Abstract
In the present study, the complete chloroplast genome of Hordeum vulgare L. var. trifurcatum was sequenced, assembled and compared with closely related species. The chloroplast genome of Hordeum vulgare L. var. trifurcatum was composed of 84 protein-coding genes (PCG), 8 ribosomal RNA (rRNA) genes, and 38 transfer RNA (tRNA) genes. The Hordeum vulgare L. var. trifurcatum chloroplast genome is 136,485 bp in size, with the GC content of 38.32%. Phylogenetic analysis based on the combined chloroplast gene dataset indicated that the Hordeum vulgare L. var. trifurcatum exhibited a close relationship with Hordeum vulgare subsp. spontaneum and Hordeum vulgare subsp. vulgare.
Keywords: Tibetan hulless barley, Hordeum, chloroplast genome, phylogenetic analysis, molecular marker
Hordeum vulgare L. var. trifurcatum, belonging to the Poaceae family (Soreng et al. 2015; Saarela et al. 2018), is one of the staple foods for Tibetans and an important livestock feed in the Tibetan Plateau (Zeng et al. 2015; Huang et al. 2020). The highland barley (Hordeum vulgare L. var. trifurcatum) shows good environmental tolerance, which can be successfully planted in high altitude, low temperature, and high salinity places (Walia et al. 2006; El-Esawi et al. 2018; Elsawy et al. 2018). In addition, the highland barley also showed good antioxidant activity (Asif et al. 2020). Thus, Hordeum vulgare L. var. trifurcatum is a promising nutritious food source and traditional Chinese medicine grows widely in plateau, which concerning by more and more researchers, just like the Fagopyrum tataricum (L.) Gaertn (Song et al. 2016; Xiang et al. 2016; Xiang, Ma, et al. 2019; Xiang, Song, et al. 2019) and Stellera chamaejasme L. (Ren et al. 2019, 2021a, 2021b). The genus Hordeum comprises more than 30 species. Some varieties are also found in this genus (Malysheva-Otto et al. 2006; Forsberg et al. 2019; Hagenblad and Morales 2020; Kumar et al. 2020). Organelle genomes have been widely used in study of taxonomy, evolution and genetics (Wang et al. 2016; Yang et al. 2019; Wang et al. 2020; Li, He, et al. 2020). However, no complete chloroplast genome of Hordeum vulgare L. var. trifurcatum was reported to date, which limits its breeding and application (Su et al. 2020).
The specimen (Hordeum vulgare L. var. trifurcatum) used for chloroplast genome assembly was collected from Qinghai, China (101.97 E; 53.70 N). A specimen was deposited at Collection Center of Chengdu University (Y. Ren, renyuanhang@cdu.edu.cn) under the voucher number ZQK_R1. The chloroplast genome of Hordeum vulgare L. var. trifurcatum was sequenced and assembled according to methods previously described (Li, Ren, et al. 2020). First, we extracted the total genomic DNA of Hordeum vulgare L. var. trifurcatum using a Plant DNA Kit (D3485-00, Omega Bio-Tek, Norcross, GA, USA). Then we purified the genomic DNA using a Gel Extraction Kit (Omega Bio-Tek, Norcross, GA, USA). The purified DNA was stored in Chengdu University (No. DNA_ ZQK_R1). Sequencing libraries of Hordeum vulgare L. var. trifurcatum was constructed using a NEBNext® Ultra™ II DNA Library Prep Kit (NEB, Beijing, China). We conducted the whole genomic sequencing (WGS) of Hordeum vulgare L. var. trifurcatum using the Illumina HiSeq 2500 Platform (Illumina, SanDiego, CA). The chloroplast genome of Hordeum vulgare L. var. trifurcatum was initially assembled using SPAdes v3.11.0 (Bankevich et al. 2012). The chloroplast sequences obtained in the above steps were used as seed sequences to assemble the complete chloroplast genome of Hordeum vulgare L. var. trifurcatum using NOVOPlasty v4.3.1 with a k-mer size of 35 (Dierckxsens et al. 2017). Approximately 1.10 million reads were assembled into a complete circular chloroplast genome. The average chloroplast sequence coverage was 2,168 ×. The complete chloroplast genome of Hordeum vulgare L. var. trifurcatum was annotated by GeSeq (Tillich et al. 2017) using the chloroplast genome of Hordeum vulgare subsp. spontaneum as the reference (Bdolach et al. 2019).
