Abstract
Experiments were conducted to measure biodegradation of polychlorinated biphenyl (PCB) congeners contained in mixture Aroclor 1248 and congeners present in wastewater lagoon sediment contaminated decades earlier at Altavista, Virginia. A well-characterized strain of aerobic PCB-degrading bacteria, Paraburkholderia xenovorans LB400 was incubated in laboratory bioreactors with PCB-contaminated sediment collected at the site. The experiments evaluated strain LB400’s ability to degrade PCBs in absence of sediment and in PCB-contaminated sediment slurry. In absence of sediment, LB400 transformed 76% of Aroclor 1248 within seven days, spanning all homolog groups present in the mixture. In sediment slurry, only mono- and di-chlorinated PCB congeners were transformed. These results show that LB400 is capable of rapidly biodegrading most PCB congeners when they are freely dissolved in liquid but cannot degrade PCB congeners having three or more chlorine substituents in sediment slurry. Finally, using GC/MS-MS triple quadrupole spectrometry, this work distinguishes between physical (sorption to cells) and biological removal mechanisms, illuminates the process by which microorganisms with LB400-type congener specificity can selectively transform lower-chlorinated congeners over time, and makes direct comparisons to other studies where individual congener data is reported.
Keywords: Polychlorinated biphenyls, Paraburkholderia xenovorans LB400, Lab bioreactor experiments, Bioremediation, Biodegradation, Bioavailability, Bioaccesibility, Biotransformation
1. Introduction
1.1. Biodegradation & bioavailability of PCBs
PCBs are a group of chlorinated hydrocarbons whose structural foundation is two aromatic rings that are linked by a single carbon-carbon bond with a total of 209 possible congeners. PCB congeners are subdivided into 10 homolog groups depending on the total number of chlorine substituents (Erickson, 1997). PCBs exist in soil and sediment as mixtures of high molecular weight (HMW) and low molecular weight (LMW) congeners. Complete mineralization of these complex mixtures can be achieved via biodegradation – the process by which microorganisms use their natural metabolic capabilities to break down relatively large compounds into smaller molecules to use for energy and growth.
PCB biodegradation often requires two sequential or concurrent microbial processes: (1) anaerobic reductive dechlorination and (2) aerobic oxidation (Abramowicz, 1995; Field and Sierra-Alvarez, 2008; Meggo et al., 2013). Anaerobic reductive dechlorination transforms HMW congeners to LMW congeners by removing chlorine substituents without destroying the biphenyl ring structure of the PCB molecule (Brown et al., 1987; Van Dort and Bedard, 1991). LMW congeners having four or fewer chlorine substituents are much more amenable to aerobic oxidation (Quensen III et al., 1988). Oxidation of PCB congeners proceeds by the upper and lower bph pathways mediated by enzymes expressed by the bph gene cluster (Passatore et al., 2014). The upper bph pathway biodegradation reaction begins when 2,3-dioxygenase (bphAEFG) enzymes introduce molecular oxygen into 2,3- or 3,4- sites along the aromatic ring (Haddock and Gibson, 1995). The aromatic ring is subsequently cleaved by dihydroxybiphenyl dioxygenase (DHBD; bphC) to yield upper pathway transformation products (Furukawa, 2000; Haddock et al., 1995). These products enter the lower bph pathway where they are further broken down to smaller compounds by other enzymes, which can be completely mineralized after entering the Krebs cycle (Dai et al., 2002; Martínez et al., 2007; Pieper and Seeger, 2008).
Lack of PCB bioavailability has been identified as a key parameter which prevents efficient bioremediation or natural attenuation of environmental PCB contamination (Ehlers and Luthy, 2003; Semple et al., 2004). Microorganisms are unable to biodegrade PCBs when PCB molecules are irreversibly (or slowly reversibly) bound to organic material in soil or sediment (Di Toro et al., 1982; Luthy et al., 1997; Ren et al., 2018; Semple et al., 2007). The degree to which PCBs are irreversibly or slowly reversibly bound to soil or sediment particles depends heavily on the total organic carbon (TOC) content or the fraction that can be defined as black carbon (BC)(Cipullo et al., 2018; Lohmann et al., 2005; Werner et al., 2010; Zhu et al., 2016). This work compares Paraburkholderia xenovorans LB400’s ability to biodegrade a commercial PCB mixture (Aroclor 1248) when the mixture is freely dissolved (bioavailable) to LB400’s ability to degrade a similar mixture strongly bound to sediment after decades of environmental contamination. Strain LB400 was first isolated from a PCB-contaminated landfill soil in Moreau, New York and has become one of the best-characterized PCB degrading microorganisms ever studied (Bedard et al., 1986; Chain et al., 2006).
The purpose of the present work is to better understand the temporal, aerobic biodegradation process by which LB400 degrades a mixture of PCB congeners in presence and absence of sediment. Previous studies which examined the capacity of LB400 to biodegrade PCBs reported only selected congeners, certain homolog groups, or used analytical techniques where individual PCB congener peaks were poorly resolved within the entire mixture (Arnett et al., 2000; Bedard et al., 1986; Bopp, 1986; Gibson et al., 1993; Haddock et al., 1993; Payne et al., 2013, 2017, 2019; Rehmann and Daugulis, 2008a, 2008b, 2008c; Tehrani et al., 2012). This study represents a novel contribution to the scientific record by screening for all 209 congeners as 174 co-eluting peaks and making direct comparisons of biodegradation, by individual congener, to other known studies which investigated PCB biodegradation by LB400 where individual congener data was reported. Additionally, this work differentiates between physical removal (sorption) and biological transformation of PCBs within the bioreactor by comparison of live cell treatment to dead cell controls. Finally, by observation and comparison of complete PCB profiles, we demonstrate that strong sorption of PCBs to aged sediments severely limits LB400’s ability to biodegrade them – except for the least chlorinated congeners (#Cl < 3) – and briefly discuss the implications of this finding on risk assessment and remedial design.
2. Methods & materials
2.1. Site description & sampling
The sediment used in this study was taken from a PCB-contaminated emergency overflow lagoon located in Altavista, VA (37°06′52″N, 79°16′21″W). The lagoon became contaminated with PCBs prior to 1977. Sediment samples were taken from sampling location between F3 and F4 (Figure S1) using a steel headed transfer shovel. Four (4) 23 L coolers and one (1) 5 L cooler were filled with sediment, limiting headspace within containers. The coolers were chilled with icepacks, sealed with duct tape, transported back to University of Iowa laboratories, and stored in refrigerators at 4 °C prior to further analysis.
2.2. Sediment homogenization and characterization
Sediment was transferred to a 20gal drum and thoroughly homogenized using a mud mixer attached to a power drill before use in experiments. Three 1.5 kg test aliquots were transferred to red clay pots. Five sub-samples were taken from each pot. The first two sub-samples from each pot were analyzed as replicates and the last three sub-samples were combined to form a composite sample. The PCB profiles and concentrations of replicate sub-samples and composite samples from each pot were compared to confirm that sediment was properly homogenized to a degree such that further samples including sediment were representative. The average total PCB concentration (ΣPCB) was 6346 ng/g (6.346 mg/kg; 6.346 ppm; 5% RSD; n = 6). The PCB-congener profile of Altavista, Virginia sediment most closely resembled Aroclor 1248 (Cos θ = 0.89; Figure S2). Altavista sediments had slightly higher mass percent in the LMW range and at the very HMW range than the commercial mixture Aroclor 1248. These differences between the PCB profile of Altavista sediment and pure Aroclor 1248 are mostly likely due to the accumulation of lightly chlorinated (mono-, di-, and trichlorobiphenyls) resulting from reductive dechlorination of tetra- and pentachlorobiphenyls by native organohalide respiring bacteria (OHRB) within the lagoon sediment (Mattes et al., 2018).
Sediment texture characterization and physico-chemical analyses were conducted by Minnesota Valley Testing Laboratories (MVTL; www.mvtl.com; New Ulm, MN). We provided MVTL with 9 × 250 g sediment samples (triplicates representing each test aliquot from the homogenization procedure and air-dried sediment taken directly from the 20gal drum). Sediment was sandy loam consisting of 72.2% sand, 24.7% silt, and 3% clay. Soil pH was 6.4, total organic matter 4.2%, total organic carbon (TOC) 7.2%, and cation exchange capacity (CEC) 6.0 cmolc/kg. Additional soil parameters can be found in Table S1.
2.3. Bacterial strain and growth conditions
Paraburkholderia xenovorans LB400 was grown aerobically at 30 °C on a platform shaker in a 500 mL Erlenmeyer flask containing 250 mL of K1 medium and solid biphenyl crystal (5 mM; 0.19 g) as a sole carbon and energy source until mid-exponential phase (OD600 = 0.8). Cells were harvested by centrifugation (5000×g, 15 min), washed twice with sterile K1 medium, resuspended in K1 medium, and inoculated into bioreactors using a 1 mL pipette (Liang et al., 2014).
