SUMMARY
RNA helicases and E3 ubiquitin ligases mediate many critical functions in cells, but their actions have largely been studied in distinct biological contexts. Here, we uncover evolutionarily conserved rules of engagement between RNA helicases and tripartite motif (TRIM) E3 ligases that lead to their functional coordination in vertebrate innate immunity. Using cryoelectron microscopy and biochemistry, we show that RIG-I-like receptors (RLRs), viral RNA receptors with helicase domains, interact with their cognate TRIM/TRIM-like E3 ligases through similar epitopes in the helicase domains. Their interactions are avidity driven, restricting the actions of TRIM/TRIM-like proteins and consequent immune activation to RLR multimers. Mass spectrometry and phylogeny-guided biochemical analyses further reveal that similar rules of engagement may apply to diverse RNA helicases and TRIM/TRIM-like proteins. Our analyses suggest not only conserved substrates for TRIM proteins but also, unexpectedly, deep evolutionary connections between TRIM proteins and RNA helicases, linking ubiquitin and RNA biology throughout animal evolution.
In Brief
Proper immune function requires multiple layers of checks and balances. Kato et al. show a conserved mechanism by which the antiviral proteins RIG-I-like receptors collaborate with a family of E3 ligases, TRIM-like proteins, to ensure high fidelity and robustness of the antiviral immune response.
Graphical Abstract
INTRODUCTION
Innate immunity to invading pathogens involves tradeoffs between efficiency and fidelity of responses. An inefficient response to an invading pathogen can be fatal, but an inappropriate immune reaction (e.g., in the absence of a pathogen) can be equally harmful. A growing number of reports suggest that ubiquitin (Ub) modification of immune components provides a general means for fine-tuning the innate immune system (Zheng and Gao, 2019). Precise control of ubiquitination and consequent alteration of the target molecule’s stability or function allows another layer of regulation to achieve fidelity and robustness of immune responses. Acting at the final step of the three-enzyme (E1, E2, and E3) Ub transfer cascade, E3 ligases dictate ubiquitination target specificity. Tripartite motif (TRIM) and TRIM-like proteins are an emerging class of E3 ligases that play important roles in innate immunity (Ozato et al., 2008; Versteeg et al., 2014). For substrate recognition, TRIM proteins often use C-terminal Pry-Spry (PSpry) domains (Esposito et al., 2017), which can recognize distinct protein features from linear peptide sequences to three-dimensional protein structures (D’Cruz et al., 2013; Perfetto et al., 2013). Despite being present in nearly 100 human proteins, our fundamental understanding of how PSpry domains recognize substrates and enable TRIM proteins to regulate immune function is limited.
The recent discovery of the TRIM-like protein RIPLET as the E3 ligase responsible for ubiquitination of RIG-I (Cadena et al., 2019; Hayman et al., 2019; Oshiumi et al., 2013) revealed important insights into PSpry:substrate interactions. RIG-I is a conserved innate immune receptor that activates type I and III interferon (IFN) pathways upon binding to viral double-stranded RNA (dsRNA) (Hur, 2019; Yoneyama and Fujita, 2010). RIPLET is required to add K63-linked Ub chains (K63-Ubn) onto RIG-I (Cadena et al., 2019; Hayman et al., 2019; Oshiumi et al., 2013), which has been shown previously to be a key trigger for RIG-I signal activation (Jiang et al., 2012; Zeng et al., 2010). K63-Ubn promotes homo-tetramerization of RIG-I’s tandem caspase activation recruitment domain (2CARD) signaling domains, which, in turn, interact with the adaptor molecule MAVS to activate downstream signaling (Peisley et al., 2014; Wu et al., 2014). RIPLET selectively recognizes RIG-I in its filamentous oligomeric form, which assembles only upon its engagement with cognate dsRNA and does not bind monomeric RIG-I free of RNA (Cadena et al., 2019). This filament-specific recognition, mediated by the bivalency of PSpry in a dimeric architecture of RIPLET, prevents constitutive ubiquitination and aberrant activation of RIG-I in the absence of viral infection (Cadena et al., 2019).
MDA5 is a RIG-I-like receptor (RLR) that performs antiviral functions distinct from RIG-I by detecting different groups of viral RNAs (Hur, 2019; Yoneyama and Fujita, 2010). Like RIG-I, MDA5 forms filaments upon binding to cognate viral dsRNAs and activates MAVS and the IFN-inducing pathway (Peisley et al., 2011; Wu et al., 2013). Moreover, K63-Ubn also serves as a key trigger for MDA5 signaling (Jiang et al., 2012). Unlike RIG-I, however, ubiquitination of MDA5 is independent of RIPLET and instead requires the E3 ligase TRIM65 (Cadena et al., 2019; Kamanova et al., 2016; Lang et al., 2017). In this study, we found that TRIM65 recognizes MDA5 only in its filamentous state, revealing avidity-dependent RLR recognition as a common mechanism by which TRIM65 and RIPLET ensure tightly regulated antiviral signaling. We also found that RIPLET and TRIM65 bind the same epitope in the helicase domains of the cognate RLRs, which nevertheless confers highly selective substrate recognition. Furthermore, our biochemical analyses showed that a similar epitope in the helicase domain is utilized for avidity-dependent recognition by a broad range of TRIM proteins, revealing the conserved rules of engagement between TRIM proteins and helicases.
RESULTS
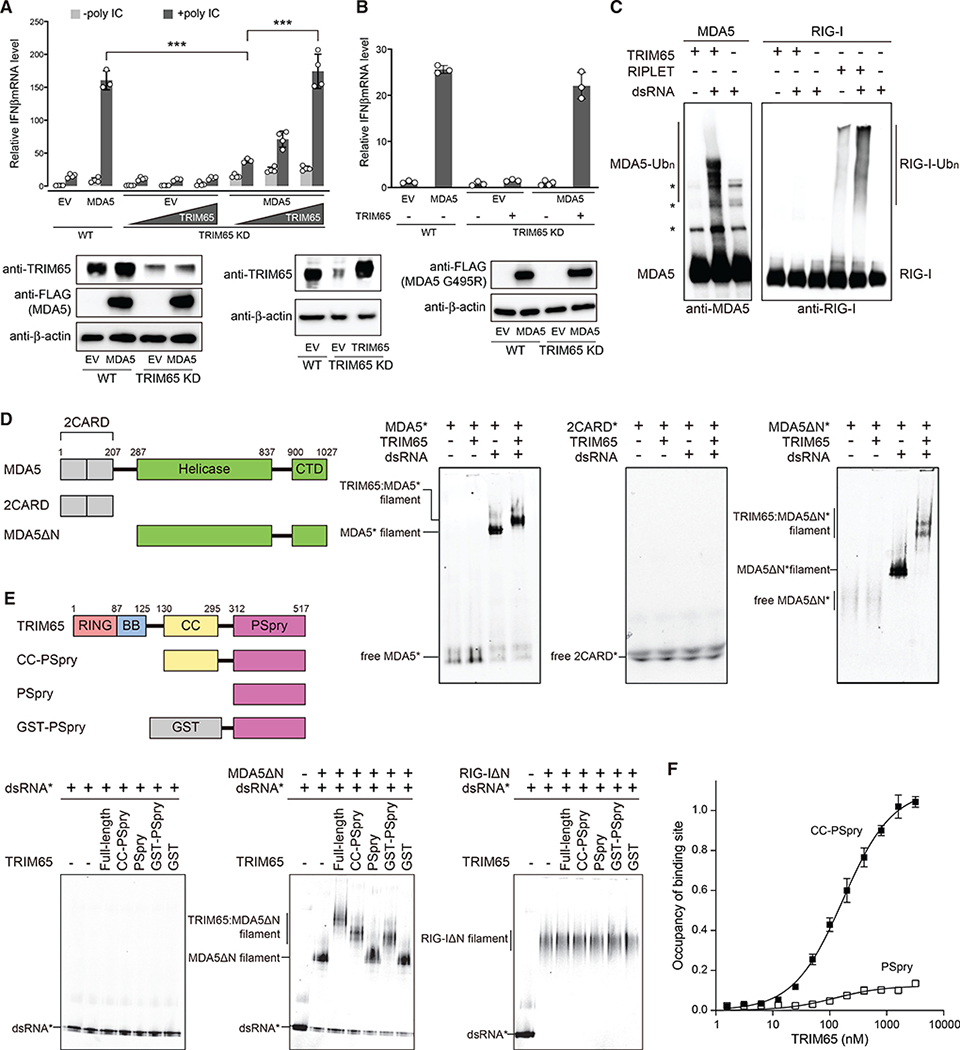
TRIM65 selectively recognizes MDA5 filaments using PSpry bivalency
We first tested the importance of TRIM65 in MDA5 signaling (Kamanova et al., 2016; Lang et al., 2017). CRISPR-mediated knockdown (KD) of TRIM65 in 293T cells impaired MDA5 signaling upon stimulation with a dsRNA mimetic, poly(I:C) (Figure 1A). Conversely, TRIM65 complementation restored MDA5 signaling, as measured by IFNβ mRNA induction (Figure 1A). TRIM65 KD did not affect IFNβ induction upon stimulation with a RIG-I-specific RNA ligand (42-bp dsRNA with a 5′-triphosphate group) or upon overexpression of MAVS or STING (Figures S1A–S1C), suggesting that TRIM65 specifically acts on the MDA5 pathway. We also examined whether the signaling activity of a gain-of-function mutant of MDA5 (G495R) relied on TRIM65. MDA5 G495R, which was identified from individuals with the auto-inflammatory disease Aicardi-Goutières syndrome (Rice et al., 2014), is known to constitutively activate MDA5 signaling because it allows spontaneous filament formation of MDA5 on endogenous dsRNAs (Ahmad et al., 2018). We found that the signaling activity of G495R also depended on TRIM65 (Figure 1B). This result not only reinforces the requirement of TRIM65 for MDA5-dependent signaling but also demonstrates that the TRIM65 requirement is independent of the source of the RNA ligand.
Figure 1. TRIM65 selectively recognizes MDA5 filaments using PSpry bivalency.
