Abstract
Despite decades of study, there are still many unanswered questions about metastasis, the process by which a localized cancer becomes a systemic disease. One of these questions is the nature of the tumor cells that give rise to metastases. Although conventional models suggest that metastases are seeded by single cells from the primary tumor, there is growing evidence that seeding requires the collective action of tumor cells traveling together in clusters. Here, we review this evidence, which comes from analysis of both experimental models and patient samples. We present a model of metastatic dissemination that highlights the activities of clusters of tumor cells that retain and require their epithelial properties.
Metastasis is a complex, multistep process that requires cancer cells to detach from the primary tumor, migrate through adjacent tissue, access and travel through the vasculature, and then survive and proliferate in distant organs (1, 2). There is an added challenge for carcinomas because the adult epithelial cells from which they arise are normally polarized and nonmotile. Metastasis therefore represents a striking divergence from their homeostatic condition. However, epithelial tissues are highly dynamic and migratory during development and tissue repair (3), and epithelial cells can acquire mesenchymal molecular features in both developmental and disease states (4).
Current therapies are not sufficiently effective in treating metastatic disease, and so it is important that we determine the cellular and molecular features of the cancer cells that “seed” new tumors in distant organs. In this Perspective, we discuss recent work that supports a role for tumor cell clusters in metastatic seeding—an alternative to the conventional view that metastases are seeded by single cells from the primary tumor.
Tumor cell clusters and polyclonal seeding of metastases
The idea that tumor cell clusters contribute to metastasis can be traced back to the 1950s, when it was reported that blood samples from cancer patients contain both single and clustered tumor cells (2) and that tumor cell clusters can rapidly traverse the lungs in animal models (5). It was later shown that tumor cell clusters were more efficient than single cells were at producing metastases when injected intravenously into mice (2, 6). However, because metastasis is known to be inefficient (2), the data from these early studies are consistent with two very different possibilities: Metastases could arise from a less efficient process of seeding by abundant single cells or from a more efficient process of seeding by rare clusters. Recent technological advances have revolutionized our ability to quantify circulating tumor cells (CTCs) and CTC clusters and to determine their molecular properties across diverse tumor types (7–10). In two such studies, the presence of CTC clusters was associated with significantly worse clinical outcomes as compared with the presence of single CTCs (8, 9). Their prognostic value is consistent with the hypothesis that tumor cell clusters make a distinct contribution to metastasis.
If the metastatic seed is a single cancer cell, then the resulting tumor will be clonal. Conversely, if the seed is a CTC cluster, then the resulting metastasis can be polyclonal from the start. Deep-sequencing analysis of tumors allows the reconstruction of evolutionary histories of cancer cell clones during metastatic progression. These evolutionary histories can be used to infer monoclonal versus polyclonal seeding. One recent study of prostate cancer patients revealed evidence for frequent polyclonal seeding from the primary tumor to secondary sites and the polyclonal spread of existing metastases to new sites in the body (11). This observation emerges in the context of an expanding literature on phenotypic and genotypic diversity within primary tumors and an increased appreciation of cooperative and competitive dynamics among cancer cell clones (12). These studies suggest that different clonal combinations in the cluster could have very different properties with respect to growth and/or response to therapy.
Polyclonal metastases could arise in two distinct ways: direct seeding by a multicellular cluster or serial accumulation of multiple single cells at a common site. Mouse models have proved valuable in distinguishing between these two mechanisms. A key feature of these experiments is the ability to establish primary tumors containing a mixture of cancer cells that express fluorescent proteins of different colors (9, 13, 14). Both single cells and cell clusters composed entirely of the same color will generate single-colored metastases (Fig. 1A). Conversely, clusters composed of more than one color will form multicolored metastases, which would provide direct evidence of their polyclonal origin (Fig. 1, A and B).
Fig. 1. Lineage analysis to identify the metastatic seeds in mouse models.
