Abstract
Most complete hydatidiform moles (CHMs) showcase an androgenetic nature of the nuclear genome. In the normal female embryo, one of the 2 X chromosomes is inactive. However, the status of X chromosome inactivation (XCI) in androgenetic CHMs remains unknown. Seventy-one androgenetic CHM tissues with the 46, XX karyotype were collected. Seventy-four normal female villi and 74 normal male villi were collected as controls. The expression of XCI markers (XIST, TSIX, and XACT) and an X-linked gene (CDX4) was detected by real-time polymerase chain reaction. Other XCI-associated genes were also examined, including the methylation status of the human androgen receptor gene (HUMARA) by methylation-specific polymerase chain reaction), and the expression of H3K27me3, USP21, and Nanog by Western blot and immunofluorescence, respectively. In addition, 126 CHMs and 63 normal female villous samples were collected for CDX4 immunohistochemical staining. The expression of XIST RNA was significantly lower, and TSIX RNA expression was significantly higher in androgenetic CHMs than that in normal female villi (both P<0.01). The expression of CDX4 mRNA in androgenetic CHMs was elevated compared with that in normal male and normal female villous samples (both P<0.01), and CDX4 protein expression was also higher than that in normal female villous samples (P<0.01). The expression of H3K27me3 was lower in androgenetic CHMs compared with that in normal female villi(P<0.01). The methylation pattern of HUMARA was found lacking in androgenetic CHMs. The expression of Nanog and UPS21 protein in androgenetic CHMs was higher than that in normal villi (both P<0.01). Both X chromosomes are active in androgenetic CHMs with the 46, XX karyotype, and the USP21-Nanog pathway may be involved in the disruption of XCI during this process.
Key Words: Complete hydatidiform moles, X chromosome inactivation, USP21, Nanog
Complete hydatidiform mole (CHM) is characterized by hydropic swelling, trophoblastic proliferation, and vascular immaturity in the stromal cells of chorionic villi. CHM is an uncommon disease in western countries, but the incidence has increased in Asian women, accounting for ∼5.0 in 1000 pregnancies in China 1. CHM has been classified into 2 types according to the genetic origin: the androgenetic CHM and biparental CHM 2. Androgenetic CHM, which arises from the fertilization of an enucleate egg and 1 or 2 haploid sperms, accounts for 90% to 95% of all CHMs. There are 2 kinds of androgenetic CHMs, the monospermic CHM, arising from the fertilization of an enucleate egg and a single haploid sperm, accounts for 70% of CHM, while the dispermic CHM, resulting from fertilization of an enucleate egg by 2 sperms, makes up 20% to 25% of CHM. Biparental CHM is quite rare and is diploid, but of biparental origin. The development of CHM has remained mysterious. Disordered genetic imprinting 3, defective angiogenesis 4, oxidative stress 5, abnormal apoptosis 6, and the dysfunction of mitochondria 7 may be involved in the development of CHM. However, all these findings failed to offer conclusive proof, and the detailed mechanisms of CHM development remain unknown.
X chromosome inactivation (XCI), is considered a dosage compensation strategy to balance sex chromosome contents between female individuals and male individuals, and is believed to be connected with the regulation of embryonic development, as well as with some diseases. The effect of XCI on recurrent miscarriage is contentious because of different inclusion criteria in the studies involved. Some studies suggested that the abnormal XCI may be associated with recurrent miscarriage, while Kaare et al. 8 found that abnormal XCI was not the cause of recurrent miscarriage. In the context of gestational trophoblastic disease, Djuric et al. 9 showed a normal XCI pattern in biparental CHM, but the status of XCI in androgenetic CHM with the 46, XX karyotype has still not been reported, although Siu et al. 10 claimed that Nanog, was an important factor in XCI 11, was overexpressed in gestational disease. Androgenetic CHM is the most common type among CHMs, and understanding its status of X chromosome inactivation may assist in better understanding of the mechanisms of the development of this disease.
