The dihydroquinoxaline moiety, with the exception of the N atom, is essentially planar with the attached phenyl ring inclined to it by 11.64 (6)° and the inner part of the methylpropanoate group nearly perpendicular to it. In the crystal, inversion dimers formed by C—H⋯O hydrogen bonds are connected into oblique stacks by π-stacking and C—H⋯π(ring) interactions.
Keywords: crystal structure, dihydroquinoxaline, hydrogen bond, π-stacking, C—H⋯π(ring)
Abstract
In the title molecule, C18H16N2O3, the dihydroquinoxaline moiety, with the exception of the N atom is essentially planar with the inner part of the methylpropanoate group (CH2—CH2—O) nearly perpendicular to it. In the crystal, inversion dimers formed by C—H⋯O hydrogen bonds are connected into oblique stacks by π-stacking and C—H⋯π(ring) interactions.
Chemical context
Quinoxaline are a class of nitrogen containing heterocyclic compounds, found in many biologically active drugs (Ramli & Essassi, 2015 ▸; Ramli et al., 2014 ▸). In addition, this heterocyclic scaffold possess anticorrosion characteristics (El Ouali et al., 2010 ▸; Zarrok et al., 2012 ▸; Tazouti et al., 2016 ▸; El Aoufir et al., 2016 ▸; Laabaissi et al., 2019 ▸). In a continuation of our recent work focused on the synthesis and biological evaluation of novel heterocyclic compounds (Guerrab et al. 2019 ▸, 2020 ▸, 2021 ▸; Abad et al., 2021a
▸,b
▸; Missioui et al. 2021 ▸) we report here the crystal structure of the title compound (Fig. 1 ▸). As with many biologically active molecules, the molecular conformation adopted may have a significant effect on its activity.
Figure 1.
The title molecule with labelling scheme and 50% probability ellipsoids. The intramolecular C—H⋯O hydrogen bonds are shown by dashed lines.
Structural commentary
The dihydroquinoxaline moiety, with the exception of N1, is planar to within 0.0186 (9) Å (r.m.s. deviation of the nine fitted atoms = 0.0116 Å). N1 lies 0.0526 (12) Å below the mean plane. The C9–C14 phenyl ring is inclined to the above plane by 11.64 (6)° while the inner part (CH2—CH2—O) of the methyl propanoate substituent is nearly perpendicular to the dihydroquinoxaline unit, as indicated by the angle of 87.34 (6)° between the N1/C15/C16/O2 and N2/C1–C8 planes. The overall conformation is determined in part by the intramolecular C5—H5⋯O3 and C14—H14⋯O1 hydrogen bonds (Table 1 ▸ and Fig. 1 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
Cg2 is the centroid of the C1–C6 benzene ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C4—H4⋯O3i | 0.974 (16) | 2.526 (16) | 3.4713 (17) | 163.6 (11) |
| C5—H5⋯O3 | 0.992 (15) | 2.592 (16) | 3.5435 (15) | 160.7 (12) |
| C14—H14⋯O1 | 0.963 (15) | 2.232 (16) | 2.8387 (16) | 120.0 (12) |
| C16—H16B⋯O3ii | 0.993 (14) | 2.553 (14) | 3.3632 (16) | 138.7 (10) |
| C18—H18A⋯Cg2iii | 0.97 (2) | 2.93 (2) | 3.7585 (17) | 144.2 (17) |
Symmetry codes: (i) -x+2, -y+1, -z+1; (ii) x-1, y, z; (iii) -x+1, -y+1, -z+1.
Supramolecular features
In the crystal, inversion dimers are formed by C4—H4⋯O3i hydrogen bonds [Table 1 ▸; symmetry code: (i) −x + 2, −y + 1, −z + 1] and are connected into oblique stacks by a combination of π-stacking interactions between the C1/C6/N1/C7/C8/N2 and C9–C14 rings [centroid–centroid distance = 3.7786 (9) Å, dihedral angle = 12.20 (6)°] and in addition C16—H16B⋯O3ii and C18—H18A⋯Cg2iii interactions [Table 1 ▸ and Fig. 2 ▸; Cg2 is the centroid of the C1–C6 ring; symmetry codes: (ii) x − 1, y, z, (iii) −x + 1, −y + 1, −z + 1]. The crystal packing also shows a C17=O3⋯Cg2 interaction [O3⋯Cg2 = 3.9578 (12) Å, C17⋯Cg2 = 3.7440 (16) Å, C17=O3⋯Cg2 = 71.04 (8)°].
Figure 2.
Perspective view of the packing. Intermolecular C—H⋯O hydrogen bonds are shown by black dashed lines while π-stacking and C—H⋯π(ring) interactions are shown, respectively, by orange and green dashed lines.