(A) Relative signaling activity of MDA5 upon poly(I:C) stimulation (0.5 μg), as measured by IFNβ mRNA induction. The signaling activity was compared in wild-type (WT) and TRIM65 KD 293T cells with and without TRIM65 complementation. Because 293T expresses endogenous MDA5 poorly, cells were transfected with MDA5-expressing vectors. EV, empty vector. For western blotting, the highest amount of TRIM65 complementation was compared with endogenous levels of TRIM65 in WT cells. See STAR methods for experimental details.
(B) Relative signaling activity of MDA5 G495R in WT and TRIM65 KD 293T cells. No exogenous RNA was introduced.
(C) In vitro ubiquitination of MDA5 and RIG-I (500 nM) using TRIM65 or RIPLET (250 nM) in the presence and absence of dsRNA (2 ng/μL). 1,012-bp and 112-bp dsRNAs were used for MDA5 and RIG-I, respectively, because the two receptors prefer different lengths of dsRNA for filament formation and immune stimulation. The Ubc13:Uev1a complex was used as E2. Reactions were analyzed by SDS-PAGE, followed by anti-MDA5 and anti-RIG-I immunoblots.
(D) Native gel mobility shift assays to monitor MDA5:TRIM65 interaction. Fluorescein-labeled (*) full-length MDA5, 2CARD, or MDA5ΔN (600 nM) were incubated with TRIM65 (300 nM) in the presence and absence of dsRNA (4 ng/μL). We used 112-bp dsRNA instead of 1,012-bp dsRNA to clearly visualize the mobility shift upon TRIM65 binding. Fluorescein fluorescence was used for gel imaging.
(E) Native gel mobility shift assays to monitor MDA5:TRIM65 interaction with TRIM65 domain mutants. Filaments of MDA5ΔN or RIG-IΔN (250 nM) were formed on Cy5-labeled (*) 112-bp dsRNA (1 ng/μL) and incubated with TRIM65 or its truncation variants (300 nM).
(F) Quantitative analysis of the mobility shift assay in Figure S1H, which showed that TRIM65 CC-PSpry has a KD of 170.6 nM. The KD could not be determined for PSpry because of inefficient mobility shift at all concentrations tested.
All data are representative of at least three independent experiments. Data in (A), (B), and (F) are mean ± SD (n = 3–4), and p values were calculated by two-tailed t test (***p < 0.001, **p < 0.005).
See Figure S1.
Reconstitution of the ubiquitination system using purified proteins in vitro showed that TRIM65 directly ubiquitinates MDA5, but not RIG-I, under equivalent conditions (Figure 1C). RIG-I, however, was efficiently ubiquitinated by RIPLET in a dsRNA-dependent manner (Figure 1C), consistent with a previous report (Cadena et al., 2019). RIPLET does not ubiquitinate MDA5 with or without dsRNA (Cadena et al., 2019). We found that the Ub chains conjugated to MDA5 by the E3 ligase TRIM65 and the E2 protein Ubc13:Uev1A were primarily K63-Ubn (Figures S1D and S1E), which are required for MDA5 signal activation (Jiang et al., 2012). Mutation or deletion of TRIM65’s RING domain, which is required for E2 binding, impaired MDA5 ubiquitination (Figure S1F). Importantly, robust MDA5 ubiquitination by TRIM65 required dsRNA (Figure 1C), which mediates MDA5 filament formation and signal activation. Native gel mobility shift assays showed that TRIM65 binds MDA5 only in the presence of dsRNA (Figure 1D, left panel), whereas it does not bind RIG-I in the presence or absence of dsRNA (Figure S1G). Because TRIM65 itself does not bind dsRNA (Figure 1E, left panel), our results suggest that the mobility shift of the MDA5 filament occurs via a direct interaction between the MDA5 filament and TRIM65.
Next we investigated which domains mediate the TRIM65: MDA5 interaction. First, we tested the 2CARD domain of MDA5 and showed that it did not bind TRIM65 in the presence or absence of dsRNA (Figure 1D, center panel). However, we found that the 2CARD-deletion mutant (MDA5ΔN), which can form filaments on dsRNA, was able to bind TRIM65 in the presence of dsRNA (Figure 1D, right panel), implicating the helicase domain and/or C-terminal domain (CTD) in the interaction. Similarly, neither the TRIM65 RING nor B-box domains were required for TRIM65 binding to MDA5 filaments, whereas the TRIM65 coiled-coil (CC) and PSpry domains were necessary (Figure 1E, center panel). The dissociation constant (KD) of CC-PSpry was 170.6 nM, whereas the KD of PSpry could not be measured because of inefficient binding (Figure 1F; Figure S1H). However, we could replace CC with an artificial dimeric fusion protein, glutathione S-transferase (GST) (Figure 1E, center panel). Considering that TRIM65 CC is a constitutive dimer (Figure S1I), this result suggests that PSpry bivalency is necessary and sufficient for MDA5 filament binding by TRIM65. Thus, not only is TRIM65 a highly specific E3 ligase for MDA5, just as RIPLET is for RIG-I (Cadena et al., 2019), but TRIM65 and RIPLET also share key aspects of substrate recognition; i.e., utilization of PSpry bivalency for selective recognition of RIG-I/MDA5 in the filamentous state.
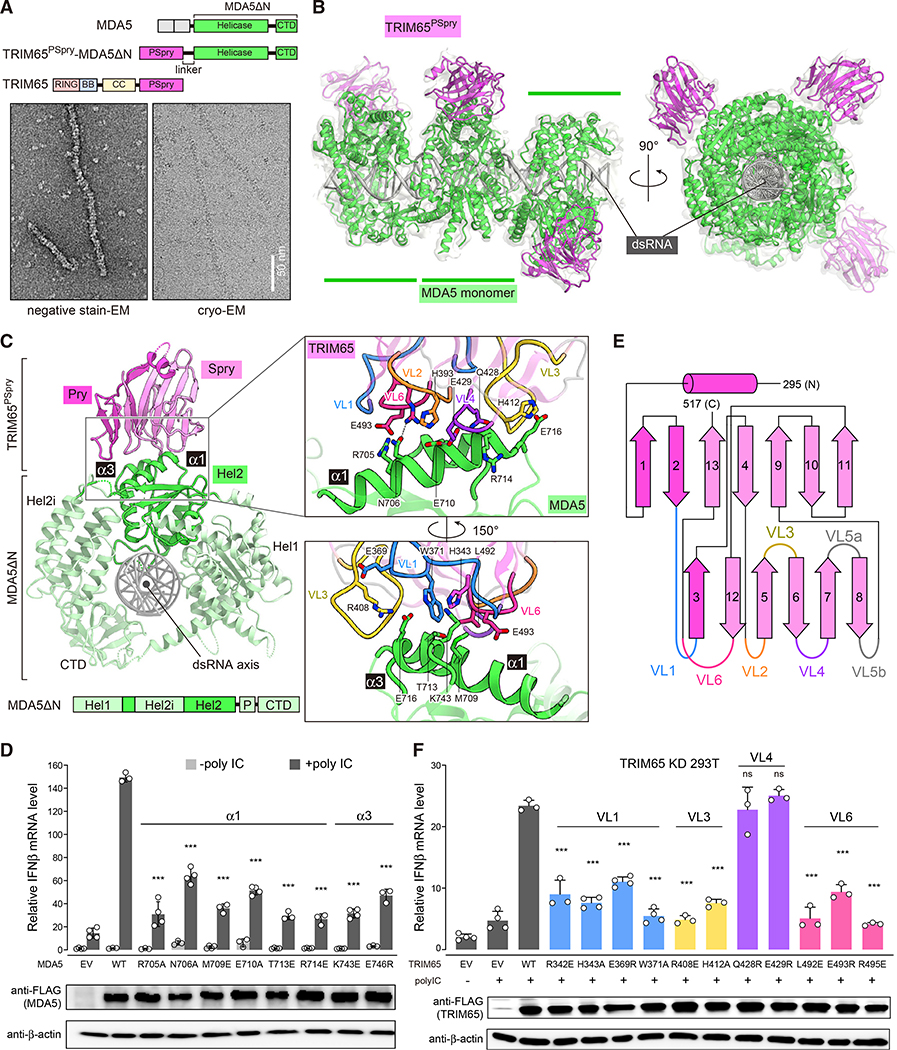
TRIM65 PSpry binds α1/α3 helices in the Hel2 domain of MDA5
We next investigated whether the functional similarity we uncovered between TRIM65:MDA5 and RIPLET:RIG-I interactions originates from a structural similarity in binding. We first determined cryoelectron microscopy (cryo-EM) structures of the MDA5 filaments on dsRNA bound by the PSpry domains from TRIM65. Because monomeric PSpry does not bind MDA5 filaments, we utilized an intramolecular fusion construct to increase the local concentration of PSpry and promote its binding to MDA5. We fused TRIM65 PSpry (TRIM65PSpry) to the N or C terminus of MDA5ΔN through a flexible 38-amino acid linker (Figure S2A). We confirmed that MDA5ΔN fused to TRIM65PSpry can form filaments on dsRNA independent of the fusion order (Figure S2B). To test whether the fused TRIM65PSpry binds to MDA5ΔN in cis, we checked for binding of the fusion constructs to GST-TRIM65PSpry in trans. Our native gel mobility shift assay showed no binding of GST-TRIM65PSpry to N and C terminus fusion filaments, suggesting that the TRIM65 binding sites in these fusion constructs are pre-occupied by TRIM65PSpry in cis (Figure S2C). For cryo-EM analysis, we chose the N-terminal fusion construct (TRIM65PSpry-MDA5ΔN) because of its superior protein stability compared with the C-terminal fusion construct.
Using fusion filaments formed on 1,012-bp dsRNA (Figure 2A), we obtained cryo-EM data and computationally extracted trimeric segments of the filaments (Table 1; STAR methods). From the resulting 15,306 trimeric segments, we reconstructed the cryo-EM map to 4.3-Å resolution (Figures 2B and 2C; Figures S2D–S2G; Table 1). We also determined the crystal structure of isolated TRIM65PSpry to aid model building (1.9 Å; Table 2). The crystal structures of MDA5ΔN (Wu et al., 2013) and TRIM65PSpry (this paper) could be fitted unambiguously in the reconstructed map (Figure 2B). Focused 3D classification and refinement of the central monomeric complex of MDA5ΔN:TRIM65PSpry improved the map quality for TRIM65PSpry (4.3 Å; Figures S2E and S2F).