In this study, we detected the expression of XCI markers (XIST, TSIX, and XACT) and an X-linked gene (CDX4), and examined other XCI-associated factors, including the methylation status of human androgen receptor gene (HUMARA), and the expressions of H3K27me3, USP21, and Nanog in androgenetic CHMs with the 46, XX karyotype. The aim of our study was to confirm the status of XCI and explain the possible association with the development of CHM.
MATERIALS AND METHODS
Sample Collection
Molar tissue samples were collected from patients treated at Women’s Hospital, School of Medicine, Zhejiang University, China. Villous tissue samples from normal early pregnancy patients were selected as controls. All normal villous tissues were donated by women who underwent surgical abortion due to unexpected pregnancy. All samples were immediately snap-frozen in liquid nitrogen and stored at −80°C for PCR and Western blot assays. Every sample was pathologically diagnosed by a senior pathologist in our hospital, and the immunohistochemical staining of p57 was performed according to a previous report 12 to confirm the diagnosis of CHM. Thereafter, gender identification was performed on the basis of the detection of sex-related gene (SRY) by PCR 13. CHM cases with the 46, XY karyotype were excluded. After gender identification, a polymorphic marker, HUMARA, was used to exclude the biparental CHMs. The HUMARA assay was performed, as previously described 14. A total of 219 fresh tissues were identified as being eligible for this study. They included 71 androgenetic CHMs with the 46, XX karyotype and 74 normal villi with 46, XX karyotype and 74 normal villi with the 46, XY karyotype. Among these, 27 CHMs with 46, XX karyotype, 30 normal villi with 46, XX karyotype, and 30 normal villi with the 46, XY karyotype, collected during April 2012 to June 2014, were used to detect the expression of XIST, TSIX, and CDX4. The remaining samples that were collected from May 2015 to December 2017 were used to detect USP21, Nanog, H3K27me3, and HUMARA-MSP. In addition, paraffin-embedded tissues, including 126 CHMs and 63 normal villi with 46, XX karyotype, collected from January 2010 to March 2012, were used for immunohistochemistry analysis of CDX4.
Sample collection was approved with the informed consent of each woman and by the institutional ethical review board of the hospital.
Real-Time Polymerase Chain Reaction
Trophoblastic tissues were washed twice with PBS to remove the blood and other impurities. Total RNA was extracted using Trizol reagent in accordance with the manufacturer’s instructions (Invitrogen, Paisley, UK). Real-time polymerase chain reaction was performed using the SYBR Premix Ex Taq PCR Kit (Takara Bio, Shiga, Japan), as previously reported 15. The relative expression of RNA was calculated using the 2−(△△Ct) method. The primers used are shown in Table 1.
TABLE 1.
The sequence of the primers
| Gene | Forward primer | Reverse primer |
|---|---|---|
| SRY | 5′AGAGAATCCCAGAATGCGAAAC3′ | 5′CTTCCGACGAGGTCGATACTT3′ |
| XIST | 5′CCATTGAAGATACCACGCTGC3′ | 5′GTCCTCAGGTCTCACATGCTCA3′ |
| TSIX | 5′GTGATCCTCACAGGACTGCAACA3′ | 5′AGCTGAGTCTTCAGCAGGTCCAA3′ |
| XACT | 5′TGCTGATGATGTCCTAAACCTG3′ | 5′AGGAGGTTTGGAGATAGGCA3′ |
| USP21 | 5′GAGCTCACTGAAGCCTTTGC3′ | 5′CCATGAGGAGCTTCAGGAAC3′ |
| Nanog | 5′AAGGTCCCGGTCAAGAAACAG3′ | 5′CTTCTGCGTCACACCATTGC3′ |
| CDX4 | 5′CGAGAAGACTGGAGCGTGTA 3′ | 5′CTGTAGTCGGTCGAGCAGAA3′ |
| GAPDH | 5′GCACCGTCAAGGCTGAGAAC3′ | 5′TGGTGAAGACGCCAGTGGA3′ |
| HUMARA | 5′GCTGTGAAGGTTGCTGTTCCTCAT3′ | 5′TCCAGAATCTGTTCCAGAGCGTGC3′ |
| HUMARA-M | 5′GCGAGCGTAGTATTTTTCGGC3′ | 5′AACCAAATAACCTATAAAACCTCTACG3′ |
| HUMARA-U | 5′GTTGTGAGTGTAGTATTTTTTGGT3′ | 5′CAAATAACCTATAAAACCTCTACA3′ |
Western Blotting Analysis
Trophoblastic tissues were lysed in RIPA buffer. The protein concentration was measured using the Bradford Protein Assay Kit (Beyotime, Shanghai, China) according to the manufacturer’s protocol. Western blot was performed as previously described 16. The membranes were incubated with primary antibodies (all from Abcam, Cambtidge, UK) against H3K27me3(ab192985, 1:1000), USP21(ab38864, 1:200), Nanog (Abcam, ab109250, 1:1000), and GAPDH (Abcam, ab8245, 1:10000). The bands were detected with FDbio-Dura ECL (FUDE, Zhejiang, China) using the Imagequant LAS400mini (GE Healthcare, Buckinghamshire, UK). Image J software (NIH, Bethesda, MD) was used to measure the gray value of each band. The signal of each band was normalized to that of GAPDH in the same membrane before comparison.