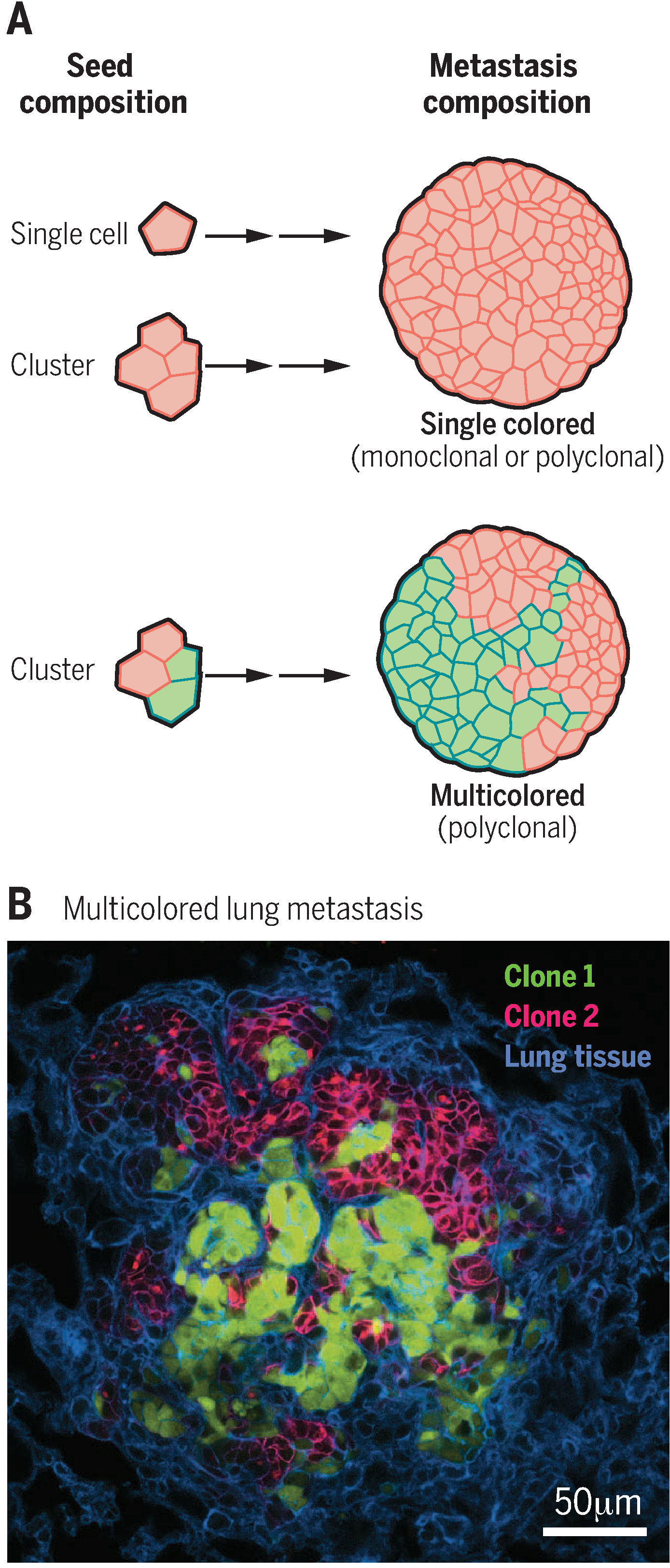
(A) A primary tumor composed of cancer cells of two different colors can generate single cells, single-colored clusters, and multicolored clusters. Single cells and single-colored clusters will generate single-colored metastases. In contrast, multicolored clusters generate multicolored metastases. The number of different colors observed in a metastasis can therefore be used to infer the cellular properties of its seed. (B) Representative example of a multicolored lung metastasis in a mouse model of breast cancer [adapted from (14)].
