Abstract
SUMOylation plays a crucial role in regulating diverse cellular processes including ribosome biogenesis. Proteomic analyses and experimental evidence showed that a number of nucleolar proteins involved in ribosome biogenesis are modified by SUMO. However, how these proteins are SUMOylated in cells is less understood. Here, we report that USP36, a nucleolar deubiquitinating enzyme (DUB), promotes nucleolar SUMOylation. Overexpression of USP36 enhances nucleolar SUMOylation, whereas its knockdown or genetic deletion reduces the levels of SUMOylation. USP36 interacts with SUMO2 and Ubc9 and directly mediates SUMOylation in cells and in vitro. We show that USP36 promotes the SUMOylation of the small nucleolar ribonucleoprotein (snoRNP) components Nop58 and Nhp2 in cells and in vitro and their binding to snoRNAs. It also promotes the SUMOylation of snoRNP components Nop56 and DKC1. Functionally, we show that knockdown of USP36 markedly impairs rRNA processing and translation. Thus, USP36 promotes snoRNP group SUMOylation and is critical for ribosome biogenesis and protein translation.
Keywords: deubiquitinating enzyme, ribosome biogenesis, snoRNP, SUMOylation, USP36
Subject Categories: Post-translational Modifications, Proteolysis & Proteomics; Protein Biosynthesis & Quality Control
The deubiquitinase USP36 also promotes protein SUMOylation and is essential for ribosome biogenesis and protein translation.
Introduction
SUMOylation, a posttranslational modification of proteins by small ubiquitin‐like modifiers (SUMOs), plays a crucial role in the regulation of diverse cellular processes, including transcription, chromatin dynamics, genome maintenance, DNA repair, RNA splicing and processing, cell cycle control, and metabolism and is essential for normal cell growth and animal development (Bergink & Jentsch, 2009; Finkbeiner et al, 2011; Jentsch & Psakhye, 2013; Chymkowitch et al, 2015; Nuro‐Gyina & Parvin, 2015; Sarangi & Zhao, 2015; Zhao, 2018). Mammals express three main SUMO isoforms: SUMO2 and SUMO3 are 97% identical (referred to as SUMO2/3) and both are 45% identical to SUMO1. SUMOylation is ATP‐dependent and occurs through sequential reactions involving a heterodimeric SUMO‐activating enzyme SAE1/SAE2 (E1), a single SUMO‐conjugating enzyme Ubc9 (E2) and one of a few SUMO ligases (E3) (Muller et al, 2001; Geiss‐Friedlander & Melchior, 2007; Jentsch & Psakhye, 2013). Ubc9 transfers SUMO to substrate acceptor lysine (Lys, K) residues, which is facilitated by SUMO E3s, via an isopeptide linkage (Muller et al, 2001; Geiss‐Friedlander & Melchior, 2007; Jentsch & Psakhye, 2013). The SUMO acceptor Lys is often present within a conserved ΨKxE motif, where Ψ is a large hydrophobic amino acid and x is any amino acid (Rodriguez et al, 2001; Sampson et al, 2001; Geiss‐Friedlander & Melchior, 2007; Gareau & Lima, 2010). While there are over 600 ubiquitin E3s in human (Deshaies & Joazeiro, 2009; Metzger et al, 2012), only a small number of bona fide SUMO E3 have been identified, including the SP‐RING family members PIAS1, PIAS3, PIASxα, PIASxβ, PIASy (Kahyo et al, 2001; Sachdev et al, 2001; Nakagawa & Yokosawa, 2002; Nishida & Yasuda, 2002; Schmidt & Muller, 2002), and Nse2 (Andrews et al, 2005; Potts & Yu, 2005; Berkholz et al, 2014), the nuclear pore Ran Binding Protein 2 (RanBP2) (Pichler et al, 2002; Reverter & Lima, 2005), and the ZNF451 family proteins (ZNF451‐1, ZNF451‐2, ZNF451‐3) (Cappadocia et al, 2015; Eisenhardt et al, 2015; Varejao et al, 2020). In addition, a few other proteins have been shown to possess SUMO E3 activity such as Pc2, TRIM28, and SLX4 (Kagey et al, 2003; Ivanov et al, 2007; Guervilly et al, 2015). SUMO modification is also highly dynamic and reversible. Removal of SUMO from substrates (deSUMOylation) is catalyzed by a group of deSUMOylating enzymes or SUMO proteases, including SENP1‐3 and SENP5‐7, DESI‐1, DESI‐2, and USPL1 (Hickey et al, 2012; Kunz et al, 2018).
SUMOylation also plays an important role in ribosome biogenesis, a multi‐step cellular process for making the ribosome in eukaryotes, requiring synthesis of ribosomal RNA (rRNA) and ribosomal proteins, rRNA processing, the assembly of the mature ribosome subunits in the nucleolus and transport into the cytoplasm, as well as the participation of many accessory factors (Rodnina & Wintermeyer, 2009; Kressler et al, 2017; Tomecki et al, 2017). Impairment of ribosome biogenesis is associated with a group of diseases called ribosomopathies whereas aberrant over‐activation of ribosome biogenesis is tightly linked to human cancers (Mills & Green, 2017; Pelletier et al, 2018). Therefore, it is crucial to understand how ribosome biogenesis is properly regulated during normal cell homeostasis. It was first found that many yeast ribosome biogenesis factors and pre‐ribosomal particles are modified by SUMO, which is critical for efficient ribosome biogenesis (Panse et al, 2006). In human, a number of nucleolar proteins involved in ribosome biogenesis, including nucleophosmin (NPM) (Tago et al, 2005; Liu et al, 2007), nucleolin (Zhang et al, 2015), and Las1L (Finkbeiner et al, 2011; Castle et al, 2012), are modified by SUMOylation. Proteomic studies found that ribosome biogenesis‐related proteins are one of the major classes of SUMOylated proteins in cells (Vertegaal et al, 2004; Matafora et al, 2009; Amente et al, 2012; Hendriks et al, 2014). Likewise, deSUMOylation is also important for ribosome biogenesis. Removal of SUMO from NPM by SENP3 is critical for 28S rRNA maturation and the subsequent nucleolar export of the 60S pre‐ribosomal subunit (Haindl et al, 2008). DeSUMOylation of Las1L by SENP3 is essential for ribosome particles partitioning from the nucleolus to cytoplasm (Finkbeiner et al, 2011; Castle et al, 2012). SUMOylation of PELP1, a component of the PELP1‐TEX10‐WDR18 complex critical for ribosome maturation, promotes the recruitment of MDN1, an AAA ATPase that removes assembly factors from the pre‐60S particles (Chen et al, 2018), to pre‐60S particles, while deSUMOylation is needed to release both MDN1 and PELP1 from pre‐ribosomes (Raman et al, 2016). Depletion of SENP3 inhibits rRNA processing reminiscent of the NPM knockdown (Yun et al, 2008). Further, Nop58 and Nhp2, components of the small nucleolar ribonucleoprotein (snoRNP) complexes responsible for rRNA 2′‐O‐ribose methylation and pseudouridylation and critical for rRNA processing (Mannoor et al, 2012; Watkins & Bohnsack, 2012; Lui & Lowe, 2013; Dupuis‐Sandoval et al, 2015), are also modified by SUMO (Matic et al, 2010; Westman et al, 2010). SUMOylation of Nop58 is required for its high‐affinity binding to snoRNAs and the nucleolar localization of snoRNAs (Westman et al, 2010). Thus, balanced levels of SUMOylation and deSUMOylation of ribosome biogenesis factors are critical for ribosome biogenesis and maturation in the nucleolus. However, how these nucleolar proteins are SUMOylated is less understood.
In this study, we identified that the ubiquitin‐specific protease USP36 mediates nucleolar SUMOylation. We show that USP36 directly interacts with SUMO2 and Ubc9, directly promotes the SUMOylation of Nop58 and Nhp2 in cells and in vitro, and enhances their binding to snoRNAs. Functionally, USP36 is required for multiple steps of rRNA processing and translation. Thus, USP36, a nucleolar deubiquitinating enzyme (DUB) (Endo et al, 2009; Sun et al, 2015), also acts to promote nucleolar SUMOylation and is critical for ribosome biogenesis.
Results
USP36 promotes SUMOylation in cells, mainly in the nucleolus
In our initial investigation of the SUMO regulation of USP36, we serendipitously observed that USP36 markedly increased the levels of protein SUMOylation in cells. We confirmed that overexpression of USP36 significantly promoted SUMO2/3‐conjugation in H1299 (Fig 1A), HeLa, and U2OS (Fig 1B) as well as other tested cell lines such as 293 (Fig EV1A) and breast cancer T47D (Fig EV1B) cells. This effect is specific for USP36 as SUMOylation was not increased by overexpression of either JOSD3, another nucleolar DUB (Fig EV1C), or the nuclear DUB USP7 (Fig EV1D). We focused on SUMO2 in this study but USP36 also promotes SUMO1 conjugation (Fig EV1E and F). Nickel (Ni2+‐NTA beads) purification methods demonstrated that overexpression of USP36 drastically enhanced SUMOylation by exogenous His‐SUMO2 in cells (Fig EV1G). To test whether endogenous USP36 also regulates the levels of protein SUMOylation in cells, we performed knockdown experiments. As shown in Fig 1C, knockdown of USP36 by two individual shRNAs drastically reduced the levels of SUMOylated species in H1299 cells. Similarly, knockdown of USP36 also reduced the levels of SUMOylated species in HeLa (Fig 1D) and U2OS (Fig 1E) cells. Thus, endogenous USP36 also promotes SUMOylation and this effect is not cell‐type specific.
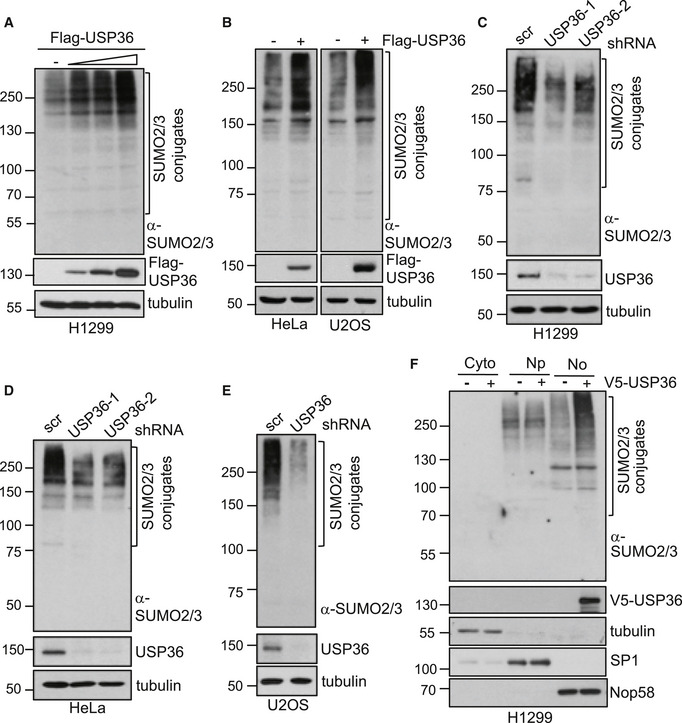
Figure 1. USP36 promotes SUMOylation in cells.
-
A, BOverexpression of USP36 promotes SUMOylation in cells. Whole cell lysates (WCL) from H1299 (A), U2OS and HeLa (B) cells transfected with control empty vector or Flag‐USP36 were assayed by IB.
-
C–EKnockdown of USP36 reduces SUMOylation in cells. H1299 (C), HeLa (D), and U2OS (E) cells infected with scrambled (scr) or USP36 shRNA encoding lentiviruses were assayed by IB.
-
FUSP36 promotes nucleolar SUMOylation. H1299 cells transfected with control or V5‐USP36 were fractionated to the cytoplasm (Cyto), nucleoplasm (Np), and nucleolus (No) fractions and assayed by IB.
Data information: SUMO2/3 conjugates are indicated in all the top panels.
Source data are available online for this figure.
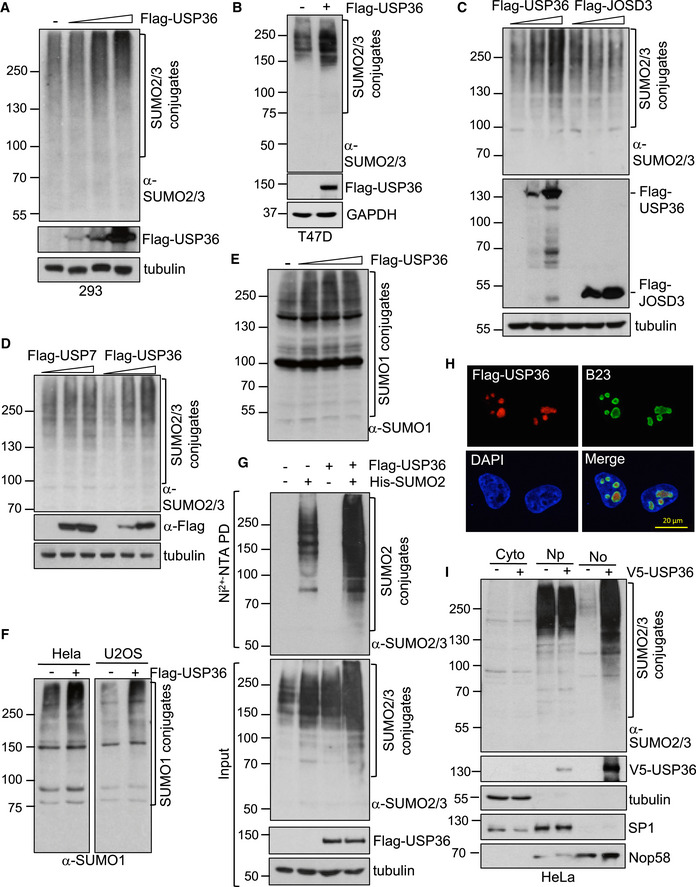
Figure EV1. USP36 promotes SUMOylation in cells.