Immunohistochemistry
Immunohistochemical staining was performed according to a previous report 12. Negative controls (substitution of primary antibody for rabbit IgG) were run simultaneously. Each sample was assigned to one of the following categories: 0 (0%–4%), 1 (5%–24%), 2 (25%–49%), 3 (50%–74%), or 4 (75%–100%). The intensity of immunostaining was graded as follows: 0, negative; 1 +, weak; 2 +, moderate or 3 +, strong. A final score was obtained by multiplying 2 scores. Scores of 0 to 4 were defined as “low expression,” scores of 5 to 8 were defined as “moderate expression,” and scores of 9 to 12 were defined as “high expression.”
Immunofluorescence Analysis
Normal villi tissues and CHM tissues were voluntarily donated by women who underwent surgical abortion. After gender identification, the trophoblastic tissue sections were immunostained with the primary antibodies (H3K27me3, Abcam, ab192985, 1:1000; USP21, Abcam, ab38864, 1:200; Nanog, Abcam, ab109250, 1:1000), as discussed before 17 and examined using confocal microscopy (Olympus, Tokyo, Japan).
HUMARA-MSP
The methylation-specific polymerase chain reaction (HUMARA-MSP) assay was used to analyze XCI in androgenetic CHM. Genomic DNA was isolated from trophoblastic tissues using the TIANamp Genomic DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. The genomic DNA was used for a bisulfite conversion procedure using the EpiTect Bisulfite Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The bisulfite-treated DNA was then amplified via polymerase chain reaction with 2 sets of primers listed in Table 1, as previously described 18. The products were examined by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Statistical Analysis
Statistical analysis was performed with the SPSS 19.0 software package (IBM, Armonk, NY). All experiments were repeated 3 times. Differences among groups were first analyzed by 1-way analysis of variance. The LSD t test was then used to further analyze the differences between each of the 2 groups. Wilcoxon rank sum test was used to analyze the difference in expression of CDX4 between CHMs and normal villi. P<0.05 was considered as statistically significant.
RESULTS
The XCI Pattern in CHM
Lower Expression of XIST and Higher Expression of TSIX in Androgenetic CHMs With the 46, XX Karyotype
Real-time polymerase chain reaction revealed that the expression of XIST RNA was significantly decreased and the expression of TSIX RNA was significantly increased in androgenetic CHMs with the 46, XX karyotype, compared with that in normal female villi (both P<0.01), while the expression of XIST RNA and TSIX RNA expression in androgenetic CHMs was similar to that in normal male villi (Fig. 1). We also detected the expression of XACT, another noncoding RNA, which can compete with XIST in the control of XCI during human early development. Although the expression of XACT was higher in androgenetic CHMs compared with that in normal villi, it was not significantly different from the levels found in the other 2 groups (P1(CHMs vs. normal female villi) =0.488; P2(CHMs vs. normal male villi)=0.381).
FIG. 1.
