Abstract
Eukaryotic cells have divided the steps of gene expression between their nucleus and cytoplasm. Protein-encoding genes generate mRNAs in the nucleus and mRNAs undergo transport to the cytoplasm for the purpose of producing proteins. Cap-binding protein (CBP)20 and its binding partner CBP80 have been thought to constitute the cap-binding complex (CBC) that is acquired co-transcriptionally by the precursors of all mRNAs. However, this principle has recently been challenged by studies of nuclear cap-binding protein 3 (NCBP3). Here we submit how NCBP3, as an alternative to CBP20, an accessory to the canonical CBP20–CBP80 CBC, and/or an RNA-binding protein – possibly in association with the exon-junction complex (EJC) – expands the capacity of cells to regulate gene expression.
Is NCBP3 a CBP, a CBC Accessory Protein, an RNA-Binding Protein, or All Three?
Evidence That NCBP3 Is an Alternative CBP
The canonical nuclear cap-binding complex (CBC) is a heterodimer comprised of the cap-binding protein (CBP)80 and CBP20, also known as nuclear cap-binding protein (NCBP)1 and NCBP2, respectively. While CBP20 directly binds the m7G cap (see Glossary) of RNA polymerase II (RNAPII)-synthesized transcripts, including the precursors of mRNAs (pre-mRNAs) and mature mRNAs, CBP80 stabilizes CBP20 binding to the cap without contacting the cap and recruits numerous cofactors that influence many nuclear and some cytoplasmic steps of mRNA metabolism (Box 1). Since its discovery in 1990 [1], studies of the CBC have been largely restricted to CBP80 because CBP80 antibody, unlike CBP20 antibody, was readily available. These studies, together with the observation that the cap-binding activity of the CBC could be recapitulated using purified recombinant CBP20 and CBP80 [2], instilled the idea that, as a rule, CBP20 and CBP80 do not function independently of one another. In 2015, Gebhardt et al. challenged this view, proposing that CBP20 function could be replaced by at least one other CBP [3]: analysis of human HeLa-cell proteins that co-purified with an m7G-capped synthetic RNA identified chromosome 17 open reading frame 85 (C17orf85), a poorly characterized RNA-binding protein, as a putative CBP. Gebhardt et al. renamed C17orf85 as NCBP3.
Box 1. Roles of the CBP20–CBP80 CBC in the Life Cycle of mRNAs.
The CBP20–CBP80 heterodimer, referred to as the canonical CBC, is acquired co-transcriptionally at the cap of RNAPII-synthesized transcripts, including but not limited to pre-mRNAs [2]. CBP80 stabilizes the direct binding of CBP20 to m7G caps and recruits numerous cofactors to support virtually every step in the mRNA life cycle [2].
The mRNA life cycle starts in the nucleus where pre-mRNAs are synthesized during a process called gene transcription. Gene transcription in higher eukaryotes is divided into highly interconnected steps that typically include chromatin loosening followed by RNAPII-mediated activities of transcription initiation, promoter-proximal pausing, transcription elongation, and transcription termination. Pre-mRNAs are processed largely co-transcriptionally in steps that include 5′-end m7G capping, which occurs coincident with promoter-proximal pausing, intron removal by splicing, and 3′-end formation by endonucleolytic cleavage coupled to the addition of a nontemplated poly(A) tail. Numerous pre-mRNA quality-control pathways have evolved to ensure that only fully processed mRNAs are exported to the cytoplasm [55]. Roles of the CBC in nuclear pre-mRNA synthesis, processing, and quality control have been reviewed recently [20,56].
Cytoplasmic ribosomes begin to translate newly synthesized mRNAs as they are exported from the nucleus to the cytoplasm, while maintaining an association with the nuclear pores. These pioneer rounds of translation, which are defined as the translation of CBP80-bound mRNAs, support an mRNA quality-control pathway called NMD. NMD prevents the production of truncated and potentially toxic proteins by mRNAs that prematurely terminate translation due to a mis-splicing event or a disease-associated genomic mutation, as two examples. Utilizing the ~5–10% of cellular mRNAs that are natural NMD targets, cells regulate the efficiency of NMD by a variety of mechanisms to adapt to differing environmental or developmental changes. We and others have reviewed CBC functions during the pioneer round of translation and NMD [40,41,57].
For the majority of cellular mRNAs not targeted for NMD, the CBC is replaced by eIF4E [58,59], a cytoplasmic CBP that supports the bulk of cellular translation [60]. Singh et al. proposed that NCBP3 acts as a transitional component during the replacement of the CBC with eIF4E at the cap of translating mRNAs [6]. Finally, eIF4E-bound mRNAs can be translationally silenced and eventually degraded via numerous mechanisms independent of the CBC [61].
Consistent with the idea that NCBP3 can replace CBP20 in a noncanonical NCBP3–CBP80 CBC (Figure 1A, panel 1), NCBP3 is sufficient to recruit CBP80 to an m7G-cap analog in vitro [4]. Like CBP20, NCBP3 directly binds m7G caps through an N-terminal RNA-recognition motif (RRM) [3,4], and its C-terminal half interacts with CBP80 in an RNase-insensitive manner [3–6] (Figure 1B). However, unlike CBP20, the affinity of NCBP3 for m7G caps (KD = 5.1 μM) is not significantly enhanced by CBP80 [4]. Moreover, while this affinity exceeds that of CBP20 alone (KD = 84 μM), it remains more than ten-times lower than that of the CBP20–CBP80 CBC (KD = 0.37 μM) [4].
Figure 1.