-
A., BUSP36 promotes SUMOylation in cells. 293 (A) and T47D (B) cells transfected with control or Flag‐USP36 plasmid were assayed by IB for SUMOylated proteins.
-
C, DUSP36 promotion of SUMOylation is specific. H1299 cells transfected with increasing amounts of Flag‐USP36, Flag‐JOSD3 (C), or Flag‐USP7 (D) were assayed by IB.
-
E, FUSP36 promotes SUMOylation by SUMO1. H1299 cells (E) or HeLa and U2OS cells (F) transfected with Flag‐USP36 were assayed by IB using anti‐SUMO1 antibody.
-
GUSP36 promotes SUMOylation by exogenous SUMO2. H1299 cells transfected with His‐SUMO2 and/or Flag‐USP36 were subjected to Ni2+‐NTA agarose beads pulldown (PD) followed by IB using anti‐SUMO2/3 antibody (top panel). WCL were also directly assayed by IB using indicated antibodies (bottom panel).
-
HNucleolar localization of USP36. HeLa cells transfected with Flag‐USP36 were stained with anti‐Flag (red) and anti‐B23 (green) antibodies followed by DAPI (blue).
-
IUSP36 promotes nucleolar SUMOylation. HeLa cells transfected with control or V5‐USP36 were fractionated to the cytoplasm (Cyto), nucleoplasm (Np), and nucleolus (No) fractions, followed by IB to detect the indicated proteins.
Source data are available online for this figure.
USP36 is mainly localized in the nucleolus (Endo et al, 2009; Sun et al, 2015) (Fig EV1H). To determine whether USP36 promotes SUMOylation in the nucleolus, we performed cell fractionation assays. As shown in Fig 1F, USP36 markedly increased the protein SUMOylation in the nucleolus, but not the nucleoplasm, in H1299 cells. Similar results were also observed in HeLa cells (Fig EV1I). Thus, USP36 mainly promotes SUMOylation of the nucleolar proteins.
The USP36 N‐terminus is necessary for promoting SUMOylation
To understand how USP36 regulates SUMOylation, we examined whether it controls the levels of the SUMO pathway enzymes, given that USP36 is a DUB (Endo et al, 2009; Sun et al, 2015). However, USP36 neither increased the levels of SUMO E1 (SAE1 and SAE2) or SUMO E2 (Ubc9; Fig EV2A), nor reduced the levels of the nuclear SUMO proteases SENP1, SENP2, SENP3, SENP5, SENP6, SENP7, and USPL1 (Fig EV2B and C), suggesting that USP36 does not promote SUMOylation by controlling the levels of the SUMO pathway enzymes. These results prompted us to test whether USP36 directly promotes SUMOylation. We then examined which region of USP36 involves in promoting SUMOylation. We transfected cells with different fragments of USP36 and found that the N‐terminal USP domain containing region (amino acids 1–420), but not the middle and C‐terminal regions, is essential for promoting SUMOylation by SUMO2/3 (Fig 2A), suggesting that this N‐terminal region might harbor SUMO E3 activity (Fig 2B).
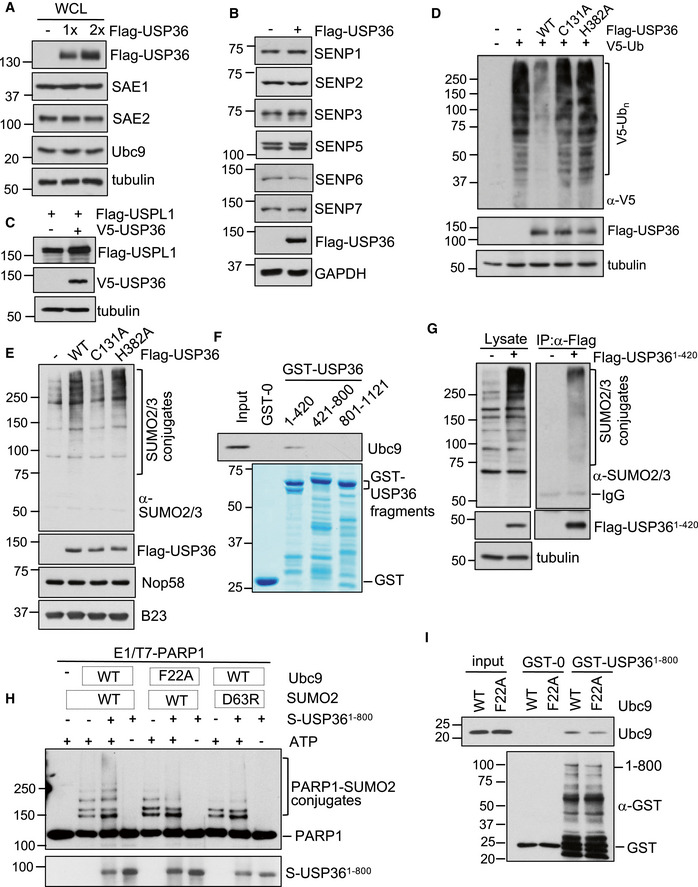
Figure EV2. USP36 neither induces the levels of SUMO E1 or E2 nor increases the levels of SUMO proteases.
-
AUSP36 does not increase the levels of SUMO E1 and E2. H1299 cells transfected with control or increased amounts of Flag‐USP36 were assayed by IB using antibodies against indicated proteins.
-
B, CUSP36 does not reduce the levels of SUMO proteases. H1299 cells transfected with Flag‐USP36 were assayed by IB using antibodies against indicated SENP proteins (B) or with Flag‐USPL1 (C) in the presence or absence of V5‐USP36 were assayed by IB.
-
DH1299 cells transfected with V5‐Ub together with Flag‐USP36 or the indicated mutants were assayed by IB to detect total ubiquitination.
-
EWT USP36 and the H382A mutant, but not the C131A mutant, promote SUMOylation in the nucleolus. H1299 cells transfected with control or the indicated Flag‐USP36 plasmids were subjected to nucleolar isolation, followed by IB. SUMO2/3 conjugates are indicated in the top panel.
-
FThe N‐terminus of USP36 interacts with Ubc9 in vitro. Purified His‐Ubc9 was incubated with GST, GST‐USP361–420, GST‐USP36421–800, or GST‐USP36801–1121. Bound protein was detected by IB. GST and GST‐fusion proteins were shown in the bottom panel by coomassie staining.
-
GThe N‐terminal USP36 binds to SUMO in cells. H1299 cells transfected with either control or Flag‐USP361–420 plasmid were assayed by IP using anti‐Flag, followed by IB with anti‐SUMO2/3 antibodies.
-
HRequirement of the SUMO–Ubc9 backside interaction for USP36's SUMO E3 activity. Recombinant T7‐PARP1 protein (0.1 μM) was incubated with SUMO E1 (50 nM, Boston Biochem), Ubc9 (50 nM, WT or the F22A mutant), SUMO2 (4 μM, WT or the D63R mutant) in the presence of USP361–800 (50 nM) and/or ATP (2.5 mM) at 30°C for 5 h and then assayed by IB.
-
IUSP36 interacts with Ubc9 in vitro. Purified His‐Ubc9 (Wt or the F22A mutant) was incubated with GST or GST‐USP361–800. Bound Ubc9 was detected by IB using anti‐Ubc9 antibody. GST and GST‐fusion proteins were detected by IB with anti‐GST.
Source data are available online for this figure.
Figure 2. The N‐terminus of USP36 is necessary for promoting SUMOylation.
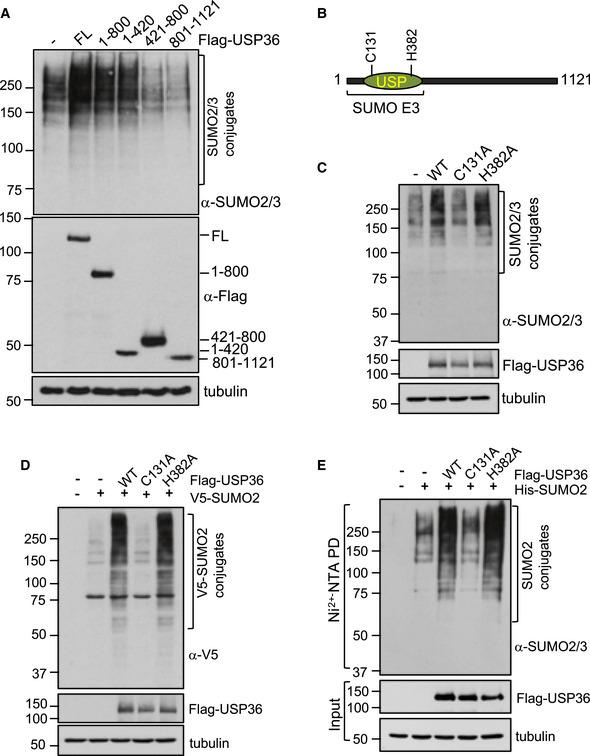
- H1299 cells were transfected with Flag‐USP36 or the indicated deletion mutants and assayed by IB. FL, full length.
- Schematic diagram of USP36 indicating that the N‐terminus of USP36, which contains the USP catalytic domain, is required for its SUMO E3 activity. Catalytic residues C131 and H382 are shown.
- H1299 cells transfected with WT USP36, the C131A, or H382A mutant were assayed by IB to detect SUMOylated proteins.
- H1299 cells transfected with WT USP36 or the indicated mutants and V5‐SUMO2 were assayed by IB using anti‐V5 antibody to detect total protein SUMOylation.
- H1299 cells transfected with WT USP36 or the indicated mutants and His‐SUMO2 were subjected to Ni2+‐NTA agarose beads pulldown (PD) followed by IB using anti‐SUMO2/3 antibody.
Source data are available online for this figure.
As this N‐terminal region contains both DUB activity and the activity to promote SUMOylation, we next asked whether the SUMO promoting activity is related to the DUB activity. To this end, we examined the effect of catalytically inactive USP36 mutants (Figs 2B and EV2D) on SUMOylation. Interestingly, mutating the DUB catalytic Cys 131 to Ala (C131A) also abolished the activity of USP36 to promote SUMOylation by either endogenous SUMO (Fig 2C) or exogenously expressed SUMO2 (Fig 2D), whereas mutating the DUB catalytic proton acceptor His 382 to Ala (H382A) promotes SUMOylation as efficiently as wild‐type (WT) USP36 (Fig 2C and D). Ni2+‐NTA pull‐down assays further confirmed that the H382A, but not the C131A, mutant enhanced SUMOylation by His‐SUMO2 in cells (Fig 2E). Further, examination of SUMOylation using purified nucleoli also confirmed that the H382A, but not the C131A, mutant promotes SUMOylation of the nucleolar proteins (Fig EV2E). Thus, the SUMO promoting activity of USP36 does not depend on its DUB activity. These results also suggest that C131 may be structurally important for the proper conformation of USP36 to mediate SUMOylation.
USP36 binds to SUMO and Ubc9 and mediates SUMOylation in vitro
Next, we examined whether USP36 indeed possesses SUMO ligase activity. SUMO ligases bind to both Ubc9 and substrates to facilitate the transfer of SUMO to substrates and also bind to SUMO (Bernier‐Villamor et al, 2002; Kagey et al, 2003; Yunus & Lima, 2009; Guervilly et al, 2015; Streich & Lima, 2016). Therefore, we first examined whether USP36 binds to Ubc9 and SUMO. Indeed, USP36 interacts with Ubc9 in cells as determined by co‐immunoprecipitation (IP) assay (Fig 3A). Glutathione S‐transferase (GST) pull‐down assays by incubating recombinant Ubc9 with GST‐fusion USP36 proteins showed that USP36 directly interacts with ubc9 in vitro (Fig 3B). The N‐terminus (aa 1–420), but not the middle and C‐terminal regions, of USP36 is also sufficient for binding to Ubc9 (Fig 3B and EV2F). To test whether USP36 interacts with SUMO, we performed co‐IP assays. As shown in Fig 3C, Flag‐USP36 binds to SUMO2/3‐conjugated proteins in cells. Also, GST‐USP36, but not GST alone, directly interacts with SUMO2, preferentially the polymeric SUMO2 chains, in vitro (Fig 3D). Again, the N‐terminus (aa 1–420), but not the middle and C‐terminal regions, binds to the SUMO2 chains in vitro (Fig 3D) and in cells (Fig EV2G). These results reveal that USP36 interacts with both Ubc9 and SUMO2 via its N‐terminus.
Figure 3. USP36 interacts with Ubc9 and SUMO and mediates SUMOylation in vitro .
