Abstract
In the last decade, the widespread use of massively parallel sequencing has considerably boosted the number of novel gene discoveries in monogenic skeletal diseases with short stature. Defects in genes playing a role in the maintenance and function of the growth plate, the site of longitudinal bone growth, are a well-known cause of skeletal diseases with short stature. However, several genes involved in extracellular matrix composition or maintenance as well as genes partaking in various biological processes have also been characterized. This review aims to describe the latest genetic findings in spondyloepiphyseal dysplasias, spondyloepimetaphyseal dysplasias, and some monogenic forms of isolated short stature. Some examples of novel genetic mechanisms leading to skeletal conditions with short stature will be described. Strategies on how to successfully characterize novel skeletal phenotypes with short stature and genetic approaches to detect and validate novel gene-disease correlations will be discussed in detail. In summary, we review the latest gene discoveries underlying skeletal diseases with short stature and emphasize the importance of characterizing novel molecular mechanisms for genetic counseling, for an optimal management of the disease, and for therapeutic innovations.
Introduction
Genetic skeletal diseases, often called skeletal dysplasias, are a heterogeneous group of heritable conditions with generalized bone and cartilage impairment caused by pathogenic variants in genes primarily affecting skeletogenesis and/or bone homeostasis (1, 2). Although the overall incidence of genetic skeletal diseases is around one in 5000 births (3), some conditions are extremely rare and can only be found in a handful of families worldwide. In the last decade, the widespread use of massively parallel sequencing (MPS) has boosted the diagnostic rate of rare skeletal diseases and has led to the identification of several novel disease loci. According to the latest Nosology of Skeletal Disorders published in 2019, in total 461 skeletal diseases have been characterized so far. These diseases have been classified into 42 groups based on a combination of radiological findings, clinical features, and underlying molecular mechanisms (2).
Genetic skeletal diseases are characterized by a broad phenotypic heterogeneity. The clinical manifestations can vary greatly among patients with the same gene defect and significantly even among those harboring the exact same pathogenic variant. This broad phenotypic and genetic heterogeneity brings challenges in establishing efficient treatments. Presently, a pharmacological treatment is only available for a limited number of bone disorders, including various genetic forms of osteoporosis, FGF23-related hypophosphatemia (MIM 193100), hypophosphatasia (MIM 146300), and achondroplasia (4, 5, 6).
Although genetic skeletal diseases most often feature short and disproportionate stature, sometimes normal or even tall stature, like in overgrowth syndromes, can be noticed. Moreover, short stature is sometimes part of a broader phenotypic spectrum that also includes other skeletal impairments, such as short extremities, skeletal deformities, high or low bone mineral density as well as extra-skeletal features, including impaired vision or hearing. The scope of this review is to describe the latest genetic discoveries underlying some severe skeletal diseases characterized by short stature.
Skeletal diseases with reduced linear growth
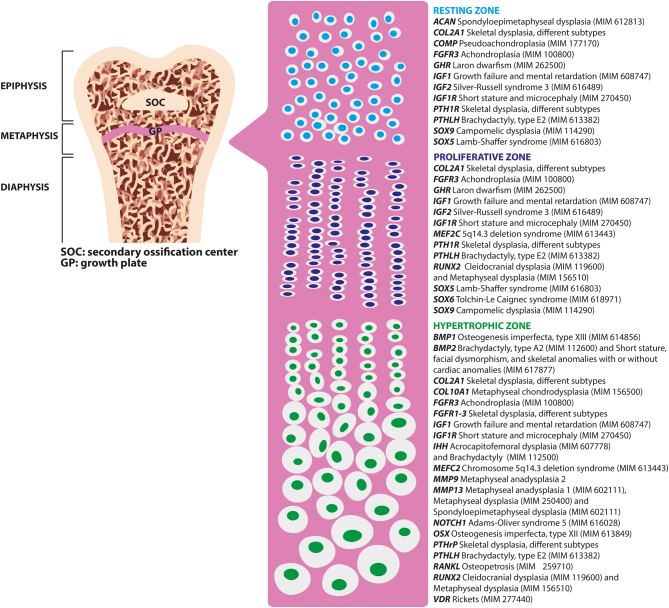
Longitudinal linear bone growth is determined by endochondral ossification, which is mediated by the growth plate, a complex cartilaginous structure located between the metaphysis and the epiphysis of the long bones (Fig. 1). From the epiphyseal end the growth plate can be subdivided into three different zones, each of which is composed of chondrocytes characterized by different size, shape, orientation, proliferative capacity, and function: (1) resting zone, (2) proliferative zone and (3) hypertrophic zone (7). The resting zone is a source of stem-like progenitor cells restoring the pool of proliferative chondrocytes (8). In the proliferative zone, the flattened chondrocytes divide longitudinally at a high rate and synthetize a large amount of ECM. Finally, in the hypertrophic zone chondrocytes stop dividing, become large in size, and start producing factors that trigger mineralization and the invasion of blood vessels, thus promoting chondrocyte apoptosis (9). Consequently, osteoblasts invade the hypertrophic zone and bone formation takes place. At the end of puberty, when linear growth no longer occurs, the growth plate fuses with the epiphysis.
Figure 1.
Structure of a long bone with major focus on the growth plate and the key genes regulating longitudinal bone growth. On the left, structure of a long bone. Pink panel: schematic representation of the chondrocytes within the three different zones of the growth plate. On the right, some key genes regulating longitudinal bone growth and list of monogenic skeletal conditions caused by pathogenic mutations in each of these genes (96, 97, 98). ACAN, aggrecan; BMP1, bone morphogenetic protein 1; COL2A1, collagen type II alpha 1 chain; COL10A1, collagen type X alpha 1 chain; FGFR1-3, fibroblast growth factor receptor 1-3; GHR, growth hormone receptor; IGF1, insulin-like growth factor 1; IGF1R, insulin-like growth factor 1 receptor; IHH, Indian hedgehog signaling molecule; MEF2C, myocyte enhancer factor 2C; MMP9/13, matrix metallopeptidase 9/13; NOTCH1, Notch receptor 1; OSX, Sp7 transcription factor; PTH1R, parathyroid hormone 1 receptor 2; PTHLH, parathyroid hormone-like hormone; RUNX2, Runt-related transcription factor 2; SOX9/5, SRY-Box transcription factor 9/5; MIM, phenotype OMIM number.
Pathogenic variants in several genes encoding factors involved in chondrocyte proliferation and differentiation as well as defects in ECM components and cell-matrix interactions are known to be responsible for different monogenic skeletal conditions often accompanied by short stature (Fig. 1). Moreover, in some conditions growth plate defect can be secondary to abnormal bone metabolism and/or mineralization, such as in hypophosphatemic rickets (MIM 307800), in which hypophosphatemia leads to impaired chondrocyte apoptosis (10).
As mentioned previously, usually disproportionate short stature is not an isolated finding, but it is part of a wider spectrum of clinical features. From 2015 to 2019, pathogenic variants in 45 novel genes have been linked to skeletal diseases (2, 11). Among these, 15 novel gene defects, which have also been included in the Online Mendelian Inheritance in Man (OMIM) database, have been linked to either spondylometaphyseal dysplasia (SMD) or spondyloepimetaphyseal dysplasia (SEMD), two diseases that are mainly characterized by severe short stature and skeletal impairments affecting the spine, metaphyses and epiphyses (only SEMD) (Table 1) (2). In addition, pathogenic variants in the gene encoding the ribosomal protein eL13 (RPL13) have been recently linked to a novel form of SEMD (Table 1) (12, 13). These newly characterized conditions are caused by pathogenic variants in genes involved in several different biological and molecular processes (Fig. 2). Below, some examples of these conditions will be described in more detail.
Table 1.
Novel genetic defects underlying SMD and SEMD.
| Gene | Protein | Function | Disease | IP | OMIM phenotype MIM # | Reference |
|---|---|---|---|---|---|---|
| AIFM1 | Apoptosis inducing factor mitochondria associated 1 | Mitochondrial protein involved in oxidative phosphorylation and redox control in healthy cells | SEMD | XLR | 300232 | Mierzewska et al. 2017 (99) |
| BGN | Biglycan | Structural component of articular cartilage. It participates in the assembly of the chondrocyte extracellular matrix and it also plays a role in cell signaling | SEMD | XLR | 300106 | Cho et al. 2016 (100) |
| COL27A1 | Collagen type XXVII alpha 1 chain | Plays a role during the calcification of cartilage and in the transition of cartilage to bone | SEMD | AR | 615155 | Gonzaga et al. 2015 (101) |
| EXTL3 | Exostosin-like glycosyltransferase 3 | Glycosyltransferase that catalyzes the transfer of N-acetylglucosamine to glycosaminoglycan chains. This reaction is important in heparin and heparan sulfate synthesis | SEMD | AR | 617425 | Volpi et al. 2017 (102) |
| FN1 | Fibronectin 1 | Glycoprotein binding cell surfaces and various compounds including collagen, fibrin, heparin, DNA, and actin | SMD | AD | 184255 | Lee et al. 2017 (17) |
| HSPA9 | Heat shock protein family A (Hsp70) member 9 | Chaperone in the mitochondria, cytoplasm, and centrosome | SEMD | AR | 616854 | Royer-Bertrand et al. 2015 (103) |
| IARS2 | Isoleucyl-TRNA synthetase 2, mitochondrial | Nuclear-encoded mitochondrial isoleucine-tRNA synthetase that catalyzes the attachment of an isoleucine residue to a cognate mt-tRNA | SEMD | AR | 616007 | Schwartzentruber et al. 2014 (104) |
| LTBP3 | Latent transforming growth factor beta binding protein 3 | Involved in TGF-beta signaling pathway | SEMD | AR | 601216 | Huckert et al. 2015 (105) |
| NANS | N-acetylneuraminate synthase | Enzyme that functions in the biosynthetic pathways of sialic acids | SEMD | AR | 610442 | van Karnebeek et al. 2016 (106) |
| PISD | Phosphatidylserine decarboxylase | Enzyme that catalyzes the conversion of phosphatidylserine to phosphatidylethanolamine in the inner mitochondrial membrane | SEMD | AR | NA | Girisha et al. 2019 (107) |
| RPL13 | Ribosomal protein eL13 | Component of the large ribosomal subunit 60S | SEMD | AD | 618728 | La Caignec et al. 2019 (12) |
| RSPRY1 | Ring finger and SPRY domain containing 1 | Glycoprotein with unknown function | SEMD | AR | 616723 | Faden et al. 2015 (108) |
| SGMS2 | Sphingomyelin synthase 2 | A major component of cell and Golgi membranes involved in the synthesis of sphingomyelin | SMD | AD | 126550 | Pekkinen et al. 2019 (13) |
| TONSL | Tonsoku like, DNA repair protein | Involved in DNA replication | SEMD | AR | 271510 | Burrage et al. 2019 (109) |
| TRIP11 | Thyroid hormone receptor interactor 11 | Predicted to play a role in the assembly and maintenance of the Golgi ribbon structure around the centrosome | SMD | AR | 184260 | Wehrle et al. 2019 (110) |
| UFSP2 | UFM1 specific peptidase 2 | Involved in protein ubiquitination | SEMD | AD | 617974; 142669 | Di Rocco et al. 2018 (111) |
AD, autosomal dominant; AR, autosomal recessive; IP, inheritance pattern; NA, not available; SMD, spondylometaphyseal dysplasia; SEMD, spondyloepimetaphyseal dysplasia; XLR, X-linked recessive.