2.4. PCB quantification
GC-MS/MS (Agilent 7890 A GC system, Agilent 7000 Triple Quad, Agilent 7693 autosampler) in multiple reaction monitoring mode (MRM) was used for identification and quantification of 209 PCBs as 174 chromatographic peaks. The GC was equipped with a Supelco SPB-Octyl capillary column (50% n-octyl, 50% methyl siloxane, 30 m × 0.25 mm ID, 0.25 μm film thicknesses) with helium as the carrier gas flowing at 0.75 mL/min and nitrogen/argon as the collision gas. The GC operated in solvent vent injection mode at the following injection conditions: initial temperature 45 °C, initial time 0.06 min, ramp 600 °C/min to inlet temperature 325 °C at 4.4 psi. The GC oven temperature program was 45 °C for 2 min, 45–75 °C at 100 °C/min and hold for 5 min, 75–150 °C at 15 °C/min and hold for 1 min, 150 to 280 at 2.5 °C/min and final hold 5 min (total run time 70.86 min). The triple quadrupole MS electron ionization source was set to 260 °C. Additional details can be found in the supporting information (Tables S3–S13).
2.5. Quality assurance & quality control (QA/QC)
Extraction efficiency, reproducibility, and accuracy was assessed using surrogate standards, replicates of method blanks, development of QC acceptance criteria (Table S2), and analysis of standard reference materials. Standard reference material purchased from the National Institute of Standards and Technology (NIST SRM, 1944, New York, New Jersey Waterway sediment; Gaithersburg, MD, USA) was analyzed in quintuple. The mean percent difference between the measured and certified values (27 congeners) was 13% ± 14% (Figure S3). Mean and standard deviation percentage recoveries of PCB14, PCB65-d5 and PCB166 were 97 ± 1%, 94 ± 4% and 114 ± 4%, respectively. According to QC criteria, any sample having a recovery less than 50% was excluded from analysis. Percentage recoveries of surrogate standards were used to correct congener mass as follows: PCB14 recovery was used to correct PCB1 to PCB39, PCB65-d5 was used to correct PCB40 to PCB127 and PCB166 was used to correct PCB128 to PCB209 (sorted by IUPAC number). PCB congener masses were corrected for surrogate recoveries less than 100%. Samples were processed in batches of five along with one method blank per batch. All materials used in sample extraction had either been triple rinsed with solvent (methanol, acetone, and hexane) or combusted overnight at 450 °C to prevent background PCB contamination. Results from the method blanks were used to determine the limit of quantification (LOQ) as the upper limit of the 95% confidence interval (average mass plus two times the standard deviation).
2.6. Statistical analysis
The concentration data set was first dichotomized at the threshold of the congener specific LOQ: concentrations of congeners below the LOQ were treated as zero. Cosine theta (cos θ) was calculated to quantitatively describe and compare PCB profiles where cos θ = 0 describes two completely different profiles and cos θ = 1 describes identical profiles (Johnson et al., 2010). GraphPad Prism’s two-sided, unpaired t-test with Welch’s correction applied was used to test for significant differences between treatment conditions and controls.
2.7. Aroclor 1248 Biodegradation Assay with Paraburkholderia xenovorans LB400 in Absence of Sediment
An experiment was conducted with LB400 cells in an aqueous solution of Aroclor 1248 to test the ability of LB400 to degrade soluble, bioavailable PCB congeners (experimental design matrix shown in Figure S4). Bioreactors were 9 mL Pyrex® test tube with PTFE-lined screw caps. Live-cell treatments and dead-cell controls were added to the bioreactors. First, 1 mL of K1 bacterial medium was transferred to the bioreactor and spiked with Aroclor 1248 in acetone (3 μL; 25 ng/mL final concentration). PCBs equilibrated with the aqueous phase by shaking on a platform shaker table at 150 rpm for 48 h prior to the addition of LB400 cells. The biodegradation reaction was started by adding 2 mL of live LB400 cells (OD600 = 0.8) suspended in K1 medium to the bioreactor. Dead-cell controls were inactivated by adding perchloric acid (20 μL; final concentration 0.7%; Bedard et al., 1986). Time-zero samples were prepared to equal volume but did not contain LB400 cells. Bioreactors were incubated at room temperature (~25 °C) for one week on a platform shaker table at 150 rpm. All treatments were prepared in quintuplet (n = 5) but two samples were lost at T = 48 h due to a faulty equipment sensor which led to the complete evaporation of one sample each in the live and dead-cell treatments resulting in n = 4 at that time point.
2.8. PCB congeners
A stock solution of Aroclor 1248 suspended in acetone was created with analytical standard obtained from AccuStandard, Inc. (New Haven, CT). The final concentration of the stock solution used to spike the bioreactors was approximately 25 mg/L (25 × 103 ng/mL). Aroclor 1248 stock solution (3 μL) was added to 1 mL K1 medium already inside the bioreactors using a 10 μL syringe for a final concentration of 25 ng/mL as Aroclor 1248. PCB congeners referenced in this work will be described by their congener number and chlorine substitution pattern, where necessary or helpful (US EPA, 2003).
PCBs were extracted from the aqueous bioreactor using a liquid-liquid extraction (LLE) with an equal volume of hexane (3 mL) added to each bioreactor. Prior to extraction, the bioreactor was spiked with surrogate standards PCB 14 (50.81 ng; 3,5-dichlorobiphenyl), deuterated PCB 65-d5 (52.5 ng; 2,3,5,6-tetrachlorobiphenyl-d5, deuterated) and PCB166 (52.56 ng; 2,3,4,4′,5,6-hexachlorobiphenyl; Cambridge Isotope Laboratories, Inc.). The bioreactor was vortexed and centrifuged. The hexane layer containing the sample extract was transferred to a TurboVap (Biotage, Uppsala, Sweden) tube. The LLE process was repeated three times. The resulting volume of solvent containing the sample extract was concentrated to approximately 1 mL using a TurboVap II Concentration Workstation (Caliper Life Sciences).
The final hexane extract was passed through a Pasteur pipette filled with 0.1 g of combusted silica gel and 1 g of acidified silica gel (2:1 silica gel:sulfuric acid by weight) and eluted with approximately 10 mL of hexane (US EPA, 1996). Samples were concentrated to approximately 1 mL and transferred to a gas chromatography vial. The final sample was spiked with internal standards deuterated PCB 30-d5 (19.6 ng; 2,4,6- trichlorobiphenyl-2′,3′,4′,5′,6′-d5, deuterated) and PCB 204 (19.6 ng; 2,2′,3,4,4′,5,6,6′-octachlorobiphenyl; Cambridge Isotope Laboratories, Inc.).
2.9. Aroclor 1248 Biodegradation Assay with Paraburkholderia xenovorans LB400 in Sediment Slurry
An experiment was conducted with LB400 cells in a biphasic solution of K1 bacterial medium and PCB-contaminated sediment slurry to test the ability of LB400 to degrade PCB congeners from the contaminated sediment gathered from the Altavista, VA field site. The sediment was not spiked with any additional PCB congeners. The biodegradation reaction was started by adding 2 mL of live LB400 cells (OD600 = 0.8) in K1 medium to the incubation vessel containing Aroclor 1248-contaminated sediment slurry (1 mL K1 medium with 0.3 g PCB-contaminated sediment; experimental design matrix shown in Figure S5). Dead-cell controls were inactivated by adding perchloric acid (20 μL; final concentration 0.7%; Bedard et al., 1986). Cells were incubated at room temperature (~25 °C) for one month (T = 28 days) on a platform shaker table at 150 rpm. All treatments were performed in quintuplet (n = 5).
The analytical method employed for sample extraction is a modification of U.S. EPA Method 3545 (US EPA, 2007). Sediment slurry samples were extracted using pressurized fluid extraction (Accelerated Solvent Extractor; Dionex ASE-200) with equal parts acetone and hexane. At the time of sampling, contents of the bioreactor were poured into a mortar and pestle containing ~7 g combusted diatomaceous earth. The mass of sediment remaining in the bioreactor after pouring was determined gravimetrically. The diatomaceous earth containing the sample was ground to a fine powder and transferred to a 33 mL ASE-200 cell containing approximately 3 g silica gel sorbent. The sample was spiked with surrogate standards PCB 14 (50.81 ng; 3,5-dichlorobiphenyl), deuterated PCB 65-d5 (52.5 ng; 2,3,5,6-tetrachlorobiphenyl-d5, deuterated) and PCB166 (52.56 ng; 2,3,4,4′,5,6-hexachlorobiphenyl; Cambridge Isotope Laboratories, Inc.). The sample extract resulting from pressurized fluid extraction was concentrated within the collection vial to approximately 1 mL and transferred to a 15 mL Pyrex® test tube with PTFE-lined screw cap.