Figure 2. TRIM65 PSpry recognizes α1/α3 of MDA5 Hel2.
(A) Top: schematic of the TRIM65PSpry-MDA5ΔN fusion construct. Bottom: representative negative-stain EM (left) and cryo-EM (right) of filaments formed by TRIM65PSpry-MDA5ΔN on 1,012-bp dsRNA.
(B) Cryo-EM map and ribbon model of the trimeric segment of the TRIM65PSpry-MDA5ΔN fusion filament. The cryo-EM map was low pass filtered at 5-A resolution and shown at 50% transparency. TRIM65PSpry, MDA5ΔN, and dsRNA are colored magenta, green, and gray, respectively. See also Figure S2 and Table 1 for details.
(C) Top view of the central monomeric complex of MDA5ΔN:TRIM65PSpry. Subdomains of MDA5ΔN are colored according to the schematic below. Note that part of Hel2 is separated by the Hel2 insertion (Hel2i) domain. “P” indicates the pincer domain. The Pry and Spry domains of TRIM65PSpry are colored magenta and pink, respectively. Right: magnified views of the interface between TRIM65PSpry and MDA5ΔN, where relevant variable loops (VLs) of TRIM65PSpry are colored according to the secondary structure schematic in Figure 2E. Key interacting residues are shown in stick models.
(D) Relative signaling activity of MDA5 with or without mutations in the TRIM65PSpry binding interface as measured by IFNβ mRNA induction. 293T cells were transfected with MDA5-expressing vectors and stimulated with poly(I:C) (0.5 μg). The amount of MDA5-expressing vector for each mutant was adjusted to ensure that the expression level is equivalent to WT.
(E) Secondary structure representation of TRIM65PSpry and definition of VLs. The Pry and Spry subdomains are distinguished by magenta and pink colors, respectively.
(F) Relative signaling activity of WT MDA5 upon poly(I:C) (0.5 μg) stimulation, as measured by IFNβ mRNA induction. TRIM65 KD 293T cells were transfected with plasmids expressing WT or mutant TRIM65. The amount of TRIM65-expressing vector for each mutant was adjusted to ensure that the expression level is equivalent to the WT. All cells were transfected with equal amounts of WT MDA5.
Data in (D) and (F) are mean ± SD (n = 3–4), and p values were calculated by comparing with the WT value using two-tailed t test (***p < 0.001; ns, not significant, p > 0.1). See Figures S2 and S3.
Table 1.
Cryo-EM data collection, refinement, and validation statistics
| MDA5-TRIM65PSpry-dsRNA | RIG-I-RIPLETPSpry-dsRNA | |||
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 105,000 | 105,000 | ||
| Voltage (kV) | 300 | 300 | ||
| Electron exposure (e−/Å2) | 76.5/72.8 | 50 | ||
| Defocus range (μm) | −1.5 to −2.5 | −0.7 to −2.5 | ||
| Pixel size (Å) | 0.85 | 0.855 | ||
| Initial particle images (no.) | 1,230,328 | 7,072,843 | ||
| Refinement | Filament | Monomer | Filament | Monomer |
| PDB ID | 7JL2 | 7JL0 | 7JL3 | 7JL1 |
| EMDB ID | EMD-22370 | EMD-22368 | EMD-22371 | EMD-22369 |
| Final particle images (no.) | 15,306 | 49,051 | 39,718 | 74,079 |
| Map resolution (Å) | 4.3 | 4.3 | 4.2 | 3.9 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Model resolution (Å) | 4.4 | 4.2 | 4.3 | 3.9 |
| FSC threshold | 0.5 | 0.5 | 0.5 | 0.5 |
| Map sharpening B factor (Å2) | −120 | −113 | −120 | −103 |
| Helical symmetry imposed | ||||
| Twist (°) | 86.941 | – | 76.1005 | – |
| Rise (Å) | 46.843 | – | 46.2706 | – |
| Model composition | ||||
| Non-hydrogen atoms | 22,122 | 7352 | 21219 | 7132 |
| Protein residues | 2,526 | 842 | 2457 | 827 |
| RNA nucleotides | 88 | 28 | 84 | 28 |
| Ligands | 12 | 4 | 12 | 4 |
| Root-mean-square deviations (RMSDs) from ideal | ||||
| Bond lengths (Å) | 0.006 | 0.008 | 0.003 | 0.010 |
| Bond angles (°) | 0.809 | 0.914 | 0.733 | 0.891 |
| Validation | ||||
| Clashscore | 18.40 | 13.08 | 11.22 | 13.12 |
| Poor rotamers (%) | 0.55 | 0.14 | 1.45 | 0.00 |
| Ramachandran plot (%) | ||||
| Favored | 94.67 | 93.29 | 97.10 | 95.65 |
| Allowed | 5.33 | 6.71 | 2.90 | 4.35 |
| Disallowed | 0.00 | 0.00 | 0.00 | 0.00 |
Table 2.
Data collection and refinement statistics
| TRIM65PSpry | |
|---|---|
| Data collection | |
| PDB ID | 7JL4 |
| Wavelength (Å) | 0.9201 |
| Space group | P32 |
| Cell dimensions | |
| a, b, c (Å) | 76.3, 76.3, 82.7 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 35.1–1.92 (1.99–1.92) |
| Rmerge | 0.079 (0.416) |
| I/σI | 13.3 (3.8) |
| Completeness (%) | 99.9 (99.8) |
| Redundancy | 5.3 (5.4) |
| CC1/2 | 0.996 (0.952) |
| Refinement | |
| Resolution (Å) | 35.1–1.92 (1.99–1.92) |
| No. of reflections | 41,053 (4,054) |
| Rwork/Rfree | 0.1682/0.2075 |
| No. of atoms | |
| Protein | 4,393 |
| Ligand/ion | 24 |
| Water | 425 |
| B-factors (Å2) | |
| Protein | 38.0 |
| Ligand/ion | 61.1 |
| Water | 48.9 |
| RMSDs from ideal | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.21 |
| Ramachandran plot (%) | |
| Favored | 97.96 |
| Allowed | 2.04 |
| Disallowed | 0 |
Values in parentheses are for the highest-resolution shell.
The structure showed that the MDA5 filament is similar to the previously reported structures of monomeric human MDA5 (Wu et al., 2013) and filamentous mouse MDA5 (Yu et al., 2018) without TRIM65 (Figure S3A), suggesting that TRIM65 binding causes little conformational change in MDA5. Each TRIM65PSpry interacts with an individual MDA5ΔN monomer with no contact across the filament interface (Figure 2B). TRIM65PSpry binds two adjacent α1 (residues 700–715) and α3 (residues 743–747) helices within a subdomain (Hel2) of the MDA5 helicase domain (Figure 2C). Mutation analysis suggests that helices α1 and α3 are important (Figure 2D). Structural comparison of MDA5 with (Wu et al., 2013) and without (Motz et al., 2013) dsRNA and in the monomeric (Wu et al., 2013) versus filamentous state with various ATP analogs (Yu et al., 2018) suggests that, although the overall helicase conformation changes upon RNA binding and ATP binding/hydrolysis, the conformations of individual Hel2 domains, especially near helices α1/α3, remain similar (Figure S3A). Consistent with these structural findings, forced dimerization of MDA5ΔN via GST fusion was sufficient to allow TRIM65 binding even in the absence of RNA, whereas cleavage of the GST tag impaired binding (Figure S3B). These data collectively suggest that the filament specificity of TRIM65 is mediated exclusively by the bivalency requirement instead of a direct readout of the filament structure.
TRIM65PSpry displays a twisted β sandwich fold (Figure 2C), which is common among the PSpry domains characterized so far (D’Cruz et al., 2013). It utilizes a cluster of variable loops (VLs) on one edge of the β sandwich for MDA5 binding (Figures 2C and 2E). The MDA5 binding surface on TRIM65PSpry is largely flat with a neutral electrostatic potential (Figure S3C), which may account for the low-affinity interaction between individual TRIM65PSpry and MDA5 molecules and the requirement for avidity. The VLs from the Pry (VL1) and Spry subdomains (VL3, VL4, and VL6) contact MDA5Hel2 (Figure 2C, inset). By mutagenesis analysis, we confirmed that VL1, VL3, and VL6 in TRIM65PSpry play important roles in MDA5 recognition, whereas residues in VL4 contacting MDA5 (e.g., Q428 and E429) may be less important (Figure 2F). Comparison with previous structures of two different PSpry domains in complex with their substrates (James et al., 2007; Woo et al., 2006; Figure S3D) suggests that, although the PSpry domains commonly utilize VLs for substrate recognition, each PSpry employs a unique set of VLs with distinct sequences and lengths for binding diverse substrates.