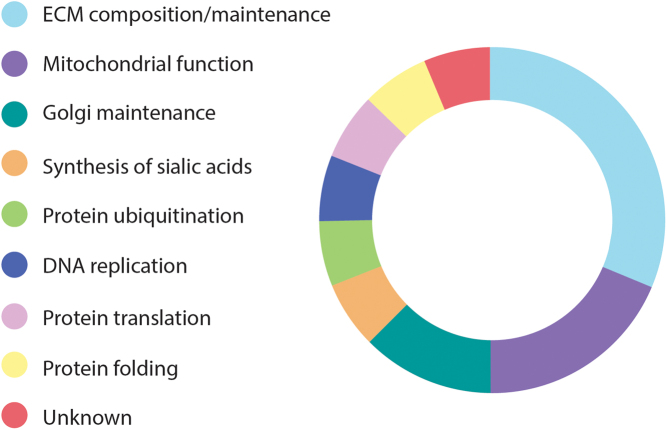
Figure 2.
Function of the 16 genes recently linked to SMD and SEMD. Most of the genes recently reported as causing SMD and SEMD play a role in the ECM (5/16), in the mitochondria (3/16) and in the Golgi apparatus (2/15). Each of the remaining genes is involved in a different biological mechanism.
Defects in fibronectin-1 cause SMD-corner fracture type
Fibronectin-1 (FN) is a dimeric glycoprotein that is abundant in several tissues and can be found either in a soluble form, like in the plasma, or as an insoluble multimeric fibrillar component of the ECM (14). FN is important for mesenchymal stem cell (MSC) differentiation and for deposition of collagen type I in the ECM (15, 16). Monoallelic pathogenic mutations in the fibronectin-1 gene (FN1) were first identified in 2017 in patients with a subtype of SMD, namely SMD with corner fractures (SMD-CF) (MIM 1842559) (17). Corner fractures are not considered as real fractures but they are key radiological findings appearing as lucent areas in the proximal metaphyses (growth plate) of tubular bones (18). SMD-CF was first described by Sutcliffe in 1966 and it was recognized as a separate entity in 1990 (19). Since then, approximately 30 families have been reported and while pathogenic variants in COL2A1 were identified in some subjects with SMD-CF, most of the patients lacked a genetic diagnosis for several decades (17, 20, 21). Nowadays, 11 different disease-causing missense variants and a single amino acid deletion in FN1 have been reported in 13 families with SMD-CF (17, 20, 21). In addition to corner fractures, patients typically feature short stature, developmental coxa vara, scoliosis, and abnormal ossification at the growth plate and secondary ossification sites.
FN is a dimer constituted by three types of modules, domains types I–III, which bind other components of the ECM, such as collagen, integrins and glycosaminoglycans as well as signaling and cell adhesion molecules (14, 22). Interestingly, the majority of the pathogenic variants that have so far been linked to SMD-CF locate within the fibronectin type-I domains. In particular, nine out of the 11 reported variants associated with SMD-CF affect cysteine residues partaking in disulfide bonds, which maintain the highly organized structure of FN. Pathogenic variants in FN1 impair the secretion of FN into the ECM in patient-derived fibroblasts as well as in HEK293 cells transfected with plasmid expressing mutant FN (17, 20). Surprisingly, pathogenic variants in FN domain type III cause another disease, named Glomerulopathy with fibronectin deposits 2 (phenotype MIM 601894). Renal dysfunction has not been reported in any patient with SMD-CF thus suggesting that the location of the pathogenic variant determines the development of either one or the other disease.
Although additional experiments are required to investigate the consequences of pathogenic variants in FN1 on the bone ECM, the critical role of FN in skeletal development and tissue maintenance is confirmed by the lethality of mice lacking FN (23).
Defects in sphingomyelin synthase 2 cause SMD
Sphingomyelin is a sphingolipid of the plasma membrane and the Golgi membranes. Monoallelic pathogenic variants in the gene encoding sphingomyelin synthase 2 (SGMS2), an enzyme that catalyzes the synthesis of sphingomyelin, have recently been identified in patients affected by a rare form of genetic osteoporosis, named Calvarial doughnut lesions with bone fragility with or without SMD (MIM 126550) (24). Thus far, only three different SGMS2 pathogenic variants in eight unrelated families have been described in the literature (24, 25). While some patients only have early-onset osteoporosis, the most severely affected patients feature severe short stature, neonatal fractures, and SMD. The variable severity of the disease is related to the type of pathogenic variant: subjects harboring the nonsense variant c.148C>T (p.Arg50*) are either healthy (25) or more mildly affected than patients with the missense changes c.185T>G (p.Ile62Ser) or c.191T>G (p.Met64Arg) (24). The absence of a bone phenotype in some patients harboring the nonsense variant suggests incomplete penetrance. The p.Arg50* change generates a truncated protein that lacks the whole membrane-spanning core domain and leads to a catalytically inactive enzyme (24). On the other hand, in cell and yeast models, the two missense variants produce a catalytically active enzyme that accumulates in the endoplasmic reticulum and synthesizes sphingomyelin at a remarkably higher rate than the wildtype enzyme. Since SMS2 is a homodimer the missense pathogenic variants might lead to a dominant-negative effect induced by the mutated SMS2 forming a heterodimer with the wildtype enzyme. Patient-derived bone biopsies reveal significant tissue-level pathology, thin cortical bone, and disturbed bone mineralization (24, 25). Pathogenic variants in two genes encoding two other enzymes involved in sphingomyelin metabolism, phosphatidylserine synthase 1 (PTDSS1) and choline-phosphate cytidylyltransferase A (PCYT1A), cause two other types of skeletal dysplasias, named Lenz-Majewski syndrome (MIM 151050) (26) and spondylometaphyseal dysplasia with cone-rod dystrophy (MIM 608940) (27).
Defects in ribosomal protein eL13 cause SEMD
In some cases, short stature and skeletal abnormalities can arise from pathogenic mutations in genes encoding ribosomal proteins (RPs), transcribing ribosomal RNAs (rRNAs) or required for ribosome biosynthesis (28). Ribosomes are vital organelles of the cells that are responsible for synthetizing proteins. Eukaryotic ribosomes (80S) are composed of two subunits: the large 60S subunit and the small 40S subunit. Human ribosomes are mainly located in the cytoplasm (either bound to the endoplasmic reticulum or floating in the aqueous part) and are formed by a complex of around 80 RPs and four rRNAs (29, 30). Recently, we and others identified inherited or de novo pathogenic variants in the gene encoding the ribosomal protein eL13 (RPL13) in altogether nine index patients with skeletal dysplasia without extra-skeletal features (12, 13, 31). The phenotype of the so far reported patients with pathogenic RPL13 variants varies greatly and it ranges from normal stature with multiple epiphyseal dysplasia in one patient (31) to disproportionate short stature, growth deficiency, broad metaphyses, and delayed ossification of the epiphyses in the majority of the other patients. Interestingly, in two families variable clinical expressivity and incomplete penetrance of the disease have been identified (13). Surprisingly, in one family the index patient died during early childhood while his mother is healthy, despite harboring the same RPL13 mutation as her child (13). Polysome profiling, carried out in both patient-derived lymphoblasts and dermal fibroblasts, shows a reduced 80S peak, thus suggesting a change in translation efficiency (12, 13). Moreover, a zebrafish model harboring a frameshift deletion within rpl13 partly recapitulates the human phenotype by featuring cartilage deformities during early stages of development (13). Although the molecular mechanisms underlying skeletal dysplasia RPL13-type are yet unknown, tissue specificity with skeletal involvement and incomplete penetrance of the disease have been found in other conditions caused by ribosomal dysfunction, collectively named ribosomopathies (28, 32). Tissue specificity suggests that ribosomes could be involved in other mechanisms beside translation (33) and RPL13 might potentially play a role in skeletogenesis.