Polar interferences and other compounds were removed by extraction with sulfuric acid. The final hexane extract was passed through a Pasteur pipette filled with 0.1 g of combusted silica gel and 1 g of acidified silica gel (2:1 silica gel:sulfuric acid by weight) and eluted with approximately 10 mL of hexane (US EPA, 1996). Samples were concentrated to approximately 1 mL and transferred to a gas chromatography vial. The final sample was spiked with internal standards deuterated PCB 30-d (19.6 ng; 2,4,6- trichlorobiphenyl-2′,3′,4′,5′,6′-d5, deuterated) and PCB 204 (19.6 ng; 2,2′,3,4,4′,5,6,6′-octachlorobiphenyl; Cambridge Isotope Laboratories, Inc.). All treatments were prepared in quintuplet (n = 5) but, due to surrogate recoveries below established QA/QC criteria (Table S2) in the time-zero measurement, two samples were excluded from the analysis, resulting in n = 3 at that time point. Additionally, one sample from the live cell treatment at the 1-month timepoint was excluded from analysis because it had low surrogate recoveries due to an instrumentation error during PCB extraction resulting in n = 4 at that time point, for that treatment.
3. Results & discussion
3.1. Aroclor 1248 biodegradation assay with Paraburkholderia xenovorans LB400 in absence of sediment
Results of an Aroclor 1248 biodegradation assay conducted in absence of sediment showed that LB400 grown on biphenyl as the sole carbon and energy source could biodegrade a wide spectrum of PCB congeners in the mixture. After one week of incubation at room temperature the live-cell treatment transformed 76% of total PCB concentration (ΣPCBs), relative to time-zero measurements (Fig. 1A). Significant differences were detected between live cell treatments and dead cell controls for all homolog groups present in the Aroclor 1248 mixture except for hexachlorobiphenyls, which make up a small fraction of the total mixture (Fig. 1B). In the dead-cell control, there was a 23% drop in PCB concentration within the first 48 h of the incubation period. Additionally, a linear relationship (r2 = 0.9072) between the relative mass abundance of an individual congener (PCBi) and its contribution to total biological removal was observed. Deviations from this relationship indicate congener preference of LB400 (Figure S6). In general, the observed magnitude of both biological and physical removal of an individual PCB congener was proportional to its relative abundance (by mass) in the Aroclor 1248 mixture. Findings from this experiment were consistent with other studies which demonstrate the broad PCB biodegradation capability of Paraburkholderia xenovorans LB400 (Table 1) but also provide new insight into how PCB biodegradation of Aroclor 1248 occurs with time (Bedard et al., 1986; Bopp, 1986; Payne et al., 2017).
Fig. 1.
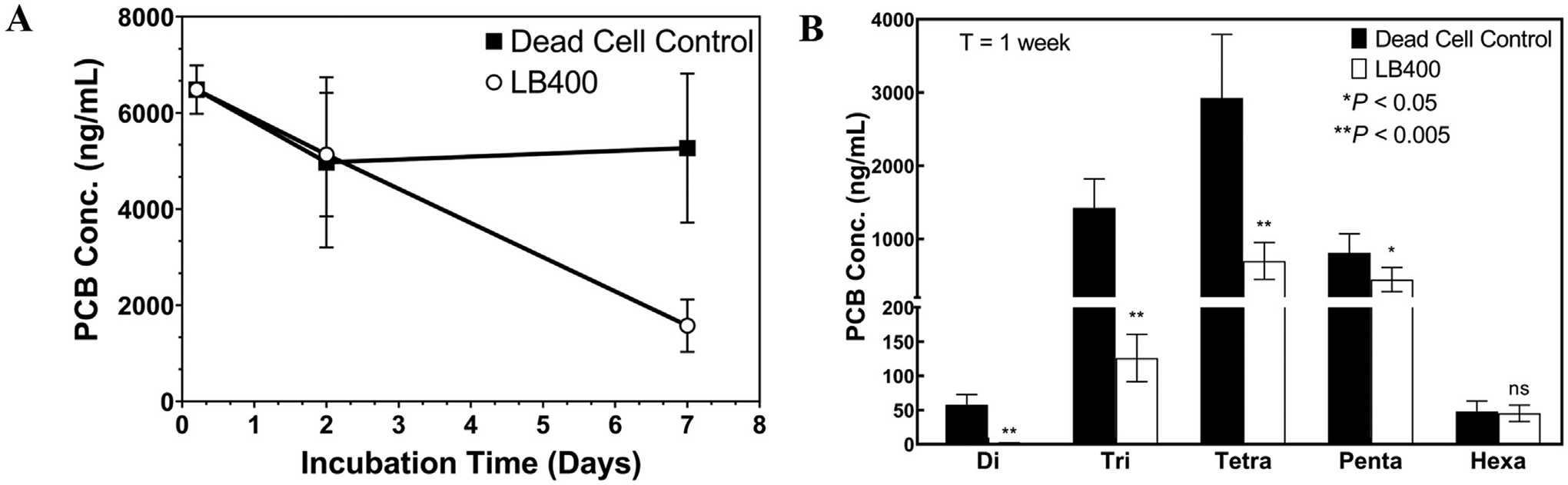
Panel A: PCB concentration in solution (ng/mL) over a 1-week incubation period. Mean and standard deviation of n = 4 replicates are shown for T = 48 h and n = 5 replicates are shown for time-zero and T = 1wk. Panel B: PCB concentration in solution (ng/mL), by homolog group at T = 1wk. Average per-congener depletion within each homolog group (%) in the live cell treatment relative to the dead cell control was as follows: Di – 93%; Tri – 88%; Tetra – 72%; Penta – 56%; Hexa – 27%. For all homolog groups, a two-sided, unpaired t-test with Welch’s correction applied was used to test for significant differences between live and dead-cell treatments. Mean and standard deviation for n = 5 replicates are shown, except for T = 48 where n = 4.
Table 1.
Comparison of PCBi congener depletion (%) by Paraburkholderia xenovorans LB400 between literature values and present work.
| Congener Description | % Depletion of PCBi | ||||||
|---|---|---|---|---|---|---|---|
| Structural Class | BZ Congener# | Substitution Pattern | Present work | Bedard et al. (1986) | Bopp et al. (1986) | Nadim et al. (1987) | Gibson et al. (1993) |
| Open 2,3 and 3,4 Sites | 4 | 2,2′ | 100 | 80–100d | 100 | 100 | 100/100e |
| 5 | 2,3 | 80–100 | 100 | 100 | 100/100 | ||
| 8 | 2,4′ | 99 | 80–100 | 100 | 100 | 100/100 | |
| 9 | 2,5 | 100 | |||||
| 10 | 2,6 | 40 | |||||
| 16 | 2,2′,3 | 99 | |||||
| 17 | 2,2′,4 | 99 | |||||
| 18 | 2,2′,5 | 99 | 80–100 | 100 | 100 | 100/100 | |
| 19a | 2,2′,6 | 74 | |||||
| 22 | 2,3,4′ | 97 | |||||
| 25 | 2,3′,4 | 97 | |||||
| 26/29b | 2,3′,5/2,4,5 | 98 | |||||
| 27 | 2,3′,6 | 98 | |||||
| 30 | 2,4,6 | 25 | |||||
| 31 | 2,4′,5 | 98 | 80–100 | 100 | 100 | 98/100 | |
| 32 | 2,4′,6 | 63 | |||||
| 33 | 2,3′,4′ | 97 | |||||
| 40 | 2,2′,3,3′ | 81 | 80–100 | 100 | 100/100 | ||
| 41 | 2,2′,3,4 | 94 | |||||
| 42 | 2,2′,3,4′ | 87 | |||||
| 43 | 2,2′,3,5 | 94 | |||||
| 44/47c | 2,2′,3,5’/2,2′,4,4′ | 89 | 80–100/80–100 | 100/100 | 100/100 | 100/81/86 | |
| 45 | 2,2′,3,6 | 66 | |||||
| 46 | 2,2′,3,6′ | 70 | |||||
| 48 | 2,2′,4,5 | 95 | |||||
| 51 | 2,2′,4,6′ | 37 | |||||
| 53 | 2,2′,5,6′ | 92 | 100 | ||||
| 56 | 2,3,3′,4′ | 92 | |||||
| 59/75 | 2,3,3′,6/2,4,4′,6 | 81 | |||||
| 63 | 2,3,4′,5 | 64 | |||||
| 64 | 2,3,4′,6 | 46 | |||||
| 70/74 | 2,3′,4′,5/2,4,4′,5 | 73 | 80–100/NA | 100/NA | 100/NA | ||
| 82 | 2,2′,3,3’,4 | 50 | |||||
| 84 | 2,2′,3,3′,6 | 67 | |||||
| 87 | 2,2′,3,4,5′ | 67 | 80–100 | 100 | 100 | 97/96 | |
| 91 | 2,2′,3,4′,6 | 41 | |||||
| 97/109/119 | 2,2′,3,4′,5’/2,3,3′,4,6/2,3′,4,4′,6 | 53 | 40–59/NA/NA | 90/NA/NA | 100/NA/0 | 50/53 | |
| 110 | 2,3,3′,4′,6 | 49 | |||||
| 129/138/163 | 2,2′,3,3′,4,5/2,2′,3,4,4 ′,5’/2,3,3′,4′,5,6 | 26 | |||||
| 132 | 2,2′,3,3′,4,6 | 25 | |||||
| 147/149 | 2,2′,3,4′,5,6/2,2′,3,4′,5′,6 | 27 | |||||
| Open 3,4 Sites | 52 | 2,2′,5,5′ | 96 | 80–100 | 100 | 100 | 100/100 |
| 54 | 2,2′,6,6′ | 0g | |||||
| 92 | 2,2′,3,5,5′ | 80 | |||||
| 95 | 2,2′,3,5′,6 | 70 | |||||
| 101 | 2,2′,4,5,5′ | 78 | 80–100 | 100 | 100 | 100/98 | |
| 103 | 2,2′,4,5,6′ | 75 | |||||
| 135 | 2,2′,3,3′,6,6′ | 35 | |||||
| Open 2,3 Sites | 15 | 4,4′ | 81 | 60–79 | 47 | 50 | 25/11 |
| 28 | 2,4,4′ | 84 | 80–100 | 98 | 100 | 89/54 | |
| 37 | 3,4,4′ | 41 | |||||
| 47 | 2,2′,4,4′ | @44/47f | 80–100 | 100 | 100 | 81/86 | |
| 60 | 2,3,4,4′ | 39 | |||||
| 66 | 2,3′,4,4′ | 48 | 40–59 | 64 | 65 | 43/34 | |
| 75 | 2,4,4′,6 | @59/75 | 0 | ||||
| 77 | 3,3′,4,4′ | 42 | 40–59 | 41 | 0 | 6/11 | |
| 85 | 2,2′,3,4,4′ | 42 | |||||
| 99 | 2,2′,4,4′,5 | 56 | |||||
| 100 | 2,2′,4,4′,6 | 0 | |||||
| 118 | 2,3′,4,4′,5 | 40 | 0 | ||||
| Blocked 2,3 & 3,4 Sites | 121 | 2,3′,4,5′,6 | 0 | ||||
| 153/168 | 2,2′,4,4′,5,5’/2,3′,4,4′,5′,6 | 30 | 40–59/NA | 59/45 | 70/NA | [41/61]/NA | |
| 155 | 2,2′,4,4′,6,6′ | 0 | |||||
PCB congeners with BZ# in bold have more than two ortho-chlorine substituents.