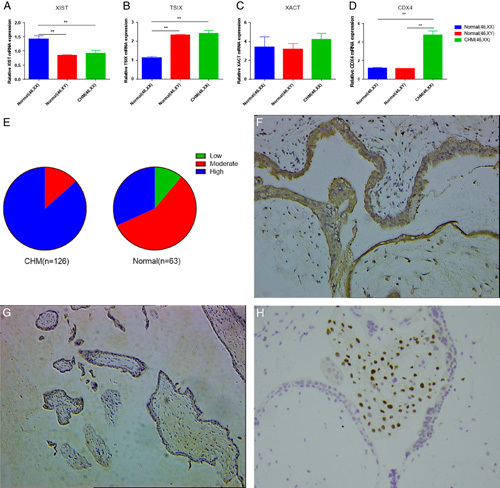
The expression of XIST, TSIX, XACT, and CDX4. (A) XIST RNA was significantly decreased in androgenetic complete hydatidiform moles (CHMs) compared with that in normal female villi (P<0.01), but was similar to that in normal male villi (P=0.573). (B) The level of TSIX RNA was significantly increased in androgenetic CHMs (P<0.01), but was similar to that in normal male villi (P=0.641). (C) XACT RNA was overexpressed in androgenetic CHMs compared with that in normal villi, but no significant difference was observed (F=0.431, P=0.651, P1=0.488, P2=0.381, P3=0.854). (D) The expression of CDX4 mRNA in androgenetic CHMs was significantly higher than that in normal female and male villi (both P<0.01). (E) CDX4 immunostaining intensity in CHMs and normal villi. (F) The immunohistochemistry of CDX4 protein in CHM (200×). (G) CDX4 immunostaining intensity in normal female villous (200×). (H) The immunohistochemistry of p57 protein in CHM (200×). Data are expressed as mean±SEM. **P<0.01. P1: androgenetic CHMs versus normal female villi, P2: androgenetic CHMs versus normal male villi, P3: normal female villi versus normal male villi.
Higher Expression of CDX4 in Androgenetic CHMs With the 46, XX Karyotype
We detected the expression of CDX4 mRNA in androgenetic CHMs with the 46, XX karyotype (n=27) and normal villi with the 46, XX (n=30) and 46, XY karyotype (n=30) by qPCR, and found that the expression of CDX4 mRNA in androgenetic CHMs was significantly higher than that in normal female and male villi (both P<0.01), as seen in Figure 1D. We further detected CDX4 protein expression in paraffin-embedded samples, including 126 CHMs and 63 normal female villi, by immunohistochemistry, and found that the expression of CDX4 protein in CHMs was significantly higher than that in normal female villi (z=−7.75, P<0.01), as shown in Figures 1E–G and Table 2.
TABLE 2.
CDX4 immunostaining intensity in CHMs and normal villi
| CDX4 expression | Low | Moderate | High | Sum |
|---|---|---|---|---|
| CHMs | 0 | 17 | 109* | 126 |
| Normal villi | 7 | 36 | 20 | 63 |
| Sum | 7 | 53 | 129 | 189 |
*Compared with normal villi (z=−7.75, P=0.000).
CHM indicates complete hydatidiform mole.
Decreased Expression of H3K27me3 in Androgenetic CHMs With the 46, XX Karyotype
We measured the levels of H3K27me3 in androgenetic CHMs with the 46, XX karyotype (n=44), and in male (n=44) and female villi (n=44) by Western blot. The levels of H3K27me3 in androgenetic CHMs were significantly lower than those in normal female villi (P<0.01), but was not significantly different from those in normal male villi (Fig. 2A). We further detected the expression and localization of H3K27me3 using immunofluorescence analysis. H3K27me3 was mainly localized in the cell nucleus (Fig. 2B), and normal female villi displayed increases in fluorescence intensity of H3K27me3 compared with androgenetic CHMs and normal male villi, consistent with the results of Western blot.
FIG. 2.