Database survey
A survey of the Cambridge Structural Database (Version 5.42, last update February 2021; Groom et al., 2016 ▸) using the search fragment II yielded 30 hits of which those most similar to the title molecule have the formula III with R = Me and R′ = CH2CO2H (DEZJAW; Missioui et al., 2018 ▸), CH2C≡CH (DUCYUW; Benzeid et al., 2009a
▸), benzyl [DUSHUV (Ramli et al., 2010b
▸) and DUSHUV01 (Ramli et al., 2018 ▸)], Et (IGANOU; Benzeid et al., 2008 ▸), CH2CH=CH2 (YUPXAJ; Ramli et al., 2010a
▸), with R = CF3 and R′ = i-Bu (DUBPUO; Wei et al., 2019 ▸), with R = Ph and R′ = CH2(cyclo-CHCH2O) (NIBXEE; Abad et al., 2018a
▸), benzyl (PUGGII; Benzeid et al., 2009b
▸), CH2CH2CH2OH (RIRBOM; Abad et al., 2018b
▸), CH2CO2Et (XEXWIJ; Abad et al., 2018c
▸), CH2CH=CH2 (YAJGEX; Benzeid et al., 2011 ▸) and with R = 3-NO2-C6H4 and R′ = benzyl (XIKHAD; Das et al., 2018 ▸).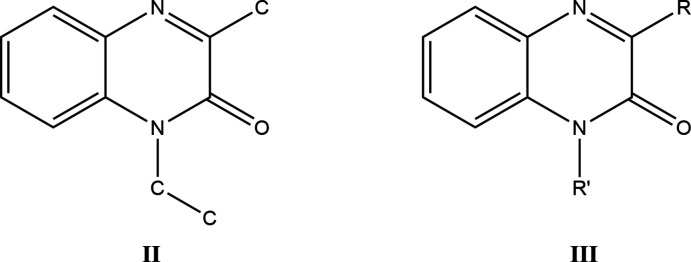
In the majority of the hits, the dihydroquinoxaline ring is essentially planar with the dihedral angle between the constituent rings being less than 1° or having the nitrogen bearing the exocyclic substituent less than 0.03 Å from the mean plane of the remaining nine atoms. Two notable exceptions are DEZJAW, where the dihedral angle between the two rings is 3.32°, and RIRBOM, where the nitrogen bearing the exocyclic substituent deviates by 0.062 Å from the plane defined by the other nine atoms.
Hirshfeld surface analysis
An effective means of probing intermolecular interactions is Hirshfeld surface analysis (McKinnon et al., 2007 ▸; Spackman & Jayatilaka, 2009 ▸), which can be conveniently carried out with Crystal Explorer 17 (Turner et al., 2017 ▸). A detailed description of the use of Crystal Explorer 17 and the plots obtained has been published (Tan et al., 2019 ▸) and will not be given here. Fig. 3 ▸ a presents front (top) and side (bottom) views of the Hirshfeld surface plotted over d norm in the range −0.1367 to 1.2965 a.u. One of the intramolecular C—H⋯O hydrogen bonds is indicated by the arrow at the left in the front view while those leading to the formation of the inversion dimers are shown by the arrows on the right of the front view. The C—H⋯π(ring) interaction and the π-stacking interactions are represented by the red spots designated by arrows in the side view. Fig. 3 ▸ b presents the same two views of the surface plotted over the shape-index. In the front view, the π-stacking interaction is evident at the center as an orange triangle surrounded by blue triangles. Fig. 3 ▸ c has the same two views of the surface plotted over the curvature index, with the flat area in the center indicating the locus of the π-stacking interaction. Fig. 4 ▸ presents fingerprint plots for all intermolecular interactions (a) and those delineated into H⋯H contacts (b, 49.4%), H⋯O/O⋯H contacts (c, 18.2%), H⋯C/C⋯H contacts (d, 17.8%) and C⋯C contacts (e, 7.2%).
Figure 3.
Front (top) and side (bottom) views of the Hirshfeld surface plotted over (a) d norm, (b) shape-index and (c) curvature.
Figure 4.
Two dimensional fingerprint plots showing (a) all intermolecular interactions and those delineated into (b) H⋯H, (c) H⋯O/O⋯H, (d) H⋯C/C⋯H and (e) C⋯C interactions.