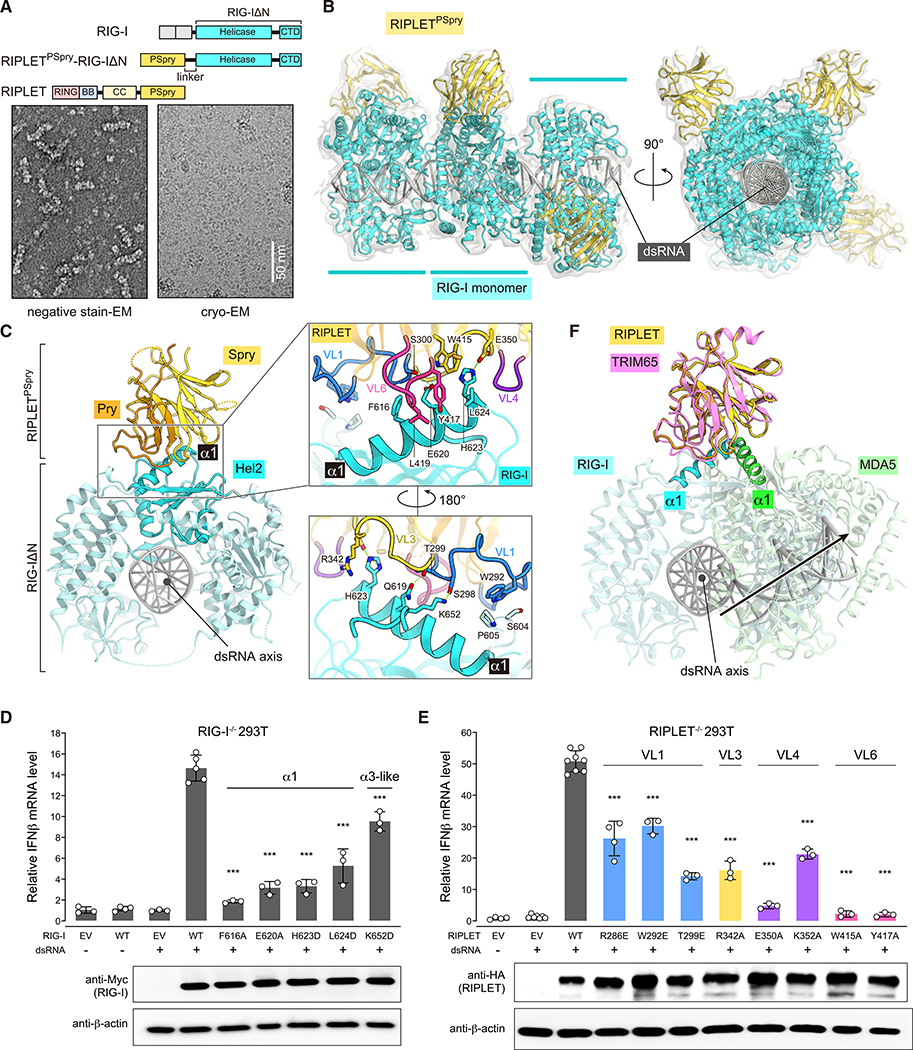
RIPLET recognizes RIG-I using a similar epitope in the helicase domain
We adopted a similar strategy to determine the cryo-EM structure of the human RIG-IΔN filament bound by RIPLETPSpry (Figures 3A and 3B; Figure S4; Table 1). The fusion filaments were formed on 112-bp dsRNA with a 5′-triphosphate group (Figure 3A), a preferred substrate for RIG-I. As with the TRIM65PSpry-MDA5ΔN filament, we first reconstructed a cryo-EM map for the trimeric segment of the RIPLETPSpry-RIG-IΔN filament (4.2 Å) and then performed focused local refinement of the central monomeric complex (3.9 Å). The maps were fitted with a previous crystal structure of human RIG-IΔN (Jiang et al., 2011) and a homology model of RIPLETPSpry (Figure 3B).
Figure 3. RIPLET PSpry recognizes RIG-I using a similar epitope as in the MDA5:TRIM65 complex, but the PSpry orientation differs.
(A) Top: schematic of the RIPLETPSpry-RIG-IΔN fusion construct. Bottom: representative negative-stain EM (left) and cryo-EM (right) of filaments formed by RIPLETPSpry-RIG-IΔN on 112-bp dsRNA.
(B) Cryo-EM map and ribbon model of the trimeric segment of the RIPLETPSpry-RIG-IΔN fusion filament. RIPLETPSpry, RIG-IΔN, and dsRNA are colored yellow, cyan, and gray, respectively.
(C) Top view of the central monomeric complex of RIG-IΔN:RIPLETPSpry. The Hel2 subdomain and the rest of RIG-IΔN are colored cyan and light cyan, respectively. The Pry and Spry domains of RIPLETPSpry are colored orange and yellow, respectively. Right: magnified view of the interface between RIPLETPSpry and RIG-IΔN, where relevant VLs are displayed according to the color code in (E). Key interacting residues are shown as stick models.
(D) Relative signaling activity of RIG-I with or without mutations in the RIPLETPSpry binding interface as measured by IFNβ mRNA induction. RIG-I−/− 293T cells (Shi et al., 2017) were transfected with RIG-I-expressing vectors and stimulated with 42-bp dsRNA with 5′-ppp (0.2 μg). The amount of RIG-I-expressing vector for each mutant was adjusted to ensure that the expression level is equivalent to the WT.
(E) Relative signaling activity of WT RIG-I upon stimulation with 5′-ppp 42-bp dsRNA (0.2 μg). RIPLET−/− 293T cells (Shi et al., 2017) were transfected with plasmids expressing WT or mutant RIPLET. The amount of RIPLET-expressing vector for each mutant was adjusted to ensure that the expression level is equivalent to the WT. All cells were transfected with an equal amount of WT RIG-I.
(F) Superposition of the MDA5ΔN:TRIM65PSpry and RIG-IΔN:RIPLETPSpry complexes by aligning the PSpry domains of TRIM65 (pink) and RIPLET (yellow). With the exception of the α1 helices, MDA5 (green) and RIG-I (cyan) proteins are shown at 50% transparency.
Data in (D) and (E) are mean ± SD (n = 3–4), and p values were calculated by comparing with the WT value using two-tailed t test (***p < 0.001). See Figures S4 and S5.
The structure of the RIG-I filament showed that the conformation of individual RIG-I molecules differs significantly from the previously reported structures of monomeric RIG-I bound to a dsRNA end (Jiang et al., 2011; Kowalinski et al., 2011; Luo et al., 2011). The CTD, which is responsible for recognition of the 5′-triphosphate group of dsRNA, is rotated away from the central axis of dsRNA to the side of the dsRNA in the RIG-I filament (Figure S5A). This leads to RIG-I adopting a ring conformation around the dsRNA stem, which closely resembles that of MDA5 in the filament (Figure S5A).
Intriguingly, the structure showed that RIPLETPSpry also binds α1 (residues 609–624) and the α3-like loop (residues 651–655) of RIG-IHel2 (Figures 3C–3D), similar to TRIM65PSpry’s recognition of α1/α3 in MDA5Hel2. RIPLETPSpry also utilizes VL1, VL3, VL4, and VL6 to contact RIG-I (Figures 3C and 3E), which induces little conformational change in Hel2 of RIG-I (Figure S5B). However, the relative orientation of RIPLETPSpry differs from that of TRIM65PSpry by ~60°, which is more evident when the two complex structures are superposed by aligning PSpry domains (Figure 3F). These divergent helicase-PSpry interaction modes are highly unexpected given the close relationship between RIG-I and MDA5 helicase domains; the overall similarity between RIPLETPSpry and TRIM65PSpry structures, including the VL conformations (Figure 3F); and the functional parallelism in signaling between the two E3:RLR cognate pairs. Our structures uniquely highlight how PSpry domains can adapt to similar epitopes in divergent ways with only minimal perturbations in VLs.
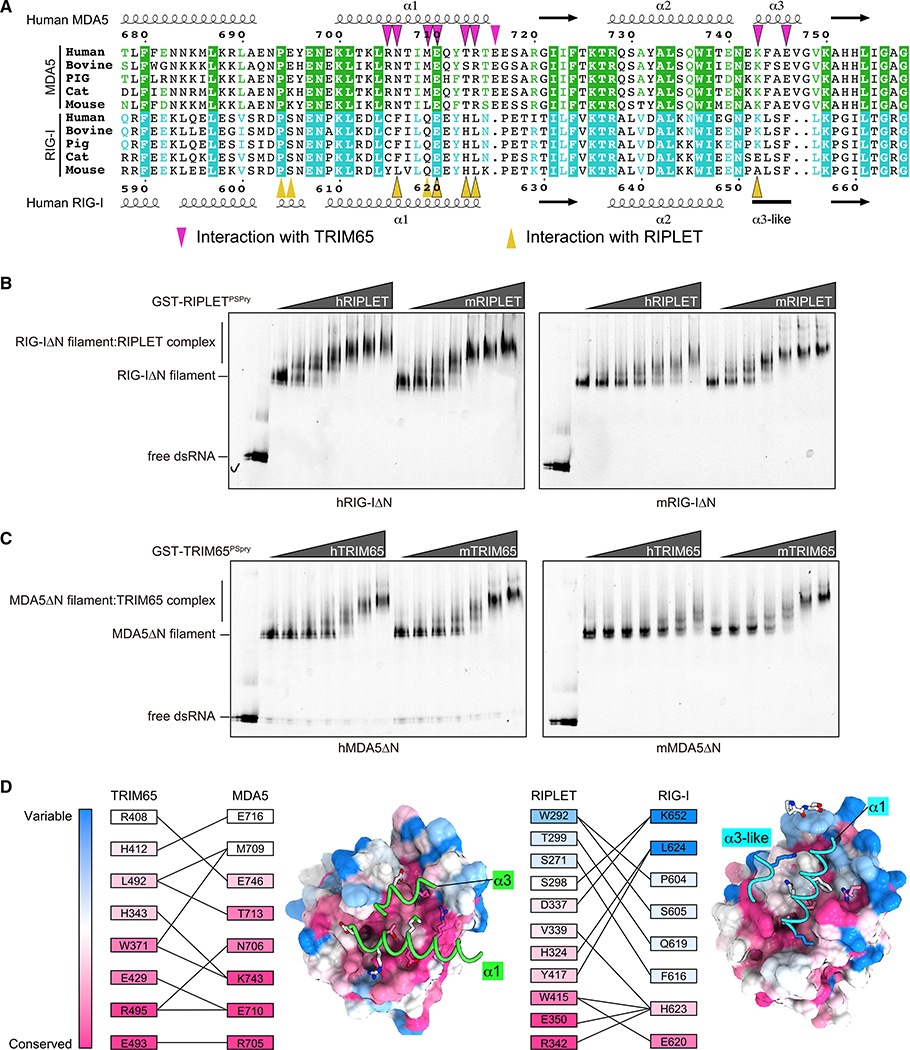
RLRs and their cognate TRIM proteins have co-evolutionary relationship
Sequence comparison among RIG-I and MDA5 homologs showed that the PSpry interface residues are relatively well conserved within RIG-I orthologs and within MDA5 orthologs but not between RIG-I and MDA5 (Figure 4A). This observation explains how RIPLET and TRIM65 achieve specificity toward RIG-I and MDA5, respectively, with little cross-reactivity. Intriguingly, some of the interface residues were not strictly conserved even among orthologs (Figure 4A; Figure S5C). As a result, there is limited cross-species reactivity between mouse and human proteins; mouse RIG-I and MDA5 are well recognized by mouse RIPLET and TRIM65, respectively, but not as well by human RIPLET and TRIM65 (Figures 4B and 4C). This is despite the fact that mouse RIPLET and TRIM65 recognize human and mouse RLRs equally well (Figures 4B and 4C). Analysis of the degree of conservation for the interface residues suggests that well-conserved residues in RIPLET/TRIM65 generally interact with well-conserved residues in RIG-I/MDA5, whereas less-conserved residues in RIPLET/TRIM65 interact with less-conserved residues in RIG-I/MDA5 (Figure 4D). These data collectively support our conclusion that interacting pairs of E3 ligases and RLRs have a co-evolutionary relationship.