Concerning the presence of short stature and skeletal abnormalities due to defects in ribosomal components, other congenital conditions with these clinical features have been described previously, including CHH, Diamond-Blackfan anemia 1 (MIM 105650), and Shwachman-Diamond syndrome (MIM 260400) (34). Moreover, common findings among ribosomopathies are bone marrow failure and anemia. Although the patients that have so far been reported as being affected by skeletal dysplasia RPL13-type do not feature hematological impairments, bone marrow dysfunction could lead to defective mesenchymal stem cell production, consequently affecting chondrogenic/osteogenic differentiation. Moreover, patients with ribosomopathies often have increased propensity to develop cancer (35, 36). Since only nine families with skeletal dysplasia due to RPL13 variants have been described so far, it is not possible to estimate the cancer risk for this type of skeletal dysplasia. Certainly, more patients need to be identified and characterized to better delineate the clinical and genetic scenario of skeletal dysplasia RPL13-type and additional functional work, possibly using chondrogenic/osteogenic cell lines and in vivo models, is required to fully explore the pathogenesis of the disease.
Mildest forms of skeletal dysplasia in patients diagnosed with idiopathic short stature
Idiopathic short stature (ISS) is defined as stature more than 2 s.d. below the mean for age and sex, when no cause for the short stature has been identified despite standard clinical and laboratory evaluation (37, 38). For diagnosis of ISS, systemic or endocrine diseases, dysmorphic syndromes, small birth size (small for gestational age, SGA), and skeletal dysplasias should be excluded (38). Height is a polygenic trait and several hits associated to height variation have been identified by genome-wide association studies so far (39, 40). However, monogenic forms of short stature have also been identified. In cohorts of ISS children, some children have pathogenic variants in genes that are associated with skeletal dysplasias and ISS phenotype, thus represents the mildest end of the spectrum of these disorders.
One well-known example is short stature homeobox (SHOX) haploinsufficiency, which causes short stature in 2–15% of children previously diagnosed with ISS (MIM 300582) (41). In addition to isolated short stature, heterozygous SHOX pathogenic variants cause Leri-Weill dyschondrosteosis (MIM 127300), a skeletal dysplasia characterized by disproportionate short stature, mesomelic limb shortening, and the Madelung deformity (42). On the other hand, biallelic pathogenic variants in SHOX cause the more severe Langer mesomelic dysplasia (MIM 249700) (43). The clinical severity of SHOX pathogenic variants varies even within family members carrying the same variant and damaging mutations of the gene encoding the retinoic acid catabolizing enzyme (CYP26C1) have been reported to act as genetic modifiers of SHOX deficiency (44). Recently, it was reported that CYP26C1 damaging mutations without SHOX deficiency can also lead to short stature (45).
Pathogenic variants in natriuretic peptide receptor 2 gene (NPR2) also cause a large variability in phenotype, ranging from acromesomelic dysplasia (MIM 602875), caused by biallelic mutations, to isolated short stature (MIM 616555). Family members who were mutation carriers were found to have significantly lower height than non-carrier family members and the general population, suggesting that heterozygous variants in NPR2 are associated with short stature (46). Subsequent studies in ISS cohorts have found heterozygous NPR2 variants in 2–6% of the patients (47, 48, 49). Variable phenotype was observed, with cases of both proportional and disproportional short stature. NPR2 is the principal receptor of natriuretic peptide C, encoded by the natriuretic peptide precursor C (NPPC). Heterozygous NPPC pathogenic variants have been identified as a cause of short stature and small hands phenotype in 0.6% of patients with short stature (50).
Pathogenic variants in the fibroblast-growth factor receptor 3 (FGFR3) are responsible for a wide range of clinical severity, from lethal thanatophoric dysplasia types I and II (MIM 187600-187601) to nonlethal achondroplasia (MIM 100800) and hypochondroplasia (MIM 146000) (51). In 2015, a FGFR3 variant was determined as a cause of proportionate short stature in one family (52), further widening the clinical spectrum. Interestingly, an analog of C-type natriuretic peptide, which promotes bone growth by inhibiting fibroblast-growth factor-mediated mitogen activated protein kinase (MAPK) activation, is presently used in phase III trials to treat achondroplasia(6).
Aggrecan (ACAN) is a proteoglycan in the extracellular matrix and its major function is to resist compression in cartilage. Homozygous or compound heterozygous pathogenic variants in ACAN cause SEMD aggrecan type (MIM 612813), while heterozygous pathogenic variants can cause either spondyloepiphyseal dysplasia Kimberley type (MIM 608361) or short stature with advanced bone age (MIM 165800) (2). Many patients with short stature and advanced bone age caused by pathogenic variants in ACAN develop early-onset osteoarthritis and degenerative disc disease (53). Midface hypoplasia, joint problems, and broad great toes have also been reported (54). Recently, subjects with ACAN haploinsufficiency were reported to have an elevated mean arm span to height ratio in childhood and adolescence, and a slightly elevated ratio until age 50 (53, 55).
Indian hedgehog signaling molecule (IHH) is known to cause acrocapitofemoral dysplasia (MIM 607778), brachydactyly type A1 (MIM 112500), syndactyly (Lueken type) and syndactyly with craniosynostosis (Fig. 1) (2). Additionally, heterozygous pathogenic IHH variants causing short stature have been identified in a cohort of patients with growth disorders (56). Short stature was mildly disproportionate in most cases and many had shortening of the middle phalanx of second and/or fifth finger. Some of the patients were born small for gestational age (SGA). Recently, heterozygous pathogenic IHH variants were identified in another cohort with short stature and/or brachydactyly, including the first patient with a complete deletion of IHH, who presented with both short stature and brachydactyly (57).
Novel molecular mechanisms underlying skeletal diseases
For a long time, it has been thought that the non-coding genome (approximately 98% of the whole genome), also named junk DNA, does not play any vital function. However, this concept has been disproved by the discovery of skeletal diseases caused by variants in the non-coding regulatory genome and two examples are discussed here.
Aberrant microRNA (miRNA) expression and processing, miRNA deletions as well as point mutations in miRNA have been associated with congenital conditions (58). In 2019, the first skeletal dysplasia caused by gain-of-function pathogenic variants in a miRNA, miRNA-140, were characterized by Grigelioniene et al. (59). The three patients from two families affected by this disease, named spondyloepiphyseal dysplasia Nishimura type (MIM 618618), mainly feature dwarfism, short limbs, and small hands and feet. All affected subjects had the same heterozygous missense variant, MIR140:NR_029681.1:n.24A>G, in the seed sequence of miRNA-140, which is highly expressed in chondrocytes and it was found to be associated with a chondrocyte-specific super-enhancer both in humans and mice (59). Mice lacking this miRNA are short and have craniofacial deformities (60). Heterozygous and homozygous knock-in mice harboring the same pathogenic variant as detected in the patients showed a more severe phenotype, including also delayed secondary ossification of tubular and carpal bones, delayed cartilage development in the larynx, trachea, anterior ribs, and decreased expression of Col10a1 (59).
In recent years, the discovery of topological associated domains (TADs), megabase-scale 3D rearrangements of the chromatin, has demonstrated that regulatory regions might be located far away (in terms of linear DNA sequence) from the gene they regulate (61). Acropectorovertebral dysplasia (MIM 102510), characterized by carpal and tarsal synostoses, syndactyly, hypodactyly and polydactyly of feet as well as spina bifida occulta arises from structural variants (SVs) disrupting a TAD domain spanning the genes WNT6, IHH, EPHA4, and PAX3 (62). This TAD disruption is likely to rewire gene-enhancer interactions, leading to abnormal gene expression. Disruption of other TAD domains might then be the cause of other skeletal diseases with unknown genetic basis.
How to identify novel phenotypes
Novel skeletal disorders and their genetic etiology can be studied by combining careful phenotyping with selected genetic strategies. Assessment includes detailed family history and information about possible consanguinity, in order to determine the possible inheritance pattern. As skeletal dysplasias may present in different family members with a varying degree of severity, a clinical evaluation of family members may be necessary to determine whether they might have a mild form of the same disease.
In a patient with short stature, accurate and multiple growth chart measurements, including SDS for height, weight, and head circumference are essential for assessing growth. It should be determined whether the patient was born with short stature and whether macrocephaly is present. Disproportionate short stature is typical in skeletal dysplasia and it can be assessed by measuring arm span, sitting height, and sitting height/height ratio. Alternatively, the upper/lower body segment ratio can be calculated and compared to references (38). Disproportionate short stature affecting the limbs can be either rhizomelic, mesomelic or acromelic depending on whether proximal (humerus and femur), middle (radius, ulna, tibia, or fibula), or distal (hand and foot) segments are shortened (63). Spinal involvement may be difficult to evaluate clinically and often requires radiographic assessment.
Evaluation of facial dysmorphic features may include evaluation of the fontanels, nasal bridge, midface, philtrum, mandible, palate, and ears (1). One should also assess upper and lower limb lengths and possible asymmetry, carrying angle of the elbow, and any anomalies in the digits (oligo-, poly-, brachy- or syndactyly). Frontal bossing, cleft palate, dental defects, clavicle aplasia or hypoplasia, scoliosis, joint hyperlaxity/contractures/dislocations, genu varum or valgum, fractures, and abnormalities in nails, hair, and skin should be examined.
Skeletal dysplasias may also be associated with various extra-skeletal manifestations, such as problems in hearing, vision, renal or respiratory system, heart defects, genital abnormalities, immune deficiency, anemia, and intestinal problems such as Hirschprung disease (1, 63, 64, 65). Further evaluation of these systems may be needed.