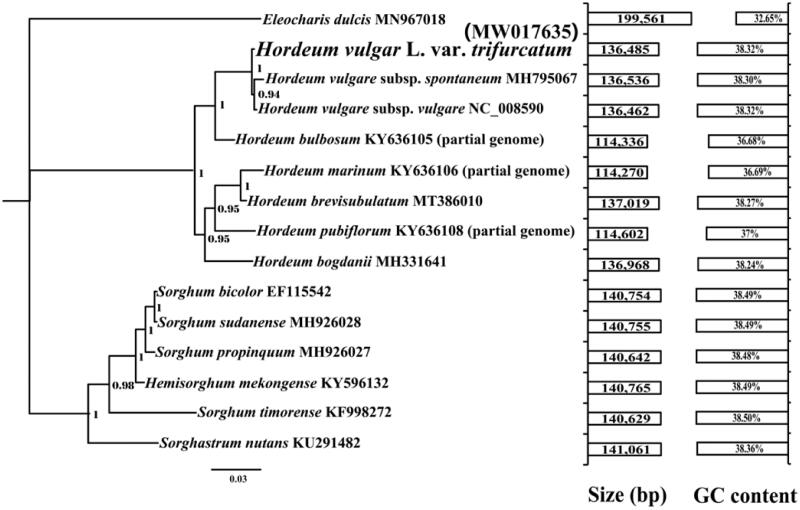
The complete chloroplast genome of Hordeum vulgare L. var. trifurcatum is 136,485 bp in length, which was larger than Hordeum vulgare subsp. vulgare (136,462 bp) (Zeng et al. 2017) and smaller than Hordeum vulgare subsp. spontaneum (136,536 bp) (Bdolach et al. 2019). The GC content of the Hordeum vulgare L. var. trifurcatum chloroplast genome is 38.32%, which is larger than that of Hordeum vulgare subsp. spontaneum (38.30%). The base compositions of the Hordeum vulgare L. var. trifurcatum chloroplast genome were as follows: A (30.93%), T (30.76%), G (19.22%) and C (19.10%). The complete chloroplast genome of Hordeum vulgare L. var. trifurcatum contains 84 protein-coding genes, 8 ribosomal RNA genes, and 38 transfer RNA (tRNA) genes. The number of protein-coding genes in Hordeum vulgare L. var. trifurcatum chloroplast genome was more than that in two subspecies (Hordeum vulgare L. var. trifurcatum and Hordeum vulgare subsp. vulgare), while the number of tRNA was less than that in the two subspecies. To investigate the phylogenetic status of Hordeum vulgare L. var. trifurcatum, we constructed a phylogenetic tree for 15 species. The protein-coding region of 13 genes conserved in the 15 species was used to construct combined a chloroplast gene set according to previous methods (Li, Xiang, et al. 2019; Wu et al. 2021). Bayesian (BI) analysis method (Li, Wu, et al. 2021) was used to construct the phylogenetic tree based on combined protein-coding genes of chloroplast genome as described by previous methods (Li, Yang, et al. 2020; Cheng et al. 2021; Li, Li, et al. 2021). MrBayes v3.2.6 (Ronquist et al. 2012) was used to construct the phylogenetic tree using Bayesian inference (BI) method. Two independent runs with four chains (three heated and one cold) each were conducted simultaneously for 2 × 106 generations. Each run was sampled every 100 generations. We assumed that stationarity had been reached when estimated sample size (ESS) was greater than 100, and the potential scale reduction factor (PSRF) approached 1.0. The first 25% samples were discarded as burn-in, and the remaining trees were used to calculate Bayesian posterior probabilities (BPP) in a 50% majority-rule consensus tree (Li, Ren, et al. 2019). According to the phylogenetic tree (Figure 1), the Hordeum vulgare L. var. trifurcatum exhibited a close relationship with Hordeum vulgare subsp. spontaneum (Bdolach et al. 2019) and Hordeum vulgare subsp. vulgare (Zeng et al. 2017).
Figure 1.
Bayesian phylogenetic analysis and comparative chloroplast genomic analysis of 15 species based on the combined protein-coding gene sets. Support values are Bayesian posterior probabilities (BPP). Accession numbers of chloroplast sequences used in the phylogenetic analysis are listed after the species names.
Funding Statement
This work was funded by Sichuan Science and Technology Program [2021YJ0477].
Disclosure statement
The authors have declared that no competing interests exist.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW017635. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA717119, SRR14082750, and SAMN18478699, respectively.