Figure 4. Conservation and co-evolution analysis of the RLR:PSpry interface.
(A) Sequence alignment of orthologs of MDA5 and RIG-I near the TRIM65/RIPLET interface. Residues involved in the interaction with TRIM65 and RIPLET are indicated with triangles Sequences were aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo), and alignment figures were generated using ESPript3 (http://espript.ibcp.fr/ESPript/ESPript).
(B) Native gel mobility shift assays of RIG-I filaments in the presence of RIPLET. RIG-I filaments were assembled by incubating Cy5-labeled 112-bp dsRNA (1 ng/μL) with human or mouse RIG-IΔN (hRIG-IΔN or mRIG-IΔN, respectively; 250 nM) in the presence of 2 mM ATP (Peisley et al., 2013). RIG-I filaments were then incubated with increasing concentrations (18–600 nM) of human or mouse RIPLETPSpry (hRIPLET or mRIPLET, respectively) fused with GST, and complex formation was analyzed by native PAGE using dsRNA fluorescence.
(C) Native gel mobility shift assay of MDA5 filaments in the presence of TRIM65. Experiments were done as in (A) except that ATP was omitted in the reaction because ATP promotes MDA5 filament disassembly (Peisley et al., 2011).
(D) Degree of conservation for interacting residues in the MDA5:TRIM65 and RIG-I:RIPLET complexes. The conservation score was calculated using vertebrate protein sequences (STAR methods) and the Consurf web server (https://consurf.tau.ac.il/). The interacting residues are arranged in the descending order of conservation score (bottom to top), and the interaction pairs are indicated by connecting lines. The conservation score was also mapped onto the structures of PSpry using the program CueMol. The PSpry domains of TRIM65/RIPLET are shown in surface representation from the equivalent viewpoints, whereas α1/α3 helices of the bound RLRs are shown in cartoon representation.
LGP2 is recognized by TRIM14 via PSpry bivalency and Hel2 epitope
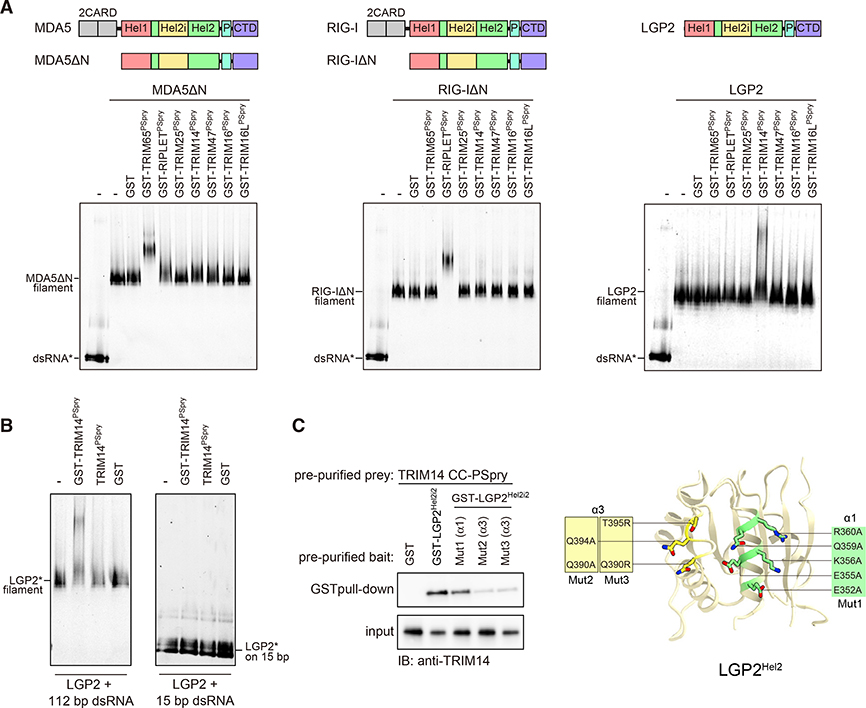
We next turned our attention to LGP2, the third member of the RLR family with a helicase domain homologous to those of RIG-I and MDA5 (Sarkar et al., 2008; Figure S6A). Like RIG-I and MDA5, LGP2 also forms filaments upon dsRNA binding (Uchikawa et al., 2016). However, unlike RIG-I and MDA5, LGP2 does not directly activate MAVS and the downstream pathway. Although some studies suggest that LGP2 has a role in modulating RIG-I/MDA5 functions (Bruns and Horvath, 2015; Venkataraman et al., 2007), other studies suggest a cell type- and ligand-dependent phenotype of LGP2 deletion (Satoh et al., 2010; Suthar et al., 2012), raising questions regarding the true functions of LGP2. Nevertheless, given the structural similarity of TRIM65:MDA5 and RIPLET:RIG-I binding, we speculated that a related TRIM protein might bind LGP2 in a similar manner. We used phylogenetic analysis to identify the human proteins most closely related to RIPLET and TRIM65 in their PSpry domains: TRIM47, TRIM25, TRIM16, TRIM16L, TRIM14, and BSPRY (Figure S6B). We were able to purify the PSpry domains of all these TRIM proteins except BSPRY as soluble GST fusions (Figure S6C). We tested these domains for their ability to interact with RIG-I, MDA5, and LGP2 filaments. Among the panel of PSpry domains tested, only TRIM65PSpry and RIPLETPSpry bound MDA5ΔN and RIG-IΔN filaments, respectively, again confirming the high selectivity of their engagement (Figure 5A). In contrast to an earlier report (Gack et al., 2007), but consistent with more recent findings (Cadena et al., 2019; Hayman et al., 2019), we found that TRIM25 does not interact with RIG-I (Figure 5A; Figure S6D).
Figure 5. LGP2 is recognized by TRIM14 via PSpry bivalency and Hel2 epitope.
(A) Native gel mobility shift assay to test interactions between RLR filaments and a panel of TRIMPSpry closely related to those of RIPLET and TRIM65 (Figure S6). RLR filaments were formed by mixing MDA5ΔN, RIG-IΔN, and LGP2 (250 nM) with Cy5-labeled (*) 112-bp dsRNA (1 ng/μL) and incubated with TRIMPSpry fused with GST (300 nM for RIG-I/MDA5 and 1.2 μM for LGP2). Cy5 fluorescence was used for gel imaging.
(B) Mobility shift assay of LGP2 (0.6 μM) bound to 112-bp or 15-bp dsRNA (2.4 ng/μL) in the presence of GST-TRIM14PSpry, TRIM14Pspry, or GST alone (2.4 μM).
(C) GST pull-down assay to examine the interaction between GST-LGP2Hel2i2 and TRIM14CC-PSpry. GST-LGP2Hel2i2 and TRIM14CC-PSpry were recombinantly expressed in E. coli and purified prior to GST pull-down. The variants (Mut1–Mut3) of LGP2Hel2i2 with mutations in the α1 and α3 helices are defined on the right. Mutated residues are mapped onto the modeled human LGP2 Hel2, which was built using the Phyre2 server (http://www.sbg.bio.ic.ac.uk) with the crystal structure of chicken LGP2 (PDB: 5JB2) as a template.
Using the same panel of GST-PSpry proteins, we also found that only TRIM14PSpry is able to specifically bind LGP2 filaments (Figure 5A). TRIM14 was linked previously to the RIG-I pathway (Tan et al., 2017; Zhou et al., 2014). Given our finding that TRIM14 directly binds LGP2 but not RIG-IΔN, we deduce that the RIG-I-stimulatory function of TRIM14 may be mediated indirectly through its interaction with LGP2. TRIM14 binding to LGP2 also requires PSpry bivalency; TRIM14PSpry without GST fusion does not bind LGP2 filaments (Figure 5B, left panel).
Furthermore, GST-TRIM14PSpry does not bind monomeric LGP2 on 15-bp dsRNA (Figure 5B, right panel). Given the evolutionary relatedness of LGP2 to RIG-I and MDA5, we next tested whether Hel2 of LGP2 is involved in TRIM14 binding. However, isolated LGP2 Hel2 was insoluble and required inclusion of the adjacent domain Hel2i to maintain solubility. Therefore, we purified LGP2 Hel2i-Hel2 (Hel2i2) fused to GST and performed GST pull-down. TRIM14CC-PSpry co-purified with GST-LGP2Hel2i2 but not with GST alone (Figure 5C). Mutations in α3 helix, but not α1, of the Hel2 domain in LGP2Hel2i2 impaired the interaction between TRIM14CC-PSpry and GST-LGP2Hel2i2 (Figure 5C). Overall, our results suggest that the avidity-driven interaction of TRIM14 with LGP2 is mediated by an epitope similar to those of RIG-I and MDA5. Our findings highlight an ancient functional and structural similarity in TRIM-mediated binding of all three RLR proteins, which last had a common ancestor prior to the origin of bony vertebrates and the IFN-based immune system ~600 million years ago (mya) (Krause and Pestka, 2005; Sarkar et al., 2008).