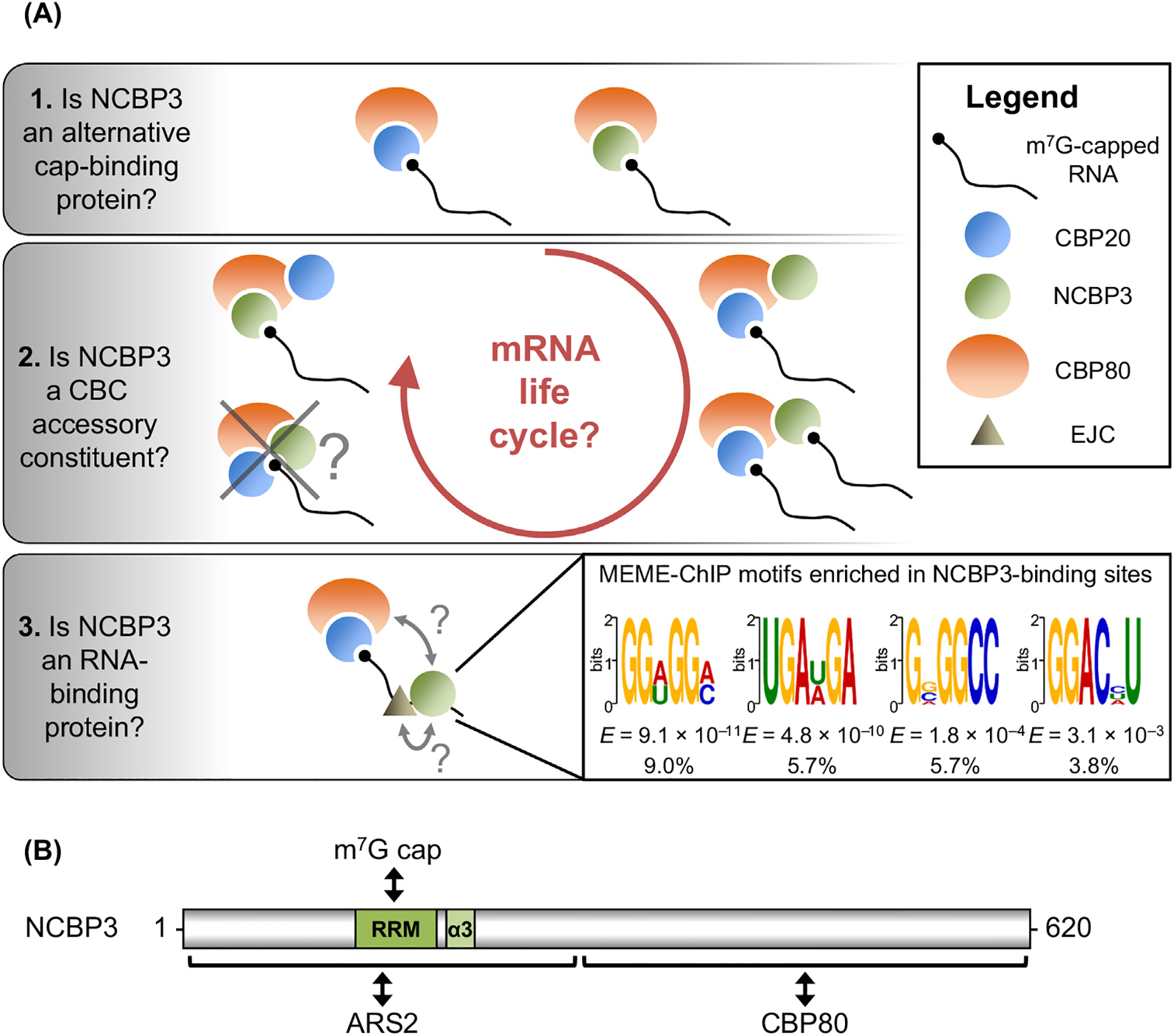
Determinants of Nuclear Cap-Binding Protein 3 (NCBP3) Functions as a Cap-Binding Protein, a Cap-Binding Complex (CBC) Accessory Protein, and an RNA-Binding Protein. (A) Panels 1–3 illustrate three possible configurations with which cellular NCBP3 may interact with pre-mRNAs and their derived mRNAs, binding directly to the m7G cap alone (Panel 1), binding together with CBP20 either directly to the m7G cap, or via CBP80 (Panel 2), and/or binding to the body of the transcript, possibly in association with exon-junction complexes (EJCs) or with the CBC (Panel 3). Any or all of these scenarios could apply to particular transcripts based on, for example, the promoter at which transcription initiates, co-transcriptional processes, or response to cellular stress. (Panel 2) We posit that, if both NCBP3 and CBP20 are bound to caps in the same complex, they are not likely to be bound to the cap of the same molecule given the established and predicted cap-binding mode of their respective RNA-recognition motifs (RRMs) (Box 2). (Panel 3) MEME-ChIP parameters: dreme dna -p /seqs-centered -n ./seqs-shuffled -norc -e 0.05 were used to define consensus binding sites. E, E-value [i.e. the number of expected hits of similar quality (score) that could be found by chance alone]. Percentages are the fractions of NCBP3-binding sites that have the corresponding MEME-ChIP motif. (B) Diagram of human NCBP3 denoting its RRM, its putative RRM-associated α3-helix, and its arsenite resistance protein 2 (ARS2)- and cap-binding protein 80 (CBP80)-interacting regions. Numbers specify amino acids.
Evidence That NCBP3 Can Coexist with CBP20 as a CBC Accessory Protein
Challenging the conclusion that NCBP3 replaces CBP20 in an alternative CBC, depletion of CBP20 abolishes the association of NCBP3 with CBP80 in HeLa cells [7]. Furthermore, in vitro work using purified recombinant proteins demonstrated that NCBP3 may form a complex with CBP20–CBP80 in both the presence and the absence of an m7GTP-cap analog and the CBC cofactor arsenite resistance protein 2 (ARS2) [4]. The model, proposed in 2018 by Schulze et al. [4], describes how ARS2 acts as a co-transcriptional platform that sorts different types of RNAs and regulates their fate through the mutually exclusive recruitment of diverse accessory constituents, including NCBP3, to m7G-cap-bound CBP20–CBP80. In this model, NCBP3 supports the productive synthesis of export-competent multiexonic mRNAs by competing for CBC binding with the RNA decay factor ZC3H18 [7]. Whether NCBP3 binds the cap in the CBP20–CBP80–ARS2–NCBP3–m7GTP complex and, if it does, whether NCBP3 binds the cap alone or with CBP20 (Figure 1A, panel 2) remains to be determined.
The existence in cells of a complex that includes NCBP3 and CBP20 is supported by the observation that CBP20 was present after the immunoprecipitation (IP) of exogenously expressed NCBP3 from human HeLa-cell lysates that were or were not treated with benzonase, an endonuclease that degrades all forms of DNA and RNA [3,7,8]. However, CBP20 was not present after IP of endogenous NCBP3 from the cytoplasmic fraction of human HEK293-cell lysates that had not been exposed to nucleases [6]. While this discrepancy may be explained by technical biases, we believe that it is more likely to reflect cell-type specificities and/or remodeling of the CBC during the mRNA lifecycle (Figure 1A and Box 1). Supporting this idea, CBP20 is ~20-times more abundant than NCBP3 in polysome fractions of HeLa S3-cell lysates [9], but it is NCBP3 and not CBP20 that is detected in polysome fractions of HEK293-cell lysates [6]. By contrast, CBP80 is readily detectable in polysome fractions of both cell types [6,9]. Thus, while CBP20 and NCBP3 may coexist in the same complex in the nucleus, the loss or acquisition of proteins associated with mRNA during its nuclear export and/or pioneer round(s) of translation (Box 1) may disrupt these complexes, resulting in translating mRNAs bound by either CBP20 or NCBP3.