Synthesis and crystallization
To a solution of 2-oxo-3-phenyl-1,2-dihydroquinoxaline (0.5 g, 2.25 mmol) in N,N-dimethylformamide (15 ml) were added 2-bromoethyl acetate (0.4 ml, 2.25 mmol), potassium carbonate (0.31 g, 2.25 mmol) and a catalytic quantity of tetra-n-butylammonium bromide. The reaction mixture was stirred at room temperature for 24 h. The solution was filtered and the solvent removed under reduced pressure. The residue thus obtained was chromatographed on a silica gel column using a hexane/ethyl acetate 9.5: 0.5 mixture as eluent. The solid obtained was recrystallized from ethanol solution to afford colorless column-like specimen of the title compound. Yield: 0.50 g, 67%; m.p. 471–473 K.
1H NMR (Bruker Avance 300 MHz, CDCl3) δ (ppm): 8.24 (d, 2H, Ar—H); 7.91 (d, 1H, Ar—H); 7.82 (m, 3H, Ar—H); 7.53 (m, 1H, Ar—H); 7.25 (m, 2H, Ar—H); 4.73 (t, 2H, O—CH2); 3.92 (t, 2H, N—CH2); 2.23 (s, 3H, OCOCH3).
13C NMR (Bruker Avance 75 MHz, CDCl3) δ (ppm):46.15 (N—CH2); 61.15 (O—CH2); 114.38, 123.82, 127.01, 127.72, 128.13, 128.96, 129.68, 130.33, 130.45,130.62 (CH—Ar); 132.78, 133.36, 135.40, 136.05, 154.24(Cq); 156.92 (C=O); 177.82 (O—C=O).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All hydrogen atoms were located from a difference electron-density map and freely refined.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C18H16N2O3 |
| M r | 308.33 |
| Crystal system, space group | Triclinic, P\overline{1} |
| Temperature (K) | 120 |
| a, b, c (Å) | 5.3518 (6), 11.6989 (14), 13.3527 (16) |
| α, β, γ (°) | 64.019 (2), 80.323 (2), 76.952 (2) |
| V (Å3) | 729.83 (15) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.10 |
| Crystal size (mm) | 0.42 × 0.18 × 0.12 |
| Data collection | |
| Diffractometer | Bruker SMART APEX CCD |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.87, 0.99 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 14193, 3909, 2929 |
| R int | 0.028 |
| (sin θ/λ)max (Å−1) | 0.688 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.049, 0.142, 1.00 |
| No. of reflections | 3909 |
| No. of parameters | 272 |
| H-atom treatment | All H-atom parameters refined |
| Δρmax, Δρmin (e Å−3) | 0.47, −0.23 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989021005247/vm2249sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021005247/vm2249Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989021005247/vm2249Isup3.cml
CCDC reference: 2062956
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
JTM thanks Tulane University for support of the Tulane Crystallography Laboratory. Author contributions are as follows. Conceptualization, YR and EME; synthesis, NA and LEG; writing (review and editing of the manuscript) CKM, JTM and YR; formal analysis, SK and JTM; crystal-structure determination, JTM; validation, JTM and YR, project administration, YR
supplementary crystallographic information