Skeletal survey is an essential tool in assessing skeletal dysplasias. A skeletal survey may include radiographs of the skull (posteroanterior and lateral), spine (anteroposterior and lateral), thorax (anteroposterior), pelvis (anteroposterior), upper limb (anteroposterior), left hand and wrist (posteroanterior) and lower limb (anteroposterior) (38). Based on localization of radiological findings, the dysplasias can be characterized as spondylo, epiphyseal, metaphyseal or diaphyseal dysplasia, or a combination of these.
Genetic approaches to variant and gene discoveries
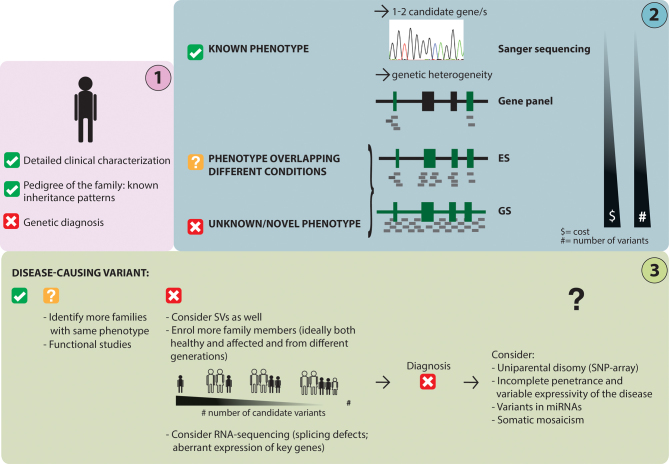
In addition to an in-depth clinical investigation, choosing the correct genetic approach to investigate a family with a rare skeletal disease with short stature is crucial for a successful diagnosis (Fig. 3).
Figure 3.
Schematic workflow of the genetic approach to identify the disease-causing variant in a patient with a skeletal disease. After an in-depth clinical characterization (step 1), the phenotype of the studied patient might overlap a known condition, several conditions or be a novel/unknown phenotype; this will determine the genetic approach/method to be chosen (step 2). Data analysis could directly lead to a genetic diagnosis, but often further investigations are needed to validate a genetic finding or to pinpoint the genetic defect (step 3). ES, exome sequencing; GS, genome sequencing; SVs, structural variants.
In last decades, the field of clinical genetics has greatly evolved thanks to the development of novel sequencing technologies and the advances in molecular biology. Before the MPS era, novel genes underlying monogenic conditions were mapped by positional cloning and linkage analysis. These methods, which allowed the identification of large regions to be prioritized by Sanger sequencing, were not only time consuming but they also required large multi-case pedigrees. Nowadays, Sanger sequencing is only used to sequence one or a couple of candidate genes/exons when a disease-gene correlation is suspected.
Since the majority of skeletal diseases is characterized by genetic heterogeneity, methods that allow to screen several loci at the same time are often required (Fig. 3). Among the different MPS methods, gene panels are designed for capturing and sequencing a certain number of candidate genes. Running a gene panel has become a faster and less expensive alternative to Sanger sequencing (66).
Alternatively, when the investigated phenotype overlaps with more than one condition and/or a long list of genes needs to be screened, exome sequencing (ES) is a more cost-effective option. ES gives the possibility not only to screen for variants in known disease-causing genes but also to search for variants in the rest of the protein-coding genes.
Finally, genome sequencing (GS), currently the most expensive method in the field, enables the sequencing of the entire genome. Some advantages of using GS instead of ES include: (1) sequencing of variants in regulatory regions (67) as well as deep intronic variants that could affect gene expression and the splicing mechanism (68), respectively, (2) sequencing of non-coding RNAs, such as miRNAs, that could regulate the expression of key genes in skeletogenesis, (3) more even coverage to improve the detection of intragenic deletions and duplications, and (4) possibility of detecting SVs, which are large chromosomal rearrangements (>50 base pairs) (69) that could disrupt known disease-causing genes. Although most of the pathogenic variants that have so far been reported to cause skeletal dysplasias are small-scale variants, sometimes SVs, including copy number variants (CNVs), inversions and translocations, can give rise to skeletal diseases (70). CNVs can also be detected using SNP and comparative genomic hybridization (CGH)-arrays, two older methods that can pinpoint large changes in DNA copies.
SNP-array, ES, or GS data can also be used to detect uniparental disomy (UPD) (71), a genetic phenomenon where both copies of a chromosome are inherited from the same parent. UPD can lead to imprinting disorders by disrupting the expression pattern of imprinted genes. For example, paternal UPD of chromosome 20 has been reported in up to 18% of the patients with pseudohypoparathyroidism type 1b (PHP1b) (72) and maternal UPD of chromosome 7 is found in approximately 5–10% of patients with Silver–Russell syndrome (SRS) (MIM 18060) (73). In addition to UPD, imprinting disorders are also caused by methylation defects, such as loss of methylation at the GNAS imprinted region in PHP1b (74) and loss of methylation at 11p15 in SRS (73).
Despite the fact that nowadays large multi-case families are not needed to pinpoint a novel disease locus, it is often advantageous to perform MPS on at least a family trio (patient and his/her parents) in order to narrow down the number of rare candidate variants.
Finally, to improve the knowledge on non-coding variants and to increase the diagnostic yield in skeletal dysplasias, a combination of ES/GS and RNA-sequencing might be needed. Transcriptome analysis in monogenic diseases does not only allow to pinpoint deep intronic variants and synonymous changes that affect the splicing mechanism but also to identify variants in regulatory regions (promoters, enhancers and UTRs) that result in aberrant gene expression (68). Since the transcriptome signatures differ in different tissues, it is crucial to perform the investigations on the affected tissue or to identify a good proxy tissue for the disease, which in the case of skeletal conditions would be bone and/or cartilage.
Recently, transcriptome analysis performed on RNA extracted from primary skin fibroblasts of patients with CHH revealed differential expression of several genes regulating the cell cycle (75). These results suggest that if the investigated gene plays a pivotal role not only for the skeleton but also for the ectoderm, skin biopsies could be used as a source material to carry out RNA-sequencing.
To summarize, it is important to choose the most appropriate genetic approach based on clinical evaluation of the patients, available resources as well as cost-effectiveness/limitations of each method.
Validation of novel gene-disease associations
In vitro studies
Once a candidate variant in a gene that has not been previously linked to disease is identified, functional studies are needed to validate this finding. As previously mentioned, an important aspect to consider when investigating genetic findings is the source of material chosen/available to investigate the disease pathogenesis. Often, bone biopsies would be the most appropriate tissue and source of cells for investigating the pathogenesis of the disease. However, obtaining a bone biopsy is an invasive procedure that is not performed unless it is required for diagnostic purposes or the patient needs to undergo elective surgery. Bone biopsies are valuable for skeletal research since they can be used to assess several parameters, such as bone microarchitecture, mineralization, and the morphology and organization of the different types of bone cells (24, 76). Moreover, bone marrow aspirates are a source of MSCs. Although several protocols for osteogenic cell line differentiation from MSCs are available, one limitation of this method is related to the limited availability of these cells from healthy donors.
If the studied gene is expressed in the ectoderm, most often skin punch biopsies obtained from patients are used to investigate the pathomolecular mechanisms leading to genetic skeletal diseases. From skin biopsies primary fibroblasts are derived and can be studied from different angles. A lot of information can be obtained for example by comparing cell proliferation, differentiation, and apoptosis as well as gene/protein expression in patients vs sex- and age-matched controls. However, it is always important to know that a defect in these cells might not necessarily reflect the true nature of the disease and, vice versa, lack of any experimental evidence in fibroblasts might not exclude that the pinpointed genetic finding has an adverse effect on bone. Despite these limitations, patient-derived fibroblasts are widely used in the field of skeletal research and have led to important discoveries. Moreover, dermal fibroblasts could be potentially reprogrammed into induced pluripotent stem cells (iPSCs) and re-differentiated into chondrocytes to study subtypes of skeletal dysplasias with short stature due to an intrinsic defect in the growth plate. However, the culture conditions allowing to differentiate iPSCs and MSCs into chondrocytes are not yet well understood (77).
Since 2012, the CRISPR-Cas9 technology (78) has transformed the field of genetic engineering by introducing the possibility of efficiently silencing a gene of interest (or introducing a specific pathogenic mutation) in commercially available cell lines (79). While this approach overcomes the problem of obtaining patient-derived osteogenic cells, the use of immortalized cell lines might introduce changes in the cells that might be incorrectly assessed as part of the phenotype induced by the introduced pathogenic variant/gene defect. Furthermore, patient-derived cells have the advantage of maintaining the whole genetic signature of each patient, thus allowing to detect the effects of potential interactions between a set of variants in the genome.
In vivo models
In order to explore the systemic effects of genetic pathogenic variants in an organism and to investigate cartilage and skeletal development, animal models are required. Genetically engineered mice (MusMusculus) have been widely used for mimicking human skeletal diseases since this species shares a high percentage of coding DNA (~85%) with humans (80) and bone development and the skeletal elements are highly conserved between these two mammalian species. Mice also undergo longitudinal bone growth and the growth plate is the structure determining cartilage production and bone apposition (81). Unlike in humans, the murine growth plate does not undergo epiphyseal fusion with sexual maturation (82). Concerning bone remodeling, it takes place in the cancellous bone, as in humans. The most striking difference between mice and humans is that murine bone lacks osteons, the structural and functional units of cortical bone (81).