References
- Asif A, Zeeshan N, Mehmood S.. 2020. Antioxidant and antiglycation activities of traditional plants and identification of bioactive compounds from extracts of Hordeum vulgare by LC-MS and GC-MS. J Food Biochem. 44(9):e13381. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bdolach E, Prusty MR, Faigenboim-Doron A, Filichkin T, Helgerson L, Schmid KJ, Greiner S, Fridman E.. 2019. Thermal plasticity of the circadian clock is under nuclear and cytoplasmic control in wild barley. Plant Cell Environ. 42(11):3105–3120. [DOI] [PubMed] [Google Scholar]
- Cheng J, Luo Q, Ren YH, Luo Z, Liao WL, Wang X, Li Q.. 2021. Panorama of intron dynamics and gene rearrangements in the phylum Basidiomycota as revealed by the complete mitochondrial genome of Turbinellus floccosus. Appl Microbiol Biotechnol. 105(5):2017–2032. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Esawi MA, Alaraidh IA, Alsahli AA, Ali HM, Alayafi AA, Witczak J, Ahmad M.. 2018. Genetic variation and alleviation of salinity stress in barley (Hordeum vulgare L.). Molecules. 23(10):2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsawy HIA, Mekawy AMM, Elhity MA, Abdel-Dayem SM, Abdelaziz MN, Assaha DVM, Ueda A, Saneoka H.. 2018. Differential responses of two Egyptian barley (Hordeum vulgare L.) cultivars to salt stress. Plant Physiol Biochem. 127:425–435. [DOI] [PubMed] [Google Scholar]
- Forsberg NEG, Leino MW, Hagenblad J.. 2019. Population structure in landrace barley (Hordeum vulgare L.) during the late 19th century crop failures in Fennoscandia. Heredity (Edinb). 123(6):733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad J, Morales J.. 2020. An evolutionary approach to the history of barley (Hordeum vulgare) cultivation in the Canary Islands. Afr Archaeol Rev. 37(4):579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Gao X, Li Y, Tian P, Nima Y, Laba Z, Ci Z, Wei X, Qu J, Guan W, et al. 2020. Content analysis of vitamins, dietary fibers and amino acids in a wide collection of barley (Hordeum vulgare L.) from Tibet, China. Bioinformation. 16(4):314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Banjarey P, Malik R, Tikle AN, Verma RPS.. 2020. Population structure and diversity assessment of barley (Hordeum vulgare L.) introduction from ICARDA. J Genet. 99(1):70. [PubMed] [Google Scholar]
- Li Q, He X, Ren Y, Xiong C, Jin X, Peng L, Huang W.. 2020. Comparative mitogenome analysis reveals mitochondrial genome differentiation in ectomycorrhizal and asymbiotic Amanita species. Front Microbiol. 11:1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li LJ, Feng HY, Tu WY, Bao ZJ, Xiong C, Wang X, Huang WL.. 2021. Characterization of the complete mitochondrial genome of Basidiomycete yeast Hannaella oryzae: intron evolution, gene rearrangement and its phylogeny. Front Microbiol. 12: 646567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ren Y, Shi X, Peng L, Zhao J, Song Y, Zhao G.. 2019. Comparative mitochondrial genome analysis of two ectomycorrhizal fungi (Rhizopogon) reveals dynamic changes of intron and phylogenetic relationships of the subphylum Agaricomycotina. Int J Mol Sci. 20(20):5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ren Y, Xiang D, Shi X, Zhao J, Peng L, Zhao G.. 2020. Comparative mitogenome analysis of two ectomycorrhizal fungi (Paxillus) reveals gene rearrangement, intron dynamics, and phylogeny of basidiomycetes. IMA Fungus. 11(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wu P, Li L, Feng H, Tu W, Bao Z, Xiong C, Gui M, Huang W.. 2021. The first eleven mitochondrial genomes from the ectomycorrhizal fungal genus (Boletus) reveal intron loss and gene rearrangement. Int J Biol Macromol. 172:560–572. [DOI] [PubMed] [Google Scholar]
- Li Q, Xiang D, Wan Y, Wu Q, Wu X, Ma C, Song Y, Zhao G, Huang W.. 2019. The complete mitochondrial genomes of five important medicinal Ganoderma species: features, evolution, and phylogeny. Int J Biol Macromol. 139:397–408. [DOI] [PubMed] [Google Scholar]
- Li Q, Yang L, Xiang D, Wan Y, Wu Q, Huang W, Zhao G.. 2020. The complete mitochondrial genomes of two model ectomycorrhizal fungi (Laccaria): features, intron dynamics and phylogenetic implications. Int J Biol Macromol. 145:974–984. [DOI] [PubMed] [Google Scholar]