Crystal data
| C18H16N2O3 | Z = 2 |
| Mr = 308.33 | F(000) = 324 |
| Triclinic, P1 | Dx = 1.403 Mg m−3 |
| a = 5.3518 (6) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 11.6989 (14) Å | Cell parameters from 5207 reflections |
| c = 13.3527 (16) Å | θ = 3.1–29.3° |
| α = 64.019 (2)° | µ = 0.10 mm−1 |
| β = 80.323 (2)° | T = 120 K |
| γ = 76.952 (2)° | Column, colourless |
| V = 729.83 (15) Å3 | 0.42 × 0.18 × 0.12 mm |
Data collection
| Bruker SMART APEX CCD diffractometer | 3909 independent reflections |
| Radiation source: fine-focus sealed tube | 2929 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.028 |
| Detector resolution: 8.3333 pixels mm-1 | θmax = 29.3°, θmin = 1.7° |
| φ and ω scans | h = −7→7 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | k = −16→16 |
| Tmin = 0.87, Tmax = 0.99 | l = −18→18 |
| 14193 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.142 | All H-atom parameters refined |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.0999P)2] where P = (Fo2 + 2Fc2)/3 |
| 3909 reflections | (Δ/σ)max < 0.001 |
| 272 parameters | Δρmax = 0.47 e Å−3 |
| 0 restraints | Δρmin = −0.23 e Å−3 |
Special details
| Experimental. The diffraction data were obtained from 3 sets of 400 frames, each of width 0.5° in ω, colllected at φ = 0.00, 90.00 and 180.00° and 2 sets of 800 frames, each of width 0.45° in φ, collected at ω = –30.00 and 210.00°. The scan time was 30 sec/frame. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.24048 (17) | 0.11347 (8) | 0.45539 (7) | 0.0287 (2) | |
| O2 | 0.42582 (17) | 0.24290 (8) | 0.68691 (6) | 0.0264 (2) | |
| O3 | 0.74296 (17) | 0.36037 (9) | 0.62940 (7) | 0.0308 (2) | |
| N1 | 0.52749 (18) | 0.24890 (9) | 0.40271 (7) | 0.0209 (2) | |
| N2 | 0.46952 (18) | 0.31557 (9) | 0.18053 (7) | 0.0205 (2) | |
| C1 | 0.6193 (2) | 0.37704 (11) | 0.20843 (9) | 0.0198 (2) | |
| C2 | 0.7488 (2) | 0.47088 (11) | 0.12307 (9) | 0.0229 (3) | |
| H2 | 0.731 (3) | 0.4919 (13) | 0.0454 (12) | 0.033 (4)* | |
| C3 | 0.9057 (2) | 0.53135 (11) | 0.14791 (10) | 0.0253 (3) | |
| H3 | 0.999 (3) | 0.5940 (13) | 0.0898 (11) | 0.025 (3)* | |
| C4 | 0.9332 (2) | 0.50106 (12) | 0.25984 (10) | 0.0240 (3) | |
| H4 | 1.043 (3) | 0.5462 (13) | 0.2757 (11) | 0.023 (3)* | |
| C5 | 0.8070 (2) | 0.40967 (11) | 0.34551 (10) | 0.0229 (3) | |
| H5 | 0.835 (3) | 0.3898 (15) | 0.4234 (13) | 0.035 (4)* | |
| C6 | 0.6527 (2) | 0.34508 (11) | 0.32082 (9) | 0.0197 (2) | |
| C7 | 0.3624 (2) | 0.19025 (11) | 0.37929 (9) | 0.0209 (2) | |
| C8 | 0.3496 (2) | 0.22703 (11) | 0.25807 (9) | 0.0193 (2) | |
| C9 | 0.2018 (2) | 0.16113 (11) | 0.22093 (9) | 0.0210 (2) | |
| C10 | 0.2376 (3) | 0.18294 (12) | 0.10782 (10) | 0.0268 (3) | |
| H10 | 0.365 (3) | 0.2385 (15) | 0.0602 (13) | 0.040 (4)* | |
| C11 | 0.1007 (3) | 0.12852 (13) | 0.06609 (10) | 0.0307 (3) | |