Three independent research groups have conducted studies of this design using different mouse models, and all have found multicolored metastases, which is consistent with the concept that polyclonal metastases can be seeded by tumor cell clusters (9, 13, 14). The first group established multicolored mammary tumors in mice and frequently observed multicolored metastases (9). When they established a single-colored tumor on one side of the mouse and a different single-colored tumor on the other side, the metastases were predominantly single-colored (9). This result suggests that aggregation of tumor cells in the blood or at the distant organ is inefficient and, therefore, that multicolored metastases arise from seeding by tumor cell clusters (9). The second group extended the lineage analysis concept to pancreatic cancer, documented polyclonal seeding by clusters in the mice, and observed strong differences in the extent to which a polyclonal seed expanded into clonal or polyclonal metastases in different organ sites (13). Studying spontaneous breast cancer in mice, the third group quantitatively related the extent of clonal mixing in the primary tumor with the frequency of detection of multicolored metastases, leading them to estimate that >97% of metastases arose from clusters (14). These three studies also provided direct evidence that clusters exhibit superior survival and colony-forming potential both in culture (14) and in vivo (9, 13, 14). Control experiments in which different color tumors were established in different locations in the mouse or different color cancer cells were injected intravenously at different times rarely or never yielded multicolored metastases. Therefore, all three studies provide data to support the concept that a multicellular cluster travels as a unit from the primary tumor to distant organs to seed polyclonal metastases (Fig. 2) (9, 13, 14).
Fig. 2. A model for metastatic spread that is based on collective dissemination of epithelial tumor cell clusters.
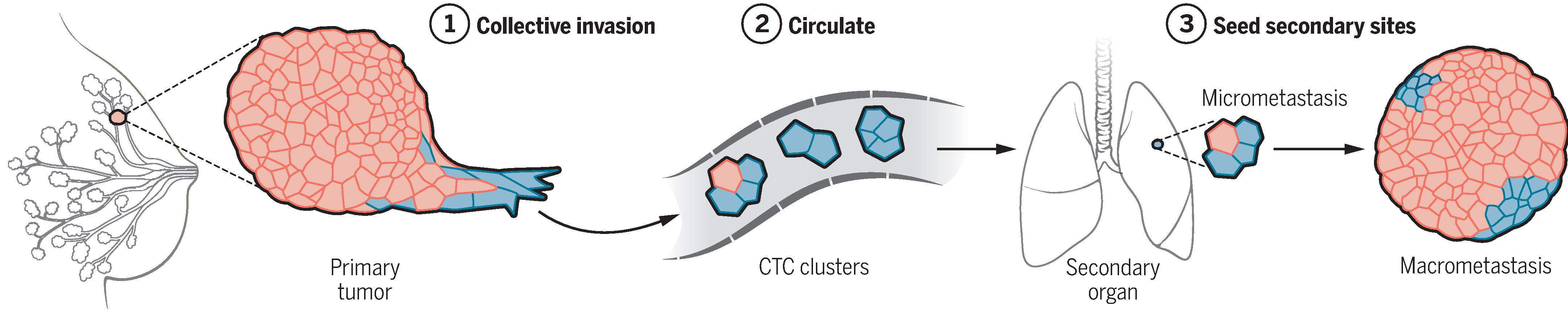
This model posits that primary tumor cells invade, circulate, and seed tumor growth at distant sites as collective units and that these activities require their expression of epithelial genes, such as K14. After arriving in the secondary organ (the lung), the predominantly K14+ seed (blue) from a primary breast cancer expands to form predominantly K14− macrometastases (red). Further details are available in (14, 16).
