Chem. Soc. Rev. 2017;46(14):4281–4298. doi: 10.1039/c7cs00055c. [DOI] [PubMed] [Google Scholar]
- Bruch R., Baaske J., Chatelle C., Meirich M., Madlener S., Weber W., Dincer C., Urban G.A. Adv. Mater. 2019;31(51) doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- CDC U.S. 2020. CDC Diagnostic Tests for COVID-19. [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shi Y., Chen Y., Yang Z., Wu H., Zhou Z., Li J., Ping J., He L., Shen H., Chen Z., Wu J., Yu Y., Zhang Y., Chen H. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112642. 112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirieleison S.M., Allen P.B., Simpson Z.B., Ellington A.D., Chen X. Nat. Chem. 2013;5(12):1000–1005. doi: 10.1038/nchem.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Bruenink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Euro Surveill. 2020;25(3):23–30. [Google Scholar]
- Dirks R.M., Pierce N.A. Proc. Natl. Acad. Sci. U.S.A. 2004;101(43):15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A., O'Connell M.R., Knight S.C., Burstein D., Cate J.H.D., Tjian R., Doudna J.A. Nature. 2016;538(7624):270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije C.A., Myhrvold C., Boehm C.K., Lin A.E., Welch N.L., Carter A., Metsky H.C., Luo C.Y., Abudayyeh O.O., Gootenberg J.S., Yozwiak N.L., Zhang F., Sabeti P.C. Mol. Cell. 2019;76(5):826–837. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y., Chen Y., Feng G., Gu Q., Wang R.R., Zhou Q., Li W. Cell Discov. 2020;6:34. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P., Barrangou R. Science. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Huang M., Zhou X., Wang H., Xing D. Anal. Chem. 2018;90(3):2193–2200. doi: 10.1021/acs.analchem.7b04542. [DOI] [PubMed] [Google Scholar]
- Huang W.E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. Microb. Biotechnol. 2020;13(4):950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. 112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Ellington A.D. Acc. Chem. Res. 2014;47(6):1825–1835. doi: 10.1021/ar500059c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. Briefings Bioinf. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Makarova K.S., Zhang F. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladha A., Saito Makoto, Kim Nam-Gyun, Woolley Ann E., Joung Julia, Woolley Ann E., Segel Michael, Barretto Robert PJ., Ranu Amardeep, Macrae Rhiannon K., Faure Guilhem, Ioannidi Eleonora I., Krajeski Rohan N., Bruneau Robert, Huang Meei-Li W., Yu Xu G., Li Jonathan Z., Walker Bruce D., Hung Deborah T., Greninger Alexander L. N. Engl. J. Med. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Ellington A.D., Chen X. Nucleic Acids Res. 2011;39(16):e110. doi: 10.1093/nar/gkr504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-Y., Cheng Q.-X., Liu J.-K., Nie X.-Q., Zhao G.-P., Wang J. Cell Res. 2018;28(4):491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-Y., Cheng Q.-X., Wang J.-M., Li X.-Y., Zhang Z.-L., Gao S., Cao R.-B., Zhao G.-P., Wang J. Cell Discov. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li S., Wang J., Liu G. Trends Biotechnol. 2019;37(7):730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Yang X., Sun S., Wang Q., Wang K., Huang J., Liu J., He L. Anal. Chem. 2013;85(16):7689–7695. doi: 10.1021/ac4001157. [DOI] [PubMed] [Google Scholar]
- Liu L., Li X., Ma J., Li Z., You L., Wang J., Wang M., Zhang X., Wang Y. Cell. 2017;170(4):714–726. doi: 10.1016/j.cell.2017.06.050. [DOI] [PubMed] [Google Scholar]
- Liu J., Zhou X., Shi H. Anal. Chem. 2018;90(3):2362–2368. doi: 10.1021/acs.analchem.7b04933. [DOI] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memon A.G., Xing Y., Zhou X., Wang R., Liu L., Zeng S., He M., Ma M. J. Hazard Mater. 2020;384 doi: 10.1016/j.jhazmat.2019.120948. [DOI] [PubMed] [Google Scholar]
- Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S.I., Kim B.-T., Maeng J.-S. J. Mol. Diagn. 2020;22(6):729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T., Athipanyasilp N., Eiamthong B., Lakkanasirorat B., Phoodokmai T., Niljianskul N., Pakotiprapha D., Chanarat S., Homchan A., Tinikul R., Kamutira P., Phiwkaow K., Soithongcharoen S., Kantiwiriyawanitch C., Pongsupasa V., Trisrivirat D., Jaroensuk J., Wongnate T., Maenpuen S., Chaiyen P., Kamnerdnakta S., Swangsri J., Chuthapisith S., Sirivatanauksorn Y., Chaimayo C., Sutthent R., Kantakamalakul W., Joung J., Ladha A., Jin X., Gootenberg J.S., Abudayyeh O.O., Zhang F., Horthongkham N., Uttamapinant C. Nat. Biomed. Eng. 2020;4:1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Seelig G., Soloveichik D., Zhang D.Y., Winfree E. Science. 2006;314(5805):1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- Shan Y., Zhou X., Huang R., Xing D. Anal. Chem. 2019;91(8):5278–5285. doi: 10.1021/acs.analchem.9b00073. [DOI] [PubMed] [Google Scholar]