| H11 | 0.131 (3) | 0.1475 (15) | −0.0164 (14) | 0.045 (4)* | |
| C12 | −0.0746 (3) | 0.05042 (12) | 0.13563 (11) | 0.0299 (3) | |
| H12 | −0.174 (3) | 0.0113 (13) | 0.1067 (11) | 0.026 (3)* | |
| C13 | −0.1069 (2) | 0.02574 (12) | 0.24773 (11) | 0.0282 (3) | |
| H13 | −0.229 (3) | −0.0255 (14) | 0.2925 (12) | 0.030 (4)* | |
| C14 | 0.0284 (2) | 0.08023 (11) | 0.29077 (10) | 0.0239 (3) | |
| H14 | −0.004 (3) | 0.0579 (14) | 0.3697 (12) | 0.030 (4)* | |
| C15 | 0.5682 (2) | 0.20405 (12) | 0.52109 (9) | 0.0224 (3) | |
| H15A | 0.751 (3) | 0.2084 (12) | 0.5233 (10) | 0.019 (3)* | |
| H15B | 0.546 (2) | 0.1157 (14) | 0.5606 (11) | 0.023 (3)* | |
| C16 | 0.3790 (2) | 0.28659 (12) | 0.57099 (9) | 0.0246 (3) | |
| H16A | 0.388 (3) | 0.3773 (14) | 0.5287 (11) | 0.027 (3)* | |
| H16B | 0.200 (3) | 0.2750 (12) | 0.5718 (10) | 0.024 (3)* | |
| C17 | 0.6177 (2) | 0.28800 (12) | 0.70394 (10) | 0.0246 (3) | |
| C18 | 0.6508 (3) | 0.23944 (15) | 0.82550 (11) | 0.0319 (3) | |
| H18A | 0.545 (4) | 0.305 (2) | 0.8477 (19) | 0.085 (7)* | |
| H18B | 0.600 (4) | 0.1579 (19) | 0.8692 (16) | 0.063 (5)* | |
| H18C | 0.829 (4) | 0.2289 (18) | 0.8386 (15) | 0.060 (5)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0367 (5) | 0.0329 (5) | 0.0186 (4) | −0.0178 (4) | 0.0010 (3) | −0.0080 (4) |
| O2 | 0.0315 (5) | 0.0335 (5) | 0.0192 (4) | −0.0123 (4) | 0.0016 (3) | −0.0136 (4) |
| O3 | 0.0309 (5) | 0.0397 (5) | 0.0282 (4) | −0.0145 (4) | 0.0022 (4) | −0.0175 (4) |
| N1 | 0.0248 (5) | 0.0241 (5) | 0.0160 (4) | −0.0082 (4) | 0.0008 (4) | −0.0094 (4) |
| N2 | 0.0230 (5) | 0.0203 (5) | 0.0195 (4) | −0.0044 (4) | −0.0001 (4) | −0.0097 (4) |
| C1 | 0.0212 (5) | 0.0209 (5) | 0.0188 (5) | −0.0040 (4) | 0.0007 (4) | −0.0102 (4) |
| C2 | 0.0267 (6) | 0.0224 (6) | 0.0191 (5) | −0.0050 (5) | 0.0012 (4) | −0.0089 (4) |
| C3 | 0.0282 (6) | 0.0221 (6) | 0.0236 (6) | −0.0081 (5) | 0.0039 (5) | −0.0080 (5) |
| C4 | 0.0239 (6) | 0.0247 (6) | 0.0280 (6) | −0.0063 (5) | −0.0005 (4) | −0.0147 (5) |
| C5 | 0.0241 (6) | 0.0256 (6) | 0.0220 (5) | −0.0051 (4) | −0.0004 (4) | −0.0127 (5) |
| C6 | 0.0203 (5) | 0.0205 (5) | 0.0185 (5) | −0.0043 (4) | 0.0017 (4) | −0.0091 (4) |
| C7 | 0.0237 (6) | 0.0216 (5) | 0.0191 (5) | −0.0063 (4) | 0.0002 (4) | −0.0097 (4) |
| C8 | 0.0209 (5) | 0.0201 (5) | 0.0178 (5) | −0.0029 (4) | −0.0013 (4) | −0.0092 (4) |
| C9 | 0.0225 (6) | 0.0200 (5) | 0.0217 (5) | −0.0020 (4) | −0.0038 (4) | −0.0097 (4) |
| C10 | 0.0338 (7) | 0.0276 (6) | 0.0223 (5) | −0.0105 (5) | −0.0018 (5) | −0.0110 (5) |
| C11 | 0.0415 (7) | 0.0316 (7) | 0.0246 (6) | −0.0096 (5) | −0.0061 (5) | −0.0141 (5) |
| C12 | 0.0322 (7) | 0.0285 (6) | 0.0373 (7) | −0.0059 (5) | −0.0088 (5) | −0.0188 (6) |
| C13 | 0.0266 (6) | 0.0269 (6) | 0.0337 (6) | −0.0085 (5) | −0.0009 (5) | −0.0137 (5) |
| C14 | 0.0235 (6) | 0.0243 (6) | 0.0246 (6) | −0.0049 (4) | −0.0009 (4) | −0.0109 (5) |