Evidence That NCBP3 Is an RNA-Binding Protein
Beyond studies establishing NCBP3 as a cap-binding or a CBC accessory protein, photoactivatable ribonucleotide-enhanced crosslinking and IP (PAR-CLIP) experiments have demonstrated that NCBP3 binds comparably with mRNA 5′ untranslated regions (UTRs) (i.e., transcribed nucleotides that reside in close proximity to the m7G cap) and to downstream coding sequences (CDSs) [10]. Arguably, while NCBP3 footprints that are not cap proximal may reflect RNA loops that connect cap-bound NCBP3 to the mRNA backbone, it is more likely that NCBP3 binds the mRNA backbone directly. Using Multiple Em for Motif Elicitation-Chromatin IP (MEME-ChIP) analyses [11], we identified four independent G-rich motifs that were significantly enriched among NCBP3-binding sites (Figure 1A, panel 3). This observation is compatible with the interesting hypothesis that NCBP3 binds m7G nucleotides in the mRNA body [7]. However, each motif constitutes less than 10% of all NCBP3-binding sites, suggesting that NCBP3 preferentially targets degenerate RNA sequences and/or secondary RNA structures, possibly by collaborating with other RNA-binding proteins or protein complexes.
One comparative analysis of CLIP experiments revealed that NCBP3 RNA-binding sites significantly overlap with binding sites for the cytoplasmic RNA ligase RTCB [12]. However, no consensus binding sequence was determined among these overlapping sites [12], and there is nothing in the literature that provides evidence for a NCBP3–RTCB complex either in cells or in vitro. Therefore, the biological relevance of NCBP3 and RTCB sharing RNA-binding sites remains to be determined (see later).
An independent analysis found that NCBP3 PAR-CLIP footprints are enriched 10–35 nucleotides upstream of mRNA exon-exon junctions [i.e., where exon-junction complexes (EJCs) reside] [7]. Consistent with this finding, NCBP3 readily associates with HeLa cell constituents of EJCs, even in the presence of RNase or benzonase [3,7,8]. Furthermore, in vitro-binding assays using purified proteins demonstrated that m7GTP-bound NCBP3 directly interacts with the EJC core, but only if the EJC has been fully assembled and locked onto RNA [7]. Thus, it is possible that the NCBP3–EJC interaction occurs after splicing and that NCBP3 can simultaneously bind the EJC and the mRNA cap or internal m7G nucleotides (Figure 1A, panel 3). Notably, NCBP3 was not co-purified with EJC core constituents in HEK293 cells [13], reinforcing the idea that NCBP3 may have different functions in different cell types.
Possible Parameters That Direct NCBP3 Binding Primarily to mRNAs
Regardless of whether it directly or indirectly binds 5′ caps or internal m7Gs, anti-NCBP3 IP of RNA followed by deep sequencing (RIP-seq) revealed that NCBP3 associates primarily with mRNAs but also, albeit to a lesser extent, with long noncoding RNAs (lncRNAs) [3]. mRNAs and lncRNAs are two types of long (i.e., ≥200–300 nucleotides) and generally polyadenylated transcripts synthesized by RNAPII. Notably, relative to mRNAs, lncRNAs contain fewer introns [14], are more often retained in the nucleus [15], and, by definition, are not thought to support protein synthesis. Unlike CBP20, NCBP3 does not significantly bind to small nuclear RNAs (snRNAs) or replication-dependent histone (RDH) mRNAs (i.e., small, intronless RNAPII-synthesized transcripts that are not polyadenylated) [3,4,16,17]. Together, these observations suggest that NCBP3 binding to RNA may depend on RNA length and/or particular RNA processes such as splicing, nuclear export, and/or translation. Experimental data support the idea that longer RNAs and splicing promote NCBP3 recruitment.
ARS2 and the RNA-binding heterogeneous nuclear ribonucleoprotein particle C (hnRNPC) most likely cooperate to promote NCBP3 recruitment to and/or stabilization at the 5′ end of long RNAPII-synthesized transcripts. In one mechanism, binding of (i) NCBP3, (ii) the snRNA CBC accessory protein phosphorylated adapter for RNA export (PHAX), and (iii) the RDH mRNA CBC accessory protein Fas-associated death domain (FADD)-like interleukin (IL)-1β-converting enzyme (FLICE) associated with a huge protein (FLASH) to the C-terminal leg domain of ARS2 are three mutually exclusive interactions [4]. In another mechanism, PHAX binding to the CBC of long RNAs is inhibited by hnRNPC, a molecular ruler around which are wrapped unstructured 200–300-nucleotide stretches [18].
Supporting the idea that splicing may promote the recruitment of NCBP3 to mRNA, NCBP3 stably associates with a reporter mRNA that derives from splicing in HeLa-cell nuclear extracts, but does not associate with a reporter mRNA that cannot undergo splicing [19]. This finding is consistent with the tight association of NCBP3 with the EJC (see earlier), which is likewise acquired as a consequence of pre-mRNA splicing.
It was also proposed that different promoters can shape the identity of CBC accessory proteins [20]. Supporting the role for additional genomic features, using ToppFun [21] we found that DNA encoding the 3′UTR of mRNAs that are preferentially bound by NCBP3 relative to CBP20 [3] are enriched in binding sites for certain transcription factors, such as ELK1 (P = 7.9 × 10−5) and NRF1 (P = 2.1 × 10−4).
Is the NCBP3 RRM and Its C-terminal α3-Helix a Versatile Platform Controlling Binding to a 5′ Cap, RNA, and/or Protein Complexes?
The RRM of NCBP3 was found to share the highest sequence homology with the RRM of poly(A)-specific ribonuclease (PARN), whose features have been used to predict the structure and function of the RRM of NCBP3 [3] (Box 2). In agreement with in silico predictions (Figure 2A), wet-bench experiments demonstrated that, like the RRM of PARN, the RRM of NCBP3 binds an m7G-cap analog using a motif located in the β2–β3 loop of the RRM β-sheet [3]. Using Iterative Threading ASSEmbly Refinement (I-TASSER) to generate a homology model of the NCBP3 RRM using previously determined structures [22], we show here that, in addition to binding the cap, the NCBP3 RRM is predicted to bind RNA through its β-sheet as well as the same β2–β3 loop residues required for cap binding (Figure 2B). These structural models suggest that NCBP3 binding to the m7G cap and NCBP3 binding to RNA are, at least to some extent, mutually exclusive.
Box 2. Structural Properties of the PARN RRM.
The RRM of PARN can bind both m7G caps and poly(A) tails, although with different affinities and through different binding strategies [24]. X-ray crystallography of a cap-bound C-terminally truncated PARN dimer demonstrated that its core RRM harbors a canonical β1–α1–β2–β3–α2–β4 structure (β, β-strand; α, α-helix) (PDB ID: 3D45) [62]. Interestingly, NMR spectroscopy of a monomeric form of an extended PARN RRM revealed a noncanonical C-terminal α-helix, referred to as the α3-helix or the αC-helix, that folds back onto the RRM β-sheet (PDB ID: 2ROK) [23]. Adding complexity to the model, X-ray crystallography of an extended PARN RRM multimer (PDB ID: 3CTR) revealed an ‘open’ β1–α1–β2–β3–α2–α3 folding, with α2 and α3 protruding from the β-sheet and α3 stacking onto the β-sheet of the dimerized RRM [63].