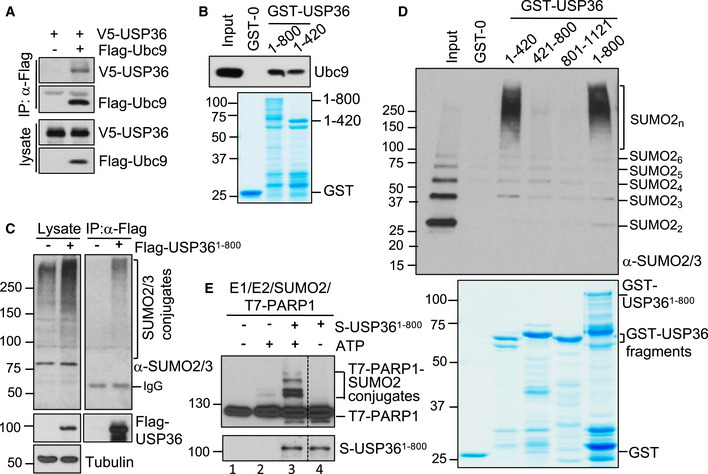
- USP36 interacts with Ubc9 in cells. H1299 cell transfected with the indicated plasmids was subjected to co‐IP with anti‐Flag antibody followed by IB.
- USP36 interacts with Ubc9 in vitro. Purified His‐Ubc9 was incubated with GST, GST‐USP361–800 or GST‐USP361–420. Bound protein was detected by IB. GST and GST‐fusion proteins were shown in the bottom panel by coomassie staining.
- USP36 interacts with SUMO in cells. H1299 cells transfected with either control or Flag‐USP361–800 were assayed by co‐IP using anti‐Flag, followed by IB with anti‐SUMO2/3 antibodies.
- USP36 interacts with SUMO2 chains in vitro. Recombinant SUMO2 chains were incubated with GST or indicated GST‐USP36 fragments. Bound proteins were assayed by IB using anti‐SUMO2/3 antibodies. Coomassie staining of the recombinant proteins was shown in bottom panel.
- USP36 SUMOylates PARP1 in vitro. Recombinant T7‐PARP1 protein (0.1 μM) was incubated with SUMO E1 (30 nM), Ubc9 (50 nM), SUMO2 (4 μM) in the presence of USP361–800 (50 nM) and/or ATP (2.5 mM) at 30°C for 5 h and then assayed by IB.
Source data are available online for this figure.
We next tested whether USP36 directly promotes SUMOylation in vitro using well‐studied SUMO target PARP1 involved in cell cycle progression (Ryu et al, 2010a). As shown in Fig 3E, in vitro SUMOylation reaction using recombinant E1 (SAE1/SAE2), E2 (Ubc9), SUMO2, and ATP resulted in marginal SUMO conjugation of PARP1 (lane 2), consistent with the notion that SUMO E1 and E2 can mediate SUMOylation in vitro, although less efficiently (Bernier‐Villamor et al, 2002; Geiss‐Friedlander & Melchior, 2007; Ryu et al, 2010a; Ryu et al, 2010b). Notably, USP36 drastically increased PARP1 SUMOylation in vitro (compared lane 3 to lane 2, Fig 3E). These results suggest that USP36 may possess a novel SUMO E3 activity. As many SUMO E3s such as PIAS1 (Eisenhardt et al, 2019) and ZNF451 (Cappadocia et al, 2015; Eisenhardt et al, 2015; Koidl et al, 2016; Eisenhardt et al, 2019) family members require a SUMO interaction with the backside of Ubc9 for their SUMO E3 activity, we tested whether this interaction could also play a role in USP36's SUMO E3. As shown in Fig EV2H, either mutating Phe 22 of Ubc9 to Ala (Ubc9F22A), which weakens the SUMO–Ubc9 backside interaction (Capili & Lima, 2007), or mutating Asp 63 of SUMO2 to Arg (SUMO2D63R), which disrupts the SUMO–Ubc9 backside interaction (Knipscheer et al, 2007), markedly inhibited USP36 activity to promote PARP1 poly‐SUMOylation. These results suggest that although USP36 directly interacts with Ubc9 or the F22A mutant (Fig EV2I), its full activity to promote poly‐SUMOylation requires the SUMO–Ubc9 backside interaction, further suggesting that USP36 acts as a SUMO E3.
USP36 promotes SUMOylation of Nop58 and Nhp2 in cells
To identify proteins whose SUMOylation is increased by USP36, we performed proteomic analysis. SUMOylated proteins were purified from cells transfected with His‐SUMO2 together with control or USP36 plasmid using Ni2+‐NTA beads purification followed by mass spectrometry analysis. Among the SUMOylated proteins including USP36 itself (Dataset EV1), we found that the number of assigned MS2 spectra of Nop58 and a number of nucleolar proteins involved in ribosome biogenesis increased in the USP36 transfected sample, (Fig EV3A). Nop58 and Nhp2, components of the box C/D and box H/ACA snoRNP complexes, respectively, were previously shown to be SUMO substrates (Westman et al, 2010), yet a SUMO E3 mediating such SUMOylation has not been reported. Therefore, we tested whether USP36 promotes the SUMOylation of Nop58 and Nhp2. Indeed, in vivo SUMOylation assays using Ni2+‐NTA pulldown showed that USP36 markedly promoted the SUMOylation of exogenous Nop58 (Fig EV3B) and Nhp2 (Fig EV3D) as well as endogenous Nop58 (Fig 4A) and Nhp2 (Fig 4B). Cell fractionation assays showed that USP36 drastically promoted the SUMOylation of both Nop58 and Nhp2 in the nucleolus (Fig 4C), consistent with its role in the nucleolar SUMOylation. We also confirmed that the two previously reported Lys residues, K467 and K487, in Nop58 and the single Lys 5 (K5) in Nhp2 were the SUMOylation sites. Mutation of the K467 and K487 to Arg in Nop58 (Nop582KR) (Fig EV3C) and K5 to R in Nhp2 (Nhp2K5R) (Fig EV3E) abrogated their SUMOylation in cells. Thus, both Nop58 and Nhp2 are the endogenous SUMO substrates of USP36. Of note, USP36 does not increase the SUMOylation of nucleolar protein NOLC1 (Fig EV3F), a previously identified SUMO substrate (Westman et al, 2010), suggesting that not all nucleolar proteins are SUMO targets of USP36. Consistently, knockdown of USP36 significantly reduced the levels of SUMOylated Nop58 (Fig 4D) and Nhp2 (Fig 4E) in cells.
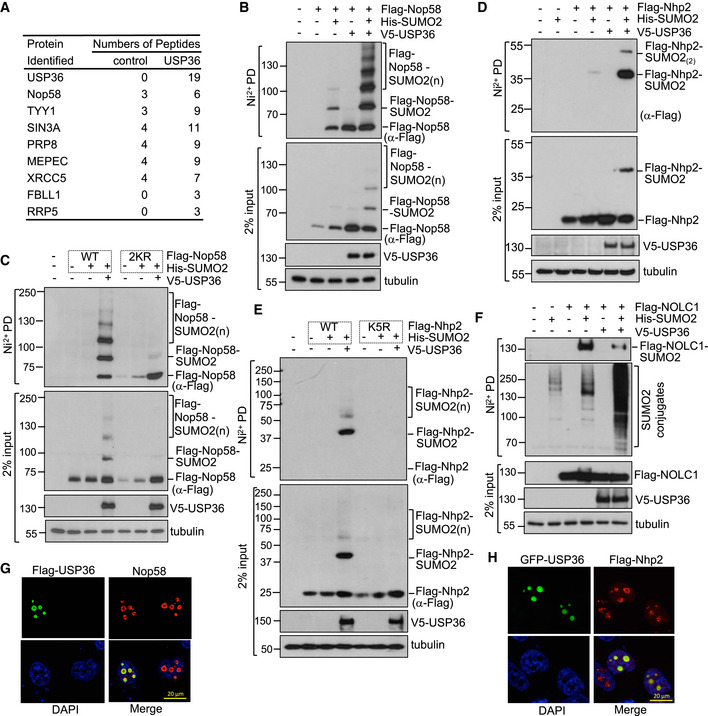
Figure EV3. USP36 promotes SUMOylation of Nop58 and Nhp2.
-
ACandidate proteins whose SUMOylation is increased by USP36. H1299 cells transfected His‐SUMO2 together with control or Flag‐USP36 plasmids were subjected to Ni2+‐NTA purification followed by LC‐MS‐MS analysis.
-
B, CUSP36 SUMOylates exogenous Nop58. H1299 cells transfected with His‐SUMO2 and/or USP36 with WT Nop58 (B) or SUMOylation‐defective mutant Nop58 (K457R/K497R, Nop582KR) (C) were subjected to Ni2+‐NTA PD followed by IB to detect SUMOylation of Nop58.
-
D, EUSP36 SUMOylates exogenous Nhp2. H1299 cells transfected with His‐SUMO2 and/or USP36 with WT Nhp2 (D) or with SUMOylation‐defective mutant Nhp2 (Nop58K5R) (E) were subjected to Ni2+‐NTA PD followed by IB to detect SUMOylation of Nph2.
-
FUSP36 does not SUMOylate nucleolar protein NOLC1. H1299 cells transfected with His‐SUMO2, Flag‐NOLC1 with or without V5‐USP36 were subjected to Ni2+‐NTA PD followed by IB to detect SUMOylation of NOLC1.
-
G, HCo‐localization of USP36 with Nop58 and Nhp2 in the nucleolus. HeLa cells were transfected with Flag‐USP36 and stained with anti‐Flag (green) and anti‐Nop58 (red) (G) or transfected with GFP‐USP36 (green) and Flag‐Nhp2 and then stained with anti‐Flag (red) (H).
Source data are available online for this figure.
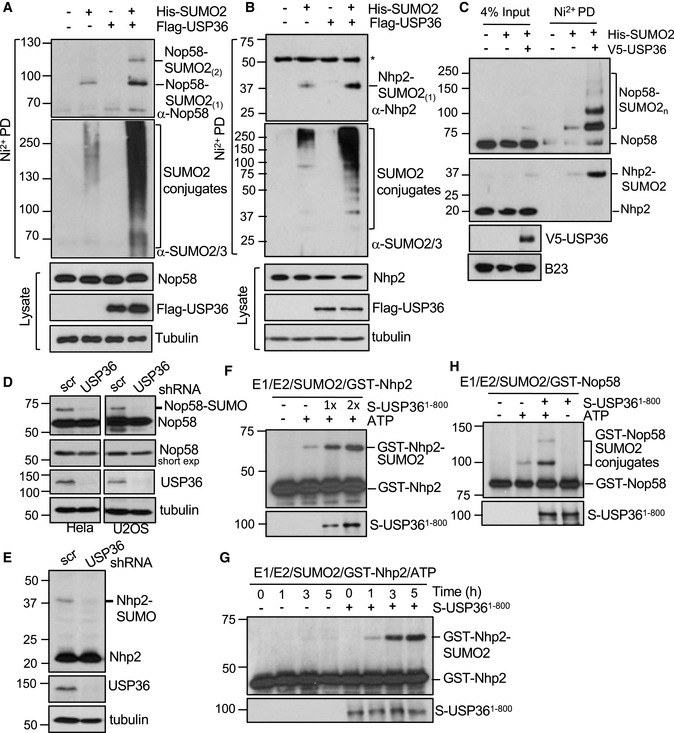
Figure 4. USP36 SUMOylates Nop58 and Nhp2 in cells and in vitro .
-
A, BH1299 cells transfected with His‐SUMO2 and/or USP36 were subjected to Ni2+‐NTA agarose beads pulldown (PD) followed by IB to detect SUMOylation of Nop58 (A) and Nhp2 (B). *indicates a non‐specific anti‐Nhp2 antibody‐reacting band.
-
CUSP36 promotes Nop58 and Nhp2 SUMOylation in the nucleolus. Nucleoli isolated from HeLa cells transfected with His‐SUMO2 and/or USP36 were subjected to Ni2+‐NTA PD, followed by IB.
-
D, EKnockdown of USP36 reduces Nop58 and Nhp2 SUMOylation in cells. Cells infected with scr or USP36 shRNA lentiviruses were assayed by IB. The SUMOylated Nop58 (D) and Nhp2 (E) are indicated.
-
F–HUSP36 SUMOylates Nhp2 and Nop58 in vitro. In vitro SUMOylation assays were performed by incubating recombinant Nhp2 (F, G) or Nop58 (H) (0.1 μM) with SUMO E1 (30 nM), Ubc9 (50 nM), SUMO2 (4 μM) in the absence or presence of USP361–800 (50 nM, 1×; 100 nM, 2×) and/or ATP (2.5 mM) at 30°C for 2 h or the indicated times. The reactions were assayed by IB using anti‐Nhp2 and anti‐Nop58 antibodies, respectively.
Source data are available online for this figure.
USP36 SUMOylates Nop58 and Nhp2 in vitro
To determine whether USP36 directly SUMOylates Nop58 and Nhp2 in vitro, we expressed and purified recombinant GST‐Nop58 and GST‐Nhp2 from bacteria for SUMOylation assays in vitro. As shown in Fig 4F, in vitro SUMOylation reaction containing recombinant E1, E2, SUMO2, and ATP resulted in marginal SUMO conjugation of Nhp2. Adding recombinant USP36 significantly promoted Nhp2 SUMOylation in vitro in dose‐dependent (Fig 4F) and time‐dependent (Fig 4G) manners. Similarly, USP36 also promotes Nop58 SUMOylation in vitro (Fig 4H). Thus, USP36 SUMOylates both Nop58 and Nhp2 in vitro.