Since generating and handling transgenic mice is expensive and time consuming, zebrafish (DanioRerio) has recently emerged as a model for studying rare skeletal diseases (83, 84, 85), primarily osteoporosis and OI (86, 87). Approximately 71% of protein-coding genes in the human genome have an ortholog in zebrafish (88). Every week zebrafish produces hundreds of eggs, which are externally fertilized, thus allowing fast and easy genetic manipulation. Moreover, zebrafish larvae are transparent, and they can be stained in whole-mount to study skeletal development and cartilage/bone mineralization. As in humans, bone development in zebrafish occurs by both intramembranous and endochondral ossification (89) and several key genes playing a role in skeletogenesis are conserved across the two species (79, 90). Despite the fact that there are also dissimilarities between human and zebrafish skeletogenesis (e.g. in zebrafish bone is not vascularized and the growth plate develops differently), several zebrafish models mimicking human skeletal disease have been generated in the last decade. Furthermore, it has been recently shown that zebrafish have structures resembling the human growth plate (91). This finding will open the possibility to better investigate diseases characterized by short stature due to impaired growth plate in this species.
In vivo models are also largely used for drug testing before a candidate undergoes clinical testing in humans. Recently, an antibody against TGF-β was tested in two different mouse models of OI (92, 93) and two chemical chaperones were proven to improve the skeletal phenotype of a zebrafish model of autosomal dominant OI due to a missense pathogenic variant in col1a1a (94).
Significance of gene discoveries
Identifying novel pathogenic mutations and novel gene defects underlying skeletal diseases is important in order to be able to provide an accurate diagnosis to the patients and to offer genetic counseling and optimized management of the disease to the families. Moreover, providing the risk of having another affected child to the parents also influences their reproductive choices. Since most of the skeletal diseases still lack a pharmacological treatment, there is necessity for exploring novel gene defects to find common disease mechanisms that can eventually lead to the development of novel effective drugs. Although orthopedic surgeries will always be required to correct skeletal deformities and severe scoliosis, pharmacotherapies might help in preventing or ameliorating some skeletal features, in particular if a diagnosis is made at an early stage. Growth hormone therapy has been shown to help in gaining height in patients with short stature due to SHOX defects, but in patients affected by other diseases, such as spondyloepiphyseal dysplasia, the skeletal impairments (e.g. scoliosis) may even worsen with treatment (95). Therefore, it is crucial to distinguish between different causes for short stature and use personalized treatment approaches. Due to the mechanistic differences in disorders leading to impaired growth, it is important, in research settings, that novel genetic findings are carefully investigated using in vitro and in vivo models to understand the pathomolecular mechanisms leading to disease.
Conclusions and future perspectives
During the past 5 years, 16 novel genetic forms of SMD and SEMD have been characterized. These successful results have been achieved not only because of the possibility to use MPS but also because of joint efforts of several experts in the field aiming to identify and carefully delineate novel skeletal phenotypes.
Although nowadays 92% of the thus far characterized skeletal conditions have a known genetic basis, it can be anticipated that in the near future the genetic defects underlying the remaining conditions will be identified. In addition, novel extremely rare skeletal phenotypes most likely continue to be characterized also in the coming years. A better knowledge about the non-coding genome and further attention on the genetic mechanisms escaping regular Mendelian inheritance, such as incomplete penetrance and somatic mosaicism, might be needed to solve the remaining cases of skeletal dysplasia with severe short stature.
Finally, the large increase in gene-disease associations that has characterized the last decade has lagged behind in in-depth characterization of the pathogenesis of these diseases. Additional work is, thus, required to explore the molecular mechanisms leading to impaired bone development and/or homeostasis and to pinpoint novel drug targets. This knowledge will eventually be applied to develop efficient therapeutic strategies to treat patients with skeletal diseases.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
Our research is financially supported by Novo Nordisk Foundation, Academy of Finland, Sigrid Jusélius Foundation, Folkhälsan Research Foundation, Biomedicum Helsinki Foundation, Vetenskapsrådet, Swedish Childhood Cancer Foundation, and the Stockholm County Council to OM.
References
- 1.Krakow D.Skeletal dysplasias. Clinics in Perinatology 2015. 42 301–19. ( 10.1016/j.clp.2015.03.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mortier GR, Cohn DH, Cormier-Daire V, Hall C, Krakow D, Mundlos S, Nishimura G, Robertson S, Sangiorgi L, Savarirayan R. et al. Nosology and classification of genetic skeletal disorders: 2019 revision. American Journal of Medical Genetics. Part A 2019. 179 2393–2419. ( 10.1002/ajmg.a.61366) [DOI] [PubMed] [Google Scholar]
- 3.Krakow D, Rimoin DL.The skeletal dysplasias. Genetics in Medicine 2010. 12 327–341. ( 10.1097/GIM.0b013e3181daae9b) [DOI] [PubMed] [Google Scholar]
- 4.Carpenter TO, Whyte MP, Imel EA, Boot AM, Hogler W, Linglart A, Padidela R, Van't Hoff W, Mao M, Chen CYet al. Burosumab therapy in children with X-linked hypophosphatemia. New England Journal of Medicine 2018. 378 1987–1998. ( 10.1056/NEJMoa1714641) [DOI] [PubMed] [Google Scholar]
- 5.Whyte MP, Madson KL, Phillips D, Reeves AL, McAlister WH, Yakimoski A, Mack KE, Hamilton K, Kagan K, Fujita KPet al. Asfotase alfa therapy for children with hypophosphatasia. JCI Insight 2016. 1 e85971. ( 10.1172/jci.insight.85971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savarirayan R, Irving M, Bacino CA, Bostwick B, Charrow J, Cormier-Daire V, Le Quan Sang KH, Dickson P, Harmatz P, Phillips Jet al. C-type natriuretic peptide analogue therapy in children with achondroplasia. New England Journal of Medicine 2019. 381 25–35. ( 10.1056/NEJMoa1813446) [DOI] [PubMed] [Google Scholar]
- 7.Brighton CT.Structure and function of the growth plate. Clinical Orthopaedics and Related Research 1978. 136 22–32. ( 10.1097/00003086-197810000-00003) [DOI] [PubMed] [Google Scholar]
- 8.Abad V, Meyers JL, Weise M, Gafni RI, Barnes KM, Nilsson O, Bacher JD, Baron J.The role of the resting zone in growth plate chondrogenesis. Endocrinology 2002. 143 1851–1857. ( 10.1210/endo.143.5.8776) [DOI] [PubMed] [Google Scholar]
- 9.Gerber HP, Vu TH, Ryan AM, Kowalski J, Werb Z, Ferrara N.VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nature Medicine 1999. 5 623–628. ( 10.1038/9467) [DOI] [PubMed] [Google Scholar]
- 10.Sabbagh Y, Carpenter TO, Demay MB.Hypophosphatemia leads to rickets by impairing caspase-mediated apoptosis of hypertrophic chondrocytes. PNAS 2005. 102 9637–9642. ( 10.1073/pnas.0502249102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonafe L, Cormier-Daire V, Hall C, Lachman R, Mortier G, Mundlos S, Nishimura G, Sangiorgi L, Savarirayan R, Sillence Det al. Nosology and classification of genetic skeletal disorders: 2015 revision. American Journal of Medical Genetics. Part A 2015. 167A 2869–2892. ( 10.1002/ajmg.a.37365) [DOI] [PubMed] [Google Scholar]
- 12.Le Caignec C, Ory B, Lamoureux F, O'Donohue MF, Orgebin E, Lindenbaum P, Teletchea S, Saby M, Hurst A, Nelson Ket al. RPL13 variants cause spondyloepimetaphyseal dysplasia with severe short stature. American Journal of Human Genetics 2019. 105 1040–1047. ( 10.1016/j.ajhg.2019.09.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costantini A, Alm JJ, Tonelli F, Valta H, Huber C, Tran AN, Daponte V, Kirova N, Kwon YU, Bae JYet al. Novel RPL13 variants and variable clinical expressivity in a human Ribosomopathy With spondyloepimetaphyseal dysplasia. Journal of Bone and Mineral Research 2021. 36 283–297. ( 10.1002/jbmr.4177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zollinger AJ, Smith ML.Fibronectin, the extracellular glue. Matrix Biology 2017. 60–61 27–37. ( 10.1016/j.matbio.2016.07.011) [DOI] [PubMed] [Google Scholar]