In all three structures, PARN RRM binding to an m7G-cap analog primarily involves a WXDD motif (W, tryptophan; X, any amino acid; D, aspartic acid), located in the β2–β3 loop, in which the tryptophan side chain stacks on the m7G of the cap. To a lesser extent [24], PARN RRM binding to the cap also involves residues in the β1–α1 loop, whose backbone and/or side chains engage in peripheral hydrogen interactions with the cap. For comparison, binding of CBP20–CBP80 to the cap involves two tyrosine residues of the CBP20 RRM, whose coplanar aromatic side chains sandwich the guanine moiety of the cap (PDB ID: 1N52) [64].
In the two structures of the C-terminally extended PARN RRM, α3 buries the RNA-recognition surface of the β-sheet in a hydrophobic core, thereby covering the RNA-binding surface that is present in canonical RRMs. This has been proposed to prevent the PARN RRM from spuriously associating with RNA [23,63], a strategy also used by the RRM of the splicing factors U1A [30,31], p14 [65], and Acinus [66], the pre-mRNA cleavage and polyadenylation factor CstF-64 [25] – and its yeast ortholog Rna15 [67] –, and the transcriptional repressor SHARP [68]. Nonetheless, the PARN RRM binds ≥10-mer oligo(A) in a mechanism that has been only partially elucidated to: (i) involve the displacement of α3 [23]; (ii) not require the WXDD m7GTP-binding motif or be inhibited by m7GTP [24]; and (iii) most probably be stabilized by a second RNA-binding region of PARN, the R3H domain [69].
Figure 2.
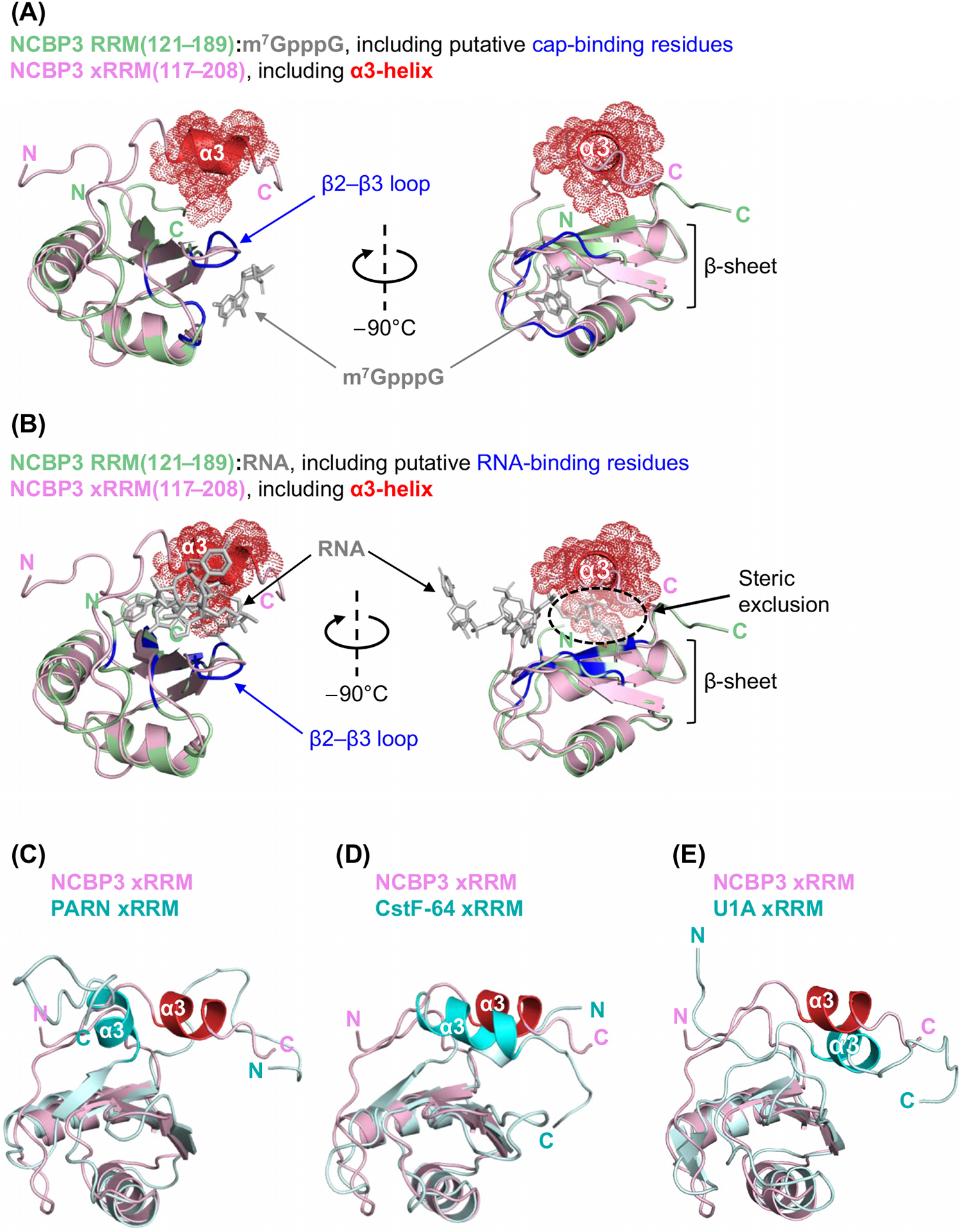
Model for Nuclear Cap-Binding Protein 3 (NCBP3) Binding to m7G Caps and RNA Bodies: Roles for the NCBP3 RNA-Recognition Motif (RRM) and Putative C-Terminal α3-Helix. (A,B) Iterative Threading ASSEmbly Refinement (I-TASSER)-predicted structure of the core NCBP3 RRM (amino acids 121–189, pale green) bound to either the cap analog m7GpppG (gray) (A) or the UUGUUU RNA hexamer (B) and superimposed on the I-TASSER-predicted structure of the NCBP3 extended RRM (xRRM) (amino acids 117–208, pale pink). The predicted α3-helix of the NCBP3 xRRM and its van der Waals surface are colored in red. Predicted cap-binding (A) and RNA-binding (B) residues are colored in dark blue. Steric incompatibility between the NCBP3 xRRM α3-helix and the UUGUUU RNA hexamer is shown by the broken-line oval (B). (C,D,E) Superimpositions of the I-TASSER-predicted structure of the NCBP3 xRRM (pale pink) and the crystal structure of the poly(A)-specific ribonuclease (PARN) xRRM (C) (pale cyan; PDB ID: 3CTR), the NMR structure of the CstF-64 xRRM (D) (pale cyan; PDB ID: 1P1T), and the RNA-free NMR structure of the U1A xRRM (E) (pale cyan; PDB ID: 1FHT). The predicted α3-helix of the NCBP3 xRRM is colored in red. The α3-helices of the PARN xRRM (C), the CstF-64 xRRM (D), and the U1A xRRM (E) are colored in bright cyan. (A–E) The N terminus (N) and C terminus (C) of each peptide is indicated, and the RRM β-sheets and α3-helices are specified.