USP36 promotes snoRNP protein group SUMOylation
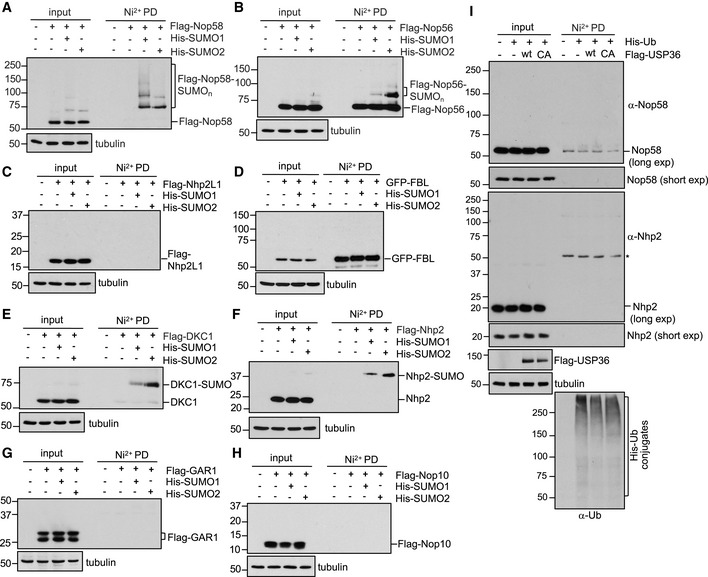
Emerging evidence suggests that SUMOylation tends to target a group of functionally and physically connected proteins called protein group SUMOylation (Psakhye & Jentsch, 2012; Jentsch & Psakhye, 2013), which allows multiple SUMO‐interacting motif (SUMO–SIM) interactions that contribute to the formation and stabilization of the multi‐protein complexes (Psakhye & Jentsch, 2012; Jentsch & Psakhye, 2013). Therefore, we examined whether USP36 promotes snoRNP protein group SUMOylation. There are two main classes of snoRNPs (Mannoor et al, 2012; Watkins & Bohnsack, 2012; Lui & Lowe, 2013; Dupuis‐Sandoval et al, 2015). Box C/D snoRNPs contain box C/D snoRNAs and four core proteins, Nop58, Nop56, Nhp2L1, and fibrillarin (FBL, a methyltransferase) and mediate rRNA 2′‐O‐ribose‐methylation, whereas box H/ACA snoRNPs contain box H/ACA snoRNAs and four other core proteins, Nhp2, Nop10, Gar1, and DKC1 (a pseudouridine synthase) and mediate rRNA pseudouridylation. These snoRNPs are assembled in the nucleoplasm in Cajal bodies and transferred into the nucleolus where they bind to pre‐rRNA and mediate rRNA modifications critical for rRNA processing (Richard et al, 2003; Watkins & Bohnsack, 2012; Dupuis‐Sandoval et al, 2015). We examined SUMOylation of all these snoRNP proteins and found that Nop56 and DKC1, but not FBL, Nhp2L1, Gar1, and Nop10, are also SUMOylated with both SUMO1 and SUMO2 in cells (Fig EV4A–H). Further, USP36 also markedly promoted the SUMOylation of Nop56 (Fig 5A) and DKC1 (Fig 5B). Thus, USP36 promotes snoRNP group SUMOylation.
Figure EV4. SUMOylation of snoRNP proteins.
-
A–HSUMOylation of snoRNP proteins. H1299 cells transfected with the indicated snoRNP proteins together with His‐SUMO1 or His‐SUMO2 were assayed by Ni2+‐NTA PD and IB with anti‐Flag (A–C, E–H) or anti‐GFP (D) antibody to detect SUMOylated snoRNP proteins.
-
IDetection of endogenous Nop58 and Nhp2 ubiquitination. H1299 cells were transfected with His‐Ub with or without Flag‐USP36 (wt or the C131A mutant) and treated with MG132 for 6 h before harvesting. The cells were then subjected to Ni2+‐NTA beads pulldown, followed by IB with anti‐Nop58 and anti‐Nhp2. The total His‐Ub conjugates are shown in the bottom. * indicates a non‐specific anti‐Nhp2 antibody‐reacting band.
Source data are available online for this figure.
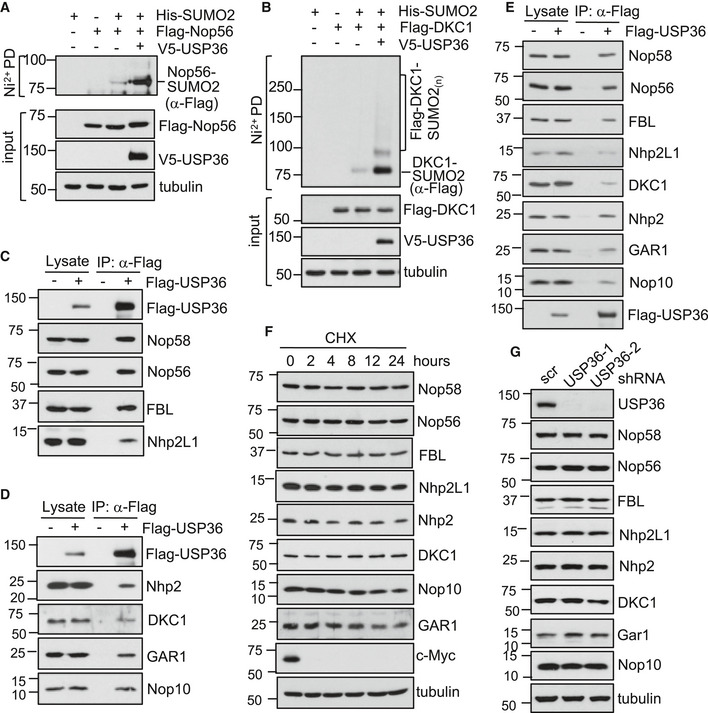
Figure 5. USP36 interacts with snoRNPs and promotes snoRNP protein group SUMOylation.
-
A, BUSP36 promotes SUMOylation of DKC1 and Nop56. H1299 cells transfected with indicated plasmids were subjected to Ni2+‐NTA PD followed by IB to detect SUMOylated Nop56 and DKC1, respectively. The protein expression was shown in the bottom of each panel.
-
C, DUSP36 interacts with both box C/D (C) and H/ACA (D) snoRNP complexes. H1299 cells transfected with empty vector or Flag‐USP36 plasmid were assayed by IP with anti‐Flag antibody, followed by IB to detect the indicated proteins.
-
EUSP36 interacts with snoRNP proteins in the nucleolus. The nucleoli purified from H1299 cells transfected with empty vector or Flag‐USP36 plasmid were assayed by co‐IP with anti‐Flag antibody, followed by IB to detect the indicated proteins.
-
FHalf‐life assays. The WCL from H1299 cells treated with 50 μg/ml cycloheximide (CHX) for indicated time were assayed by IB for the indicated snoRNP proteins.
-
GUSP36 does not change the levels of endogenous snoRNP proteins. HeLa cells infected with scrambled or the indicated USP36 shRNA lentiviruses were assayed by IB.
Source data are available online for this figure.
USP36 interacts with snoRNPs
To test whether USP36 associates with snoRNP proteins, we performed co‐IP assays. As shown in Fig 5C and D, all endogenous box C/D and box H/ACA snoRNP proteins were co‐immunoprecipitated with Flag‐USP36. Immunofluorescene (IF) staining showed that USP36 co‐localizes with Nop58 and Nhp2 in the nucleolus (Fig EV3G and H). The interaction of USP36 with snoRNP proteins in the nucleolus was also confirmed by co‐IP assays using lysates from purified nucleoli (Fig 5E). These data reveal that USP36 binds to snoRNPs and promotes snoRNP protein group SUMOylation.
USP36 does not affect the levels of snoRNP proteins
To understand how USP36 might affect snoRNP function, we first examined whether it regulates the levels of these snoRNP proteins, given that USP36 is a DUB and that SUMOylation crosstalks with protein ubiquitination (Hendriks & Vertegaal, 2016; Lamoliatte et al, 2017). We examined the half‐lives of these snoRNP proteins and found that all these snoRNP proteins are stable proteins (Fig 5F). Neither overexpression (left two lanes in Fig 5C and D) or knockdown (Fig 5G) of USP36 significantly changed the levels of endogenous snoRNP proteins. Consistently, the steady‐state levels of Nop58 and Nhp2 ubiquitination are below detectable levels (Fig EV4I), suggesting that USP36 mainly acts to promote snoRNP protein SUMOylation.
USP36 promotes the binding of Nhp2 and Nop58 to snoRNAs
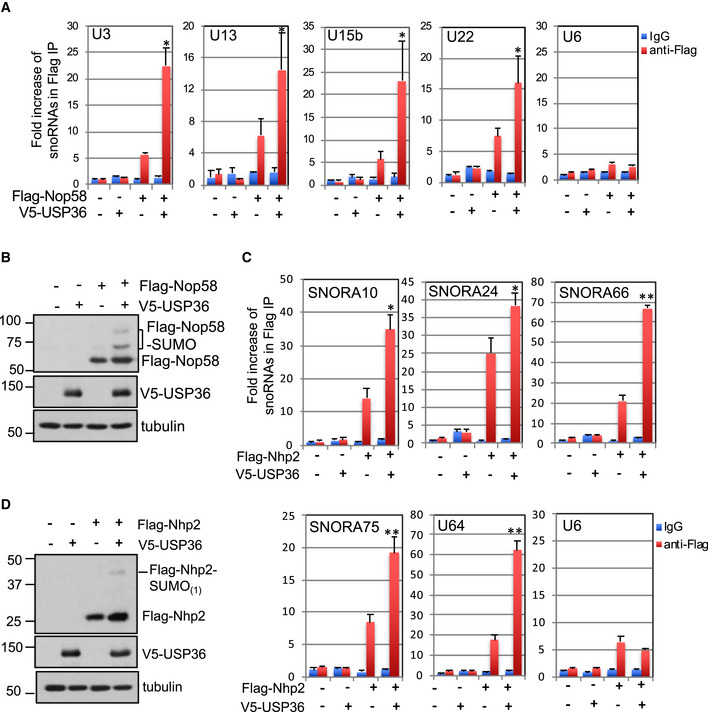
It has been previously shown that SUMOylation of Nop58 is critical for high‐affinity binding to snoRNAs (Westman et al, 2010). Thus, we next examined whether USP36‐mediated SUMOylation promotes the binding of Nop58 and Nhp2 to snoRNAs using RNA‐IP assays. As shown in Fig 6A and B, overexpression of USP36 significantly promoted the binding of Nop58 to the tested Box C/D class of snoRNAs, including U3, U13, U15b, and U22, but not control U6. Similarly, USP36 also significantly promoted the binding of Nhp2 to the tested box H/ACA class of snoRNAs, including SNORA10, SNORA24, SNORA66, SNORA75, and U64, but not U6 (Fig 6C and D), suggesting that USP36‐mediated SUMOylation plays an important role in the regulation of snoRNP association with snoRNAs and thus the snoRNP function in rRNA processing and ribosome biogenesis.
Figure 6. USP36 promotes Nop58 and Nhp2 binding to snoRNAs.
-
A, BUSP36 promotes the binding of Nop58 to box C/D snoRNAs. H1299 cells transfected with Flag‐Nop58 and V5‐USP36 individually or together were assayed by RNA‐IP with anti‐Flag or control mouse IgG, followed by RT–qPCR detection of the indicated box C/D snoRNAs (A). The expression of Nop58 and USP36 proteins is shown in (B). Shown is one representative experiment of three independent experiments. Data were presented as mean ± SD, n = 3 technical replicates. *P < 0.05, compared with Nop58 alone (Student's t‐test).
-
C, DUSP36 promotes the binding of Nhp2 to snoRNAs. H1299 cells transfected with Flag‐Nhp2 and V5‐USP36 individually or together were assayed by RNA‐IP with anti‐Flag or control mouse IgG, followed by RT–qPCR detection of the indicated box H/ACA snoRNAs (C). The expression of Nhp2 and USP36 proteins is shown in (D). Shown is one representative experiment of three independent experiments. Data were presented as mean ± SD, n = 3 technical replicates. *P < 0.05; **P < 0.01, compared with Nhp2 alone (Student's t‐test).
Source data are available online for this figure.
USP36 is critical for rRNA processing, translation, and cell growth
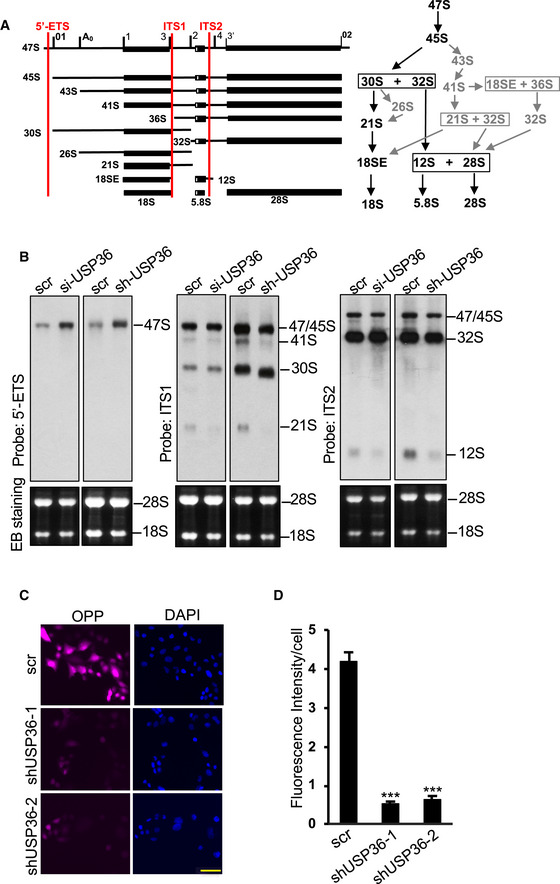
USP36 has been implicated in rRNA processing, yet the detailed role in this function has not been fully studied (Endo et al, 2009). Pre‐rRNA is processed to mature rRNAs via the major and the minor pathways as illustrated in Fig 7A. We examined the role of USP36 in rRNA processing using Northern blot with probes specifically hybridizing regions at the 5′‐external transcribed spacer (ETS), internal transcribed spacer 1 (ITS1), and ITS2 (Fig 7A) to detect different processed rRNA species. As shown in Fig 7B, knockdown of USP36 by either siRNA or shRNA lentiviruses (Sun et al, 2015) attenuated multiple steps of rRNA processing, leading to the profound accumulation of 47S and the reduction of 21S and 12S rRNA precursors. Consistently, knockdown of USP36 markedly inhibited protein translation in cells as determined by Click‐iT OPP protein synthesis assays (Fig 7C and D). Thus, USP36 is essential for ribosome biogenesis and protein translation.