- 15.Singh P, Schwarzbauer JE.Fibronectin and stem cell differentiation - lessons from chondrogenesis. Journal of Cell Science 2012. 125 3703–3712. ( 10.1242/jcs.095786) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sottile J, Hocking DC.Fibronectin polymerization regulates the composition and stability of extracellular matrix fibrils and cell-matrix adhesions. Molecular Biology of the Cell 2002. 13 3546–3559. ( 10.1091/mbc.e02-01-0048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee CS, Fu H, Baratang N, Rousseau J, Kumra H, Sutton VR, Niceta M, Ciolfi A, Yamamoto G, Bertola Det al. Mutations in fibronectin cause a subtype of Spondylometaphyseal dysplasia with “corner fractures”. American Journal of Human Genetics 2017. 101 815–823. ( 10.1016/j.ajhg.2017.09.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Currarino G, Birch JG, Herring JA.Developmental coxa vara associated with spondylometaphyseal dysplasia (DCV/SMD): "SMD-Corner Fracture Type" (DCV/SMD-CF) demonstrated in most reported cases. Pediatric Radiology 2000. 30 14–24. ( 10.1007/s002470050005) [DOI] [PubMed] [Google Scholar]
- 19.Langer LO, Jr., Brill PW, Ozonoff MB, Pauli RM, Wilson WG, Alford BA, Pavlov H, Drake DG.Spondylometaphyseal dysplasia, corner fracture type: a heritable condition associated with coxa vara. Radiology 1990. 175 761–766. ( 10.1148/radiology.175.3.2343127) [DOI] [PubMed] [Google Scholar]
- 20.Cadoff EB, Sheffer R, Wientroub S, Ovadia D, Meiner V, Schwarzbauer JE.Mechanistic insights into the cellular effects of a novel FN1 variant associated with a spondylometaphyseal dysplasia. Clinical Genetics 2018. 94 429–437. ( 10.1111/cge.13424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costantini A, Valta H, Baratang NV, Yap P, Bertola DR, Yamamoto GL, Kim CA, Chen J, Wierenga KJ, Fanning EAet al. Novel fibronectin mutations and expansion of the phenotype in spondylometaphyseal dysplasia with "corner fractures". Bone 2019. 121 163–171. ( 10.1016/j.bone.2018.12.020) [DOI] [PubMed] [Google Scholar]
- 22.Potts JR, Campbell ID.Fibronectin structure and assembly. Current Opinion in Cell Biology 1994. 6 648–655. ( 10.1016/0955-0674(9490090-6) [DOI] [PubMed] [Google Scholar]
- 23.George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO.Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 1993. 119 1079–1091. [DOI] [PubMed] [Google Scholar]
- 24.Pekkinen M, Terhal PA, Botto LD, Henning P, Makitie RE, Roschger P, Jain A, Kol M, Kjellberg MA, Paschalis EPet al. Osteoporosis and skeletal dysplasia caused by pathogenic variants in SGMS2. JCI Insight 2019. 4 e126180. ( 10.1172/jci.insight.126180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson ME, Bardai G, Veilleux LN, Glorieux FH, Rauch F.Musculoskeletal phenotype in two unrelated individuals with a recurrent nonsense variant in SGMS2. Bone 2020. 134 115261. ( 10.1016/j.bone.2020.115261) [DOI] [PubMed] [Google Scholar]
- 26.Sousa SB, Jenkins D, Chanudet E, Tasseva G, Ishida M, Anderson G, Docker J, Ryten M, Sa J, Saraiva JMet al. Gain-of-function mutations in the phosphatidylserine synthase 1 (PTDSS1) gene cause Lenz-Majewski syndrome. Nature Genetics 2014. 46 70–76. ( 10.1038/ng.2829) [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto GL, Baratela WA, Almeida TF, Lazar M, Afonso CL, Oyamada MK, Suzuki L, Oliveira LA, Ramos ES, Kim CAet al. Mutations in PCYT1A cause spondylometaphyseal dysplasia with cone-rod dystrophy. American Journal of Human Genetics 2014. 94 113–119. ( 10.1016/j.ajhg.2013.11.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trainor PA, Merrill AE.Ribosome biogenesis in skeletal development and the pathogenesis of skeletal disorders. Biochimica et Biophysica Acta 2014. 1842 769–778. ( 10.1016/j.bbadis.2013.11.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de la Cruz J, Karbstein K, Woolford JL., JrFunctions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annual Review of Biochemistry 2015. 84 93–129. ( 10.1146/annurev-biochem-060614-033917) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ben-Shem A, Jenner L, Yusupova G, Yusupov M.Crystal structure of the eukaryotic ribosome. Science 2010. 330 1203–1209. ( 10.1126/science.1194294) [DOI] [PubMed] [Google Scholar]
- 31.Reinsch B, Grand K, Lachman RS, Kim HKW, Sanchez‐Lara PA.Expanding the phenotypic spectrum of RPL13‐related skeletal dysplasia. American Journal of Medical Genetics. Part A 2020. [epub]. ( 10.1002/ajmg.a.61965) [DOI] [PubMed] [Google Scholar]
- 32.Mills EW, Green R.Ribosomopathies: there's strength in numbers. Science 2017. 358 eaan2755. ( 10.1126/science.aan2755) [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, Liao WJ, Liao JM, Liao P, Lu H.Ribosomal proteins: functions beyond the ribosome. Journal of Molecular Cell Biology 2015. 7 92–104. ( 10.1093/jmcb/mjv014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narla A, Ebert BL.Ribosomopathies: human disorders of ribosome dysfunction. Blood 2010. 115 3196–3205. ( 10.1182/blood-2009-10-178129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ajore R, Raiser D, McConkey M, Joud M, Boidol B, Mar B, Saksena G, Weinstock DM, Armstrong S, Ellis SRet al. Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations. EMBO Molecular Medicine 2017. 9 498–507. ( 10.15252/emmm.201606660) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taskinen M, Ranki A, Pukkala E, Jeskanen L, Kaitila I, Makitie O.Extended follow-up of the Finnish cartilage-hair hypoplasia cohort confirms high incidence of non-Hodgkin lymphoma and basal cell carcinoma. American Journal of Medical Genetics. Part A 2008. 146A 2370–2375. ( 10.1002/ajmg.a.32478) [DOI] [PubMed] [Google Scholar]
- 37.Murray PG, Clayton PE, Chernausek SD.A genetic approach to evaluation of short stature of undetermined cause. Lancet: Diabetes and Endocrinology 2018. 6 564–574. ( 10.1016/S2213-8587(18)30034-2) [DOI] [PubMed] [Google Scholar]
- 38.Wit JM, Clayton PE, Rogol AD, Savage MO, Saenger PH, Cohen P.Idiopathic short stature: definition, epidemiology, and diagnostic evaluation. Growth Hormone and IGF Research 2008. 18 89–110. ( 10.1016/j.ghir.2007.11.004) [DOI] [PubMed] [Google Scholar]
- 39.Visscher PM.Sizing up human height variation. Nature Genetics 2008. 40 489–490. ( 10.1038/ng0508-489) [DOI] [PubMed] [Google Scholar]
- 40.Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PMet al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Human Molecular Genetics 2018. 27 3641–3649. ( 10.1093/hmg/ddy271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Binder G.Short stature due to SHOX deficiency: genotype, phenotype, and therapy. Hormone Research in Paediatrics 2011. 75 81–89. ( 10.1159/000324105) [DOI] [PubMed] [Google Scholar]
- 42.Jorge AA, Souza SC, Nishi MY, Billerbeck AE, Liborio DC, Kim CA, Arnhold IJ, Mendonca BB.SHOX mutations in idiopathic short stature and Leri-Weill dyschondrosteosis: frequency and phenotypic variability. Clinical Endocrinology 2007. 66 130–135. ( 10.1111/j.1365-2265.2006.02698.x) [DOI] [PubMed] [Google Scholar]
- 43.Bunyan DJ, Baker KR, Harvey JF, Thomas NS.Diagnostic screening identifies a wide range of mutations involving the SHOX gene, including a common 47.5 kb deletion 160 kb downstream with a variable phenotypic effect. American Journal of Medical Genetics. Part A 2013. 161A 1329–1338. ( 10.1002/ajmg.a.35919) [DOI] [PubMed] [Google Scholar]
- 44.Montalbano A, Juergensen L, Roeth R, Weiss B, Fukami M, Fricke-Otto S, Binder G, Ogata T, Decker E, Nuernberg Get al. Retinoic acid catabolizing enzyme CYP26C1 is a genetic modifier in SHOX deficiency. EMBO Molecular Medicine 2016. 8 1455–1469. ( 10.15252/emmm.201606623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montalbano A, Juergensen L, Fukami M, Thiel CT, Hauer NH, Roeth R, Weiss B, Naiki Y, Ogata T, Hassel Det al. Functional missense and splicing variants in the retinoic acid catabolizing enzyme CYP26C1 in idiopathic short stature. European Journal of Human Genetics 2018. 26 1113–1120. ( 10.1038/s41431-018-0148-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olney RC, Bukulmez H, Bartels CF, Prickett TC, Espiner EA, Potter LR, Warman ML.Heterozygous mutations in natriuretic peptide receptor-B (NPR2) are associated with short stature. Journal of Clinical Endocrinology and Metabolism 2006. 91 1229–1232. ( 10.1210/jc.2005-1949) [DOI] [PubMed] [Google Scholar]
- 47.Vasques GA, Amano N, Docko AJ, Funari MF, Quedas EP, Nishi MY, Arnhold IJ, Hasegawa T, Jorge AA.Heterozygous mutations in natriuretic peptide receptor-B (NPR2) gene as a cause of short stature in patients initially classified as idiopathic short stature. Journal of Clinical Endocrinology and Metabolism 2013. 98 E1636–E1644. ( 10.1210/jc.2013-2142) [DOI] [PubMed] [Google Scholar]
- 48.Amano N, Mukai T, Ito Y, Narumi S, Tanaka T, Yokoya S, Ogata T, Hasegawa T.Identification and functional characterization of two novel NPR2 mutations in Japanese patients with short stature. Journal of Clinical Endocrinology and Metabolism 2014. 99 E713–E718. ( 10.1210/jc.2013-3525) [DOI] [PubMed] [Google Scholar]
- 49.Hwang IT, Mizuno Y, Amano N, Lee HJ, Shim YS, Nam HK, Rhie YJ, Yang S, Lee KH, Hasegawa Tet al. Role of NPR2 mutation in idiopathic short stature: identification of two novel mutations. Molecular Genetics and Genomic Medicine 2020. 8 e1146. ( 10.1002/mgg3.1146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hisado-Oliva A, Ruzafa-Martin A, Sentchordi L, Funari MFA, Bezanilla-Lopez C, Alonso-Bernaldez M, Barraza-Garcia J, Rodriguez-Zabala M, Lerario AM, Benito-Sanz Set al. Mutations in C-natriuretic peptide (NPPC): a novel cause of autosomal dominant short stature. Genetics in Medicine 2018. 20 91–97. ( 10.1038/gim.2017.66) [DOI] [PubMed] [Google Scholar]