PCB congeners separated with a forward slash co-elute with each other.
PCB congeners underlined or in italics belong to the corresponding structural class separate from the PCB congener(s) it co-elutes with i.e. PCB congeners that are underlined belong to the structural class with open 2,3 sites and those in italics belong to that with blocked 2,3 and 3,4 sites.
No specific values were given by Bedard et al. (1986), only approximate ranges.
Gibson et al. (1993) assayed the biodegradation ability of LB400 using both whole cells and cell extracts from LB400. Depletion percentage by whole cells is given first, followed by that of cell extracts.
Since some congeners that are both present in the Aroclor 1248 mixture co-elute in the analytical method used in the present work, a determination cannot be made about exactly how much of either congener was depleted but the ‘@’ symbol references the first entry in the table for the co-eluting congeners.
Zero values indicate that none of the PCB congener was depleted whereas a blank space indicates the congener was not detected in the corresponding work. Congeners not detected in the present work are not present in Aroclor 1248 but were screened for.
Biodegradation of the Aroclor 1248 mixture was observed on a congener-by-congener basis throughout the 1-week incubation period (Fig. 2A–C). Di- and trichlorinated congeners with the highest relative abundance (by mass) were the preferred substrate of LB400 over the first 48 h of the incubation period (Fig. 2A). Similarly, the most abundant tetrachlorobiphenyls PCB 44/47, 48, 49, and 52 were partially transformed during the first 48 h. To illustrate how LB400 transformed HMW congeners after preferentially depleting LMW congeners, we subtracted PCBi removal after 48 h on a congener-by-congener basis from total PCBi removal after 1-week (PCBi RemovalT=1wk − PCBi RemovalT=48h; Fig. 2B). By observation of Fig. 2A and B, we reason that transformation of pentachlorobiphenyls during the first 48 h was competitively inhibited because LB400 preferentially degraded di-, tri-, and a select few tetrachlorobiphenyls. In addition, the 23% drop in PCB concentration displayed during the first 48 h in the dead cell control was due to rapid sorption of PCB congeners to inactivated cell material. In the dead cell control, removal of individual congeners occurred proportionally to each congener’s relative abundance in the mixture and regardless of molecular weight (Fig. 2A and B). In the live cell treatment, strain LB400 acted upon all congeners and demonstrated at least 20% transformation for each one, up to and including hexachlorobiphenyls (Fig. 2C; Table 1). The biodegradation pattern observed in the present work is consistent with reports that LB400 has broad congener specificity which encompasses congeners belonging to separate structural classes but is limited in its ability to degrade double-para-substituted congeners (Table 1; Bedard et al., 1986; Bopp, 1986; Gibson et al., 1993; Nadim et al., 1987). Most double-para-substituted congeners belong to the final two structural classes shown in Table 1.
Fig. 2. –
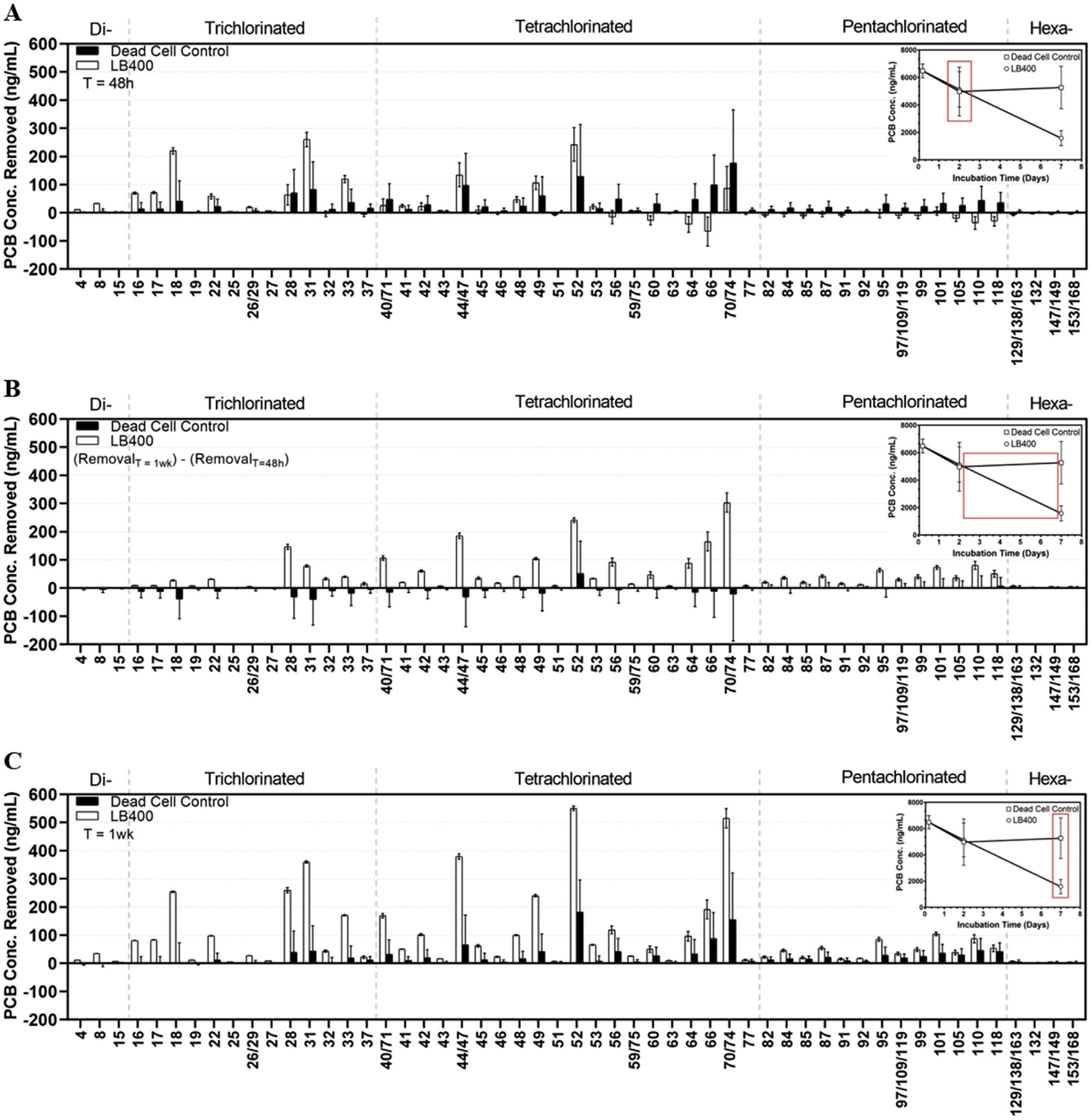
Panel A: PCB removal from K1 medium, by individual congener, (ng/ml) over 48 h. In the live-cell treatment (white bars), LB400 demonstrates a clear preference for degradation and removal of the most abundant di-, tri-, and a select few tetrachlorbiphenyls (from PCB4 to PCB52) during the first 48 h of the incubation period, whereas pentachlorobiphenyls remain unaffected. In the dead-cell control (black bars), removal across the wide-range of congeners is assumed to be due to rapid sorption to inactivated cell material. Inset displays the time interval reflecting the congener-by-congener removal. Panel B: PCB removal from K1 medium, by individual congener (ng/ml) between T = 48 h and T = 1wk. At this time point, the live-cell treatment, LB400 transformed the most abundant di- and trichlorobiphenyls and shifted to tetra- and pentachlorinated congeners through PCB 118. In the dead-cell control, highly hydrophobic (pentachlorinated) PCBs were bound tightly to dead cell membranes with no further removal, whereas some more moderately chlorinated (tri-, and tetrachlorobiphenyls) became re-associated with the aqueous phase. Panel C: PCB removal, by individual congener, (ng/mL) after 1-week incubation in liquid K1 medium. LB400 was effective at removing di-, tri-, tetra-, and pentachlorinated congeners with limited ability to transform those belonging to the hexa-chlorinated homolog groups. Hexachlorobiphenyls make up a small portion of the Aroclor 1248 mixture. Mean and standard deviation for 5 replicates are shown for all time points except T = 48 h.