| C15 | 0.0265 (6) | 0.0247 (6) | 0.0170 (5) | −0.0068 (5) | −0.0007 (4) | −0.0089 (4) |
| C16 | 0.0254 (6) | 0.0309 (6) | 0.0202 (5) | −0.0069 (5) | −0.0011 (4) | −0.0122 (5) |
| C17 | 0.0245 (6) | 0.0294 (6) | 0.0241 (5) | −0.0037 (5) | −0.0003 (4) | −0.0161 (5) |
| C18 | 0.0388 (8) | 0.0373 (8) | 0.0229 (6) | −0.0034 (6) | −0.0053 (5) | −0.0159 (6) |
Geometric parameters (Å, º)
| O1—C7 | 1.2275 (13) | C9—C14 | 1.3995 (16) |
| O2—C17 | 1.3467 (14) | C9—C10 | 1.4036 (15) |
| O2—C16 | 1.4489 (13) | C10—C11 | 1.3828 (17) |
| O3—C17 | 1.2043 (14) | C10—H10 | 0.990 (16) |
| N1—C7 | 1.3800 (14) | C11—C12 | 1.3884 (19) |
| N1—C6 | 1.3882 (14) | C11—H11 | 1.016 (16) |
| N1—C15 | 1.4692 (13) | C12—C13 | 1.3822 (18) |
| N2—C8 | 1.3016 (14) | C12—H12 | 0.988 (14) |
| N2—C1 | 1.3763 (14) | C13—C14 | 1.3894 (17) |
| C1—C2 | 1.4022 (15) | C13—H13 | 0.934 (15) |
| C1—C6 | 1.4114 (14) | C14—H14 | 0.962 (14) |
| C2—C3 | 1.3721 (17) | C15—C16 | 1.5155 (17) |
| C2—H2 | 0.972 (15) | C15—H15A | 0.996 (13) |
| C3—C4 | 1.4027 (16) | C15—H15B | 0.958 (14) |
| C3—H3 | 0.962 (13) | C16—H16A | 0.967 (14) |
| C4—C5 | 1.3795 (16) | C16—H16B | 0.993 (14) |
| C4—H4 | 0.973 (14) | C17—C18 | 1.4940 (16) |
| C5—C6 | 1.3975 (16) | C18—H18A | 0.97 (2) |
| C5—H5 | 0.992 (15) | C18—H18B | 0.95 (2) |
| C7—C8 | 1.4911 (14) | C18—H18C | 0.97 (2) |
| C8—C9 | 1.4861 (15) | ||
| C17—O2—C16 | 115.33 (9) | C9—C10—H10 | 117.0 (9) |
| C7—N1—C6 | 123.19 (9) | C10—C11—C12 | 120.53 (12) |
| C7—N1—C15 | 116.52 (9) | C10—C11—H11 | 118.3 (9) |
| C6—N1—C15 | 120.28 (9) | C12—C11—H11 | 121.2 (9) |
| C8—N2—C1 | 120.49 (9) | C13—C12—C11 | 119.10 (11) |
| N2—C1—C2 | 119.20 (9) | C13—C12—H12 | 119.6 (8) |
| N2—C1—C6 | 121.67 (10) | C11—C12—H12 | 121.3 (8) |
| C2—C1—C6 | 119.10 (10) | C12—C13—C14 | 121.02 (12) |
| C3—C2—C1 | 120.73 (10) | C12—C13—H13 | 117.5 (9) |
| C3—C2—H2 | 119.6 (8) | C14—C13—H13 | 121.4 (9) |
| C1—C2—H2 | 119.7 (8) | C13—C14—C9 | 120.32 (11) |
| C2—C3—C4 | 119.78 (11) | C13—C14—H14 | 116.0 (9) |
| C2—C3—H3 | 121.2 (8) | C9—C14—H14 | 123.7 (9) |
| C4—C3—H3 | 119.0 (8) | N1—C15—C16 | 110.08 (9) |
| C5—C4—C3 | 120.72 (11) | N1—C15—H15A | 106.2 (7) |
| C5—C4—H4 | 120.7 (8) | C16—C15—H15A | 112.9 (7) |
| C3—C4—H4 | 118.5 (8) | N1—C15—H15B | 109.3 (8) |
| C4—C5—C6 | 119.78 (10) | C16—C15—H15B | 110.1 (8) |
| C4—C5—H5 | 118.0 (9) | H15A—C15—H15B | 108.2 (11) |
| C6—C5—H5 | 122.2 (9) | O2—C16—C15 | 109.30 (10) |
| N1—C6—C5 | 122.87 (9) | O2—C16—H16A | 111.5 (8) |
| N1—C6—C1 | 117.29 (10) | C15—C16—H16A | 111.6 (8) |
| C5—C6—C1 | 119.84 (10) | O2—C16—H16B | 105.9 (7) |
| O1—C7—N1 | 120.36 (10) | C15—C16—H16B | 110.3 (7) |
| O1—C7—C8 | 124.70 (10) | H16A—C16—H16B | 108.0 (11) |
| N1—C7—C8 | 114.94 (9) | O3—C17—O2 | 123.41 (10) |
| N2—C8—C9 | 117.09 (9) | O3—C17—C18 | 124.81 (11) |
| N2—C8—C7 | 122.12 (10) | O2—C17—C18 | 111.77 (10) |