- Shen J., Zhou X., Shan Y., Yue H., Huang R., Hu J., Xing D. Nat. Commun. 2020;11:267. doi: 10.1038/s41467-019-14135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Zhou Y., Ye J., Al-Maskri A.A.A., Kang Y., Zeng S., Cai S. J. Pharm. Anal. 2020;10(2):97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., Zhang F., Koonin E.V. Mol. Cell. 2015;60(3):385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., Abudayyeh O.O., Gootenberg J.S., Makarova K.S., Wolf Y.I., Severinov K., Zhang F., Koonin E.V. Nat. Rev. Microbiol. 2017;15(3):169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Wenjie, Zhao Xiang, Ma Xuejun, Wang Wenling, Niu Peihua, Xu Wenbo, Gao George F., Wu1 Guizhen. China CDC Weekly. 2020;2:61–62. [PMC free article] [PubMed] [Google Scholar]
- Tian T., Shu B., Jiang Y., Ye M., Liu L., Guo Z., Han Z., Wang Z., Zhou X. ACS Nano. 2021;15(1):1167–1178. doi: 10.1021/acsnano.0c08165. [DOI] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Muenker M.C., Moore A.J., Klein J., Lu P., Lu-Culligan A., Jiang X., Kim D.J., Kudo E., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Tokuyama M., Venkataraman A., Weizman O.-E., Wong P., Yang Y., Cheemarla N.R., White E.B., Lapidus S., Earnest R., Geng B., Vijayakumar P., Odio C., Fournier J., Bermejo S., Farhadian S., Dela Cruz C.S., Iwasaki A., Ko A.I., Landry M.L., Foxman E.F., Grubaugh N.D. Nat. Microbiol. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Lu C.H., Willner I. Chem. Rev. 2014;114(5):2881–2941. doi: 10.1021/cr400354z. [DOI] [PubMed] [Google Scholar]
- Wang R., Xiang Y., Zhou X., Liu L.-h., Shi H. Biosens. Bioelectron. 2015;66:11–18. doi: 10.1016/j.bios.2014.10.079. [DOI] [PubMed] [Google Scholar]
- Wang R., Zhou X., Zhu X., Yang C., Liu L., Shi H. ACS Sens. 2017;2(2):257–262. doi: 10.1021/acssensors.6b00746. [DOI] [PubMed] [Google Scholar]
- Wang R., Zhu X., Xing Y., Memon A.G., Shi H., Zhou X. ACS Appl. Mater. Interfaces. 2019;11(40):36444–36448. doi: 10.1021/acsami.9b14452. [DOI] [PubMed] [Google Scholar]
- Wang M., Zhang R., Li J. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112430. 112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., Zhao M., Qian F., Yu H., Liu Z., Ni T., Zheng Y., Wang Y. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112766. 112766-112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO., 2020. https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/.
- Wolfe B.R., Porubsky N.J., Zadeh J.N., Dirks R.M., Pierce N.A. J. Am. Chem. Soc. 2017;139(8):3134–3144. doi: 10.1021/jacs.6b12693. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Chen X., Zhang M., Wang X., Chen Y., Qian C., Wu J., Xu J. Trac. Trends Anal. Chem. 2021;135 [Google Scholar]
- Xiong D., Dai W., Gong J., Li G., Liu N., Wu W., Pan J., Chen C., Jiao Y., Deng H., Ye J., Zhang X., Huang H., Li Q., Xue L., Zhang X., Tang G. PLoS Biol. 2020;18(12) doi: 10.1371/journal.pbio.3000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong E., Jiang L., Tian T., Hu M., Yue H., Huang M., Lin W., Jiang Y., Zhu D., Zhou X. Angew Chem. Int. Ed. Engl. 2021;60(10):5307–5315. doi: 10.1002/anie.202014506. [DOI] [PubMed] [Google Scholar]
- Zadeh J.N., Steenberg C.D., Bois J.S., Wolfe B.R., Pierce M.B., Khan A.R., Dirks R.M., Pierce N.A. J. Comput. Chem. 2011;32(1):170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- Zhang D.Y., Seelig G. Nat. Chem. 2011;3(2):103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- Zhang K., Lv S., Zhou Q., Tang D. Sensor. Actuator. B Chem. 2020;307 [Google Scholar]
- Zhang N., Wang L., Deng X., Liang R., Su M., He C., Hu L., Su Y., Ren J., Yu F., Du L., Jiang S. J. Med. Virol. 2020;92(4):408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Hu L., Ying L., Zhao Z., Chu P.K., Yu X.-F. Nat. Commun. 2018;9:5012. doi: 10.1038/s41467-018-07324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel C. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J., Fu L., Xu M., Zhou Q., Chen G., Tang D. Biosens. Bioelectron. 2013;45:52–57. doi: 10.1016/j.bios.2013.01.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.