LB400’s limited ability to degrade double-para-substituted congeners results from extremely slight differences in the amino acid sequences of LB400’s large subunit, relative to other PCB-degraders. These slight differences in amino acid sequence are also the reason for dramatic differences in substrate specificity, compared to other microorganisms with lesser PCB biodegradation capability, such as Pseudomonas furukawaii KF707 (Erickson and Mondello, 1992, 1993; Haddock and Gibson, 1995; Mondello et al., 1997). The dioxygenases of LB400 and KF707 share >98% sequence similarity and their large subunits differ by only 20 amino acids (Haddock et al., 1995; Mondello et al., 1997). Strains with LB400-type specificity can degrade a broad range of congeners due to relaxed regiospecificity of where their terminal oxygenase (bphA) inserts molecular oxygen to the aromatic ring structure of a PCB molecule (Arnett et al., 2000). That is, microorganisms with LB400-type specificity can initiate dioxygenase attack at an unchlorinated 2,3 or 3,4 site of a PCB congener but are relatively weak against di-para-substituted congeners (Bedard et al., 1986; Mondello et al., 1997). In contrast, strains with KF707-type specificity degrade a much narrower range of PCBs but have enhanced ability to degrade di-para-substituted congeners. For example, PCB 15 (4,4′-CB) is poorly degraded by LB400 compared to strains with KF707-type specificity (Mondello et al., 1997). Interestingly, LB400 degrades 2,4,4′-CB and 2,4,2′,4′-CB (PCBs 28 and 47, respectively) much more effectively than other di-para-substituted congeners, suggesting that the additional ortho-chlorine assists with enzymatic binding to the dichlorinated ring (Table 1; Bedard et al., 1987, 1986; Erickson and Mondello, 1993; Gibson et al., 1993; Mondello et al., 1997).
To further observe the difficulty with which LB400 degrades double-para-substituted congeners we compared the transformation of two tetrachlorinated congeners that are abundant in Aroclor 1248: PCB 52 (2,2′,5,5′-CB, 7.37% by mass) and PCB 66 (2,3′,4,4′-CB, 5.69% by mass; Frame, 1997; Koh, 2015). PCB 52 is a congener that is notoriously difficult to degrade for microorganisms without LB400-type specificity because it only has unchlorinated 3,4 sites available for enzymatic binding. Most microorganisms without LB400-type specificity can only attack PCB congeners at unchlorinated 2,3 sites (Bedard et al., 1987; Mondello et al., 1997). PCB 52 was one of the few tetrachlorinated congeners significantly degraded by LB400 within the first 48 h of the incubation period (Fig. 2A). Significantly greater (p < 0.05) biological transformation of PCB 52 was observed relative to PCB 66 even though the difference in relative abundance in Aroclor 1248 between the two congeners was small (1.68%, by mass). Presumably, each congener should display removal proportional to its relative abundance in the mixture. A linear relationship (r2 = 0.9072) was observed by plotting the relative mass abundance of PCBi and its contribution to total biological removal (Figure S6). PCB 52 falls above the best-fit line whereas PCB 66 falls below which indicates LB400’s preference for PCB 52 and aversion for double-para-substituted PCB 66. It may be possible to overcome LB400’s limited capacity to biodegrade double-para-substituted congeners in a bioremediation scenario by using bioaugmentation consortia containing strains with both LB400-type and KF707-type congener specificity.
It is unclear whether LB400 used PCBs for growth based on the results of this study. The Aroclor 1248 mixture was not 100% degraded perhaps due to the brevity of the 7 d exposure period or the possible accumulation of transformation byproducts such as chlorobenzoic acids (CBAs) and/or 2-hydroxy-6-oxo-6-phenyl-hexa-2,4-dienoic acids (HOPDAs, or meta-cleavage products) in the bioreactor, which can interfere with the bph pathway (Adebusoye et al., 2008; Guilbeault et al., 1994; Seah et al., 2000; Sondossi et al., 1992). Percent depletion of PCBi is shown in Table 1. Table 1 represents the most detailed dataset of individual PCB congener biodegradation by LB400 to date (Bako et al., 2020). Direct comparisons are made between the present work and past studies that have reported individual congener data in biodegradation assays using LB400.
4. Aroclor 1248 biodegradation assay with Paraburkholderia xenovorans LB400 in sediment slurry
Results of a biodegradation assay in presence of PCB-contaminated sediment slurry show that LB400 could not significantly biodegrade PCB congeners with more than two chlorine substituents that are bound to sediment gathered from Altavista, VA. After one month of incubation (T = 28 days) at room temperature, the live-cell treatment did not significantly transform the total PCB concentration (ΣPCBs), relative to time-zero measurements or dead cell controls (Fig. 3A). Significant differences were detected between live cell treatments and dead cell controls only in the mono- and dichlorinated homolog groups (Fig. 3B). In the dead-cell treatment, there was a detected increase in PCB concentration over the course of the incubation period which could be attributed to sediment sampling heterogeneity between time-zero and subsequent samples. Findings from this experiment were consistent with the literature that indicates microorganisms capable of PCB biodegradation are unable to do so when congeners are irreversibly or slowly reversibly bound to soil or sediment particles (Cipullo et al., 2018; Ghosh et al., 2000; Harkness et al., 1993; Luthy et al., 1997; Ren et al., 2018).
Fig. 3.
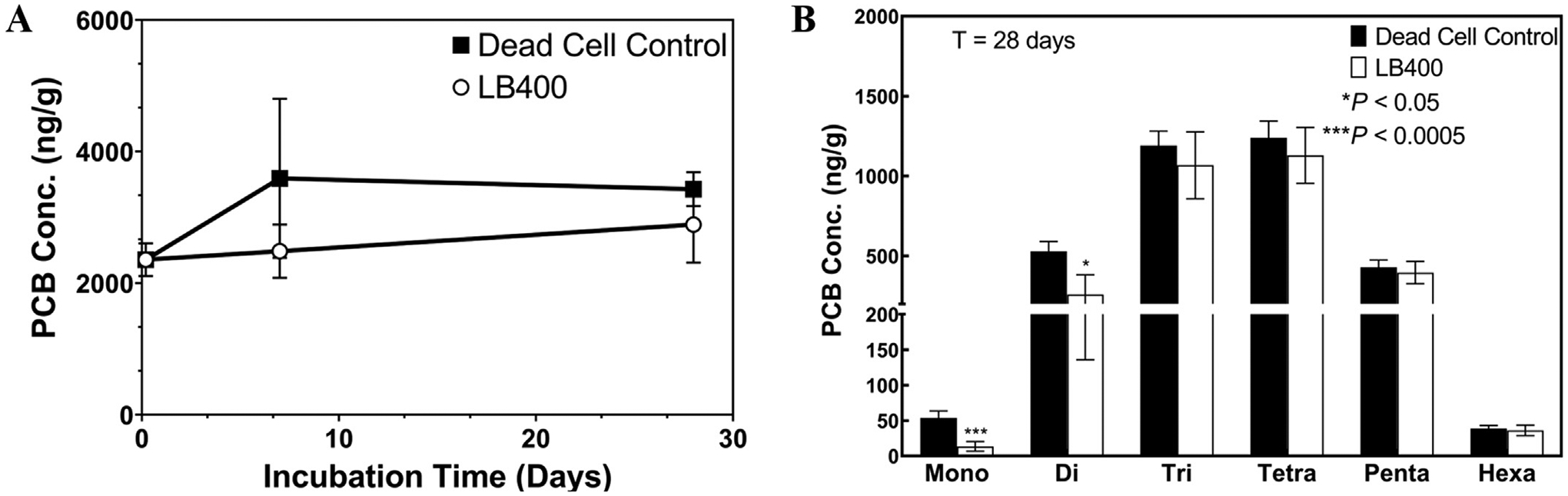
Panel A: PCB concentration (ng/g sediment) over a 28-day incubation period. Mean and standard deviation of n = 5 replicates shown. Panel B: PCB concentration (ng/g), by homolog group at T = 28 days. Significant differences (p < 0.05) were detected between the live cell treatment and dead-cell control for only mono- and dichlorinated homolog groups. For all homolog groups, a two-sided, unpaired t-test with Welch’s correction applied was used to test for significant differences. Mean and standard deviation from n = 5 replicates shown for dead cell control. Mean and standard deviation from n = 3 replicates shown for both the dead cell control and LB400 at T = 0 days. Mean and standard deviation from n = 4 replicates shown for LB400 at T = 28 days.