Importantly, the RRM of PARN is extended by a noncanonical α-helix situated C-terminal to the RRM (Box 2). This α-helix is referred to as the α3-helix, and it covers the RRM β-sheet, supposedly to prevent spurious binding to uncapped RNA [23,24]. This configuration and inhibitory mode of binding to nontarget RNAs is reminiscent of other RRM-containing proteins (Box 2). Using I-TASSER to model the extended NCBP3 RRM sequence likewise revealed a short α-helix situated C-terminal to the RRM that folded onto the RRM β-sheet (Figures 1B and 2A–E). As expected, the alignment of our modeled structures indicates that the position of the NCBP3 α3-helix is compatible with binding to caps (Figure 2A) but not RNA (Figure 2B). Notably, the putative α3-helix of NCBP3, rather than being orientated like the α3-helix of PARN (Figure 2C), is predicted to align better with that of CstF-64 (Figure 2D) and U1A (Figure 2E). However, the length and thus the accompanying flexibility of the loop residing N-terminal to the NCBP3 α3-helix may confound these predictions.
On binding of the CstF-64 RRM to G/U-rich RNA, the α3-helix unfolds and extends into a hinge domain that mediates binding to the cleavage factor CstF-77 [25]. Conversely, binding of CstF-77 to the CstF-64 hinge domain results in rearrangement of the α3-helix [26] and promotes RNA binding [27]. Thus, by homology, one hypothesis is that the NCBP3 α3-helix plays a pivotal role in the combinatorial binding of NCBP3 to degenerate RNA motifs and to cofactors such as ARS2, which binds the N-terminal half of NCBP3 that encompasses the RRM (Figure 1B) [4].
In contrast to CstF-64, the α3-helix of U1A is somehow flexible and often found at least partially unfolded in the absence of RNA [28,29]. Binding of the U1A RRM to its RNA target U1 hairpin II results in the reorientation and stabilization of the α3-helix, which increases RNA-binding specificity and stabilizes the U1A–RNA complex [28–31]. Thus, a second hypothesis is that the NCBP3 α3-helix, following the reorientation allowed by its flexible N-terminal loop, stabilizes the binding to conserved RNA sequences and/or structures. In summary, we propose that the RRM of NCBP3 together with its noncanonical α3-helix functions as a regulatable platform controlling NCBP3 binding to the m7G cap, RNA, and proteins, including the CBC constituent ARS2.
Does NCBP3 Control Multiple Aspects of mRNA Metabolism?
Possible Functions of NCBP3 in (Pre-)mRNA Processing
In various cell-types, NCBP3 physically associates in an RNase-insensitive manner with the core spliceosome constituent U1 small nuclear ribonucleoprotein particle (snRNP) [32]. NCBP3 binding to U1 snRNP may be mediated by the U1A [32] and/or LUC7L2 [33] U1 snRNP subunits or via association with the U2AF2 subunit of U2 snRNP [8]. NCBP3 also readily co-purifies with the multifunctional transcription and export (TREX) protein complex in the presence and absence of benzonase [3,7,8]. Both U1 snRNP and TREX constituents exhibit roles in pre-mRNA splicing and 3′-end processing [32,34], implicating NCBP3 in these functionally interdependent co-transcriptional processes [35].
On the one hand, both CBP20 and CBP80 have been reported to facilitate first-intron splicing through diverse mechanisms, one of which involves U1 snRNP recruitment and/or stabilization at 5′ splice sites [20]. The findings that equimolar amounts of CBP20 and CBP80 are detected together with the human splicing machinery [36] and that ARS2 associates with U1 snRNP [32] suggest that NCBP3 would function in pre-mRNA splicing as a CBC accessory protein. Notably, depletion of NCBP3 in HeLa cells did not affect those EJC-dependent alternative splicing events that were tested [7], suggesting that NCBP3 binding to EJCs might function only in other processes, such as mRNA export and possibly mRNA translation (see later).
On the other hand, CBP80 – possibly together with CBP20 – cooperates with ARS2 and a yet-to-be-identified cofactor, which we hypothesize could be NCBP3, to inhibit the selection of cryptic (e.g., first-intron localized) polyadenylation sites of pre-mRNAs [20].
As noted above, a significant fraction of the mRNA-binding sites of NCBP3 and the RNA ligase RTCB overlap [12]. This observation suggests that NCBP3 could also function in cytoplasmic mRNA splicing, a poorly elucidated stress-associated pathway in which the RNA re-ligation step involves RTCB [37].
Evidence That NCBP3 Promotes Nuclear mRNA Export
Co-depletion of both CBP20 and NCBP3 was required to phenocopy defects in the nuclear export of bulk mRNAs observed after CBP80 knockdown [3]. While the mechanism whereby CBP20 and NCBP3 could functionally replace one another is unclear [3], it was recently demonstrated that NCBP3 cooperates with TREX to promote mRNA export [7]. Interestingly, the TREX Aly/REF subunit directly binds CBP80 but not CBP20 [38] to support what has been assumed to be CBP80–CBP20-dependent mRNA export (i.e., the canonical Aly/REF–TREX mRNA export pathway) [2]. The EJC has also been proposed to promote mRNA export by serving as a binding platform for TREX [39]. Thus, NCBP3 may support mRNA export by facilitating TREX recruitment to an alternative NCBP3–CBP80 CBC and/or to NCBP3-associated EJCs.
Evidence That NCBP3 Promotes mRNA Translation
Once in the cytoplasm, unless the mRNAs are degraded by nonsense-mediated mRNA decay (NMD), the CBC is rapidly replaced by the eukaryotic translation initiation factor (eIF) 4E, another CBP that typically supports the bulk of mRNA translation (Box 1). While it is clear that CBP80 supports pioneer rounds of translation and NMD, as well as the steady-state translation of some mRNAs (e.g., RDH mRNAs [40,41]), little is known about roles for CBP20 or NCBP3 in these processes.
Studies that used NMD as a readout for pioneer rounds of translation in human HeLa cells or the fungus Neurospora crassa confirm a role for CBP20 in these processes [42–44]. CBP20 contributions to mRNA translation may depend on its direct interaction with eIF4G [42], which mediates CBP80-dependent initiation of translation [41].