Figure 7. USP36 is critical for rRNA processing and translation.
-
ADiagram of the pre‐rRNA processing showing the pre‐rRNA processing intermediate products and the location of probes (5′‐ETS, ITS1 and ITS2) used in Northern blot analysis (left) as well as the major (black) and minor (gray) processing pathways (right).
-
BUSP36 is required for rRNA processing. HeLa cells transfected with scr or USP36 siRNA (left panels) or infected with scr or USP36 shRNA lentiviruses (right panels) were assayed for rRNA processing by Northern blot using 5′‐ ETS, ITS1, and ITS2 probes as indicated.
-
C, DKnockdown of USP36 inhibits translation. HeLa cells infected with scr or USP36 shRNA lentiviruses were incubated with O‐propargyl‐puromycin (OPP) followed by Click‐iT OPP protein synthesis assays. Representative images (C) and quantification from three independent experiments (D). Data were presented as mean ± SD, n = 3 biological replicates. ***P < 0.001, compared with scr control (Student's t‐test). Scale bar = 50 μm.
Source data are available online for this figure.
To further test the role of USP36 in SUMOylation, translation, and cell growth, we generated immortalized USP36 +/+ ; Cre‐ER and USP36lox / lox; Cre‐ER mouse embryonic fibroblast (MEF) cells. Treatment of USP36lox / lox; Cre‐ER MEF cells with 4‐hydroxytamoxifen (4‐OHT) reduced protein SUMOylation (Fig EV5A) and markedly inhibited protein translation (Fig EV5B and C) and cell proliferation (Fig EV5D and E). The inhibition of protein translation upon depletion of mouse USP36 was significantly rescued by lentiviral‐mediated expression of WT human USP36, but not its DUB catalytically inactive C131A or H382A mutants (Fig EV5C and E). The efficient deletion of endogenous mouse USP36 and the exogenous expression of human USP36 were confirmed by reverse transcriptase‐quantitative polymerase chain reaction (RT–qPCR) (Fig EV5D) and immunoblot (Fig EV5C), respectively. Since the H382A mutant retains SUMO promoting activity, our results suggest that the role of USP36's SUMO promoting activity of the H382A mutant is likely overshadowed by the essential role of its DUB activity in translation and cell growth.
Figure EV5. Deletion of the USP36 gene inhibits translation and cell growth in MEF cells.
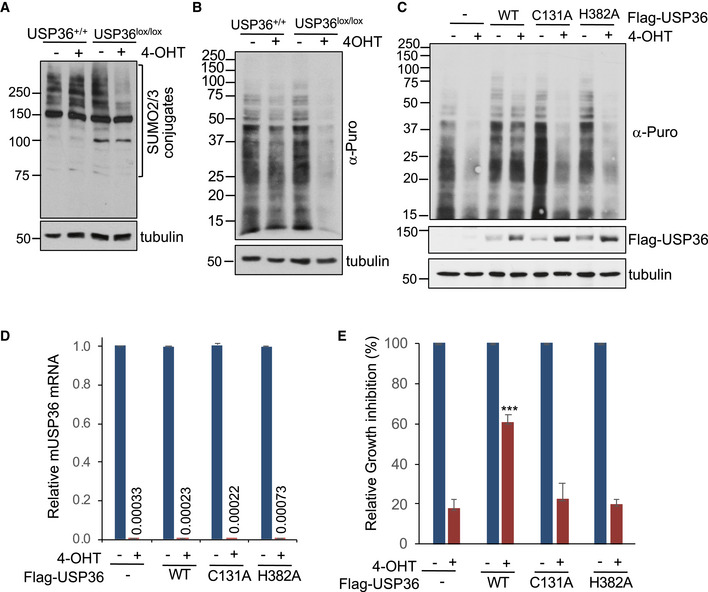
-
ADeletion of USP36 inhibits SUMOylation in MEF cells. USP36 +/+ ;Cre‐ER and USP36lox / lox;Cre‐ER MEF cells were treated with 1 mM 4‐OHT or vehicle control for 72 h and assayed for total SUMOylation by IB using anti‐SUMO2/3 antibodies.
-
BDeletion of USP36 inhibits translation in MEF cells. USP36 +/+ ;Cre‐ER and USP36lox / lox;Cre‐ER MEF cells were treated with 1 mM 4‐OHT or vehicle control for 72 h. The cells were then labeled with 10 μg/ml puromycin for 10 min followed by detection of total protein translation by IB using anti‐puromycin antibody.
-
C, DWT human UPS36 (hUSP36), but not its C131A and H382A mutants, rescues the inhibition of translation by the deletion of endogenous mouse USP36 (mUSP36). USP36lox / lox;Cre‐ER MEF cells stably expressing control, WT hUSP36, C131A, or the H382A mutant were treated with 1 mM 4‐OHT or vehicle control for 72 h. The cells were then labeled with 10 μg/ml puromycin for 10 min followed by detection of total protein translation by IB using anti‐puromycin antibody. The expression of exogenous Flag‐hUSP36 was detected by IB with anti‐Flag antibody (C). The depletion of endogenous mUSP36 mRNA was detected by RT–qPCR (D). The numbers are average ratios of mUSP36 mRNA expression in cells treated with 4‐OHT/ that in cells treated with vehicle control. Data were presented as mean ± SD, n = 3 biological replicates.
-
EWT human UPS36, but not its C131A and H382A mutants, rescues the cell growth inhibition by the deletion of endogenous mUSP36. USP36lox / lox;Cre‐ER MEF cells stably expressing control, Flag‐hUSP36 (WT, C131A, or the H382A mutant) using lentiviral expression were treated with 1 mM 4‐OHT or vehicle control and cultured for 96 h. The percentile inhibition of cell proliferation was determined by cell counting using Countess II (Life Technologies) and compared cells treated with 4‐OHT to cells with vehicle control. Data were presented as mean ± SD, n = 4 biological replicates. ***P < 0.001; compared with empty vector control infected cells treated with 4‐OHT (Student's t‐test).
Source data are available online for this figure.
Discussion
USP36 is a member of ubiquitin‐specific protease family that removes ubiquitin from ubiquitinated substrates. The main function of USP36 is so far attributed to its nucleolar role in ribosome biogenesis by deubiquitinating and stabilizing several proteins critical for ribosome biogenesis, including NPM (Endo et al, 2009), DHX33 (Fraile et al, 2018) and the yeast RNA Pol I subunit Rpa190 (human RPA194) (Richardson et al, 2012), as well as c‐Myc (Sun et al, 2015), a master regulator of ribosome biogenesis (van Riggelen et al, 2010). In addition, USP36 has been shown to deubiquitinate H2B (DeVine et al, 2018), regulate autophagy (Taillebourg et al, 2012), and play a role in oxidative stress response by deubiquitinating mitochondria protein SOD2 (Kim et al, 2011). Like many nucleolar proteins critical for ribosome biogenesis (Newton et al, 2003; Grisendi et al, 2005; Zhu et al, 2006; Blomen et al, 2015), USP36 knockout is embryonic lethal in mice (Fraile et al, 2018) and ablation of USP36 expression in various cell lines impairs cell growth and proliferation (Sun et al, 2015; Fraile et al, 2018). However, the detailed mechanism underlying the essential role for USP36 in ribosome biogenesis and cell growth is not well understood. Our finding here showing that USP36 promotes snoRNP protein group SUMOylation thus reveals a novel mechanism for its role in ribosome biogenesis.
Ribosome biogenesis‐related proteins are among the major group of functionally interconnected SUMO targets in cells (Hendriks & Vertegaal, 2016), and SUMOylation plays a key role in ribosome biogenesis in the nucleolus (Panse et al, 2006; Finkbeiner et al, 2011; Raman et al, 2016). Yet, how these proteins are SUMOylated in the nucleolus remains to be understood. In this study, we show that USP36 associates with both box C/D and Box H/ACA snoRNPs and promotes SUMOylation of two proteins in each complex (Nop58 and Nop56, Nhp2 and DKC1, respectively), demonstrating that USP36 promotes snoRNP group SUMOylation. Our results support the recently emerged view that SUMOylation tends to modify multiple proteins in functional protein complexes such as DNA damage repair proteins (Psakhye & Jentsch, 2012), telomeres (Potts & Yu, 2007; Ferreira et al, 2011; Hang et al, 2011), and ribosome biogenesis‐related proteins (Panse et al, 2006; Finkbeiner et al, 2011; Castle et al, 2012). Interestingly, the snoRNPs are quite stable and neither USP36 overexpression nor its knockdown significantly affected the levels of snoRNP proteins in cells. Consistently, endogenous Nop58 and Nhp2 do not undergo significant ubiquitination (Fig EV4I), Thus, USP36 mainly functions to mediate Nop58 and Nhp2 SUMOylation to regulate their activity, but not levels. It is possible that under normal conditions, USP36 acts mainly to SUMOylate nucleolar proteins and regulate their function, whereas its DUB activity may play a role in maintaining these proteins assembled in multi‐protein complexes in their deubiquitinated state in the nucleolus. SUMOylation of Nop58 has been shown to be essential for its binding to snoRNAs (Westman et al, 2010). Consistently, we show that USP36 augments the binding of Nop58 and Nhp2 to respective snoRNAs. Therefore, USP36 might be essential for high‐affinity binding to snoRNAs in the nucleolus by promoting snoRNP group SUMOylation and thus critical for snoRNP biogenesis, rRNA modifications, and rRNA processing. Indeed, knockdown of USP36 markedly impairs multiple steps of rRNA processing and protein translation (Fig 7).
Mechanistically, we show that USP36 may possess a novel SUMO ligase activity. SUMO ligases act as scaffolds to facilitate the transfer of SUMO from charged Ubc9 to the substrates requiring binding to Ubc9 (E2), SUMO, and substrates (Bernier‐Villamor et al, 2002; Yunus & Lima, 2009; Guervilly et al, 2015; Streich & Lima, 2016; Pichler et al, 2017; Varejao et al, 2020). We show that USP36 binds to both Ubc9 (E2) and its substrates Nop58 and Nhp2. It also interacts with SUMO2 in cells and in vitro. Importantly, USP36 directly promotes SUMOylation of multiple substrates including Nop58, Nhp2, and PARP1 in vitro using recombinant proteins (Figs 3E and 4F–H). Thus, our data suggest that USP36 acts like a SUMO ligase to mediate nucleolar protein SUMOylation. Biochemical and structural analyses have revealed the action mechanism underlying the three families of bona fide SUMO E3s. SP‐RING family SUMO E3s utilize the SP‐RING domain similar to ubiquitin E3 RING domains and adjacent SP C‐terminal domain (SP‐CTD) containing SIM‐like motifs (Yunus & Lima, 2009; Gareau & Lima, 2010). The RanBP2 SUMO E3 activity was mapped to an internal repeat (IR)1‐M‐IR2 region (Pichler et al, 2002; Pichler et al, 2004), whereas the ZNF451 family E3s utilize their N‐terminal tandem SIM region called SIM1‐M‐SIM2 motif (Cappadocia et al, 2015; Eisenhardt et al, 2015). These SUMO E3 domains mediate interaction with SUMO and Ubc9 and promote the transfer of SUMO from the SUMO charged Ubc9 to the substrates. Of note, while RanBP2 directly interacts with the backside of Ubc9 (Eisenhardt et al, 2019), many SUMO E3s such as PIAS1 (Eisenhardt et al, 2019) and ZNF451 (Cappadocia et al, 2015; Eisenhardt et al, 2015; Koidl et al, 2016; Eisenhardt et al, 2019) family members require the SUMO–Ubc9 backside interaction for their SUMO E3 activity. We show that the full SUMO promoting activity of USP36 also requires the SUMO–Ubc9 backside interaction, as disruption of the interaction by either the SUMO2D63R or the Ubc9F22A mutant suppressed the activity of USP36 to promote poly‐SUMOylation, further supporting USP36 as a SUMO E3, although it remains possible that USP36 acts as a cofactor to enhance SUMOylation (Eisenhardt et al, 2019). Intriguingly, both the DUB and SUMO promoting activities are located to the N‐terminal region of USP36. Yet, the SUMO promoting activity is independently of its DUB activity as the H382A mutant lacking DUB function retains the SUMO promoting activity (Fig 2). Therefore, we can differentiate the two activities by this single point mutation. As mutating the DUB catalytic residue C131 also abolished USP36's SUMO promoting activity, C131 might be structurally required for the SUMO E3. Future structural studies would clarify whether USP36 is a bona fide SUMO E3, the mechanisms underlying the potential switches between its DUB and SUMO E3 activities, and aid in finding USP36 mutants that specifically inactivate SUMO E3, but not DUB. Such mutants would be critical for elucidating the SUMO E3‐specific function in ribosome biogenesis and cell growth, given that USP36H382A failed to rescue the inhibition of protein translation and cell growth by the deletion of endogenous USP36 (Fig EV5).