Fig. 4 indicates transformation of PCB 1 (2-chlorobiphenyl), PCB 4 (2,2′-dichlorobiphenyl), and PCB 8 (2,4′-dichlorobiphenyl) in the live cell treatment. No biodegradation of double-para-substituted PCB 15 (4,4′-dichlorobiphenyl) was detected. In the dead cell control, there were detected increases in PCBi concentration for nearly all congeners likely due to slight sediment sampling heterogeneity between time zero samples and T = 28 days samples. Results of this experiment show that LB400 is unable to degrade HMW PCB congeners in Altavista sediment because they are likely irreversibly or slowly-reversibly bound to sediment particles. LB400 still significantly (p < 0.05) biodegraded congeners that are most likely to become airborne (mono- and dichlorobiphenyls) – partially disrupting a significant pathway of human exposure to PCBs (Awad et al., 2016; Herkert et al., 2018; Jahnke and Hornbuckle, 2019;Kvasnicka et al., 2019; Marek et al., 2017; (Martinez et al., 2017), 2019; Shanahan et al., 2015). Volatilization of PCBs to the atmosphere has long been recognized as a significant mass transport process (Achman et al., 1993; Hafner and Hites, 2003; Jeremiason et al., 1994; Totten et al., 2001). As such, the inhalation exposure pathway of PCBs has been included as a major factor when making risk-based decisions regarding remediation. New methods for measuring PCB concentrations in air, over time, offer new insight into the inhalation exposure pathway which has traditionally been considered secondary to the consumption of PCB-contaminated foods ((Martinez et al., 2017), 2019).
Fig. 4.
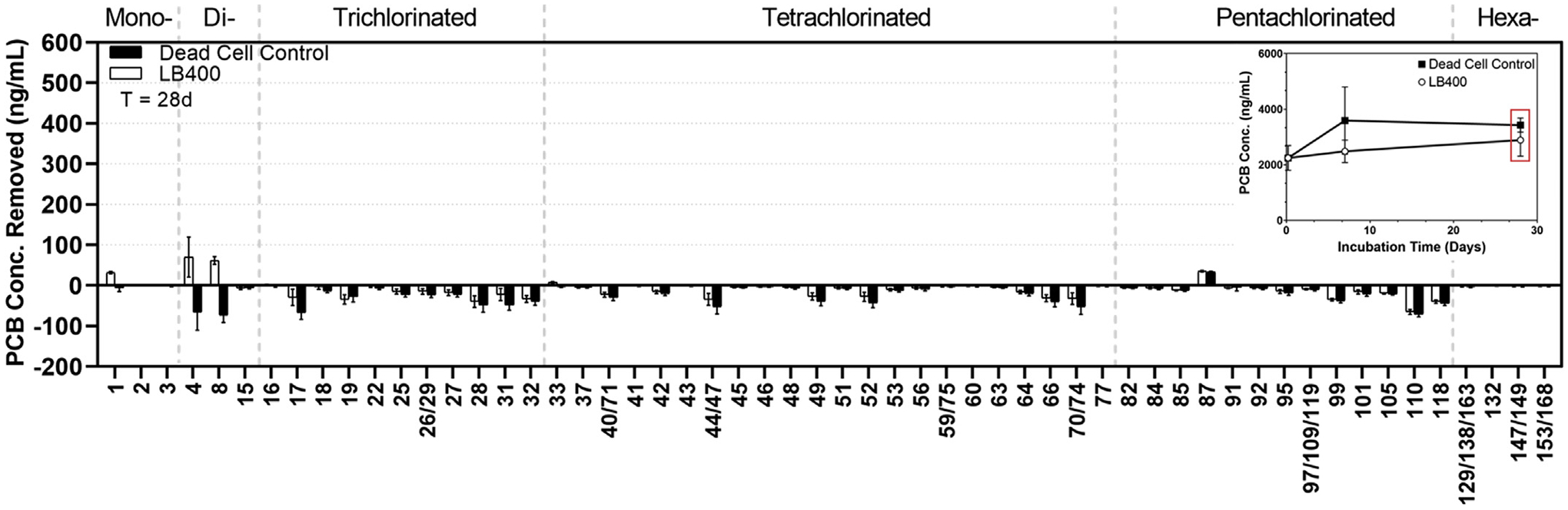
PCB removal, by individual congener, due to biodegradation (ng/g) after 28-day incubation in liquid K1 medium. LB400 was only able to partially transform mono- and dichlorinated congeners. Inset shows the time point reflected by the congener-by-congener breakdown. Mean and standard deviation for 5 replicates for are shown. Although we screened for 209 congeners, only a condensed profile is shown for ease of profile comparison.
Together, these two biodegradation assays clearly display how LB400 microorganisms possess a high degree of PCB biodegradation capacity but cannot use their ability to fully remediate PCB-contaminated environments unless PCBs are soluble and/or bioavailable to a greater extent. Substrate availability and cell viability also become limiting factors due to slow desorption kinetics of PCBs in the sediment environment (Ren et al., 2018; Semple et al., 2004, 2007).
5. Conclusion
For the first time by use of GC-MS/MS QQQ, we show that biodegradation by LB400 in aged, contaminated sediments is limited to degradation of the mono- and dichlorinated PCB congeners out of the entire 209 possible due to strong sorption to aged sediment. Mono- and dichlorobiphenyls are the most volatile substrates in the congener profile, so bioaugmentation with LB400 should help to decrease aquatic concentrations and airborne flux of these toxic LMW-PCBs thereby limiting inhalation exposure in communities where PCB contamination of nearby waterways is common (Ortega-Calvo et al., 2015). Our findings are also important because they inform strategies for bioaugmentation/biostimulation of legacy sediments. To biodegrade the highly chlorinated PCBs with LB400, the highly sorbed-compounds must be made more soluble and bioavailable to microorganisms through amendments such as biosurfactants, solvents, and/or novel sorbent materials.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Environmental Health Sciences (NIEHS) grant #P42ES013661, United States.. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Andres Martinez for advice and assistance in data analysis and visualization; Reid Simmer for preparing Figure S2 in ArcGIS; Jess Ewald and Patrick Richards for their guidance in maintaining living cell cultures and as manuscript preparation; as well as Brian Westra and Marina Zhang for assistance in preparing and depositing the underlying dataset to the Iowa Research Online (IRO) data repository.
Role of funding source
This work was supported by the National Institutes of Environmental Health Sciences (NIEHS) grant #P42ES013661. The funding sponsor did not have any role in study design; in collection, analysis, and/or interpretation of data; in writing this report; and/or in the decision to submit this article for publication.
Footnotes
This paper has been recommended for acceptance by Eddy Y. Zeng.
Data availability statement
The underlying data for this work (Bako et al., 2020) has been deposited in the Iowa Research Online (IRO) institutional data repository for future reuse under an Open Data Commons Attribution License (ODC-By). There are no registration or fee requirements to download the underlying dataset for this work.
Declaration of competing interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envpol.2020.116364.