Supporting a role for the alternative NCBP3–CBP80 CBC in mRNA translation, CBP80 associates with NCBP3 but not with CBP20 in HEK293-cell polysomes [6]. However, NCBP3 knockdown failed to significantly reduce the polysome association of JUND mRNA [6], whose translation appears to be CBP80 dependent [45]. Nonetheless, following treatment with a potent innate immune stimulator, Ncbp3-gene knockout (KO) in mouse embryonic fibroblasts decreased the expression of proinflammatory cytokines while not affecting their mRNA levels [46]. Thus, a role for NCBP3 in mRNA translation may depend on the target mRNA and/or the cellular context (e.g., cell type and stress); otherwise, NCBP3 and CBP20 may be functionally redundant as described above for mRNA export. Notably, both internal m7G nucleotides [47] and the EJC stimulate mRNA translation [48]. Therefore, a role for NCBP3 in mRNA translation may also rely on its hypothetical binding to internal m7G nucleotides and/or on its ability to associate with the EJC.
NCBP3 Is an Antiviral Protein and a Putative Oncogenic Protein
Ncbp3-gene KO (Ncbp3−/−) mice deriving from heterozygote crosses (Ncbp3+/− × Ncbp3+/−) exhibit lower birth rates and reduced body weight compared with their wild-type littermates [46]. Nevertheless, the current literature indicates that NCBP3 is a nonessential protein under basal conditions (i.e., in the absence of stress). Co-depletion studies using unstressed HeLa cells or mouse embryonic fibroblasts showed that NCBP3 can be functionally replaced by either CBP20 or accessory EJC constituents to support mRNA export and cell growth [3,7]. However, NCBP3 may become critical for the maintenance of cellular homeostasis under stressful conditions, such as viral infection and cancer.
In support of this idea, loss of NCBP3 dramatically increases the growth of a broad range of RNA viruses, including vesicular stomatitis virus variant M2 (VSV-M2), Semliki Forest virus (SFV), encephalomyocarditis virus (EMCV), and influenza A virus (IAV) [3,46]. Furthermore, genetic depletion of NCBP3 in mice induces mortality on infection by IAV [46]. These phenotypes were attributed to the inability of NCBP3-depleted cells to properly execute the innate immune response [46]. Notably, NCBP3 protein levels were not induced by the infection of five different cell types with any one of 20 viruses from nine virus classes [49], and the ability of NCBP3 to bind mRNAs was unchanged on infection of HEK293 cells by the Sindbis RNA virus (SINV) [50]. Therefore, the molecular mechanism by which NCBP3 controls the innate immune response may depend on the activation of accessory proteins.
A role for NCBP3 as an oncogenic protein derives from the finding that NCBP3–CBP80-dependent upregulation of the E3 ubiquitin ligase Cullin 4B (CUL4B) promotes lung adenocarcinoma progression [51]. Interestingly, the CBC and CUL4 genes are often hijacked by viruses to augment viral replication and survival [52–54], raising the possibly of an NCBP3–CBP80–CUL4 axis acting at the interface of virology and oncology.
Concluding Remarks
The scientific community has believed for at least 25 years that CBP20 and CBP80 constitute the heterodimer at the cap of all nascent pre-mRNAs and their derived mRNAs. This simplistic view was recently challenged by studies that brought to light a poorly characterized protein, C17orf85, which was renamed NCBP3. Despite the relatively low number of publications focusing on this protein, it is already clear that NCBP3 is a multifaceted regulator of gene expression. NCBP3 seems to exist in possibly mutually exclusive states: (i) bound to the cap, where it would replace CBP20 in an alternative NCBP3–CBP80 CBC; (ii) associated with the canonical CBP20–CBP80 CBC and ARS2; or (iii) bound to RNA either directly or together with either RNA-binding proteins or RNA-binding complexes such as the EJC (Figure 1A). We propose that the RRM of NCBP3 and its extended α3-helix play a pivotal role in the transition of NCBP3 between these three states (Figure 2 and Box 2).
Just as the canonical CBC is directly implicated in numerous steps of gene expression (Box 1), NCBP3 is also likely to regulate pre-mRNA splicing, pre-mRNA 3′-end processing, mRNA nuclear export, and mRNA translation. Whereas many questions pertaining to its molecular functions remain unanswered (see Outstanding Questions), it appears that NCBP3 becomes functionally critical under pathological conditions that include viral infection [3,46] and cancer [51].
Outstanding Questions.
Does NCBP3 binding to a cap analog reflect its ability to bind 5′ caps and/or internal m7Gs?
How do cells benefit from having NCBP3 instead of – or in addition to – CBP20 at the m7G caps of mRNAs?
What regulates the preferential recruitment of either NCBP3 or CBP20 to the cap of mRNAs?
Is NCBP3 and CBP20 binding to m7G caps dynamic and regulated? In other words, can there be a precursor–product relationship between NCBP3–CBP80 and CBP20–CBP80 or vice versa?
Does the addition of NCBP3 increase the affinity of the canonical CBP20–CBP80 CBC for m7G caps?
Similar to CBP80 increasing CBP20 binding to caps, are there proteins that enhance the m7G-binding ability of NCBP3?
Does NCBP3 bind m7G caps or cap-bound CBP20–CBP80 and, simultaneously, the EJC?
What gene, transcript, or cellular features determine whether NCBP3 regulates the processing, nuclear export, and/or translation of a transcript?
What are good cell lines and animal models for further studies of NCBP3?
Does the NCBP3 RRM form multiple structures and, if it does, how are they regulated?
How, if at all, is NBCP3 regulated by post-translational modifications?
Notably, the co-depletion of CBP20 and NCBP3 does not inhibit cell growth to the same extent as does the depletion of CBP80 alone [3]. This raises the exciting possibility that, in addition to CBP20 and NCBP3, there exist other, yet-to-be-discovered CBP80 binding partners at 5′ caps.
Highlights.
Conventional views of nuclear cap-binding proteins (NCBPs) have recently been challenged. There exist different opinions on whether NCBP3 is an alternative to cap-binding protein (CBP)20 at the 5′ m7G cap of pre-mRNAs, binds to CBP80 and arsenite resistance protein 2 (ARS2) in a cap-bound CBP20–CBP80–ARS2 complex, and/or binds to RNA – possibly in association with exon-junction complexes.
We predict that mutually exclusive binding of NCBP3 to m7G caps, the cap-binding complex (CBC), and/or RNA is regulated by NCBP3’s noncanonical RNA-recognition motif (RRM) and a putative α3-helix that resides distal to the RRM.