| C9—C8—C7 | 120.78 (9) | C17—C18—H18A | 104.9 (13) |
| C14—C9—C10 | 118.14 (10) | C17—C18—H18B | 113.1 (11) |
| C14—C9—C8 | 124.43 (10) | H18A—C18—H18B | 110.9 (18) |
| C10—C9—C8 | 117.43 (10) | C17—C18—H18C | 111.5 (11) |
| C11—C10—C9 | 120.87 (11) | H18A—C18—H18C | 110.2 (17) |
| C11—C10—H10 | 122.2 (9) | H18B—C18—H18C | 106.3 (16) |
| C8—N2—C1—C2 | 179.20 (10) | O1—C7—C8—N2 | 175.20 (11) |
| C8—N2—C1—C6 | 1.29 (17) | N1—C7—C8—N2 | −5.32 (16) |
| N2—C1—C2—C3 | −178.09 (10) | O1—C7—C8—C9 | −6.07 (18) |
| C6—C1—C2—C3 | −0.13 (17) | N1—C7—C8—C9 | 173.42 (9) |
| C1—C2—C3—C4 | −1.29 (18) | N2—C8—C9—C14 | −168.77 (10) |
| C2—C3—C4—C5 | 0.91 (18) | C7—C8—C9—C14 | 12.44 (17) |
| C3—C4—C5—C6 | 0.92 (18) | N2—C8—C9—C10 | 10.61 (16) |
| C7—N1—C6—C5 | 175.87 (10) | C7—C8—C9—C10 | −168.19 (10) |
| C15—N1—C6—C5 | −4.42 (16) | C14—C9—C10—C11 | 1.58 (18) |
| C7—N1—C6—C1 | −4.13 (16) | C8—C9—C10—C11 | −177.84 (11) |
| C15—N1—C6—C1 | 175.58 (10) | C9—C10—C11—C12 | −0.4 (2) |
| C4—C5—C6—N1 | 177.66 (10) | C10—C11—C12—C13 | −1.2 (2) |
| C4—C5—C6—C1 | −2.34 (17) | C11—C12—C13—C14 | 1.52 (19) |
| N2—C1—C6—N1 | −0.14 (16) | C12—C13—C14—C9 | −0.28 (19) |
| C2—C1—C6—N1 | −178.05 (10) | C10—C9—C14—C13 | −1.26 (17) |
| N2—C1—C6—C5 | 179.86 (10) | C8—C9—C14—C13 | 178.11 (10) |
| C2—C1—C6—C5 | 1.95 (16) | C7—N1—C15—C16 | −92.28 (12) |
| C6—N1—C7—O1 | −173.92 (10) | C6—N1—C15—C16 | 88.00 (12) |
| C15—N1—C7—O1 | 6.36 (16) | C17—O2—C16—C15 | 82.16 (12) |
| C6—N1—C7—C8 | 6.57 (15) | N1—C15—C16—O2 | −178.70 (9) |
| C15—N1—C7—C8 | −173.15 (9) | C16—O2—C17—O3 | 0.52 (16) |
| C1—N2—C8—C9 | −177.24 (9) | C16—O2—C17—C18 | 179.37 (10) |
| C1—N2—C8—C7 | 1.54 (17) |
Hydrogen-bond geometry (Å, º)
Cg2 is the centroid of the C1–C6 benzene ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C4—H4···O3i | 0.974 (16) | 2.526 (16) | 3.4713 (17) | 163.6 (11) |
| C5—H5···O3 | 0.992 (15) | 2.592 (16) | 3.5435 (15) | 160.7 (12) |
| C14—H14···O1 | 0.963 (15) | 2.232 (16) | 2.8387 (16) | 120.0 (12) |
| C16—H16B···O3ii | 0.993 (14) | 2.553 (14) | 3.3632 (16) | 138.7 (10) |
| C18—H18A···Cg2iii | 0.97 (2) | 2.93 (2) | 3.7585 (17) | 144.2 (17) |
Symmetry codes: (i) −x+2, −y+1, −z+1; (ii) x−1, y, z; (iii) −x+1, −y+1, −z+1.
References
- Abad, N., Ferfra, S., Essassi, E. M., Mague, J. T. & Ramli, Y. (2021b). Z. Kristallogr. New Cryst. Struct. 236, 173–175.
- Abad, N., El Bakri, Y., Sebhaoui, J., Ramli, Y., Essassi, E. M. & Mague, J. T. (2018a). IUCrData, 3, x180610.
- Abad, N., El Bakri, Y., Sebhaoui, J., Ramli, Y., Essassi, E. M. & Mague, J. T. (2018c). IUCrData, 3, x180519.
- Abad, N., Ramli, Y., Lahmidi, S., El Hafi, Y., Essassi, E. M. & Mague, J. T. (2018b). IUCrData, 3, x181633.
- Abad, N., Sallam, H. H., Al-Ostoot, F. H., Khamees, H. A., Al-horaibi, S. A., Khanum, S. A., Madegowda, M., Hafi, M. E., Mague, J. T., Essassi, E. M. & Ramli, Y. (2021a). J. Mol. Struct. 1232, 130004.