References
- Abramowicz DA, 1995. Aerobic and anaerobic PCB biodegradation in the environment. Environ. Health Perspect 103, 97. 10.2307/3432489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achman DR, Hornbuckle KC, Eisenreich SJ, 1993. Volatilization of polychlorinated biphenyls from green bay, lake michigian. Environ. Sci. Technol 27, 75–87. 10.1021/es00038a007. [DOI] [Google Scholar]
- Adebusoye SA, Picardal FW, Ilori MO, Amund OO, 2008. Influence of chlorobenzoic acids on the growth and degradation potentials of PCB-degrading microorganisms. World J. Microbiol. Biotechnol 24, 1203–1208. 10.1007/s11274-007-9594-1. [DOI] [Google Scholar]
- Arnett CM, Parales JV, Haddock JD, 2000. Influence of chlorine substituents on rates of oxidation of chlorinated biphenyls by the biphenyl dioxygenase of burkholderia sp. strain LB400. Appl. Environ. Microbiol 66, 2928–2933. 10.1128/AEM.66.7.2928-2933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad AM, Martinez A, Marek RF, Hornbuckle KC, 2016. Occurrence and distribution of two hydroxylated polychlorinated biphenyl congeners in chicago air. Environ. Sci. Technol. Lett 3, 47–51. 10.1021/acs.estlett.5b00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bako CM, Mattes TE, Marek RF, Hornbuckle KC, Schnoor JL, 2020. Dataset describing biodegradation of individual polychlorinated biphenyl congeners (PCBs) by Paraburkholderia xenovorans LB400 in presence and absence of sediment slurry. Iowa Res. Online V1 10.25820/data.006135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard DL, Haberl ML, May RJ, Brennan MJ, 1987. Evidence for novel mechanisms of polychlorinated biphenyl metabolism in Alcaligenes eutrophus H850. Appl. Environ. Microbiol 53, 1103–1112. 10.1128/AEM.53.5.1103-1112.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard DL, Unterman R, Bopp LH, Brennan MJ, Haberl ML, Johnson C, 1986. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl. Environ. Microbiol 51, 761–768. 10.1128/AEM.51.4.761-768.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp LH, 1986. Degradation of highly chlorinated PCBs by Pseudomonas strain LB400. J. Ind. Microbiol 1, 23–29. 10.1007/BF01569413. [DOI] [Google Scholar]
- Brown JF, Bedard DL, Brennan MJ, Carnahan JC, Feng H, Wagner RE, 1987. Polychlorinated biphenyl dechlorination in aquatic sediments. Science 84 236, 709–712. 10.1126/science.236.4802.709. [DOI] [PubMed] [Google Scholar]
- Chain PSG, Denef VJ, Konstantinidis KT, Vergez LM, Agullo L, Reyes VL, Hauser L, Cordova M, Gomez L, Gonzalez M, Land M, Lao V, Larimer F, LiPuma JJ, Mahenthiralingam E, Malfatti SA, Marx CJ, Parnell JJ, Ramette A, Richardson P, Seeger M, Smith D, Spilker T, Sul WJ, Tsoi TV, Ulrich LE, Zhulin IB, Tiedje JM, 2006. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. Unit. States Am 103, 15280–15287. 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipullo S, Prpich G, Campo P, Coulon F, 2018. Assessing bioavailability of complex chemical mixtures in contaminated soils: progress made and research needs. Sci. Total Environ 615, 708–723. 10.1016/j.scitotenv.2017.09.321. [DOI] [PubMed] [Google Scholar]
- Dai S, Vaillancourt FH, Maaroufi H, Drouin NM, Neau DB, Snieckus V, Bolin JT, Eltis LD, 2002. Identification and analysis of a bottleneck in PCB biodegradation. Nat. Struct. Biol 9, 934–939. 10.1038/nsb866. [DOI] [PubMed] [Google Scholar]
- Di Toro DM, Horzempa LM, Casey MM, Richardson W, 1982. Reversible and resistant components of PCB adsorption-desorption: adsorbent concentration effects. J. Great Lake. Res 8, 336–349. 10.1016/S0380-1330(82)71976-8. [DOI] [Google Scholar]
- Ehlers LJ, Luthy RG, 2003. Peer reviewed: contaminant bioavailability in soil and sediment. Environ. Sci. Technol 37, 295A–302A. 10.1021/es032524f. [DOI] [PubMed] [Google Scholar]
- Erickson BD, Mondello FJ, 1993. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl. Environ. Microbiol 59, 3858–3862. 10.1128/AEM.59.11.3858-3862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson BD, Mondello FJ, 1992. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J. Bacteriol 174, 2903–2912. 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson MD, 1997. Analytical Chemistry of PCBs, second ed. CRC Press, Taylor & Francis Group, New York. [Google Scholar]
- Field JA, Sierra-Alvarez R, 2008. Microbial transformation and degradation of polychlorinated biphenyls. Environ. Pollut 155, 1–12. 10.1016/j.envpol.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Frame GM, 1997. A collaborative study of 209 PCB congeners and 6 Aroclors on 20 different HRGC columns: 1. Retention and coelution database. Fresenius’ J. Anal. Chem 357, 701–713. 10.1007/s002160050237. [DOI] [Google Scholar]
- Furukawa K, 2000. Biochemical and genetic bases of microbial degradation of polychlorinated biphenyls (PCBs). J. Gen. Appl. Microbiol 46, 283–296. 10.2323/jgam.46.283. [DOI] [PubMed] [Google Scholar]
- Ghosh U, Weber AS, Jensen JN, Smith JR, 2000. Relationship between PCB desorption equilibrium, kinetics, and availability during land biotreatment. Environ. Sci. Technol 34, 2542–2548. 10.1021/es9905389. [DOI] [Google Scholar]
- Gibson DT, Cruden DL, Haddock JD, Zylstra GJ, Brand JM, 1993. Oxidation of polychlorinated biphenyls by Pseudomonas sp. strain LB400 and Pseudomonas pseudoalcaligenes KF707. J. Bacteriol 175, 4561–4564. 10.1128/JB.175.14.4561-4564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbeault B, Sondossi M, Ahmad D, Sylvestre M, 1994. Factors affecting the enhancement of PCB degradative ability of soil microbial populations. Int. Biodeterior. Biodegrad 33, 73–91. 10.1016/0964-8305(94)90056-6. [DOI] [Google Scholar]
- Haddock JD, Gibson DT, 1995. Purification and characterization of the oxygenase component of biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J. Bacteriol 177, 5834–5839. 10.1128/JB.177.20.5834-5839.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddock JD, Horton JR, Gibson DT, 1995. Dihydroxylation and dechlorination of chlorinated biphenyls by purified biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J. Bacteriol 177, 20–26. 10.1128/jb.177.1.20-26.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddock JD, Nadim LM, Gibson DT, 1993. Oxidation of biphenyl by a multicomponent enzyme system from Pseudomonas sp. strain LB400. J. Bacteriol 175, 395–400. 10.1128/jb.175.2.395-400.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner WD, Hites RA, 2003. Potential sources of pesticides, PCBs, and PAHs to the atmosphere of the Great Lakes. Environ. Sci. Technol 37, 3764–3773. 10.1021/es034021f. [DOI] [PubMed] [Google Scholar]
- Harkness MR, McDermott JB, Abramowicz DA, Salvo JJ, Flanagan WP, Stephens ML, Mondello FJ, May RJ, Lobos JH, Carroll KM, Brennan MJ, Bracco AA, Fish KM, Warner GL, Wilson PR, Dietrich DK, Lin DT, Morgan CB, Gately WL, 1993. In situ stimulation of aerobic PCB biodegradation in Hudson River sediments. Science 80 (259), 503–507. 10.1126/science.8424172. [DOI] [PubMed] [Google Scholar]
- Herkert NJ, Jahnke JC, Hornbuckle KC, 2018. Emissions of tetrachlorobiphenyls (PCBs 47, 51, and 68) from polymer resin on kitchen cabinets as a non-aroclor source to residential air. Environ. Sci. Technol 52, 5154–5160. 10.1021/acs.est.8b00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke JC, Hornbuckle KC, 2019. PCB emissions from paint colorants. Environ.Sci. Technol 53 (9), 5187–5194. 10.1021/acs.est.9b01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeremiason JD, Hornbuckle KC, Eisenreich SJ, 1994. PCBs in lake superior, 1978–1992: decreases in water concentrations reflect loss by volatilization. Environ. Sci. Technol 28, 903–914. 10.1021/es00054a023. [DOI] [PubMed] [Google Scholar]
- Johnson GW, Quensen JFI, Chiarenzelli JR, Hamilton CM, 2010. Polychlorinated biphenyls. In: Morrison R, Murphy B (Eds.), Environmental Forensics: Contaminant Specific Guide. Elsevier Science & Technology; Academic Press, pp. 188–221. [Google Scholar]
- Koh WX, 2015. Polychlorinated Biphenyls and Their Hydroxylated Metabolites in Human Serum from Urban and Rural Cohorts in the United States. ProQuest Diss. Theses University of Iowa. 10.17077/etd.xbjjeo6a. [DOI] [Google Scholar]
- Kvasnicka J, Stylianou KS, Nguyen VK, Huang L, Chiu WA, Burton GA, Semrau J, Jolliet O, 2019. Human health benefits from fish consumption vs. Risks from inhalation exposures associated with contaminated sediment remediation: dredging of the hudson river. Environ. Health Perspect 127 10.1289/EHP5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Meggo R, Hu D, Schnoor JL, Mattes TE, 2014. Enhanced polychlorinated biphenyl removal in a switchgrass rhizosphere by bioaugmentation with Burkholderia xenovorans LB400. Ecol. Eng 71, 215–222. 10.1016/j.ecoleng.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann R, Macfarlane JK, Gschwend PM, 2005. Importance of black carbon to sorption of native PAHs, PCBs, and PCDDs in Boston and New York Harbor sediments. Environ. Sci. Technol 39, 141–148. 10.1021/es049424+. [DOI] [PubMed] [Google Scholar]