Distinct TRIM proteins recognize common epitopes in diverse helicases using PSpry bivalency
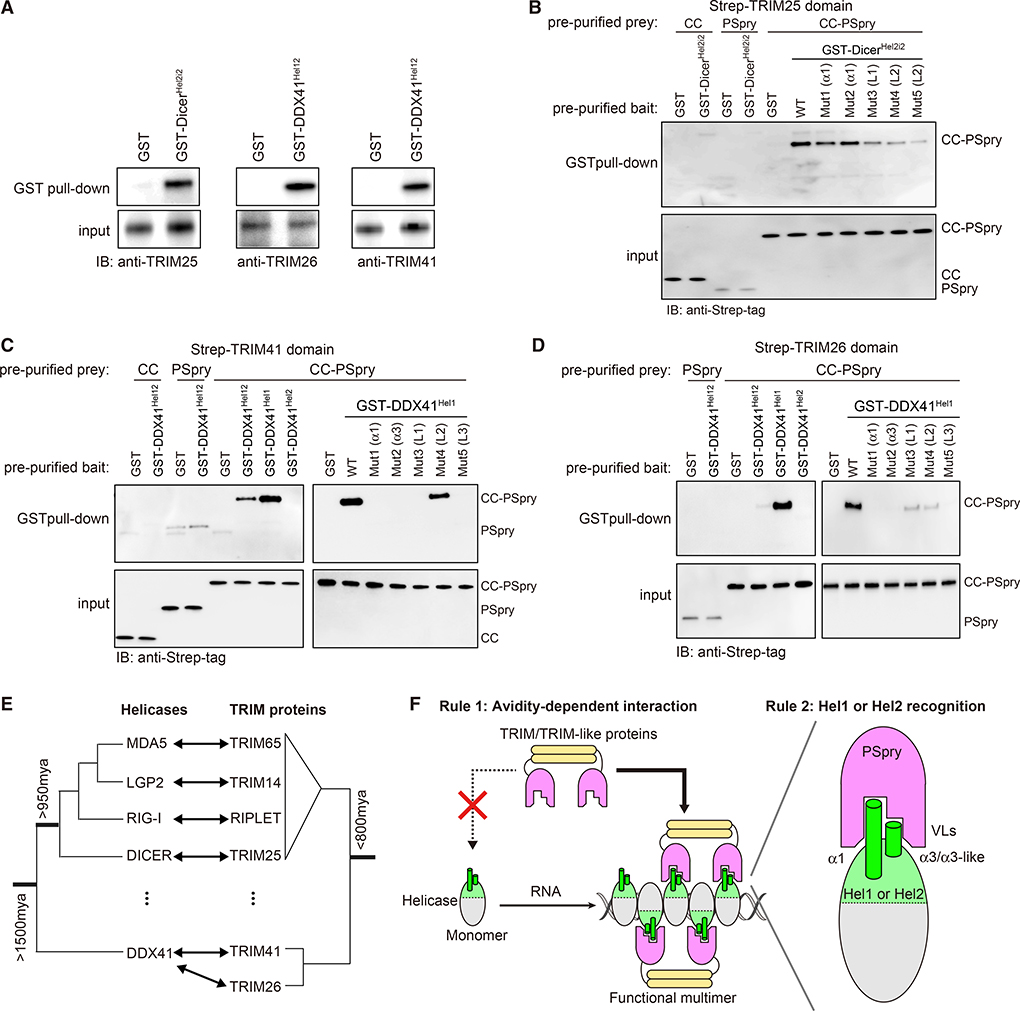
We next asked whether the helicase:PSpry interaction mode we identified with RLRs is more widespread among distantly related helicases. We investigated Dicer, which contains a helicase domain similar to those of RLRs (Kim et al., 2009) and, together with RLRs, belongs to the dsRNA-activated ATPases (DRAs) (Luo et al., 2013). Dicer is a conserved ribonuclease involved in RNA interference, which is considered one of the primary antiviral mechanisms that existed prior to the birth of the IFN system (Aguado et al., 2017; Maillard et al., 2019). To identify TRIM/TRIM-like proteins that interact with DicerHel2, we mixed purified GST-DicerHel2i2 with 293T lysate and subjected it to GST pull-down. We conjectured that GST fusion would mimic helicase dimerization and allow identification of avidity-dependent interactors. Co-purified proteins were analyzed by mass-spectrometry and/or western blot analyses. Putative interaction partners containing PSpry domains were then recombinantly purified and reanalyzed for direct interactions with GST-DicerHel2i2. We found that TRIM25 co-purifies with GST-DicerHel2i2 from 293T lysate but not with GST (Figure 6A). The interaction is direct because TRIM25CC-PSpry and GST-DicerHel2i2 pre-purified from E. coli are also co-isolated by GST pull-down (Figure 6B). Neither TRIM25PSpry nor TRIM25CC alone can bind GST-DicerHel2i2 (Figure 6B), consistent with the importance of PSpry bivalency for their interactions. To examine whether the Hel2 regions equivalent to α1 and α3 of RLRs are involved in the interaction with TRIM25CC-PSpry, we made mutations in and around the α1 helix and α3-like loop, which include linker residues (designated L1 and L2; Figure S6E) that are uniquely present in Dicer but not RLRs. Although the importance of the α3-like loop could not be tested because of insolubility of the mutants, L1 adjacent to the α3-like loop and L2 adjacent to the α1 helix were found to be important for TRIM25 binding (Figure 6B). These results suggest that TRIM25 binds Dicer by recognizing a similar region of Hel2 as in RLRs, again using PSpry bivalency. Thus, the mode of TRIM-helicase interaction we have uncovered significantly preceded the origin of the IFN-based immune system (Krause and Pestka, 2005).
Figure 6. Multiple TRIM proteins recognize common epitopes in helicases beyond RLRs.
(A) GST pull-down assays to identify interacting TRIM partners of Dicer and DDX41 helicase domains. GST-fused Hel2i-Hel2 of Dicer (GST-DicerHel2i2) and Hel1-Hel2 of DDX41 (GST-DDX41Hel12) were recombinantly purified and mixed with 293T lysate for GST pull-down.
(B) GST pull-down assay to examine direct interaction between GST-DicerHel2i2 and TRIM25. GST-DicerHel2i2 and TRIM25 (CC, Pspry, or CC-PSpry) were recombinantly expressed in E. coli and purified prior to GST pull-down. The variants (Mut1–Mut5) of DicerHel2i2 with mutations in α1 or L1 and L2 are defined in Figure S6E.
(C and D) GST pull-down assays to examine direct interaction between GST-DDX41Hel12 and TRIM41CC-PSpry (C) or TRIM26CC-PSpry (D). GST-DDX41 (Hel12, Hel1, or Hel2) and TRIM41 or TRIM26 (CC, Pspry, or CC-PSpry) were recombinantly expressed in E. coli and purified prior to GST pull-down. TRIM26 CC was insoluble and, thus, not included in the GST pull-down. The mutations (Mut1–Mut5) in DDX41Hel1 are defined in Figure S6E.
(E) Schematic phylogenetic trees of helicases and TRIM/TRIM-like proteins discussed in this study. See Figures S6A and S6B for full trees.
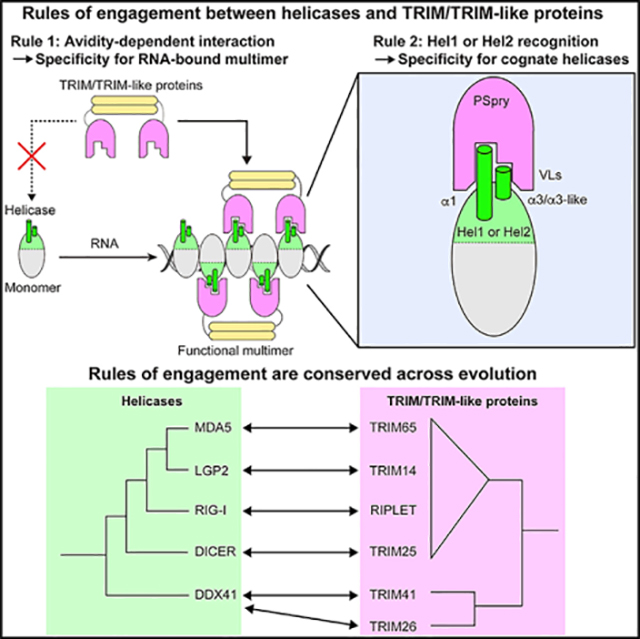
(F) Cartoon summarizing the two rules of engagement between helicases and TRIM proteins. Rule 1: PSpry bivalency allows TRIMs to specifically recognize the cognate helicases in the multimeric form versus monomeric form. This restricts the actions of TRIMs to the helicases in complex with certain RNAs that can induce helicase multimerization. Multimerization can be in the form of filaments (as for RLRs) or other structures shaped by the conformation of the bound RNAs. Rule 2: individual PSpry recognizes regions in or near α1/α3 of Hel1 or Hel2. Utilization of the common epitope by a wide range of helicases for their interaction with PSpry suggests an ancient evolutionary relationship between helicase and PSpry domains.
See also Figure S6.
Finally, we investigated an even more divergent helicase, DDX41. Although DDX41 belongs to the same superfamily of helicases as Dicer and RLRs (Singleton et al., 2007), it is a DEAD-box helicase rather than a DRA (Luo et al., 2013) and diverged from DRAs more than 1,500 mya (Figure 6E). DDX41 has been known previously to be involved in splicing, translation, and many other cellular RNA processes (Jiang et al., 2017; Peters et al., 2017; Polprasert et al., 2015) but has been shown recently to participate in foreign DNA sensing, albeit through a poorly understood mechanism (Parvatiyar et al., 2012; Quynh et al., 2015; Zhang et al., 2011). DDX41 does not contain a Hel2i domain but harbors Hel1 and Hel2, which share the RecA-like fold (Omura et al., 2016). Given the structural similarity between Hel1 and Hel2, we expanded our analysis to search for partners of DDX41 Hel1 as well as Hel2. We purified DDX41 Hel1-Hel2 fused with GST (GST-DDX41Hel12) and subjected it to the same pull-down and mass spectrometry analysis as GST-DicerHel2i2. We found that two TRIM proteins, TRIM26 and TRIM41, specifically co-purified with GST-DDX41Hel12 from 293T lysate (Figure 6A). Using pre-purified proteins, we showed that these interactions are direct, dependent on PSpry bivalency, and mediated by regions in DDX41 Hel1 equivalent to α1/α3 of Hel2 (Figures 6C and 6D). Our findings thus suggest that the rules of PSpry engagement we identified with RLRs and Dicer are not restricted to DRAs but shared with other helicases that diverged close to the origin of eukaryotes nearly a billion years ago (Figure 6E).