NCBP3 may regulate various steps of RNA metabolism: mRNA export from the nucleus, mRNA translation, and possibly pre-mRNA splicing, pre-mRNA 3′-end formation, and noncanonical splicing of cytoplasmic mRNAs.
NCBP3 function appears to become critical when cells are stressed, such as during viral infection and when cells are transformed to become cancer cells.
Acknowledgments
This work was supported by an American Heart Association (AHA) Post-doctoral Fellowship 18POST33960339 to X.R. and by National Institutes of Health (NIH) grant R01 GM059514 to L.E.M. We thank the Maquat laboratory members Rina Nagao for comments and Elizabeth Abshire for help with structural modeling.
Glossary
- α-Helix
secondary protein structure in which hydrogen bonds are formed between the backbone of amino acids located three or four residues apart to form a righthand helix
- β-Sheet
secondary protein structure comprising β-strands connected laterally by at least two or three backbone hydrogen bonds to form a pleated sheet
- β-Strand
polypeptide of three to ten amino acids having a backbone in an extended configuration
- Chromatin
protein-packaged genomic DNA of eukaryotic cells
- Cytoplasmic mRNA splicing
stress-induced process of removing an internal fragment of a cytoplasmic mRNA associated with the endoplasmic reticulum, involving, at least in the case of the XBP1 mRNA, endonucleolytic cleavage by IRE1 followed by ligation by RTCB
- Exon-junction complex (EJC)
complex of proteins deposited ~20–24 nucleotides upstream of mRNA exon-exon junctions as a consequence of pre-mRNA splicing (i.e., intron removal)
- Immunoprecipitation (IP)
method to isolate a protein, often complexed with other proteins, using an antibody specific to that protein
- Long noncoding RNA (lncRNA)
≥200–300-nucleotide polyadenylated RNAPII-synthesized transcript that is computationally defined as not encoding protein
- Loop
flexible protein region that fails to form an α-helix or a β-strand
- m7G cap
inverted 7-methyl guanosine linked via a 5′–5′ triphosphate bridge to the first transcribed nucleotide (N) to form m7G(5′)ppp(5′)N
- mRNA
transcript derived from pre-mRNA processing in the nucleus to support protein synthesis in the cytoplasm
- NMR
type of spectroscopy that generates a magnetic field around an atom in a molecule to change its resonance frequency, providing the electronic structure of its constituent groups
- Photoactivatable ribonucleotide-enhanced crosslinking and immunoprecipitation (PAR-CLIP)
method to define direct cellular protein-binding sites on RNA dependent on the protein directly contacting the photoactivatable nucleotide (e.g., 4-thiouridine)
- Polysomes
mRNAs associated with multiple 80S ribosomes
- Precursor of mRNA (pre-mRNA)
RNAPII-synthesized transcript that undergoes co-transcriptional capping at its 5′ end, splicing to remove introns, and cleavage and polyadenylation of its 3′ end to generate an mRNA
- RNA immunoprecipitation coupled to RNA-seq (RIP-seq)
method that, unlike CLIP variations, identifies RNA molecules that bind not only directly but also indirectly with a protein of interest
- RNA polymerase II (RNAPII)
550-kDa complex of 12 protein subunits that uses DNA as a template to synthesize pre-mRNAs, lncRNAs, most snRNAs, and primary miRNAs
- RNA recognition motif (RRM)
RNA- or protein-binding domain typically comprising four antiparallel β-strands and two α-helices arranged in a β-α-β-β-α-β fold
- Small nuclear RNA (snRNA)
RNAPII-synthesized RNA acquiring an m7G cap that (with the exception of U6 snRNA), after export to the cytoplasm, is converted to an m2,2,7G(5′)ppp(5′)N trimethyl cap prior to reimport into the nucleus for function in pre-mRNA splicing
- Untranslated region (UTR)
mRNA sequences situated either upstream (5′UTR) or downstream (3′UTR) of the protein-coding region
- X-ray crystallography
3D production of electron density deriving from the diffraction of an X-ray beam to determine the atomic structure of the molecules that have been crystallized
References
- 1.Ohno M et al. (1990) A nuclear cap binding protein from HeLa cells. Nucleic Acids Res. 18, 6989–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonatopoulos-Pournatzis T and Cowling VH (2014) Cap-binding complex (CBC). Biochem. J 457, 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gebhardt A et al. (2015) mRNA export through an additional cap-binding complex consisting of NCBP1 and NCBP3. Nat. Commun 6, 8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulze WM et al. (2018) Structural analysis of human ARS2 as a platform for co-transcriptional RNA sorting. Nat. Commun 9, 1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegele A et al. (2012) Dynamic protein–protein interaction wiring of the human spliceosome. Mol. Cell 45, 567–580 [DOI] [PubMed] [Google Scholar]
- 6.Singh G et al. (2020) The mRNA encoding the JUND tumor suppressor detains nuclear RNA-binding proteins to assemble polysomes that are unaffected by mTOR. J. Biol. Chem 295, 7763–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dou Y et al. (2020) NCBP3 positively impacts mRNA biogenesis. Nucleic Acids Res, gkaa744 Published online September 22, 2020 10.1093/nar/gkaa744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dou Y et al. (2020) Affinity proteomic dissection of the human nuclear cap-binding complex interactome. bioRxiv Published online April 21, 2020. 10.1101/2020.04.20.048470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aviner R et al. (2017) Proteomic analysis of polyribosomes identifies splicing factors as potential regulators of translation during mitosis. Nucleic Acids Res. 45, 5945–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baltz AG et al. (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 [DOI] [PubMed] [Google Scholar]
- 11.Machanick P and Bailey TL (2011) MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li YE et al. (2017) Identification of high-confidence RNA regulatory elements by combinatorial classification of RNA-protein binding sites. Genome Biol. 18, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh G et al. (2012) The cellular EJC interactome reveals higher-order mRNP structure and an EJC–SR protein nexus. Cell 151, 750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derrien T et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palazzo AF and Lee ES (2018) Sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front. Genet 9, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guiro J and Murphy S (2017) Regulation of expression of human RNA polymerase II-transcribed snRNA genes. Open Biol. 7, 170073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzluff WF et al. (2008) Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet 9, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCloskey A et al. (2012) hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science 335, 1643–1646 [DOI] [PubMed] [Google Scholar]
- 19.Merz C et al. (2007) Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA 13, 116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rambout X and Maquat LE (2020) The nuclear cap-binding complex as choreographer of gene transcription and pre-mRNA processing. Genes Dev. 34, 1113–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J et al. (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J et al. (2015) The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagata T et al. (2008) The RRM domain of poly(A)-specific ribonuclease has a noncanonical binding site for mRNA cap analog recognition. Nucleic Acids Res. 36, 4754–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nilsson P et al. (2007) A multifunctional RNA recognition motif in poly(A)-specific ribonuclease with cap and poly(A) binding properties. J. Biol. Chem 282, 32902–32911 [DOI] [PubMed] [Google Scholar]