As USP36 increases the total levels of nucleolar SUMOylation (Figs 1E and EV1I), it is conceivable that USP36 may promote SUMOylation of other nucleolar proteins associated with ribosome biogenesis and act as a central regulatory hub for the nucleolar SUMOylation to regulate ribosome biogenesis and translation. Further proteomic analyses are warranted to identify additional USP36 SUMO targets in the nucleolus. As hyperactivation of ribosome biogenesis is tightly linked to human cancers (Mills & Green, 2017; Pelletier et al, 2018) and USP36 itself is overexpressed in various human cancers (Sun et al, 2015), USP36 could be a promising therapeutic target in human cancers.
Materials and Methods
Plasmids and recombinant proteins
V5‐tagged USP36 plasmid were described previously (Sun et al, 2015). The full‐length USP36 cDNAs (WT and the C131A mutant) were also cloned into the pcDNA3‐2Flag vector to generate Flag‐tagged USP36 plasmids. Flag‐USP361–420, Flag‐USP36421–800, Flag‐USP36801–1121, and Flag‐USP361–800 deletion mutants were also described (Sun et al, 2015). The above cDNAs were also cloned into pGEX‐4T.1 vector (GE Healthcare) to express GST‐USP361–420, GST‐USP36421–800, GST‐USP36801–1121, GST‐USP361–800 fusion proteins. His‐tagged SUMO1, SUMO2, and SUMO3 were cloned by PCR into pcDNA3‐His vector. Flag‐USPL1 was described (Sun et al, 2018). Nhp2 cDNA was amplified from HeLa cDNA and cloned into pcDNA3‐2Flag to generate Flag‐Nhp2 plasmid. Flag‐Nhp2K5R was generated by site‐directed mutagenesis using QuikChange Kit (Agilent). Flag‐Nop58 (WT and K467R/K487R (2KR) mutant) plasmids were cloned into pcDNA3‐2Flag vector by PCR using pcDNA3‐His‐Nop58 (WT and 2KR mutant) (provided by Dr. Angus I. Lamond, University of Dundee, UK) (Westman et al, 2010) as templates. Nhp2 and Nop58 cDNAs were also subcloned into pGEX‐4T.1 to generate GST‐Nhp2 and GST‐Nop58 plasmids for recombinant proteins expression in bacteria. USP361–800 cDNA was subcloned into pET30a vector for bacterial expression of His‐tagged proteins. Flag‐Nop56, Flag‐Nhp2L1, Flag‐DKC1, Flag‐GAR1, Flag‐Nop10, and Flag‐Ubc9 plasmids were cloned into pcDNA3‐2Flag vector by PCR. Lentiviral vectors expressing Flag‐USP36, pLenti4‐Flag‐USP36WT, and pLenti4‐Flag‐USP36C131A were described previously (Sun et al, 2015). pLenti4‐Flag‐USP36H382A was generated by site‐directed mutagenesis.
All the plasmids were confirmed by sequencing.
Recombinant proteins were expressed in Escherichia coli (BL21) by induction with Isopropyl β‐d‐1‐thiogalactopyranoside (IPTG) and purified using glutathione agarose for GST‐fusion proteins and Ni2+‐NTA agarose beads for His‐tagged proteins. Recombinant SUMO E1 (SAE1/SAE2), Ubc9, and T7‐tagged PARP1 were kindly provided by Dr. Yoshi (Ryu & Azuma, 2010; Ryu et al, 2010a). Ubc9 and SUMO2 cDNAs were also subcloned into the pProxEX HTa vector (Life technologies) to generate pProxEX‐His‐Ubc9 and pProxEX‐His‐SUMO2 vectors. His‐Ubc9F22A and His‐SUMO2D63R mutant plasmids were generated by site‐directed mutagenesis. The expression and purification of these proteins were described (Ryu & Azuma, 2010; Ryu et al, 2010a; Sun et al, 2015).
Antibodies and reagents
Anti‐Flag (M2, F3165, Sigma), anti‐V5 (R960‐25, Life technologies), anti‐Nop58 (A302‐719A, Bethyl), anti‐Nhp2 (ab180498, abcam), anti‐B23 (19‐7288, Zymed), anti‐Nop56 (A302‐720, Bethyl), anti‐FBL (sc‐25397, Santa Cruz Biotech), anti‐Nhp2L1 (A304‐030A, Bethyl), anti‐DKC1 (ab64667, abcam), anti‐Gar1 (A12748, ABclonal), anti‐Nop10 (ab134902, abcam), anti‐GST (A00865, GenScript), anti‐Ubc9 (4918, Cell Signaling), anti‐SAE1 (13585, Cell Signaling), anti‐SAE2 (A302‐926A, Bethyl), anti‐T7‐HRP (69048, Millipore), and anti‐puromycin (clone 12D10, MABE343, EMD Millipore) antibodies were purchased. Rabbit polyclonal anti‐SUMO1 and anti‐SUMO2/3 antibodies were provided by our collaborator Dr. Yoshiaki Azuma (University of Kansas). Rabbit anti‐USP36 serum was provided by Dr. Masayuki Komada (Tokyo Institute of Technology, Japan) (Endo et al, 2009; Sun et al, 2015). Puromycin (Invitrogen) and 4‐OHT (Sigma) were purchased.
Cell culture, transfection, immunoblot (IB), and co‐immunoprecipitation (co‐IP) analyses
Human H1299, HeLa, U2OS, and 293 cells (from ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin and 0.1 mg/ml streptomycin at 37°C in a 5% CO2 humidified atmosphere. Cells transfection were performed using TransIT®‐LT1 (Mirus Bio Corporation) or Lipofectamine 2000 (Life technologies) reagents following the manufacturers' protocol. Cells were harvested at 36–48 h post‐transfection and lysed in lysis buffer consisting of 50 mM Tris–HCl (pH 8.0), 0.5% Nonidet P‐40, 1 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM DTT, 1 µg/ml pepstatin A, and 1 mM leupeptin. Equal amounts of cell lysates were used for IB analysis. Co‐IP was conducted as described previously (Sun et al, 2015; Sun et al, 2018). Bound proteins were detected by IB using antibodies indicated in figure legends.
Gene knockdown by RNA interference
Lentiviral vectors encoding shRNAs against USP36 were purchased (Open Biosystems). The shRNA sequences are 5′‐GCGGTCAGTCAGGATGCTATT‐3′ (shRNA‐1, used for all experiments, except where indicated) and 5′‐CGTCCGTATATGTCCCAGAAT‐3′ (shRNA‐2). The plasmids were transfected with VSVG, pLP1, pLP2 plasmids into 293FT cells using calcium chloride (Promega). The viruses were then used to infect cells in the presence of polybrene (6 µg/ml). The cells were harvested at 72 h post‐transduction for IB analysis. For siRNA‐mediated USP36 knockdown, the 21‐nucleotide siRNA duplexes with a 3′ dTdT overhang were synthesized by Dharmacon Inc (Lafayette, CO). The target sequence for USP36 is 5′‐TGTCCTGAGTGGAGAGAAT‐3′. The control scrambled RNA sequence has been described previously (Sun et al, 2012). These siRNA duplexes (100 nM) were introduced into cells using Lipofectamine 2000 (Life technologies) following the manufacturer's protocol.
In vivo SUMOylation assays
In vivo SUMOylation assays under denaturing conditions was conducted using a Ni2+‐NTA pull‐down method. Briefly, cells transfected with His‐SUMO and indicated plasmids were lysed in 8 M urea lysis buffer containing 100 mM NaH2PO4, 10 mM Tris–HCl pH8.0, 250 mM NaCl, 5% Glycerol, 0.1% Triton X‐100, 10 mM imidazole and 10 mM β‐mercaptoethanol (β‐ME), followed by brief sonication to reduce viscosity. The cell lysates were then incubated with Ni2+‐NTA agarose beads for 4 h at room temperature. The beads were collected by spinning at 750 g for 5 min, washed with 8 M urea lysis buffer containing 20 mM imidazole, and eluted in elution buffer containing 150 mM Tris–HCl (pH 6.7), 150 mM NaCl, 10% Glycerol, 5% SDS, 300 mM imidazole, and 0.72 M β‐ME. The elution was boiled in 1× SDS sample buffer and assayed by IB.
In vitro SUMOylation assays
In vitro SUMOylation assays were carried out as described previously with minor modification (Ryu & Azuma, 2010; Ryu et al, 2010a). In brief, reaction buffer consists of 100 mM NaCl, 5 mM MgCl2, 20 mM HEPES (pH 7.6), 5% glycerol, and 1 mM DTT. Unless otherwise specified, the in vitro reactions contained SUMO E1 heterodimer, Ubc9 (E2), USP361–800, processed form of SUMO2, ATP, and substrates at the final concentration of 30 nM, 50 nM, 50 nM, 4 μM, 2.5 mM, and 0.1 μM, respectively. The reactions were incubated at 30°C for 2 h or different times as indicated and stopped by adding an equal volume of 2× SDS sample buffer, followed by IB.
Nucleolar fractionation
Nucleolar fractionation was performed as described previously (Challagundla et al, 2011; Sun et al, 2015). Briefly, freshly harvested cells were washed with 1× PBS, resuspended in hypotonic buffer A (10 mM HEPES pH7.8, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT) in the presence of PMSF and incubated for 10 min on ice. The cells were homogenized using B pestle douncer followed by spinning down at 228 g for 5 min at 4°C. The supernatant (cytoplasmic fraction) was supplemented with 1/10 volume of buffer B (0.3 M Tris–HCl pH 7.8, 1.4 M KCl, 30 mM MgCl2). The nuclear pellets were washed with buffer A and then resuspended in buffer S1 (0.25 M sucrose, 10 mM MgCl2), layered over buffer S2 (0.35 M sucrose, 0.5 mM MgCl2), and centrifuged at 1,430 g for 10 min at 4°C. The pelleted nuclei were resuspended in buffer S2 with PMSF and sonicated using a microtip probe at power setting of 50%. The sonicated nuclei were then layered over buffer S3 containing 0.88 M sucrose and 0.5 mM MgCl2 and centrifuged at 3,000 g for 10 min at 4°C. The pellet contained purified nucleoli and the supernatant represented the nucleoplasm. The nucleoli were then lysed in high salt RIPA buffer containing 50 mM Tris pH 7.5, 500 mM NaCl, 1% Nonidet P‐40, 0.5% deoxycholate, and protease inhibitors in the presence of 80 U/ml DNase I on ice for 15–30 min. The lysates were then added with 2× volume of RIPA buffer without salt, left on ice for an additional 10 min, followed by centrifugation at maximal speed for 15 min. The supernatant was collected as soluble nucleolar fraction for IP analysis (Sun et al, 2015).
Glutathione S‐transferase‐fusion protein association assays
Poly‐SUMO2 chains were purchased from Boston Biochem. GST‐fusion protein–protein association assays were conducted as described (Tatham et al, 2008; Guervilly et al, 2015). Briefly, SUMO2 chains (200 ng) were incubated with the glutathione Sepharose 4B beads (Sigma) containing 1.5 μg of GST or GST‐USP36 fragments in SUMO binding buffer containing 50 mM Tris–HCl (pH 7.4), 250 mM NaCl, 0.1% NP‐40, 2 mM DTT, and 5% glycerol at 4°C for 2 h. After wash, bound proteins were analyzed using IB with anti‐SUMO2 and anti‐GST antibodies.
Mass spectrometry
H1299 cells transfected with His‐SUMO2 with or without Flag‐USP36 were lysed in 8 M urea lysis buffer, and total SUMOylated proteins were purified using the Ni2+‐NTA agarose bead pull‐down method as described above. The eluted proteins were analyzed by mass spectrometry to identify the SUMOylated proteins. Briefly, samples were run into a SDS–PAGE gel for 6 min, the gel stained, protein bands at the top of the gel excised, reduction, and alkylation of cysteines performed using dithiothreitol and iodoacetamide, respectively, and proteins in‐gel trypsinized as previously described (Ritchie et al, 2015). Dried peptides were then dissolved in 5% formic acid, injected onto a 75 µm × 250 mm nano C18 column via a peptide trap, and data‐dependent MS/MS data for peptides were acquired using a LTQ Velos linear ion trap (Thermo Scientific) as before (Ritchie et al, 2015), except using a 7.5–30% acetonitrile gradient over 195 min. MS/MS data were then searched using SEQUEST vs 28 (rev. 12) against a human database containing both Swissprot and TrEMBL sequences, plus 179 common contaminates and the reversed forms of all those proteins (269,858 total sequences). A static modification +57 was specified on Cys, and a dynamic modification of +16 on Met. Average parent ion mass tolerance was 2.5 Da, and monoisotopic fragment ion mass tolerance was 1.0 Da, with trypsin specified as the enzyme. A linear discriminant transformation was then used to improve the identification sensitivity from the SEQUEST analysis (Keller et al, 2002; Elias & Gygi, 2007; Wilmarth et al, 2009), score histograms of forward and reverse sequence entries generated, and matches to forward sequence proteins filtered as previously described (Ritchie et al, 2015), except a protein false discovery rate under 1% was used. After removal of contaminates, this resulted in the identification of 203 proteins. Estimation of protein abundance differences in the His‐SUMO2 pulldowns with or without Flag‐USP36 was performed using the numbers of assigned MS/MS spectra to each protein across the two samples.