- Benzeid, H., Bouhfid, R., Massip, S., Leger, J. M. & Essassi, E. M. (2011). Acta Cryst. E67, o2990. [DOI] [PMC free article] [PubMed]
- Benzeid, H., Ramli, Y., Vendier, L., Essassi, E. M. & Ng, S. W. (2009a). Acta Cryst. E65, o2196. [DOI] [PMC free article] [PubMed]
- Benzeid, H., Saffon, N., Garrigues, B., Essassi, E. M. & Ng, S. W. (2009b). Acta Cryst. E65, o2685. [DOI] [PMC free article] [PubMed]
- Benzeid, H., Vendier, L., Ramli, Y., Garrigues, B. & Essassi, E. M. (2008). Acta Cryst. E64, o2234. [DOI] [PMC free article] [PubMed]
- Brandenburg, K. & Putz, H. (2012). DIAMOND, Crystal Impact GbR, Bonn, Germany.
- Bruker (2016). APEX3, SADABS and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Das, D. K., Pampana, V. K. K. & Hwang, K. C. (2018). Chem. Sci. 9, 7318–7326. [DOI] [PMC free article] [PubMed]
- El Aoufir, Y., Lgaz, H., Bourazmi, H., Kerroum, Y., Ramli, Y., Guenbour, A., Salghi, R., El-Hajjaji, F., Hammouti, B. & Oudda, H. (2016). J. Mater. Environ. Sci. 7, 4330–4347.
- El Ouali, I., Hammouti, B., Aouniti, A., Ramli, Y., Azougagh, M., Essassi, E. M. & Bouachrine, M. (2010). J. Mater. Envir. Sci. 1, 1–8.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Guerrab, W., Chung, I.-M., Kansiz, S., Mague, J. T., Dege, N., Taoufik, J., Salghi, R., Ali, I. H., Chung, I. M., Lgaz, H. & Ramli, Y. (2019). J. Mol. Struct. 1197, 127630.
- Guerrab, W., Lgaz, H., Kansiz, S., Mague, J. T., Dege, N., Ansar, M., Marzouki, R., Taoufik, J., Ali, I. H., Chung, I. M. & Ramli, Y. (2020). J. Mol. Struct. 1205, 127630.
- Guerrab, W., Missioui, M., Zaoui, Y., Mague, J. T. & Ramli, Y. (2021). Z. Kristallogr. New Cryst. Struct. 236, 133–134.
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Laabaissi, T., Benhiba, F., Rouifi, Z., Allali, M., Missioui, M., Ourrak, K., Oudda, H., Ramli, Y., Warad, I. & Zarrouk, A. (2019). Int. J. Corros. Scale Inhib. 8, 241–256.
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- Missioui, M., El Fal, M., Taoufik, J., Essassi, E. M., Mague, J. T. & Ramli, Y. (2018). IUCRData 3, x180882.
- Missioui, M., Mortada, S., Guerrab, W., Serdaroğlu, G., Kaya, S., Mague, J. T., Essassi, E. M., Faouzi, M. E. A. & Ramli, Y. (2021). J. Mol. Struct. In the press.
- Ramli, Y., El Bakri, Y., El Ghayati, L., Essassi, E. M. & Mague, J. T. (2018). IUCrData 3, x180390.
- Ramli, Y. & Essassi, E. M. (2015). Adv. Chem. Res. 27, 109–160.
- Ramli, Y., Moussaif, A., Karrouchi, K. & Essassi, E. M. (2014). J. Chem. Article ID 563406, 1–21.
- Ramli, Y., Moussaif, A., Zouihri, H., Lazar, S. & Essassi, E. M. (2010b). Acta Cryst. E66, o1922. [DOI] [PMC free article] [PubMed]
- Ramli, Y., Slimani, R., Zouihri, H., Lazar, S. & Essassi, E. M. (2010a). Acta Cryst. E66, o1767. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Tan, S. L., Jotani, M. M. & Tiekink, E. R. T. (2019). Acta Cryst. E75, 308–318. [DOI] [PMC free article] [PubMed]
- Tazouti, A., Galai, M., Touir, R., Touhami, M. E., Zarrouk, A., Ramli, Y., Saraçoğlu, M., Kaya, S., Kandemirli, F. & Kaya, C. (2016). J. Mol. Liq. 221, 815–832.
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). Crystal Explorer 17. The University of Western Australia.
- Wei, Z., Qi, S., Xu, Y., Liu, H., Wu, J., Li, H., Xia, C. & Duan, G. (2019). Adv. Synth. Catal. 361, 5490–5498.
- Zarrok, H., Zarrouk, A., Salghi, R., Oudda, H., Hammouti, B., Ebn Touhami, M., Bouachrine, M. & Pucci, O. H. (2012). Electrochim. Acta, 30, 405–417.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989021005247/vm2249sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021005247/vm2249Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989021005247/vm2249Isup3.cml
CCDC reference: 2062956
Additional supporting information: crystallographic information; 3D view; checkCIF report