- 25.Pérez Cañadillas JM and Varani G (2003) Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 22, 2821–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grozdanov PN et al. (2018) The structural basis of CstF-77 modulation of cleavage and polyadenylation through stimulation of CstF-64 activity. Nucleic Acids Res. 46, 12022–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W et al. (2018) Reconstitution of the CstF complex unveils a regulatory role for CstF-50 in recognition of 3′-end processing signals. Nucleic Acids Res. 46, 493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurisaki I et al. (2014) Combined mechanism of conformational selection and induced fit in U1A–RNA molecular recognition. Biochemistry 53, 3646–3657 [DOI] [PubMed] [Google Scholar]
- 29.Han Z et al. (2019) Interpreting the dynamics of binding interactions of snRNA and U1A using a coarse-grained model. Biophys. J 116, 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avis JM et al. (1996) Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J. Mol. Biol 257, 398–411 [DOI] [PubMed] [Google Scholar]
- 31.Law MJ et al. (2013) The role of the C-terminal helix of U1A protein in the interaction with U1hpII RNA. Nucleic Acids Res. 41, 7092–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.So BR et al. (2019) A complex of U1 snRNP with cleavage and polyadenylation factors controls telescripting, regulating mRNA transcription in human cells. Mol. Cell 76, 590–599.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huttlin EL et al. (2017) Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heath CG et al. (2016) The role of TREX in gene expression and disease. Biochem. J 473, 2911–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaida D (2016) The reciprocal regulation between splicing and 3′-end processing. Wiley Interdiscip. Rev. RNA 7, 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt C et al. (2014) Mass spectrometry-based relative quantification of proteins in precatalytic and catalytically active spliceosomes by metabolic labeling (SILAC), chemical labeling (iTRAQ), and label-free spectral count. RNA 20, 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filipowicz W (2014) Making ends meet: a role of RNA ligase RTCB in unfolded protein response. EMBO J. 33, 2887–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng H et al. (2006) Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127, 1389–1400 [DOI] [PubMed] [Google Scholar]
- 39.Le Hir H et al. (2001) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20, 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryu I and Kim YK (2017) Translation initiation mediated by nuclear cap-binding protein complex. BMB Rep. 50, 186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maquat LE et al. (2010) CBP80-promoted mRNP rearrangements during the pioneer round of translation, nonsensemediated mRNA decay, and thereafter. Cold Spring Harb. Symp. Quant. Biol 75, 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lejeune F et al. (2004) eIF4G is required for the pioneer round of translation in mammalian cells. Nat. Struct. Mol. Biol 11, 992–1000 [DOI] [PubMed] [Google Scholar]
- 43.Hosoda N et al. (2005) CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol 12, 893–901 [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y and Sachs MS (2015) Control of mRNA stability in fungi by NMD, EJC and CBC factors through 3′UTR introns. Genetics 200, 1133–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Short JD and Pfarr CM (2002) Translational regulation of the JunD messenger RNA. J. Biol. Chem 277, 32697–32705 [DOI] [PubMed] [Google Scholar]
- 46.Gebhardt A et al. (2019) The alternative cap-binding complex is required for antiviral defense in vivo. PLoS Pathog. 15, e1008155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang LS et al. (2019) Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol. Cell 74, 1304–1316.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boehm V and Gehring NH (2016) Exon junction complexes: supervising the gene expression assembly line. Trends Genet. 32, 724–735 [DOI] [PubMed] [Google Scholar]
- 49.Hubel P et al. (2019) A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nat. Immunol 20, 493–502 [DOI] [PubMed] [Google Scholar]
- 50.García-Moreno M et al. (2019) System-wide profiling of RNA-binding proteins uncovers key regulators of virus infection. Mol. Cell 74, 196–211 e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H et al. (2019) NCBP1 promotes the development of lung adenocarcinoma through up-regulation of CUL4B. J. Cell. Mol. Med 23, 6965–6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hannah J and Zhou P (2015) Distinct and overlapping functions of the Cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene 573, 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García-de-Gracia F et al. (2019) CBP80/20-dependent translation initiation factor (CTIF) inhibits HIV-1 Gag synthesis by targeting the function of the viral protein Rev. bioRxiv Published online July 22, 2019. 10.1101/710137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruiz JC et al. (2019) Kaposi’s sarcoma-associated herpesvirus ORF57 protein protects viral transcripts from specific nuclear RNA decay pathways by preventing hMTR4 recruitment. PLoS Pathog. 15, e1007596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolin SL and Maquat LE (2019) Cellular RNA surveillance in health and disease. Science 366, 822–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rambout X et al. (2020) Transcriptional coactivator PGC-1α binding to newly synthesized RNA via CBP80: a nexus for co- and posttranscriptional gene regulation. Cold Spring Harb. Symp. Quant. Biol 84, 040212. [DOI] [PubMed] [Google Scholar]
- 57.Kurosaki T et al. (2019) Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol 20, 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trcek T et al. (2013) Temporal and spatial characterization of nonsense-mediated mRNA decay. Genes Dev. 27, 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lejeune F et al. (2002) The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 21, 3536–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pelletier J and Sonenberg N (2019) The organizing principles of eukaryotic ribosome recruitment. Annu. Rev. Biochem 88, 307–335 [DOI] [PubMed] [Google Scholar]
- 61.Ivanov P et al. (2019) Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol 11, a32813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu M et al. (2009) Structural basis of m7GpppG binding to poly(A)-specific ribonuclease. Structure 17, 276–286 [DOI] [PubMed] [Google Scholar]
- 63.Monecke T et al. (2008) Crystal structure of the RRM domain of poly(A)-specific ribonuclease reveals a novel m7G-cap-binding mode. J. Mol. Biol 382, 827–834 [DOI] [PubMed] [Google Scholar]
- 64.Calero G et al. (2002) Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol 9, 912–917 [DOI] [PubMed] [Google Scholar]
- 65.Schellenberg MJ et al. (2006) Crystal structure of a core spliceosomal protein interface. Proc. Natl. Acad. Sci. U. S. A 103, 1266–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fernandes H et al. (2018) Crystal structure of human Acinus RNA recognition motif domain. PeerJ 6, e5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pancevac C et al. (2010) Structure of the Rna15 RRM–RNA complex reveals the molecular basis of GU specificity in transcriptional 3′-end processing factors. Nucleic Acids Res. 38, 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arieti F et al. (2014) The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs. Nucleic Acids Res. 42, 6742–6752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Virtanen A et al. (2013) Poly(A)-specific ribonuclease (PARN): an allosterically regulated, processive and mRNA cap-interacting deadenylase. Crit. Rev. Biochem. Mol. Biol 48, 192–209 [DOI] [PubMed] [Google Scholar]