- 51.Geister KA, Camper SA.Advances in skeletal dysplasia genetics. Annual Review of Genomics and Human Genetics 2015. 16 199–227. ( 10.1146/annurev-genom-090314-045904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kant SG, Cervenkova I, Balek L, Trantirek L, Santen GW, de Vries MC, van Duyvenvoorde HA, van der Wielen MJ, Verkerk AJ, Uitterlinden AGet al. A novel variant of FGFR3 causes proportionate short stature. European Journal of Endocrinology / European Federation of Endocrine Societies 2015. 172 763–770. ( 10.1530/EJE-14-0945) [DOI] [PubMed] [Google Scholar]
- 53.Gkourogianni A, Andrew M, Tyzinski L, Crocker M, Douglas J, Dunbar N, Fairchild J, Funari MF, Heath KE, Jorge AAet al. Clinical characterization of patients With autosomal dominant short stature due to aggrecan mutations. Journal of Clinical Endocrinology and Metabolism 2017. 102 460–469. ( 10.1210/jc.2016-3313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Steen M, Pfundt R, Maas SJWH, Bakker-van Waarde WM, Odink RJ, Hokken-Koelega ACS.ACAN gene mutations in short children born SGA and response to growth hormone treatment. Journal of Clinical Endocrinology and Metabolism 2017. 102 1458–1467. ( 10.1210/jc.2016-2941) [DOI] [PubMed] [Google Scholar]
- 55.Gervera W, Gkourogianni A, Dauberd A, Nilsson O, Witf J.Arm span and its relation to height in a 2- to 17-year-old reference population and heterozygous carriers of ACAN variants. Hormone Research in Paediatrics 2020. 93 212–212. ( 10.1159/000509867) [DOI] [PubMed] [Google Scholar]
- 56.Vasques GA, Funari MFA, Ferreira FM, Aza-Carmona M, Sentchordi-Montane L, Barraza-Garcia J, Lerario AM, Yamamoto GL, Naslavsky MS, Duarte YAOet al. IHH gene mutations causing short stature With nonspecific skeletal abnormalities and response to growth hormone therapy. Journal of Clinical Endocrinology and Metabolism 2018. 103 604–614. ( 10.1210/jc.2017-02026) [DOI] [PubMed] [Google Scholar]
- 57.Sentchordi-Montane L, Benito-Sanz S, Aza-Carmona M, Pereda A, Parron-Pajares M, de la Torre C, Vasques GA, Funari MFA, Travessa AM, Dias Pet al. Clinical and molecular description of 16 families With heterozygous IHH variants. Journal of Clinical Endocrinology and Metabolism 2020. 105 2654–2666. ( 10.1210/clinem/dgaa218) [DOI] [PubMed] [Google Scholar]
- 58.Esteller M.Non-coding RNAs in human disease. Nature Reviews: Genetics 2011. 12 861–874. ( 10.1038/nrg3074) [DOI] [PubMed] [Google Scholar]
- 59.Grigelioniene G, Suzuki HI, Taylan F, Mirzamohammadi F, Borochowitz ZU, Ayturk UM, Tzur S, Horemuzova E, Lindstrand A, Weis MAet al. Gain-of-function mutation of microRNA-140 in human skeletal dysplasia. Nature Medicine 2019. 25 583–590. ( 10.1038/s41591-019-0353-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakamura Y, Inloes JB, Katagiri T, Kobayashi T.Chondrocyte-specific MicroRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Molecular and Cellular Biology 2011. 31 3019–3028. ( 10.1128/MCB.05178-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TKet al. Topologically associating domains are stable units of replication-timing regulation. Nature 2014. 515 402–405. ( 10.1038/nature13986) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova Ret al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 2015. 161 1012–1025. ( 10.1016/j.cell.2015.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cho SY, Jin DK.Guidelines for genetic skeletal dysplasias for pediatricians. Annals of Pediatric Endocrinology and Metabolism 2015. 20 187–191. ( 10.6065/apem.2015.20.4.187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tunkel D, Alade Y, Kerbavaz R, Smith B, Rose-Hardison D, Hoover-Fong J.Hearing loss in skeletal dysplasia patients. American Journal of Medical Genetics. Part A 2012. 158A 1551–1555. ( 10.1002/ajmg.a.35373) [DOI] [PubMed] [Google Scholar]
- 65.Makitie O, Kaitila I.Cartilage-hair hypoplasia--clinical manifestations in 108 Finnish patients. European Journal of Pediatrics 1993. 152 211–217. ( 10.1007/BF01956147) [DOI] [PubMed] [Google Scholar]
- 66.Bean LJH, Funke B, Carlston CM, Gannon JL, Kantarci S, Krock BL, Zhang S, Bayrak-Toydemir P.ACMG Laboratory Quality Assurance Committee. Diagnostic gene sequencing panels: from design to report—a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genetics in Medicine 2020. 22 453–461. ( 10.1038/s41436-019-0666-z) [DOI] [PubMed] [Google Scholar]
- 67.Ma M, Ru Y, Chuang LS, Hsu NY, Shi LS, Hakenberg J, Cheng WY, Uzilov A, Ding W, Glicksberg BSet al. Disease-associated variants in different categories of disease located in distinct regulatory elements. BMC Genomics 2015. 16(Supplement 8) S3. ( 10.1186/1471-2164-16-S8-S3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonorazky HD, Naumenko S, Ramani AK, Nelakuditi V, Mashouri P, Wang P, Kao D, Ohri K, Viththiyapaskaran S, Tarnopolsky MAet al. Expanding the boundaries of RNA sequencing as a diagnostic tool for rare Mendelian disease. American Journal of Human Genetics 2019. 104 466–483. ( 10.1016/j.ajhg.2019.01.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tattini L, D'Aurizio R, Magi A.Detection of genomic structural variants from next-generation sequencing data. Frontiers in Bioengineering and Biotechnology 2015. 3 92. ( 10.3389/fbioe.2015.00092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lindstrand A, Eisfeldt J, Pettersson M, Carvalho CMB, Kvarnung M, Grigelioniene G, Anderlid BM, Bjerin O, Gustavsson P, Hammarsjo Aet al. From cytogenetics to cytogenomics: whole-genome sequencing as a first-line test comprehensively captures the diverse spectrum of disease-causing genetic variation underlying intellectual disability. Genome Medicine 2019. 11 68. ( 10.1186/s13073-019-0675-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.King DA, Fitzgerald TW, Miller R, Canham N, Clayton-Smith J, Johnson D, Mansour S, Stewart F, Vasudevan P, Hurles MEet al. A novel method for detecting uniparental disomy from trio genotypes identifies a significant excess in children with developmental disorders. Genome Research 2014. 24 673–687. ( 10.1101/gr.160465.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Colson C, Decamp M, Gruchy N, Coudray N, Ballandonne C, Bracquemart C, Molin A, Mittre H, Takatani R, Juppner Het al. High frequency of paternal iso or heterodisomy at chromosome 20 associated with sporadic pseudohypoparathyroidism 1B. Bone 2019. 123 145–152. ( 10.1016/j.bone.2019.03.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wakeling EL, Brioude F, Lokulo-Sodipe O, O'Connell SM, Salem J, Bliek J, Canton AP, Chrzanowska KH, Davies JH, Dias RPet al. Diagnosis and management of Silver-Russell syndrome: first international consensus statement. Nature Reviews: Endocrinology 2017. 13 105–124. ( 10.1038/nrendo.2016.138) [DOI] [PubMed] [Google Scholar]
- 74.Mantovani G, Spada A, Elli FM.Pseudohypoparathyroidism and Gsalpha-cAMP-linked disorders: current view and open issues. Nature Reviews: Endocrinology 2016. 12 347–356. ( 10.1038/nrendo.2016.52) [DOI] [PubMed] [Google Scholar]
- 75.Vakkilainen S, Skoog T, Einarsdottir E, Middleton A, Pekkinen M, Ohman T, Katayama S, Krjutskov K, Kovanen PE, Varjosalo Met al. The human long non-coding RNA gene RMRP has pleiotropic effects and regulates cell-cycle progression at G2. Scientific Reports 2019. 9 13758. ( 10.1038/s41598-019-50334-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kampe AJ, Costantini A, Levy-Shraga Y, Zeitlin L, Roschger P, Taylan F, Lindstrand A, Paschalis EP, Gamsjaeger S, Raas-Rothschild Aet al. PLS3 deletions lead to severe spinal osteoporosis and disturbed bone matrix mineralization. Journal of Bone and Mineral Research 2017. 32 2394–2404. ( 10.1002/jbmr.3233) [DOI] [PubMed] [Google Scholar]