Epithelial properties of the metastatic seed
The observations discussed above raise several intriguing questions, including how tumor cell clusters escape from the primary tumor and what their molecular properties are as they transit to the distant site. We will focus on breast cancer, where there has been substantial progress in understanding the molecular properties of tumor cells as they invade, disseminate, and travel to distant organs. Carcinomas can invade the adjacent tissue in a collective fashion, as groups of cancers cells that maintain at least some aspects of epithelial cell-cell adhesion (3). A recent three-dimensional analysis of the stromal border of human breast tumors detected frequent collective invasion and virtually no single cancer cells (15). These collective invasion strands could release small clusters of cancer cells to seed the metastatic process. A simple starting point for validating this model is to test for molecular similarities between cancer cells in collective invasion strands and at later stages of metastatic spread. The cancer cells leading collective invasion were shown to have common molecular properties across subtypes of breast cancer, in both mouse models and in human breast tumors (16). These invasive cancer cells were characterized by their expression of molecular markers of basal epithelial differentiation, including cytokeratin 14 (K14), K5, P-cadherin, and p63 (16). Cells in this K14+ invasive state were observed in all stages of disseminative spread—collective invasion, local dissemination, CTC cluster, and micrometastasis—but were rare in the primary tumor and in macrometastases (14). These data suggest that there are distinct epithelial molecular programs driving tumor cell growth and dissemination (Fig. 2). Gene expression in these invasive leader cells resembles that of a basal phenotype mammary stem cell (14). This is consistent with single-cell transcriptomic analysis of human metastatic breast cancer cells (17), which showed that disseminated cancer cells share a basal stem cell gene expression profile in micrometastases but give rise to macrometastases composed largely of luminal phenotype cancer cells (17). Together, these studies reveal that changes in epithelial differentiation state accompany metastatic dissemination.
Epithelial cells are characterized by specific intercellular adhesion complexes. These junctions are often thought to impede motility; thus, it is conceptually attractive to think of metastasis as involving a transient or permanent loss of epithelial features, through a process such as the epithelial-to-mesenchymal transition (EMT) (4). However, recent studies have presented an alternative model in which epithelial adhesion complexes play an important role in metastatic tumor cells. These studies revealed that metastatic breast cancer cells retain the expression of epithelial cytoskeletal and adhesion genes such as K5, K8, K14, E-cadherin, and P-cadherin (14, 16). This concept is supported by the observation of the desmosomal adhesion protein plakoglobin in CTC clusters (9) and in the requirement for plakoglobin (9), E-cadherin (18), and K14 (14) in various experimental models of breast cancer metastasis. Knockdown of plakoglobin induced disaggregation of cancer cell clusters and compromised their efficiency in metastasis formation (9), whereas loss of K14 led to loss of a broader program of metastasis-related genes, such as AdamTS1, CARD10, and tenascin C (14). Furthermore, the highly invasive K14+ cells in a mouse mammary tumor were enriched for the genes that constitute both a major epithelial cell-cell adhesion complex, the desmosome, and a major epithelial cell-matrix adhesion complex, the hemidesmosome (14). Thus, the epithelial cell adhesion machinery can be retained in disseminating cell clusters and can contribute to the metastatic process.
The role of epithelial genes in metastasis has precedent in developmental systems; migratory clusters of epithelial cells use mechanical signals through E-cadherin to coordinate their direction sensing and collective migration (19). Further studies are warranted to determine whether cancer cell clusters possess analogous migratory advantages during metastasis or whether their superior metastatic potential is solely attributable to enhanced survival. The retention of epithelial features in breast cancer clusters is also consistent with a recent demonstration that breast cancer cells do not require EMT to accomplish metastasis (20). Although the data clearly support the concept that epithelial-phenotype clusters can seed polyclonal metastases, it seems unlikely that this is the only mechanism of metastatic spread. Mesenchymal CTCs and CTC clusters exist in human patients, and the relative frequency of epithelial- versus mesenchymal-phenotype CTCs within a patient can shift with disease progression and cancer therapy (10). It remains an important future challenge to distinguish the relative frequency with which epithelial-phenotype versus mesenchymal-phenotype tumor cell clusters function as metastatic seeds.