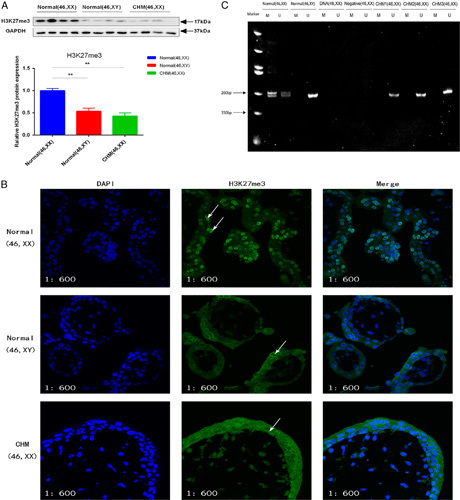
Levels of H3K27me3 and the result of HUMARA-MSP. (A) The level of H3K27me3 was decreased in androgenetic complete hydatidiform moles (CHMs) and in normal male villi compared with that in normal female villi (P<0.01), but there was no difference of the level of H3K27me3 between androgenetic CHMs and normal male villi (P=0.268). (B) Normal female villi showed intense H3K27me3 staining in cell nucleus (arrows). CHMs and normal male villi showed moderate H3K27me3 staining, and mostly in the cytoplasm (arrows); 600× (C) Size of the PCR products of the HUMARA gene was ∼200 bp. U, PCR products amplified with HUMARA-U primers; M, PCR products amplified with HUMARA-M primers. Data are expressed as mean±SEM. **P<0.01.
Lack of HUMARA Methylation in Androgenetic CHMs With the 46, XX Karyotype
We detected the methylation status of XCI CpG sites in exon1 of the HUMARA X chromosome gene in androgenetic CHMs using HUMARA-MSP. After electrophoresis, the number of bands in each lane indicated the genetic origins of samples. Specifically, 1 band meant that the sample only contained androgenetic genetic material, while 2 bands indicated that the samples had biparental genetic origin. If the products amplified with HUMARA-M primers existed, there was an inactive X chromosome in the sample, and vice versa. The lengths of the PCR products from the CAG repeat region in the HUMARA was ∼200 bp. The products amplified with HUMARA-U primers showed 2 bands in normal female villi, while there was only 1 band in normal male villi and androgenetic CHMs. In addition, the products amplified with HUMAR-M primers showed 2 bands in normal female villi, but no bands were evident in normal male villi and androgenetic CHMs (Fig. 2C). These results indicated that there was no inactive X chromosome in androgenetic CHM. In other words, both X chromosomes were active in androgenetic CHM with the 46, XX karyotype.
The Expression of USP21 and Nanog in Androgenetic CHMs With 46, XX Karyotype
Measurement of the expression of Nanog and USP21 protein in androgenetic CHMs with the 46, XX karyotype (n=44), in male villi (n=44), and in female villi (n=44) by Western blot revealed the higher expression of Nanog and USP21 protein in androgenetic CHMs than that in normal villi (both P<0.01) (Figs. 3A, B). Furthermore, we examined the localization of USP21 and Nanog by immunofluorescence analysis. We found that USP21 was colocalized with Nanog in villous tissues (Fig. 3C). Meanwhile, the fluorescence intensity of USP21 and Nanog was higher in androgenetic CHMs than in normal villi.
FIG. 3.
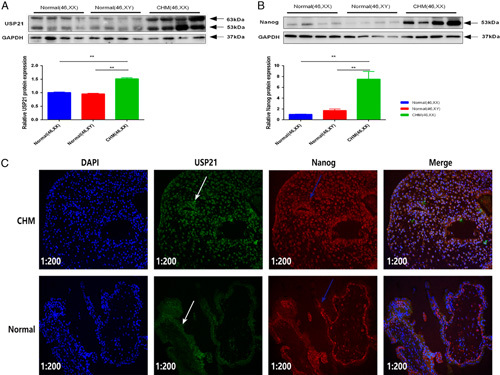
Expression of USP21 and Nanog. (A and B) USP21 and Nanog protein were overexpressed in androgenetic complete hydatidiform moles (CHMs), and their levels were compared with that in normal villi (both P<0.01). (C) Normal villi showed moderate USP21 (white arrows) and Nanog (blue arrows) expressions, which were mostly localized in the cytoplasm. Androgenetic CHMs displayed intense nuclear expression of USP21 and Nanog. Immunofluorescence analysis also showed that USP21 colocalized with Nanog in androgenetic CHMs (200×). Data are expressed as mean±SEM. **P<0.01.