- 77.Boeuf S, Richter W.Chondrogenesis of mesenchymal stem cells: role of tissue source and inducing factors. Stem Cell Research and Therapy 2010. 1 31. ( 10.1186/scrt31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E.A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012. 337 816–821. ( 10.1126/science.1225829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu N, Liu B, Du H, Zhao S, Li Y, Cheng X, Wang S, Lin J, Zhou J.Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study, et al. The Progress of CRISPR/Cas9-Mediated Gene Editing in Generating Mouse/Zebrafish Models of Human Skeletal Diseases. Computational and Structural Biotechnology Journal 2019. 17 954–962. ( 10.1016/j.csbj.2019.06.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Batzoglou S, Pachter L, Mesirov JP, Berger B, Lander ES.Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Research 2000. 10 950–958. ( 10.1101/gr.10.7.950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jilka RL.The relevance of mouse models for investigating age-related bone loss in humans. Journals of Gerontology: Series A, Biological Sciences and Medical Sciences 2013. 68 1209–1217. ( 10.1093/gerona/glt046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Börjesson AE, Windahl SH, Karimian E, Eriksson EE, Lagerquist MK, Engdahl C, Antal MC, Krust A, Chambon P, Sävendahl Let al. The role of estrogen receptor-α and its activation function-1 for growth plate closure in female mice. American Journal of Physiology-Endocrinology and Metabolism 2012. 302 E1381–E1389. ( 10.1152/ajpendo.00646.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Busse B, Galloway JL, Gray RS, Harris MP, Kwon RY.Zebrafish: an emerging model for orthopedic research. Journal of Orthopaedic Research 2020. 38 925–936. ( 10.1002/jor.24539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jao LE, Wente SR, Chen W.Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. PNAS 2013. 110 13904–13909. ( 10.1073/pnas.1308335110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bergen DJM, Kague E, Hammond CL.Zebrafish as an emerging model for osteoporosis: a primary testing platform for screening new osteo-active compounds. Frontiers in Endocrinology (Lausanne) 2019. 10 6. ( 10.3389/fendo.2019.00006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gistelinck C, Kwon RY, Malfait F, Symoens S, Harris MP, Henke K, Hawkins MB, Fisher S, Sips P, Guillemyn Bet al. Zebrafish type I collagen mutants faithfully recapitulate human type I collagenopathies. PNAS 2018. 115 E8037–E8046. ( 10.1073/pnas.1722200115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fiedler IAK, Schmidt FN, Wolfel EM, Plumeyer C, Milovanovic P, Gioia R, Tonelli F, Bale HA, Jahn K, Besio Ret al. Severely impaired bone material quality in Chihuahua zebrafish resembles classical dominant human osteogenesis imperfecta. Journal of Bone and Mineral Research 2018. 33 1489–1499. ( 10.1002/jbmr.3445) [DOI] [PubMed] [Google Scholar]
- 88.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews Let al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013. 496 498–503. ( 10.1038/nature12111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Apschner A, Schulte-Merker S, Witten PE.Not all bones are created equal – using zebrafish and other teleost species in osteogenesis research. Methods in Cell Biology 2011. 105 239–255. ( 10.1016/B978-0-12-381320-6.00010-2) [DOI] [PubMed] [Google Scholar]
- 90.Witten PE, Harris MP, Huysseune A, Winkler C.Small teleost fish provide new insights into human skeletal diseases. Methods in Cell Biology 2017. 138 321–346. ( 10.1016/bs.mcb.2016.09.001) [DOI] [PubMed] [Google Scholar]
- 91.Heubel BP, Bredesen CA, Schilling TF, Le Pabic P.Endochondral growth zone pattern and activity in the zebrafish pharyngeal skeleton. Developmental Dynamics 2021. 250 74–87. ( 10.1002/dvdy.241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grafe I, Yang T, Alexander S, Homan EP, Lietman C, Jiang MM, Bertin T, Munivez E, Chen Y, Dawson Bet al. Excessive transforming growth factor-beta signaling is a common mechanism in osteogenesis imperfecta. Nature Medicine 2014. 20 670–675. ( 10.1038/nm.3544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tauer JT, Abdullah S, Rauch F.Effect of anti-TGF-beta treatment in a mouse model of severe osteogenesis imperfecta. Journal of Bone and Mineral Research 2019. 34 207–214. ( 10.1002/jbmr.3617) [DOI] [PubMed] [Google Scholar]
- 94.Gioia R, Tonelli F, Ceppi I, Biggiogera M, Leikin S, Fisher S, Tenedini E, Yorgan TA, Schinke T, Tian Ket al. The chaperone activity of 4PBA ameliorates the skeletal phenotype of Chihuahua, a zebrafish model for dominant osteogenesis imperfecta. Human Molecular Genetics 2017. 26 2897–2911. ( 10.1093/hmg/ddx171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kanazawa H, Tanaka H, Inoue M, Yamanaka Y, Namba N, Seino Y.Efficacy of growth hormone therapy for patients with skeletal dysplasia. Journal of Bone and Mineral Metabolism 2003. 21 307–310. ( 10.1007/s00774-003-0425-7) [DOI] [PubMed] [Google Scholar]
- 96.Kozhemyakina E, Lassar AB, Zelzer E.A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development 2015. 142 817–831. ( 10.1242/dev.105536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Voller T, Cameron P, Watson J, Phadnis J.The growth plate: anatomy and disorders. Orthopaedics and Trauma 2020. 34 135–140. ( 10.1016/j.mporth.2020.03.006) [DOI] [Google Scholar]
- 98.Samsa WE, Zhou X, Zhou G.Signaling pathways regulating cartilage growth plate formation and activity. Seminars in Cell and Developmental Biology 2017. 62 3–15. ( 10.1016/j.semcdb.2016.07.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mierzewska H, Rydzanicz M, Bieganski T, Kosinska J, Mierzewska-Schmidt M, Lugowska A, Pollak A, Stawinski P, Walczak A, Kedra A. et al. Spondyloepimetaphyseal dysplasia with neurodegeneration associated with AIFM1 mutation - a novel phenotype of the mitochondrial disease. Clinical Genetics 2017. 91 30–37. ( 10.1111/cge.12792) [DOI] [PubMed] [Google Scholar]
- 100.Cho SY, Bae JS, Kim NKD, Forzano F, Girisha KM, Baldo C, Faravelli F, Cho TJ, Kim D, Lee KY. et al. BGN mutations in X-linked spondyloepimetaphyseal dysplasia. American Journal of Human Genetics 2016. 98 1243–1248. ( 10.1016/j.ajhg.2016.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gonzaga-Jauregui C, Gamble CN, Yuan B, Penney S, Jhangiani S, Muzny DM, Gibbs RA, Lupski JR, Hecht JT. Mutations in COL27A1 cause Steel syndrome and suggest a founder mutation effect in the Puerto Rican population. European Journal of Human Genetics 2015. 23 342–346. ( 10.1038/ejhg.2014.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Volpi S, Yamazaki Y, Brauer PM, van Rooijen E, Hayashida A, Slavotinek A, Sun Kuehn H, Di Rocco M, Rivolta C, Bortolomai I, EXTL3 mutations cause skeletal dysplasia, immune deficiency, and developmental delay. Journal of Experimental Medicine 2017. 214 623–627. ( 10.1084/jem.20161525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Royer-Bertrand B, Castillo-Taucher S, Moreno-Salinas R, Cho TJ, Chae JH, Choi M, Kim OH, Dikoglu E, Campos-Xavier B, Girardi E, et al. Mutations in the heat-shock protein A9 (HSPA9) gene cause the EVEN-PLUS syndrome of congenital malformations and skeletal dysplasia. Scientific Reports 2015. 5 17154. ( 10.1038/srep17154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schwartzentruber J, Buhas D, Majewski J, Sasarman F, Papillon-Cavanagh S, Thiffault I, Sheldon KM, Massicotte C, Patry L, Simon M, et al. Mutation in the nuclear-encoded mitochondrial isoleucyl-tRNA synthetase IARS2 in patients with cataracts, growth hormone deficiency with short stature, partial sensorineural deafness, and peripheral neuropathy or with Leigh syndrome. Human Mutation 2014. 35 1285–1289. ( 10.1002/humu.22629) [DOI] [PubMed] [Google Scholar]
- 105.Huckert M, Stoetzel C, Morkmued S, Laugel-Haushalter V, Geoffroy V, Muller J, Clauss F, Prasad MK, Obry F, Raymond JL, et al. Mutations in the latent TGF-beta binding protein 3 (LTBP3) gene cause brachyolmia with amelogenesis imperfecta. Human Molecular Genetics 2015. 24 3038–3049. ( 10.1093/hmg/ddv053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van Karnebeek CD, Bonafe L, Wen XY, Tarailo-Graovac M, Balzano S, Royer-Bertrand B, Ashikov A, Garavelli L, Mammi I, Turolla L, et al. NANS-mediated synthesis of sialic acid is required for brain and skeletal development. Nature Genetics 2016. 48 777––784.. ( 10.1038/ng.3578) [DOI] [PubMed] [Google Scholar]
- 107.Girisha KM, von Elsner L, Neethukrishna K, Muranjan M, Shukla A, Bhavani GS, Nishimura G, Kutsche K, Mortier G. The homozygous variant c.797G>A/p.(Cys266Tyr) in PISD is associated with a Spondyloepimetaphyseal dysplasia with large epiphyses and disturbed mitochondrial function. Human Mutation 2019. 40 299–309. ( 10.1002/humu.23693) [DOI] [PubMed] [Google Scholar]
- 108.Faden M, AlZahrani F, Mendoza-Londono R, Dupuis L, Hartley T, Kannu P, Raiman JA, Howard A, Qin W, Tetreault M, et al. Identification of a recognizable progressive skeletal dysplasia caused by RSPRY1 mutations. American Journal of Human Genetics 2015. 97 608––615.. ( 10.1016/j.ajhg.2015.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Burrage LC, Reynolds JJ, Baratang NV, Phillips JB, Wegner J, McFarquhar A, Higgs MR, Christiansen AE, Lanza DG, Seavitt JR, et al. Bi-allelic variants in TONSL cause SPONASTRIME dysplasia and a spectrum of skeletal dysplasia phenotypes. American Journal of Human Genetics 2019. 104 422–438. ( 10.1016/j.ajhg.2019.01.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wehrle A, Witkos TM, Unger S, Schneider J, Follit JA, Hermann J, Welting T, Fano V, Hietala M, Vatanavicharn N, et al. Hypomorphic mutations of TRIP11 cause odontochondrodysplasia. JCI Insight 2019. 4 e124701. ( 10.1172/jci.insight.124701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Di Rocco M, Rusmini M, Caroli F, Madeo A, Bertamino M, Marre-Brunenghi G, Ceccherini I. Novel spondyloepimetaphyseal dysplasia due to UFSP2 gene mutation. Clinical Genetics 2018. 93 671––674.. ( 10.1111/cge.13134) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a