Outlook
A number of studies in both preclinical models and patient samples have converged on the theme that tumor cells can metastasize collectively as clusters. Cancer cell clusters can retain and require epithelial gene expression and can transition between distinct epithelial differentiation states to accomplish the proliferative versus migratory components of metastasis.
At least three questions need to be addressed in this rapidly evolving field. First, what is the biological basis of metastatic seeding by tumor cell clusters? The answer to this question will require a deeper understanding of the mechanisms driving the enhanced metastatic efficiency of clusters relative to single cells and molecular insights into how cell clusters coordinate their migration and differentiation through the many steps of metastatic spread. It will also be important to understand how clusters interface with stromal populations in the primary tumor, in the circulation, and in distant organs (1). Second, are the cellular and molecular properties of the metastatic seed the same across different tumor types and metastatic sites, or do they vary? Addressing this question will require lineage analysis techniques to quantify the fraction of single-cell versus multicellular seeds and extensive CTC and CTC cluster analyses in order to determine their molecular properties. Single-cell sequencing and proteomics will also be necessary to characterize the genotypic and phenotypic heterogeneity of the cancer cells within these clusters, both in the circulation and as they expand in different organ sites (12). Last, what are the therapeutic implications of tumor cell clusters? Clusters present a potential challenge because they could contain tumor cells with different drug-resistance properties. Cluster organization itself may also influence therapeutic response. On a more positive note, tumor cell clusters also present potential opportunities for novel therapeutic strategies that target either their multicellular organization or the changes in epithelial differentiation that enable their spread through the body.
ACKNOWLEDGMENTS
We thank S. Marshall and P. Tran for critical comments on the manuscript and B. Moore for the image in Fig. 1B. K.J.C. is supported by a Burroughs Wellcome Fund Career Award for Medical Scientists. A.J.E. is supported by a Research Scholar Grant (RSG-12–141-01-CSM) from the American Cancer Society, funds from the NIH/National Cancer Institute (P30 CA006973), funds from the Cindy Rosencrans Fund for Triple Negative Breast Cancer Research, a Research Leadership Award from the Metastatic Breast Cancer Network, and an award from The Pink Agenda and The Breast Cancer Research Foundation. We apologize to the authors whose work we could not cite because of space constraints. The authors are coinventors on a patent application (U.S. 14/276,099) filed by The Johns Hopkins University that is related to the use of K14 as a biomarker for identifying patients at elevated risk of metastasis.
REFERENCES AND NOTES
- 1.Massagué J, Obenauf AC, Nature 529, 298–306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talmadge JE, Fidler IJ, Cancer Res. 70, 5649–5669 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedl P, Gilmour D, Nat. Rev. Mol. Cell Biol 10, 445–457 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Ye X, Weinberg RA, Trends Cell Biol. 25, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeidman I, Buss JM, Cancer Res. 12, 731–733 (1952). [PubMed] [Google Scholar]
- 6.Liotta LA, Saidel MG, Kleinerman J, Cancer Res. 36, 889–894 (1976). [PubMed] [Google Scholar]
- 7.Cho EH et al. , Phys. Biol 9, 016001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou JM et al. , J. Clin. Oncol 30, 525–532 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Aceto N et al. , Cell 158, 1110–1122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu M et al. , Science 339, 580–584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gundem G et al. , Nature 520, 353–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabassum DP, Polyak K, Nat. Rev. Cancer 15, 473–483 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Maddipati R, Stanger BZ, Cancer Discov. 5, 1086–1097 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung KJ et al. , Proc. Natl. Acad. Sci. U.S.A 113, E854–E863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bronsert P et al. , J. Pathol 234, 410–422 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Cheung KJ, Gabrielson E, Werb Z, Ewald AJ, Cell 155, 1639–1651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawson DA et al. , Nature 526, 131–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H et al. , Cancer Cell 27, 193–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai D et al. , Cell 157, 1146–1159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer KR et al. , Nature 527, 472–476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]