Immunofluorescence staining
Cells were fixed and stained with monoclonal anti‐B23 and polyclonal anti‐Nop58 or anti‐Nhp2 antibodies followed by staining with Alexa Fluor 488 (green) goat anti‐mouse antibody and Alexa Fluor 546 (red) goat anti‐rabbit antibody (Life technologies) as well as DAPI for DNA staining. Stained cells were analyzed under a Leica inverted fluorescence microscope.
RNA‐IP
Cells were lysed in HNTG buffer containing 20 mM HEPES pH 7.9, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 10% glycerol, 1% Triton X‐100 in the presence of EDTA‐free complete protease inhibitor cocktail (Roche), and 20 U/ml RNase inhibitor (Invitrogen) for 30 min, briefly sonicated, and centrifuged at 15,000 g for 10 min at 4°C. The supernatants were pre‐cleared with protein G beads for 30 min, followed by incubation with anti‐Flag (M2)‐conjugated beads (Sigma) or control IgG coated beads for 4 h at 4°C. After wash with lysis buffer for four times, the beads were suspended in 100 μl NT2 buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% NP‐40) containing 10 U DNase I and incubate at 37°C for 10 min. RNAs were then extracted using TRIzol‐reagent (Life technologies) and subjected to reverse transcription (RT) using iScript cDNA synthesis kit (Bio‐Rad), followed by qPCR analysis.
Reverse transcriptase‐quantitative polymerase chain reaction
Quantitative real‐time PCR was performed on an ABI StepOne™ real‐time PCR system (Applied Biosystems) using SYBR Green Mix (Thermo Fisher Scientific). All reactions were carried out in triplicate. Relative gene expression was calculated using the ΔCτ method following the manufacturer's instruction. The primers used were as follows: 5′‐CTCGCTTCGGCAGCACA‐3′ and 5′‐AACGCTTCACGAATTTGCGT‐3′ for U6; 5′‐CGTGTAGAGCACCGAAAACC‐3′ and 5′‐CACTCAGACCGCGTTCTCTC‐3′ for U3; 5′‐TTCATGAGCGTGATGATTGG‐3′ and 5′‐GTAATGTGCCCACGTCGTAA‐3′ for U13; 5′‐CAGTGATGACACGATGACGA‐3′ and 5′‐GGACACTTCTGCCAAAGGAA‐3′ for U15b; 5′‐TTTCACATGTCTTACTCTCTGTCC‐3′ and 5′‐CCTCAGACAGTTCCTTCTGGA‐3′ for U22; 5′‐TCTCAGCTCCGCTTAACCA‐3′ and 5′‐CGTGCATTAGGAGAGCCTTT‐3′ for SNORA10; 5′‐TCCATGTATCTTTGGGACCTG‐3′ and 5′‐TGGTGACAGCTTTGCCAATA‐3′ for SNORA24; 5′‐GCAAACTCGATCACTAGCTCTG‐3′ and 5′‐ACTTTTGCAAACCTGGTTCC‐3′ for SNORA66; 5′‐TCACCCGTGTGACTTTCGTA‐3′ and 5′‐ACTTTTGCAAACCTGGTTCC‐3′ for U64; 5′‐TCTTCTCATTGAGCTCCTTTCTG‐3′ and 5′‐TTCTTCTCGTGCGAATCCAT‐3′ for SNORA75; 5′‐CCATTGATGCGATGCAGAAG‐3′ and 5′‐ TGTCTGCCGATCCAACTTAGC‐3′ for mouse USP36; 5′‐ CATGGCCTTCCGTGTTCCTA‐3′ and 5′‐CCTGCTTCACCACCTTCTTGAT‐3′ for mouse GAPDH.
Translation
Translation in individual cells was measured by using Click‐iT® Plus OPP Protein Synthesis Assay Kit (Invitrogen) following the manufacturer's protocol. Briefly, cells were incubated with 20 μM O‐propargyl‐puromycin (OPP) for 30 min, fixed in 3.7% formaldehyde in PBS for 15 min, and then permeabilized in 0.5% Triton X‐100 in PBS for 15 min at room temperature. The cells were then incubated with Click‐iT® Plus OPP reaction cocktail at room temperature for 30 min for “click” reaction between the Alexa Fluor® picolyl azide and the alkyne OPP. After wash, the cells were counterstained with NuclearMask™ Blue Stain, mounted with antifade mountant (Life technologies), and imaged by fluorescence microscope (Zeiss Apotome) for the fluorescent labeled newly synthesized peptides. The intensity of the fluorescent OPP stain in single cells was quantified by image J. For detecting global protein translation, Cells were pre‐treated with 10 μg/ml puromycin (Invitrogen) for 10 min for puromycylation of nascent peptides. Cell lysates were then assayed by IB using anti‐puromycin as previously described (Schmidt et al, 2009; David et al, 2012).
Northern blot
rRNA processing was examined using non‐radioactive Northern blot method as described (Tafforeau et al, 2013; Wu et al, 2013; Sharma et al, 2015). Briefly, a total of 5 μg of total RNAs was loaded onto agarose denaturing gels (6% formaldehyde/1.2% agarose in HEPES‐EDTA buffer) and electrophoresed for 4 h at 75 V. After wash, gels were transferred to nylon membranes by capillarity overnight in 10× saline sodium citrate (SSC). Membranes were UV cross‐linked (120 mJ/cm2), followed by prehybridization in 50% formamide, 5× SSPE, 5× Denhardt's solution, 1% w/v SDS, 200 μg/ml fish sperm DNA solution (Sigma) for 1 h at 65°C. The digoxigenin‐labeled oligonucleotide probe was then added and incubated for 1 h at 65°C and then at 37°C overnight. The sequence of Northern blot probes are as follows: 5′‐ CGGAGGCCCAACCTCTCCGACGACAGGTCGCCAGAGGACAGCGTGTCAGC‐3′ (5′‐ETS); 5′‐CCTCGCCCTCCGGGCTCCGGGCTCCGTTAATGATC‐3′ (ITS1); 5′‐ CTGCGAGGGAACCCCCAGCCGCGCA‐3′ (ITS2) (Tafforeau et al, 2013). After washed with 2× SSC, membranes were blocked in 1× blocking buffer (Roche) for 0.5 h at room temperature and incubated with anti‐digoxigenin antibody (Roche, 1:10,000 dilution) for 0.5 h at room temperature, followed by washing steps (twice, each 15 min) with washing buffer (0.1 M maleic acid, 0.15 M NaCl at pH 7.5, 0.3% Tween 20 [v/v]). After equilibration in detection buffer (0.1 M Tris, 0.1 M NaCl at pH 9.5), membranes were incubated with chemiluminescent substrate CDP star ® (Roche, 1:200 dilution) at room temperature for 10 min and then exposed to films.
Generation of mouse embryonic fibroblast cell lines
ES cell clones containing the knockout‐first (targeted) USP36 alleles (USP36tm1a) were purchased from the European Mutant Mouse Archive (http://www.knockoutmouse.org/martsearch/project/25050) (Testa et al, 2004). The USP36tm1a allele generated through homologous recombination contains a β‐gal‐neo expression cassette inserted in intron 5 of the USP36 gene. The advantage of this targeting is that removal of the β‐gal‐neo cassette by Flp converts the targeted allele into a conditional allele (USP36tm1c, named USP36lox in this study) such that Cre recombination removes exon 6, leading to deletion of most of the USP36 protein (1,098 amino acids), possibly making a truncated peptide containing only the N‐terminal 218 amino acids (deletion allele, USP36tm1d). The ES cells were used to generate chimeric mice as described (Fedorov et al, 1997). The chimeric mice were then crossed with C57BL/6 mice to generate USP36tm1a /+ mice. Heterozygous USP36tm1a /+ mice were subsequently crossed with C57BL/6a mice expressing a flipase recombinase, Tg(ACTFLPe) (Flp mice, Jackson laboratory stock no: 003800), which recognizes the FRT sites and excises the β‐gal‐neo expression cassette, to generate conditional USP36lox /+ and USP36lox / lox mice. The USP36lox / lox mice were crossed with the inducible Tg(UbC‐cre/ERT2) Cre expressing mice (Jackson laboratory stock no: 007001(Ruzankina et al, 2007) to generate USP36 lox /+ ; Cre‐ER mice. All mice are C57BL/6 background. Mice were handled in accordance with the OHSU Institutional Animal Care and Use Committee (IACUC). Primary MEF cells were isolated from E13.5 embryos resulting from the USP36 lox /+ ; Cre‐ER x USP36 lox /+ ; Cre‐ER crossings. Briefly, single embryos were separated by removing placental and any maternal tissues and then dissected by removing head as well as heart and liver red tissues. The heads were used for genotyping. Embryos were minced with razor blade in 1ml 0.05% trypsin and incubated in incubator at 37°C for 30 min. DMEM medium supplemented with 15% FBS, and 50 U/ml penicillin and 0.1 mg/ml streptomycin were added to quench trypsin followed by culture for 2–3 days to allow cells to grow to confluency. The resulting USP36 +/+ and USP36lox / lox MEFs were then immortalized by SV40 T antigen.
Statistical analysis
Unless otherwise stated in the figure legend, all the statistical differences were analyzed using a standard Student's t‐test analysis from three independent experiments. P < 0.05 was considered statistically significant.
Author contributions
M‐SD and HR conceived the project and wrote the manuscript. M‐SD supervised the research. HR, X‐XS, YC, YL, XW, H‐MZ, RSD, JK, LD, LMF, and M‐SD performed the experiments. YA provided key reagents. RCS provided scientific input for the project.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Dataset EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We thank Dr. Lionel Tafforeau (University of Mons, Belgium) and Dr. Jingyan Wu (Stanford University, San Francisco, USA) for sharing their Northern blot protocol and expertise and Dr. Masayuki Komada (Tokyo Institute of Technology, Japan) for providing reagents. We gratefully acknowledge OHSU Transgenic Core for generating the USP36 knockout mice and Mr. John Klimek for assistance with the mass spectrometry analysis. We thank the members from the Dai and Sears laboratories for active discussion. This work was supported by grants from NIH (R01 CA160474 and R01 GM130604 to M.‐S.D. and R01 CA186241 to M.‐S.D. and R.C.S.). Proteomic analysis was partially supported by NIH grants P30 EY010572, P30 CA069533, and S10 RR025571.
EMBO reports (2021) 22: e50684.
Data availability
The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD022987 (https://www.ebi.ac.uk/pride/archive/projects/PXD022987).