DISCUSSION
We discovered two molecular principles that govern interactions between TRIM/TRIM-like proteins and RNA helicases (Figure 6F). The first principle is avidity-dependent interaction; many TRIM/TRIM-like proteins engage with the cognate helicases only in a multimeric state. The importance of this principle is best illustrated with TRIM65 and RIPLET, which, respectively, recognize filamentous MDA5 and RIG-I in the presence of foreign dsRNAs but not in the monomeric resting state. This enables tight control of MDA5/RIG-I-mediated immune signaling and ensures activation only upon viral infection. This avidity-dependent interaction also appears to apply to other helicases (LGP2, Dicer, and DDX41). We identified TRIM partners for these helicases and showed that their interactions are also dependent on PSpry bivalency. LGP2 forms filamentous oligomers along dsRNA, like MDA5 and RIG-I. Exactly how and when Dicer and DDX41 form multimers is still unclear. Given the similarity between the helicase domains of Dicer and RLRs, Dicer may form filamentous oligomers on long dsRNA. Alternatively, multiple Dicer and DDX41 molecules may be bridged indirectly by certain RNAs, allowing bivalent binding of cognate TRIM proteins (Figure 6F). Regardless of the mechanism for helicase multimerization, our findings suggest avidity-dependent binding as a general rule by which TRIM/TRIM-like proteins regulate their engagement with helicases. Considering that helicase multimerization can be dependent on RNA conformation and structure, as is the case for RIG-I and MDA5, our findings also raise an intriguing possibility that TRIM/TRIM-like proteins may function as a new type of indirect RNA sensor by monitoring helicase multimerization.
The second principle we uncovered is that regions of Hel1 or Hel2 near α1/α3 are commonly recognized by a broad range of TRIM/TRIM-like proteins harboring PSpry domains (Figure 6F). Our cryo-EM structures of the TRIM65:MDA5 and RIPLET:RIG-I complexes revealed that TRIM65 and RIPLET PSpry domains recognize α1/α3 of cognate helicases but adopt two distinct orientations. Remarkably, our analysis of LGP2, Dicer, and DDX41 and their TRIM partners suggests that these interactions also use similar epitopes in the helicase domains. These observations raise questions regarding the evolutionary origins of this seemingly widespread utilization of common epitopes in Hel1/Hel2 domains of helicases by TRIM proteins. Phylogenetic analysis suggests that DDX41, Dicer, and the ancestral RLR helicases diverged from one another significantly earlier than their partner TRIMs (Figure 6E), so helicase divergence could not have driven concurrent TRIM evolution. It is possible that multiple TRIMs evolved independently to recognize the Hel1/Hel2 epitope region, perhaps because it allows sufficient specificity and accessibility when the helicase domain multimerizes. Given the low likelihood that convergent evolution would arrive at such similar binding determinants, we favor the alternate hypothesis that recognition of Hel1 or Hel2 epitopes might have been a property of an ancestral PSpry-containing TRIM protein. This ability may have been preserved in some descendant extant TRIM/TRIM-like proteins, whereas other lineages may have diverged to recognize non-helicase substrates. Regardless of whether the multiple TRIM-helicase interactions we have uncovered have resulted from convergent or divergent evolution, the recurrence of this interaction in multiple branches of TRIMs and helicases suggests widespread existence of such interactions and their co-option by different helicases to acquire Ub-dependent or Ub-independent regulatory functions of TRIMs. Given that the functions and mechanisms of many helicases and TRIM proteins remain poorly understood, our findings may provide a new mechanistic link between the two protein families in innate immunity and beyond.
Limitations
Key outstanding questions are the biological roles of the interactions between LGP2, Dicer, and DDX41 and their cognate TRIM proteins. More specifically, it remains to be investigated exactly in what biological context these interactions occur and how the interactions alter the functions of the helicases or TRIM proteins. In addition to functional studies, more detailed structural analyses of additional helicase:TRIM complexes will help further validate and generalize the rules of engagement we put forward in this paper.
STAR⋆METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sun Hur (sun.hur@crystal.harvard.edu)
Materials availability
All plasmids generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The cryo-EM maps included in this study have been deposited in the Electron Microscopy Data Bank with accession codes EMDB: EMD-22368, EMD-22369, EMD-22370 and EMD-22371. The atomic coordinates have been deposited in the Protein Data Bank with accession codes PDB: 7JL0, 7JL1, 7JL2, 7JL3, and 7JL4.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HEK293T cells
Cells were maintained in DMEM (High glucose, L-glutamine, Pyruvate) with 10% fetal bovine serum, 1% penicillin/streptomycin.
METHOD DETAILS
Material Preparation
Plasmids
Mammalian expression plasmids for MDA5 and RIPLET were described previously (Cadena et al., 2019; Wu et al., 2013). Human Trim65 was inserted in pFLAG-CMV4 vector between HindIII and ClaI sites for mammalian expression. Myc tagged Human Rig-I was inserted between KpnI and NotI sites of pcDNΑ3.1. Mammalian expression plasmid for MAVS has been described previously (Wu et al., 2014) and pcDNA4 expressing STING V155M variant was a kind gift from P. Kranzusch, Harvard Medical School. Baculovirus expression construct for LGP2 and TRIM65CC-PSpry (residues 125–517) were cloned into the modified pFastBac1vector (Invitrogen), in which His6-Strep-tag and HRV 3C protease recognition site were inserted between the polyhedrin promoter and BamHI site. For bacterial expression of MDA5, the gene encoding MDA5, MDA5ΔN (residues 287–1025), helicase (residues 287–837) and α1 (residues 689–718) were inserted into pE-SUMO vector (LifeSensors) or the modified pGEX-6P-1 vector (GE healthcare) with N-terminal His6-tag. Bacterial expression plasmid for RIG-I was described previously (Cadena et al., 2019). The gene encoding RIG-IΔN (residues 204–925) was inserted into the pE-SUMO vector. For the plasmids of TRIM65PSpry-MDA5ΔN, MDA5ΔN-TRIM65PSpry and RIPLETPSpry-RIG-IΔN, the two DNA fragments of PSpry (TRIM65 or RIPLET) and RLH (MDA5ΔN or RIG-IΔN) were cloned into pE-SUMO vector using In-Fusion HD (Clontech) together with a gene encoding a flexible linker comprised of 38 residues. All Hel2i2 constructs for MDA5 (residues 532–837), LGP2 (211–487) and Dicer (243–610) and GST-PSpry constructs for TRIM65 (295–517), RIPLET (249–442), TRIM14 (238–442), TRIM16 (346–564), and TRIM47 (401–638) were cloned into the modified pGEX-6P-1 vector with N-terminal His6-tag. All DDX41 constructs, Hel12 (155–578), Hel1 (155–401) and Hel2 (402–578) were cloned into the modified pGEX-6P-1 vector with N-terminal His6-tag. TRIM14CC-PSpry (residues 59–442) was cloned into the pE-SUMO vector. All bacterial expression constructs for TRIM25, TRIM26 and TRIM41 were cloned into the modified pE-SUMO vector, in which His6-Strep-tag and HRV 3C protease recognition site were inserted after the region encoding SUMO. For expression construct of TRIM65PSpry used for crystallization, the gene encoding the residues 312–504 of TRIM65 were cloned into the modified pMal-c2x vector (NEB), in which N-terminal His6-tag and the HRV 3C protease recognition site were inserted into 5′-upstream a maltose binding protein sequence and a multiple cloning site, respectively. All mutations were introduced by a PCR-based method, and the sequences were confirmed by DNA sequencing.
RNAs
Double-stranded RNAs (dsRNAs) used in this study were prepared by in vitro T7 transcription as described previously (Peisley et al., 2011). The templates for RNA synthesis were generated by PCR amplification. The sequences of 42, 112 and 1012 bp dsRNAs were taken from the first 30, 100 and 1000 bp of the MDA5 gene, respectively, flanked by 5’-gggaga and 5’-tctccc. The sequence for 15 bp dsRNAs was 5’-agggcccgggaugcu. The two complementary RNA strands were co-transcribed, and the duplex was purified using 8.5% acrylamide (112 bp) or 6% acrylamide (1012 bp) gel electrophoresis. RNA was gel-extracted using the Elutrap electroelution kit (Whatman), ethanol precipitated, and stored in 20 mM HEPES pH 7.0. For 112 bp dsRNA with a 5-nt 5′ overhang (5′ovg 112 bp dsRNA), 5-nt sequence (GGGTT) was inserted between T7 promoter sequence and the MDA5 gene of the template DNA transcribed for the negative strand RNA, and the two complementary RNA strands were transcribed and purified separately. The purified negative strand RNA was treated with Calf intestinal alkaline phosphatase (CIP, NEB) to remove 5′-triphosphate moiety, and was re-purified using QIAquick Nucleotide removal kit (QIAGEN). The dephosphorylated RNA was annealed with the 112-nt positive strand RNA in 50 mM NaCl and 10 mM EDTA. Qualities of RNAs were analyzed by 1X TBE polyacrylamide gel and the RNA duplex integrity was established using Acridine Orange staining (Sigma Aldrich) (Mu et al., 2018). For 3′-labeling with Cy5, 112 bp dsRNA was oxidized with 0.1 M sodium meta-periodate (Pierce) overnight in 0.1 M NaOAc pH 5.4. The reaction was quenched with 250 mM KCl, buffer exchanged using Zaba desalting columns (Thermo Fischer) into 0.1 M NaOAc pH 5.4 and further incubated with 0.1 M Cy5-Monohydrazide (GE Healthcare) for 4–6 hr at RT.
Cell lines
The TRIM65-KD cells were generated using the method described previously (Chavez et al., 2015). The TRIM65-specific guide RNA sequences (Forward: TGCCGAGCGCCTCAAGCGCG; Reverse: CGCGCTTGAGGCGCTCGGCA) were inserted in pSB700 vector (Addgene) using BsmBI site. The plasmid was then transfected in HEK293T cells in a 6-well plate along with psPAX2 (Addgene) and pMD2.G (Addgene) packaging vectors in a ratio of 5:2.5:1. The lentiviral particles were harvested 48 h post-transfection and filtered using a 0.45 μ filter. The virus was then transduced into Cas9-stable HEK293T cells (generous gift from G. Church, Harvard Medical School) and allowed to grow in the presence of 3 μg/ml puromycin, 2.5 μg/ml blasticidin and 10 μg/ml hygromycin. For parental WT cells, virus derived from pSB700 empty vector was used and the rest of the procedure was the same as that for knockdown cells. 7 days after transduction, monoclonal cell masses were picked and screened for knockdown of TRIM65 using western blot analysis. RIG-I−/−, RIPLET−/− and their respective parental WT HEK293T cell lines were obtained from the Hou lab (Shi et al., 2017).