- Malysheva-Otto LV, Ganal MW, Roder MS.. 2006. Analysis of molecular diversity, population structure and linkage disequilibrium in a worldwide survey of cultivated barley germplasm (Hordeum vulgare L.). BMC Genet. 7(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Li Q, Lu L, Jin H, Tao K, Hou T.. 2021a. Isochamaejasmin induces toxic effects on Helicoverpa zea via DNA damage and mitochondria-associated apoptosis. Pest Manag Sci. 77(1):557–567. [DOI] [PubMed] [Google Scholar]
- Ren Y, Li Q, Lu L, Jin H, Tao K, Hou T.. 2021b. Toxicity and physiological actions of biflavones on potassium current in insect neuronal cells. Pestic Biochem Physiol. 171:104735. [DOI] [PubMed] [Google Scholar]
- Ren Y, Mu Y, Yue Y, Jin H, Tao K, Hou T.. 2019. Neochamaejasmin A extracted from Stellera chamaejasme L. induces apoptosis involving mitochondrial dysfunction and oxidative stress in Sf9 cells. Pestic Biochem Physiol. 157:169–177. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarela JM, Burke SV, Wysocki WP, Barrett MD, Clark LG, Craine JM, Peterson PM, Soreng RJ, Vorontsova MS, Duvall MR.. 2018. A 250 plastome phylogeny of the grass family (Poaceae): topological support under different data partitions. PeerJ. 6 :e4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Xiang DB, Yan L, Song Y, Zhao G, Wang YH, Zhang BL.. 2016. Changes in seed growth, levels and distribution of flavonoids during tartary buckwheat seed development. Plant Prod Sci. 19(4):518–527. [Google Scholar]
- Soreng RJ, Peterson PM, Romaschenko K, Davidse G, Zuloaga FO, Judziewicz EJ, Filgueiras TS, Davis JI, Morrone O.. 2015. A worldwide phylogenetic classification of the Poaceae (Gramineae). J Syst Evol. 53(2):117–137. [Google Scholar]
- Su X, Zhao J, Wang Z.. 2020. The complete chloroplast genome of Hordeum brevisubulatum. Mitochondrial DNA B Resour. 5(3):2988–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, Wilson C, Wahid A, Condamine P, Cui X, Close TJ.. 2006. Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Funct Integr Genomics. 6(2):143–156. [DOI] [PubMed] [Google Scholar]
- Wang X, Song A, Wang F, Chen M, Li X, Li Q, Liu N.. 2020. The 206 kbp mitochondrial genome of Phanerochaete carnosa reveals dynamics of introns, accumulation of repeat sequences and plasmid-derived genes. Int J Biol Macromol. 162:209–219. [DOI] [PubMed] [Google Scholar]
- Wang YL, Wei ZX, Xu QJ, Zeng XQ, Yuan HJ, Tang YW, Tashi N.. 2016. The complete mitochondrial genome of Tibetan hulless barley. Mitochondrial DNA B Resour. 1(1):430–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Bao Z, Tu W, Li L, Xiong C, Jin X, Li P, Gui M, Huang W, Li Q.. 2021. The mitogenomes of two saprophytic Boletales species (Coniophora) reveals intron dynamics and accumulation of plasmid-derived and non-conserved genes. Comput Struct Biotechnol J. 19:401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang DB, Ma CR, Song Y, Wu Q, Wu XY, Sun YX, Zhao G, Wan Y.. 2019. Post-anthesis photosynthetic properties provide insights into yield potential of tartary buckwheat cultivars. Agronomy-Basel. 9(3):149. [Google Scholar]
- Xiang DB, Song Y, Wu Q, Ma CR, Zhao JL, Wan Y, Zhao G.. 2019. Relationship between stem characteristics and lodging resistance of Tartary buckwheat (Fagopyrum tataricum). Plant Prod Sci. 22(2):202–210. [Google Scholar]
- Xiang DB, Zhao G, Wan Y, Tan ML, Song C, Song Y.. 2016. Effect of planting density on lodging-related morphology, lodging rate, and yield of tartary buckwheat (Fagopyrum tataricum). Plant Prod Sci. 19(4):479–488. [Google Scholar]
- Yang LX, Li Q, Zhao G.. 2019. Characterization of the complete chloroplast genome of Chenopodium sp. (Caryophyllales: Chenopodiaceae). Mitochondrial DNA B Resour. 4(2):2574–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng QX, Yuan JH, Wang LY, Xu JQ, Nyima T.. 2017. The complete chloroplast genome of Tibetan hulless barley. Mitochondrial DNA A DNA Mapp Seq Anal. 28(3):324–325. [DOI] [PubMed] [Google Scholar]
- Zeng X, Long H, Wang Z, Zhao S, Tang Y, Huang Z, Wang Y, Xu Q, Mao L, Deng G, et al. 2015. The draft genome of Tibetan hulless barley reveals adaptive patterns to the high stressful Tibetan Plateau. Proc Natl Acad Sci U S A. 112(4):1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW017635. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA717119, SRR14082750, and SAMN18478699, respectively.