DISCUSSION
Androgenetic CHM is the main kind of CHM, and is prone to undergoing malignant transformation. However, the mechanisms involved in the development of this kind of CHMs remain unclear. X chromosome inactivation is a very important process during the development of normal female mammals. Failure of X chromosome inactivation blocks normal embryogenesis shortly after implantation. Although it was confirmed that biparental CHM possessed a normal XCI pattern, the status of XCI in androgenetic CHM with the 46, XX karyotype has been unknown. Here, for the first time, we find that loss of X chromosome inactivation may have something to do with the development of androgenetic CHM.
Many methods have been established to evaluate the XCI pattern according to the characteristics of the inactive X chromosome and the mechanisms of XCI. Compared with the active X chromosome, the inactive X chromosome showed many characteristics of constitutive heterochromatin 19, including maintenance of the condensed state throughout interphase, late DNA replication, decreased abundance of histone acetylation, lower methylation level of histone H3 lysine-4, and higher methylation level of histone H3 lysine-9 and H3 lysine-27. These characteristics have been used to evaluate the XCI pattern 20,21. It has also been confirmed that XIST is important in the process of XCI. Thus, the detection of XIST RNA expression and XIST RNA coating in cis using fluorescence in situ hybridization has been widely used to examine the XCI pattern 22. Here, we monitored the expression of XCI markers, including XIST, TSIX, and H3K27me3, and using HUMARA-MSP to elucidate the status of XCI in androgenetic CHMs. The levels of XIST RNA were significantly decreased, while those of TSIX RNA were significantly increased in androgenetic CHMs compared with those in normal female villi, but the levels of both XIST and TSIX RNA were similar to those in normal male villi. The levels of H3K27me3 in androgenetic CHMs were significantly lower than those in normal female villi, but were similar to those in normal male villi. Moreover, there was a lack of HUMARA methylation in CHMs. The collective results suggested that both the X chromosomes were activated in androgenetic CHMs with the 46, XX karyotype. Previous studies demonstrated that the X-linked CDX4 gene is essential for the development of the villi in a variety of ways, including interaction with the Wnt pathway and enhancing the function of CDX2 23–26. The latter can cooperate with SOX2 and GATA3 and prompt the proliferation of trophoblastic cells during trophectoderm formation. In our study, CDX4 was overexpressed in androgenetic CHMs compared with normal villi. Thus, our results suggest that androgenetic CHM lacks the normal mechanics of XCI, and the loss of XCI may contribute to the development of androgenetic CHM by inducing the overexpression of CDX4.
Ubiquitination played a role in the process of XCI by stabilizing many factors 27,28. USP21, a wildly explored deubiquitinase, has been confirmed to have the ability to deubiquitinate and stabilize Nanog 29, a star molecule involved in the process of XCI. Nanog can regulate XCI in many ways, which include regulating the expression of XIST by binding to its first intron 11, occupying approximately one-third of the PRC2-occupied genes, which are closely related with embryonic development 30, and interacting with many chromatin-remodeling factors, such as SWI-SNF 31 and NuRD 32 complexes. In this study, the expressions of Nanog and UPS21 protein were both higher in androgenetic CHMs than those in normal villi, which was consistent with the previous description of Nanog overexpression in gestational trophoblastic disease 10. Meanwhile, we found USP21 colocalized with Nanog in villous tissues. Thus, our results hinted that USP21 may cause the disruption of XCI in androgenetic CHM via stabilization of Nanog.