- Luthy GR, Ainken RG, Brusseau LM, Cunningham DS, Gschwend MP, Pignatello JJ, Reinhard M, Traina JS, Weber JWJ, Westall CJ, 1997. Sequestration of hydrophobic organic contaminants by geosorbents. Environ. Sci. Technol 31, 3341–3347. 10.1021/es970512m. [DOI] [Google Scholar]
- Marek RF, Thorne PS, Herkert NJ, Awad AM, Hornbuckle KC, 2017. Airborne PCBs and OH-PCBs inside and outside urban and rural U.S. Schools. Environ. Sci. Technol 51, 7853–7860. 10.1021/acs.est.7b01910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Awad AM, Herkert NJ, Hornbuckle KC, 2019. Determination of PCB fluxes from Indiana Harbor and Ship Canal using dual-deployed air and water passive samplers. Environ. Pollut 244, 469–476. 10.1016/j.envpol.2018.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Hadnott BN, Awad AM, Herkert NJ, Tomsho K, Basra K, Scammell MK, Heiger-Bernays W, Hornbuckle KC, 2017. Release of airborne polychlorinated biphenyls from new bedford harbor results in elevated concentrations in the surrounding air. Environ. Sci. Technol. Lett 4, 127–131. 10.1021/acs.estlett.7b00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, Agulló L, Hernández M, Seeger M, 2007. Chlorobenzoate inhibits growth and induces stress proteins in the PCB-degrading bacterium Burkholderia xenovorans LB400. Arch. Microbiol 188, 289–297. 10.1007/s00203-007-0247-4. [DOI] [PubMed] [Google Scholar]
- Mattes TE, Ewald JM, Liang Y, Martinez A, Awad A, Richards P, Hornbuckle KC, Schnoor JL, 2018. PCB dechlorination hotspots and reductive dehalogenase genes in sediments from a contaminated wastewater lagoon. Environ. Sci. Pollut. Res 25, 16376–16388. 10.1007/s11356-017-9872-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meggo RE, Schnoor JL, Hu D, 2013. Dechlorination of PCBs in the rhizosphere of switchgrass and poplar. Environ. Pollut 178, 312–321. 10.1016/j.envpol.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondello FJ, Turcich MP, Lobos JH, Erickson BD, 1997. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl. Environ. Microbiol 63, 3096–3103. 10.1128/AEM.63.8.3096-3103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadim LM, Schocken MJ, Higson FK, Gibson DT, 1987. Bacterial oxidation of polychlorinated biphenyls. In: Proceedings of the 13th Annual Research Symposium on Land Disposal, Remedial Action, Incineration, and Treatment of Hazardous Waste. EPA/600/9–87/015 U.S. Environmental Protection Agency, Cincinnati, Ohio, pp. 395–402. [Google Scholar]
- Ortega-Calvo J-J, Harmsen J, Parsons JR, Semple KT, Aitken MD, Ajao C, Eadsforth C, Galay-Burgos M, Naidu R, Oliver R, Peijnenburg WJGM, Römbke J, Streck G, Versonnen B, 2015. From bioavailability science to regulation of organic chemicals. Environ. Sci. Technol 49, 10255–10264. 10.1021/acs.est.5b02412. [DOI] [PubMed] [Google Scholar]
- Passatore L, Rossetti S, Juwarkar AA, Massacci A, 2014. Phytoremediation and bioremediation of polychlorinated biphenyls (PCBs): state of knowledge and research perspectives. J. Hazard Mater 278, 189–202. 10.1016/j.jhazmat.2014.05.051. [DOI] [PubMed] [Google Scholar]
- Payne RB, Fagervold SK, May HD, Sowers KR, 2013. Remediation of polychlorinated biphenyl impacted sediment by concurrent bioaugmentation with anaerobic halorespiring and aerobic degrading bacteria. Environ. Sci. Technol 47, 3807–3815. 10.1021/es304372t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RB, Ghosh U, May HD, Marshall CW, Sowers KR, 2019. A pilot-scale field study: in situ treatment of PCB-impacted sediments with bioamended activated carbon. Environ. Sci. Technol 53, 2626–2634. 10.1021/acs.est.8b05019. [DOI] [PubMed] [Google Scholar]
- Payne RB, Ghosh U, May HD, Marshall CW, Sowers KR, 2017. Mesocosm studies on the efficacy of bioamended activated carbon for treating PCB-impacted sediment. Environ. Sci. Technol 51, 10691–10699. 10.1021/acs.est.7b01935. [DOI] [PubMed] [Google Scholar]
- Pieper DH, Seeger M, 2008. Bacterial metabolism of polychlorinated biphenyls. J. Mol. Microbiol. Biotechnol 15, 121–138. 10.1159/000121325. [DOI] [PubMed] [Google Scholar]
- Quensen III JF, Tiedje JM, Boyd SA, 1988. Reductive dechlorination of polychlorinated biphenyls by anaerobic microorganisms from sediments. Science 80 (242), 752–754. 10.1126/science.242.4879.752. [DOI] [PubMed] [Google Scholar]
- Rehmann L, Daugulis AJ, 2008a. Biodegradation of PCBs in two-phase partitioning bioreactors following solid extraction from soil. Biotechnol. Bioeng 99, 1273–1280. 10.1002/bit.21674. [DOI] [PubMed] [Google Scholar]
- Rehmann L, Daugulis AJ, 2008b. Bioavailability of PCBs in biphasic bioreactors. Biochem. Eng. J 38, 219–225. 10.1016/j.bej.2007.07.004. [DOI] [Google Scholar]
- Rehmann L, Daugulis AJ, 2008c. Enhancement of PCB degradation by Burkholderia xenovorans LB400 in biphasic systems by manipulating culture conditions. Biotechnol. Bioeng 99, 521–528. 10.1002/bit.21610. [DOI] [PubMed] [Google Scholar]
- Ren X, Zeng G, Tang L, Wang J, Wan J, Liu Y, Yu J, Yi H, Ye S, Deng R, 2018. Sorption, transport and biodegradation – an insight into bioavailability of persistent organic pollutants in soil. Sci. Total Environ 610 (611), 1154–1163. 10.1016/j.scitotenv.2017.08.089. [DOI] [PubMed] [Google Scholar]
- Seah SYK, Labbé G, Nerdinger S, Johnson MR, Snieckus V, Eltis LD, 2000. Identification of a serine hydrolase as a key determinant in the microbial degradation of polychlorinated biphenyls. J. Biol. Chem 275, 15701–15708. 10.1074/jbc.275.21.15701. [DOI] [PubMed] [Google Scholar]
- Semple KT, Doick KJ, Jones KC, Burauel P, Craven A, Harms H, 2004. Peer reviewed: defining bioavailability and bioaccessibility of contaminated soil and sediment is complicated. Environ. Sci. Technol 38, 228A–231A. 10.1021/es040548w. [DOI] [PubMed] [Google Scholar]
- Semple KT, Doick KJ, Wick LY, Harms H, 2007. Microbial interactions with organic contaminants in soil: definitions, processes and measurement. Environ. Pollut 150, 166–176. 10.1016/j.envpol.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Shanahan CE, Spak SN, Martinez A, Hornbuckle KC, 2015. Inventory of PCBs in chicago and opportunities for reduction in airborne emissions and human exposure. Environ. Sci. Technol 49, 13878–13888. 10.1021/acs.est.5b00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondossi M, Sylvestre M, Ahmad D, 1992. Effects of chlorobenzoate transformation on the Pseudomonas testosteroni biphenyl and chlorobiphenyl degradation pathway. Appl. Environ. Microbiol 58, 485–495. 10.1128/AEM.58.2.485-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani R, Lyv MM, Kaveh R, Schnoor JL, Van Aken B, 2012. Biodegradation of mono-hydroxylated PCBs by Burkholderia xenovorans. Biotechnol. Lett 34, 2247–2252. 10.1007/s10529-012-1037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totten LA, Brunciak PA, Gigliotti CL, Dachs J, Glenn IV TR, Nelson ED, Eisenreich SJ, 2001. Dynamic air-water exchange of polychlorinated biphenyls in the New York-New Jersey harbor estuary. Environ. Sci. Technol 35, 3834–3840. 10.1021/es010791k. [DOI] [PubMed] [Google Scholar]
- US EPA, 2007. Method 3545a Pressurized Fluid Extraction (PFE), Method 3545A. Washington, D.C. [Google Scholar]
- US EPA, 2003. Table of Polychlorinated Biphenyl (PCB) Congeners. EPA. United States Environ. Prot. Agency [WWW Document] https://www.epa.gov/pcbs/table-polychlorinated-biphenyl-pcb-congeners. (Accessed 29 August 2020).
- US EPA, 1996. Method 3630C: Silica Gel Cleanup. Washington, D.C. [Google Scholar]
- Van Dort HM, Bedard DL, 1991. Reductive ortho and meta dechlorination of a polychlorinated biphenyl congener by anaerobic microorganisms. Appl. Environ. Microbiol 57, 1576–1578. 10.1128/AEM.57.5.1576-1578.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner D, Hale SE, Ghosh U, Luthy RG, 2010. Polychlorinated biphenyl sorption and availability in field-contaminated sediments. Environ. Sci. Technol 44, 2809–2815. 10.1021/es902325t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Wu S, Xia X, Lu X, Zhang X, Xia N, Liu T, 2016. Effects of carbonaceous materials on microbial bioavailability of 2,2,4,4-tetrabromodiphenyl ether (BDE-47) in sediments. J. Hazard Mater 312, 216–223. 10.1016/j.jhazmat.2016.03.065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