References
- Amente S, Lavadera ML, Palo GD, Majello B (2012) SUMO‐activating SAE1 transcription is positively regulated by Myc. Am J Cancer Res 2: 330–334 [PMC free article] [PubMed] [Google Scholar]
- Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ (2005) Nse2, a component of the Smc5‐6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol 25: 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Jentsch S (2009) Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458: 461–467 [DOI] [PubMed] [Google Scholar]
- Berkholz J, Michalick L, Munz B (2014) The E3 SUMO ligase Nse2 regulates sumoylation and nuclear‐to‐cytoplasmic translocation of skNAC‐Smyd1 in myogenesis. J Cell Sci 127: 3794–3804 [DOI] [PubMed] [Google Scholar]
- Bernier‐Villamor V, Sampson DA, Matunis MJ, Lima CD (2002) Structural basis for E2‐mediated SUMO conjugation revealed by a complex between ubiquitin‐conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 [DOI] [PubMed] [Google Scholar]
- Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A et al (2015) Gene essentiality and synthetic lethality in haploid human cells. Science 350: 1092–1096 [DOI] [PubMed] [Google Scholar]
- Capili AD, Lima CD (2007) Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2‐Ubl interaction. J Mol Biol 369: 608–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadocia L, Pichler A, Lima CD (2015) Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat Struct Mol Biol 22: 968–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle CD, Cassimere EK, Denicourt C (2012) LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol Biol Cell 23: 716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challagundla KB, Sun XX, Zhang X, DeVine T, Zhang Q, Sears RC, Dai MS (2011) Ribosomal protein L11 recruits miR‐24/miRISC to repress c‐Myc expression in response to ribosomal stress. Mol Cell Biol 31: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Suzuki H, Kobayashi Y, Wang AC, DiMaio F, Kawashima SA, Walz T, Kapoor TM (2018) Structural insights into Mdn1, an essential AAA protein required for ribosome biogenesis. Cell 175: 822–834.e818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chymkowitch P, Nguea PA, Enserink JM (2015) SUMO‐regulated transcription: challenging the dogma. BioEssays 37: 1095–1105 [DOI] [PubMed] [Google Scholar]
- David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, Bennink JR, Yewdell JW (2012) Nuclear translation visualized by ribosome‐bound nascent chain puromycylation. J Cell Biol 197: 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- DeVine T, Sears RC, Dai MS (2018) The ubiquitin‐specific protease USP36 is a conserved histone H2B deubiquitinase. Biochem Biophys Res Commun 495: 2363–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis‐Sandoval F, Poirier M, Scott MS (2015) The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA 6: 381–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhardt N, Chaugule VK, Koidl S, Droescher M, Dogan E, Rettich J, Sutinen P, Imanishi SY, Hofmann K, Palvimo JJ et al (2015) A new vertebrate SUMO enzyme family reveals insights into SUMO‐chain assembly. Nat Struct Mol Biol 22: 959–967 [DOI] [PubMed] [Google Scholar]
- Eisenhardt N, Ilic D, Nagamalleswari E, Pichler A (2019) Biochemical characterization of SUMO‐conjugating enzymes by in vitro sumoylation assays. Methods Enzymol 618: 167–185 [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP (2007) Target‐decoy search strategy for increased confidence in large‐scale protein identifications by mass spectrometry. Nat Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Endo A, Matsumoto M, Inada T, Yamamoto A, Nakayama KI, Kitamura N, Komada M (2009) Nucleolar structure and function are regulated by the deubiquitylating enzyme USP36. J Cell Sci 122: 678–686 [DOI] [PubMed] [Google Scholar]
- Fedorov LM, Haegel‐Kronenberger H, Hirchenhain J (1997) A comparison of the germline potential of differently aged ES cell lines and their transfected descendants. Transgenic Res 6: 223–231 [DOI] [PubMed] [Google Scholar]
- Ferreira HC, Luke B, Schober H, Kalck V, Lingner J, Gasser SM (2011) The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat Cell Biol 13: 867–874 [DOI] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Muller S (2011) The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J 30: 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile JM, Campos‐Iglesias D, Rodriguez F, Astudillo A, Vilarrasa‐Blasi R, Verdaguer‐Dot N, Prado MA, Paulo JA, Gygi SP, Martin‐Subero JI et al (2018) Loss of the deubiquitinase USP36 destabilizes the RNA helicase DHX33 and causes preimplantation lethality in mice. J Biol Chem 293: 2183–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss‐Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP (2005) Role of nucleophosmin in embryonic development and tumorigenesis. Nature 437: 147–153 [DOI] [PubMed] [Google Scholar]
- Guervilly JH, Takedachi A, Naim V, Scaglione S, Chawhan C, Lovera Y, Despras E, Kuraoka I, Kannouche P, Rosselli F et al (2015) The SLX4 complex is a SUMO E3 ligase that impacts on replication stress outcome and genome stability. Mol Cell 57: 123–137 [DOI] [PubMed] [Google Scholar]
- Haindl M, Harasim T, Eick D, Muller S (2008) The nucleolar SUMO‐specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep 9: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang LE, Liu X, Cheung I, Yang Y, Zhao X (2011) SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat Struct Mol Biol 18: 920–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, D'Souza RC, Yang B, Verlaan‐de Vries M, Mann M, Vertegaal AC (2014) Uncovering global SUMOylation signaling networks in a site‐specific manner. Nat Struct Mol Biol 21: 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, Vertegaal AC (2016) A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol 17: 581–595 [DOI] [PubMed] [Google Scholar]
- Hickey CM, Wilson NR, Hochstrasser M (2012) Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 13: 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG et al (2007) PHD domain‐mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 28: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, Psakhye I (2013) Control of nuclear activities by substrate‐selective and protein‐group SUMOylation. Annu Rev Genet 47: 167–186 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, Wotton D (2003) The polycomb protein Pc2 is a SUMO E3. Cell 113: 127–137 [DOI] [PubMed] [Google Scholar]
- Kahyo T, Nishida T, Yasuda H (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell 8: 713–718 [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kim MS, Ramakrishna S, Lim KH, Kim JH, Baek KH (2011) Protein stability of mitochondrial superoxide dismutase SOD2 is regulated by USP36. J Cell Biochem 112: 498–508 [DOI] [PubMed] [Google Scholar]
- Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK (2007) Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J 26: 2797–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koidl S, Eisenhardt N, Fatouros C, Droescher M, Chaugule VK, Pichler A (2016) The SUMO2/3 specific E3 ligase ZNF451‐1 regulates PML stability. Int J Biochem Cell Biol 79: 478–487 [DOI] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J (2017) A puzzle of life: crafting ribosomal subunits. Trends Biochem Sci 42: 640–654 [DOI] [PubMed] [Google Scholar]
- Kunz K, Piller T, Muller S (2018) SUMO‐specific proteases and isopeptidases of the SENP family at a glance. J Cell Sci 131: jcs211904 [DOI] [PubMed] [Google Scholar]
- Lamoliatte F, McManus FP, Maarifi G, Chelbi‐Alix MK, Thibault P (2017) Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat Commun 8: 14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Liu Z, Jang SW, Ma Z, Shinmura K, Kang S, Dong S, Chen J, Fukasawa K, Ye K (2007) Sumoylation of nucleophosmin/B23 regulates its subcellular localization, mediating cell proliferation and survival. Proc Natl Acad Sci USA 104: 9679–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui L, Lowe T (2013) Small nucleolar RNAs and RNA‐guided post‐transcriptional modification. Essays Biochem 54: 53–77 [DOI] [PubMed] [Google Scholar]
- Mannoor K, Liao J, Jiang F (2012) Small nucleolar RNAs in cancer. Biochim Biophys Acta 1826: 121–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matafora V, D'Amato A, Mori S, Blasi F, Bachi A (2009) Proteomics analysis of nucleolar SUMO‐1 target proteins upon proteasome inhibition. Mol Cell Proteomics 8: 2243–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, Gnad F, Mann M, Vertegaal AC (2010) Site‐specific identification of SUMO‐2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell 39: 641–652 [DOI] [PubMed] [Google Scholar]
- Metzger MB, Hristova VA, Weissman AM (2012) HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci 125: 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Green R (2017) Ribosomopathies: there's strength in numbers. Science 358: eaan2755 [DOI] [PubMed] [Google Scholar]
- Muller S, Hoege C, Pyrowolakis G, Jentsch S (2001) SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol 2: 202–210 [DOI] [PubMed] [Google Scholar]
- Nakagawa K, Yokosawa H (2002) PIAS3 induces SUMO‐1 modification and transcriptional repression of IRF‐1. FEBS Lett 530: 204–208 [DOI] [PubMed] [Google Scholar]
- Newton K, Petfalski E, Tollervey D, Caceres JF (2003) Fibrillarin is essential for early development and required for accumulation of an intron‐encoded small nucleolar RNA in the mouse. Mol Cell Biol 23: 8519–8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida T, Yasuda H (2002) PIAS1 and PIASxalpha function as SUMO‐E3 ligases toward androgen receptor and repress androgen receptor‐dependent transcription. J Biol Chem 277: 41311–41317 [DOI] [PubMed] [Google Scholar]
- Nuro‐Gyina PK, Parvin JD (2015) Roles for SUMO in pre‐mRNA processing. Wiley Interdiscip Rev RNA 7: 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG, Kressler D, Pauli A, Petfalski E, Gnadig M, Tollervey D, Hurt E (2006) Formation and nuclear export of preribosomes are functionally linked to the small‐ubiquitin‐related modifier pathway. Traffic 7: 1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Thomas G, Volarevic S (2018) Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 18: 51–63 [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109–120 [DOI] [PubMed] [Google Scholar]
- Pichler A, Knipscheer P, Saitoh H, Sixma TK, Melchior F (2004) The RanBP2 SUMO E3 ligase is neither HECT‐ nor RING‐type. Nat Struct Mol Biol 11: 984–991 [DOI] [PubMed] [Google Scholar]
- Pichler A, Fatouros C, Lee H, Eisenhardt N (2017) SUMO conjugation ‐ a mechanistic view. Biomol Concepts 8: 13–36 [DOI] [PubMed] [Google Scholar]
- Potts PR, Yu H (2005) Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol 25: 7021–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Yu H (2007) The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere‐binding proteins. Nat Struct Mol Biol 14: 581–590 [DOI] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S (2012) Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151: 807–820 [DOI] [PubMed] [Google Scholar]
- Raman N, Weir E, Muller S (2016) The AAA ATPase MDN1 acts as a SUMO‐targeted regulator in mammalian pre‐ribosome remodeling. Mol Cell 64: 607–615 [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD (2005) Insights into E3 ligase activity revealed by a SUMO‐RanGAP1‐Ubc9‐Nup358 complex. Nature 435: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T (2003) A common sequence motif determines the Cajal body‐specific localization of box H/ACA scaRNAs. EMBO J 22: 4283–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson LA, Reed BJ, Charette JM, Freed EF, Fredrickson EK, Locke MN, Baserga SJ, Gardner RG (2012) A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell Rep 2: 372–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW (2010) MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309 [DOI] [PubMed] [Google Scholar]
- Ritchie C, Cylinder I, Platt EJ, Barklis E (2015) Analysis of HIV‐1 Gag protein interactions via biotin ligase tagging. J Virol 89: 3988–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W (2009) Recent mechanistic insights into eukaryotic ribosomes. Curr Opin Cell Biol 21: 435–443 [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT (2001) SUMO‐1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 276: 12654–12659 [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon‐Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ (2007) Deletion of the developmentally essential gene ATR in adult mice leads to age‐related phenotypes and stem cell loss. Cell Stem Cell 1: 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Al‐Ani G, Deckert K, Kirkpatrick D, Gygi SP, Dasso M, Azuma Y (2010a) PIASy mediates SUMO‐2/3 conjugation of poly(ADP‐ribose) polymerase 1 (PARP1) on mitotic chromosomes. J Biol Chem 285: 14415–14423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Azuma Y (2010) Rod/Zw10 complex is required for PIASy‐dependent centromeric SUMOylation. J Biol Chem 285: 32576–32585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Furuta M, Kirkpatrick D, Gygi SP, Azuma Y (2010b) PIASy‐dependent SUMOylation regulates DNA topoisomerase IIalpha activity. J Cell Biol 191: 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R (2001) PIASy, a nuclear matrix‐associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev 15: 3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson DA, Wang M, Matunis MJ (2001) The small ubiquitin‐like modifier‐1 (SUMO‐1) consensus sequence mediates Ubc9 binding and is essential for SUMO‐1 modification. J Biol Chem 276: 21664–21669 [DOI] [PubMed] [Google Scholar]
- Sarangi P, Zhao X (2015) SUMO‐mediated regulation of DNA damage repair and responses. Trends Biochem Sci 40: 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Muller S (2002) Members of the PIAS family act as SUMO ligases for c‐Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA 99: 2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6: 275–277 [DOI] [PubMed] [Google Scholar]
- Sharma S, Langhendries JL, Watzinger P, Kotter P, Entian KD, Lafontaine DL (2015) Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res 43: 2242–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streich FC Jr, Lima CD (2016) Capturing a substrate in an activated RING E3/E2‐SUMO complex. Nature 536: 304–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Challagundla KB, Dai MS (2012) Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J 31: 576–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, He X, Yin L, Komada M, Sears RC, Dai MS (2015) The nucleolar ubiquitin‐specific protease USP36 deubiquitinates and stabilizes c‐Myc. Proc Natl Acad Sci USA 112: 3734–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Chen Y, Su Y, Wang X, Chauhan KM, Liang J, Daniel CJ, Sears RC, Dai MS (2018) SUMO protease SENP1 deSUMOylates and stabilizes c‐Myc. Proc Natl Acad Sci USA 115: 10983–10988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL (2013) The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre‐rRNA processing factors. Mol Cell 51: 539–551 [DOI] [PubMed] [Google Scholar]
- Tago K, Chiocca S, Sherr CJ (2005) Sumoylation induced by the Arf tumor suppressor: a p53‐independent function. Proc Natl Acad Sci USA 102: 7689–7694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillebourg E, Gregoire I, Viargues P, Jacomin AC, Thevenon D, Faure M, Fauvarque MO (2012) The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins. Autophagy 8: 767–779 [DOI] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT (2008) RNF4 is a poly‐SUMO‐specific E3 ubiquitin ligase required for arsenic‐induced PML degradation. Nat Cell Biol 10: 538–546 [DOI] [PubMed] [Google Scholar]
- Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF (2004) A reliable lacZ expression reporter cassette for multipurpose, knockout‐first alleles. Genesis 38: 151–158 [DOI] [PubMed] [Google Scholar]
- Tomecki R, Sikorski PJ, Zakrzewska‐Placzek M (2017) Comparison of preribosomal RNA processing pathways in yeast, plant and human cells ‐ focus on coordinated action of endo‐ and exoribonucleases. FEBS Lett 591: 1801–1850 [DOI] [PubMed] [Google Scholar]
- Varejao N, Lascorz J, Li Y, Reverter D (2020) Molecular mechanisms in SUMO conjugation. Biochem Soc Trans 48: 123–135 [DOI] [PubMed] [Google Scholar]
- Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M, Lamond AI (2004) A proteomic study of SUMO‐2 target proteins. J Biol Chem 279: 33791–33798 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT (2012) The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414 [DOI] [PubMed] [Google Scholar]
- Westman BJ, Verheggen C, Hutten S, Lam YW, Bertrand E, Lamond AI (2010) A proteomic screen for nucleolar SUMO targets shows SUMOylation modulates the function of Nop5/Nop58. Mol Cell 39: 618–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmarth PA, Riviere MA, David LL (2009) Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J Ocul Biol Dis Infor 2: 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang HY, Hopper AK (2013) A rapid and sensitive non‐radioactive method applicable for genome‐wide analysis of Saccharomyces cerevisiae genes involved in small RNA biology. Yeast 30: 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, Dasso M (2008) Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol 183: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus AA, Lima CD (2009) Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol Cell 35: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Liang Y, Xie Q, Gao G, Wei J, Huang H, Li J, Gao J, Huang C (2015) A novel post‐translational modification of nucleolin, SUMOylation at Lys‐294, mediates arsenite‐induced cell death by regulating gadd45alpha mRNA stability. J Biol Chem 290: 4784–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X (2018) SUMO‐mediated regulation of nuclear functions and signaling processes. Mol Cell 71: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Yasumoto H, Tsai RY (2006) Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol Cell Biol 26: 9279–9290 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Dataset EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Data Availability Statement
The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD022987 (https://www.ebi.ac.uk/pride/archive/projects/PXD022987).