However, there are some shortages in our study. First, early complete moles often present with minimal amount of chorionic villi and frequently admixed with large quantity of gestational endometrium in a curettage specimen; hence, no matter how careful we are, it is very difficult to totally avoid the maternal tissue contamination, which to some extent, may lower the possibility of our results, which show that 2 X chromosomes are both active in CHMs. Second, androgenetic CHMs with 46, XX karyotype can be further divided into 2 categories, monospermic CHM and dispermic CHM, and, although they are all of androgenetic origin, they still have some difference, which to some extent has some effect on our results. Third, due to the inaccessibility of the CHM cell lines, all our research studies have been carried out on tissues. As a result, although we have found that 2 X chromosomes may all be active in androgenetic CHMs, and the expressions of USP21 and Nanog are higher in androgenetic CHMs, we still do not know whether they are the cause or the result of androgenetic CHMs. Further research studies are needed in order to better understand the mechanisms involved in the initiation and development of CHMs.
CONCLUSIONS
To our knowledge, these findings, for the first time, show that both X chromosomes are active in androgenetic CHMs with the 46, XX karyotype, and that the USP21-Nanog pathway may be involved in the disruption of XCI during this process. These findings may be relevant as one of the mechanisms involved in the development of androgenetic CHM.
Acknowledgments
The authors are grateful for the funding supported by the key research and development program of Zhejiang province, China.
Footnotes
The authors consider that the first 2 authors should be regarded as joint first authors: X.C. and Y.M.
This study was approved by the Ethics Committee of Women’s Hospital, School of Medicine, Zhejiang University. All patients provided written informed consent for tissue sampling used for scientific research before surgical treatment.
X. Chen, Y.M., and L.W.: designed the study, collected the samples, performed the experiments, carried out statistical analysis and drafted the manuscript. Y.Y.: participated in the sample collection and statistical analysis. X.Z. participated in the sample collection and re-reviewed all the studied specimens at the time of this study. X. Cheng, X.X., and W.L.: conceived the study, participated in its design and coordination, and revised the final manuscript.
Supported by the key research and development program of Zhejiang province, China (Grant No: 2019C03010).
The authors declare no conflict of interest.
REFERENCES
- 1. Song HZ, Wu PC. Hydatidiform mole in China—a preliminary survey of incidence on more than 3 million women. B World Health Organ 1987;65:507–11. [PMC free article] [PubMed] [Google Scholar]
- 2. Fisher RA, Hodges MD. Genomic imprinting in gestational trophoblastic disease—a review. Placenta 2003:S111–S8. [DOI] [PubMed] [Google Scholar]
- 3. Fisher RA, Hodges MD, Rees HC, et al. The maternally transcribed gene p57(KIP2) (CDNK1C) is abnormally expressed in both androgenetic and biparental complete hydatidiform moles. Hum Mol Genet 2002;11:3267–72. [DOI] [PubMed] [Google Scholar]
- 4. Lisman BAM, Boer K, Bleker OP, et al. Vasculogenesis in complete and partial hydatidiform mole pregnancies studied with CD34 immunohistochemistry. Hum Reprod 2005;20:2334–9. [DOI] [PubMed] [Google Scholar]
- 5. Muneyyrici-Delale O, Nacharaju VL, Sidell J, et al. 11beta-Hydroxysteroid dehydrogenase activity in pregnancies complicated by hydatidiform mole. Am J Reprod Immunol 2006;55:415–9. [DOI] [PubMed] [Google Scholar]
- 6. Sharp AN, Heazell AEP, Crocker IP, et al. Placental apoptosis in health and disease. Am J Reprod Immunol 2010;64:159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Durand S, Dumur C, Flury A, et al. Altered mitochondrial gene expression in human gestational trophoblastic diseases. Placenta 2001;64:220–6. [DOI] [PubMed] [Google Scholar]
- 8. Kaare M, Painter JN, Ulander VM, et al. Sex chromosome characteristics and recurrent miscarriage. Fertil Steril 2008;90:2328–33. [DOI] [PubMed] [Google Scholar]
- 9. Djuric U, El-Maarri O, Lamb B, et al. Familial molar tissues due to mutations in the inflammatory gene, NALP7, have normal postzygotic DNA methylation. Hum Genet 2006;120:390–5. [DOI] [PubMed] [Google Scholar]
- 10. Siu MK, Wong ES, Chan HY, et al. Overexpression of NANOG in gestational trophoblastic diseases: effect on apoptosis, cell invasion, and clinical outcome. Am J Pathol 2008;173:1165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Navarro P, Chambers I, Karwacki-Neisius V, et al. Molecular coupling of Xist regulation and pluripotency. Science 2008;321:1693–5. [DOI] [PubMed] [Google Scholar]
- 12. Liang Y, Zhang X, Chen X, et al. Diagnostic value of progesterone receptor, p16, p53 and pHH3 expression in uterine atypical leiomyoma. Int J Clin Exp Pathol 2015;8:7196–202. [PMC free article] [PubMed] [Google Scholar]
- 13. Xiang J, Li Z, Wan Q, et al. A qPCR method to characterize the sex type of the cell strains from rats. Biosci Biotechnol Biochem 2016;80:1917–24. [DOI] [PubMed] [Google Scholar]
- 14. Allen RC, Zoghbi HY, Moseley AB, et al. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992;51:1229–39. [PMC free article] [PubMed] [Google Scholar]
- 15. Ye F, Jiao J, Zhou C, et al. Nucleotide excision repair gene subunit XPD is highly expressed in cervical squamous cell carcinoma. Pathol Oncol Res 2012;18:969–75. [DOI] [PubMed] [Google Scholar]
- 16. Hu D, Zhou J, Wang F, et al. HPV-16 E6/E7 promotes cell migration and invasion in cervical cancer via regulating cadherin switch in vitro and in vivo. Arch Gynecol Obstet 2015;292:1345–54. [DOI] [PubMed] [Google Scholar]
- 17. Soni S, Rath G, Prasad CP, et al. Fas-FasL system in molar pregnancy. Am J Reprod Immunol 2011;65:512–20. [DOI] [PubMed] [Google Scholar]
- 18. Andoh-Noda T, Akamatsu W, Miyake K, et al. Differential X chromosome inactivation patterns during the propagation of human induced pluripotent stem cells. Keio J Med 2017;66:1–8. [DOI] [PubMed] [Google Scholar]
- 19. Panning B, Dausman J, Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell 1997;90:907–16. [DOI] [PubMed] [Google Scholar]
- 20. Cotton AM, Lam L, Affleck JG, et al. Chromosome-wide DNA methylation analysis predicts human tissue-specific X inactivation. Hum Genet 2011;130:187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Plath K, Fang J, Mlynarczyk-Evans SK, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003;300:131–5. [DOI] [PubMed] [Google Scholar]
- 22. Panova AV, Nekrasov ED, Lagarkova MA, et al. Late replication of the inactive X chromosome is independent of the compactness of chromosome territory in human pluripotent stem cells. Acta Naturae 2013;5:54–61. [PMC free article] [PubMed] [Google Scholar]
- 23. Beck F, Erler T, Russell A, et al. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn 1995;204:219–27. [DOI] [PubMed] [Google Scholar]
- 24. Savory JGA, Mansfield M, St Louis C, et al. Cdx4 is a Cdx2 target gene. Mech Develop 2011;128:41–48. [DOI] [PubMed] [Google Scholar]
- 25. Keramari M, Razavi J, Ingman KA, et al. Sox2 is essential for formation of trophectoderm in the preimplantation embryo. Plos One 2010;5:e13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ralston A, Cox BJ, Nishioka N, et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 2010;137:395–403. [DOI] [PubMed] [Google Scholar]
- 27. Gontan C, Achame EM, Demmers J, et al. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature 2012;485:386–90. [DOI] [PubMed] [Google Scholar]
- 28. Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 2006;75:243–69. [DOI] [PubMed] [Google Scholar]
- 29. Jin J, Liu J, Chen C, et al. The deubiquitinase USP21 maintains the stemness of mouse embryonic stem cells via stabilization of Nanog. Nat Commun 2016;7:13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006;125:301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fry CJ, Peterson CL. Chromatin remodeling enzymes: who’s on first? Curr Biol 2001;11:R185–197. [DOI] [PubMed] [Google Scholar]
- 32. Liang J, Wan M, Zhang Y, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 2008;10:731–9. [DOI] [PubMed] [Google Scholar]