Protein expression and purification
TRIM65 protein was expressed in HEK293T cells using polyethylenimine (PEI, Polysciences). The harvested cells were lysed by Dounce homogenizer in hypotonic buffer (10 mM KCl, 10 mM Tris pH7.5 and 1.5 mM MgCl2) supplemented with 1 mM PMSF and mammalian protease inhibitor cocktail (G-Biosciences). The lysed cells were centrifuged, and the supernatant was applied to anti-FLAG M2 agarose beads (Sigma-Aldrich). The protein was eluted with 3X FLAG peptide (Sigma-Aldrich), and buffer was exchanged with 50 mM HEPES pH7.5, 150 mM NaCl and 2 mM DTT using Zeba desalting columns, 40 kDa molecular-weight cutoff (Thermo Scientific) to remove the 3X FLAG peptide. LGP2 protein was expressed in Sf9 cells using the Bac-to-Bac baculovirus expression systems (Invitrogen). Sf9 cells were infected in HyClone SFX Insect cell culture media (GE Healthcare) for 48 h at 27°C. The harvested cells were lysed by sonication and centrifuged. The supernatant was applied to Ni-NTA agarose (QIAGEN) and the eluted protein was treated with HRV 3C protease at 4°C overnight to cleave the N-terminal tag. The protein was further purified by chromatography on HiTrap Heparin (GE Healthcare) and Superdex 200 Increase 10/300 (GE Healthcare) columns. For TRIM65CC-PSpry (residues 125–517), expression and lysis were done in the same manner as LGP2, while protein purification was done by chromatography on Strep-Tactin Superflow (IBA). The eluted protein was treated with HRV 3C protease, and further purified by size-exclusion chromatography on Superdex 200 Increase 10/300 columns. All other proteins were expressed in Escherichia coli Rosetta2 (DE3) (Novagen) by inducing with 0.25 mM isopropyl β-D-thiogalactopyranoside (IPTG) at 18°C overnight. Cells were lysed by high-pressure homogenization using an Emulsiflex C3 (Avestin). The N-terminally His6-SUMO-tagged MDA5, RIG-I, and its variants were purified by chromatography on Ni-NTA agarose. The eluted proteins were incubated with Ulp1 protease at 4°C for 6 h to cleave the His6-SUMO-tag. The proteins were further purified by chromatography on HiTrap Heparin and Superdex 200 Increase 10/300 columns. All His6-GST-tagged proteins were purified by a combination of chromatography on Ni-NTA agarose and HiTrap Heparin/HiTrap Q (GE Healthcare) columns, followed by size-exclusion chromatography on Superdex 200 Increase 10/300. The CC-PSpry domains for TRIM14 (residues 59–442), TRIM25 (residues 189–630), TRIM26 (residues 134–539) and TRIM41 (residues 258–630) were expressed as a fusion protein with N-terminal His6-SUMO-tag, and purified by a combination of chromatography on Ni-NTA agarose, Resource Q/Resource S (GE Healthcare) and Superdex 200 Increase 10/300 columns. The CC domains (TRIM25: residues 189–362, TRIM26: residues 134–270 and TRIM41: residues 258–391) and the PSpry domains (TRIM25: residues 447–630, TRIM26: residues 271–539 and TRIM41: residues 392–630) were purified by a combination of chromatography on Ni-NTA agarose and Superdex 200 Increase 10/300 columns. TRIM65PSpry (residues 295–517) and its crystallization construct (312–504, C356L, C407S and C421S) were expressed as a fusion protein with N-terminal His6-maltose binding protein (MBP)-tag. The proteins were purified by Ni-NTA agarose and treated with HRV 3C protease to cleave the His6-MBP-tag. The cleaved proteins were further purified by Resource S and Superdex 75 Increase 10/300 (GE Healthcare) columns. For N-terminal fluorescent labeling of MDA5, TRIM65 or LGP2 and their variants, the proteins (0.5–2 mg/ml) were incubated with 100 μM peptide (LPETGG) conjugated with fluorescein (Anaspec) and 1 mg/ml S. aureus sortase A (a gift from H. Ploegh, MIT) (Antos et al., 2009) at RT for 4 h away from light, followed by size-exclusion chromatography on Superdex 200 Increase 10/300 to remove sortase A. All purified proteins were frozen in liquid nitrogen and stored at −80°C until further use.
Antibodies
The antibodies used for immunoblots were: Anti-FLAG-HRP (SIGMA A8592), Anti-HA (Cell Signaling 3724S), Anti-myc (Cell Signaling 2278S), Anti-TRIM65 (Invitrogen PA5–54459), Anti-RIG-I (Enzo ALX-210–932-C100), Anti-STING (Cell Signaling 13647S), Anti-Actin (Cell Signaling 8457L), Anti-MDA5 (Enzo ALX-210–935-C100), Anti-TRIM14 (Abcam ab185349, LS Bio C110434), Anti-TRIM25 (BD Biosciences 610570), Anti-TRIM26 (Santa Cruz sc-393832), Anti-TRIM41 (Abcam ab111580), Anti-Strep-tag (Invitrogen MA5–17283), Anti-mouse-IgG-HRP (GE Healthcare NA931V) and Anti-rabbit-IgG-HRP (Cell Signaling 7074P2).
RT-qPCR
HEK293T cells were grown in 48-well plates in Dulbecco’s modified Eagle medium (Cellgro) supplemented with 10% fetal calf serum. The cells were transfected with indicated amounts of plasmid (MDA5 WT = 5 ng; MDA5 G495R = 10 ng; TRIM65 = 5–50 ng; RIG-I WT = 10 ng; RIPLET = 10 ng; MAVS = 25 ng; STING V155M = 750 ng) at around 70%–80% confluency using Lipofectamine 2000 (Thermo Fisher) following manufacturer’s protocol. The plasmid dose for all point mutants were optimized using western blot analysis to match the wild-type expression levels. Similarly, for experiments requiring comparison of WT and KD cells, the plasmid dose was optimized to get similar expression levels across cell-lines. For experiments requiring RNA stimulation, the media was changed 6 h after the first transfection and the cells were then additionally transfected with 500 ng polyI:C (Invivogen) or 200 ng 42 bp dsRNA. The cells were lysed ~20 h after stimulation and the whole cell RNA was extracted with TRIzol reagent (Thermo Fisher) using Direct-zol RNA Miniprep kit (Zymoresearch) followed by cDNA synthesis using High Capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacture’s instruction. The cDNA was subjected to real-time PCR using a set of gene specific primers and SYBR Green Master Mix (Applied Biosystems) in the StepOne Real-Time PCR Systems (Applied Biosystems). The interferon signaling activity was quantified by measuring the increase in levels of IFNβ mRNA relative to the change in levels of 18S rRNA. Average values and standard deviations were calculated using Microsoft excel. Each datapoint was derived from 3–4 independent experiments and the P values were calculated using the two-tailed unpaired Student’s t test in Microsoft excel. The P values are denoted by ** (p < 0.005) and ***(p < 0.001).
Ubiquitination assay
Ubiquitination assay was performed as described previously (Cadena et al., 2019). Briefly, purified MDA5 or RIG-I (0.5 μM) was first incubated with 1012 bp or 112 bp dsRNA (2 ng/μl) respectively, in 20 mM HEPES pH 7.5, 50 mM NaCl, 1.5 mM MgCl2, 5 mM ATP and 2 mM DTT at RT for 30 min. The protein-RNA complex was then further mixed with 20 μM ubiquitin, 1 μM mE1, 5 μM Ubc13, 2.5 μM Uev1A and 0.25 μM TRIM65, and the mixture was incubated at 37°C for 30 min. The reaction was quenched with SDS loading buffer and analyzed on SDS-PAGE followed by anti-MDA5 or anti-RIG-I western blot.
Native gel-shift assay
For experiments requiring dsRNA, 112 bp dsRNA was used unless mentioned otherwise. For detection purpose, either Cy5-labeled dsRNA or fluorescein-labeled protein was used (as mentioned in the relevant experiments). For experiments where Cy5-labeled dsRNA was used, MDA5 (or other RLHs) and/or their variants at 0.25 μM were first incubated with dsRNA (1 ng/μl) for 30 min in 20 mM HEPES pH 7.5, 100 mM NaCl, 1.5 mM MgCl2 and 2 mM DTT at RT with or without 2 mM ATP. This was followed by the addition of the TRIM proteins (TRIM65, RIPLET, TRIM14, TRIM16, TRIM25 or TRIM47) and/or their variants (as mentioned in the experiment). The mixture was further incubated at RT for 10 min and the samples were then analyzed on Bis-Tris native PAGE (Life Technologies). For experiments where fluorescin-labeled TRIM65 CC-PSpry was used, GST-tagged MDA5 variants (1.6 μM) were directly incubated with fluorescin-labeled TRIM65 CC-PSpry (0.8 μM) for 15 min at RT before analyzing on native PAGE. For experiments where fluorescin-labeled MDA5 was used, fluorescin-labeled MDA5 or its variants (0.6 μM) was incubated with TRIM65 (0.3 μM) in the presence or absence of 112 bp dsRNA (8 ng/μl) for 30 min, and the mixture was analyzed on native PAGE. The gels were imaged using either Cy5 or fluorescin fluorescence in iBright FL1000 (Invitrogen).
Multi-Angle Light Scattering (MALS)
The molecular masses of TRIM65CC-PSpry and TRIM65PSpry were determined by MALS using a Superdex 200 Increase 10/300 column (GE Healthcare) attached to MiniDAWN detector (Wyatt Technology) in Phosphate Buffered Saline (PBS, Corning) buffer containing 2 mM DTT and the data were analyzed using ASTRA7.3.1 software (Wyatt Technology).
Pull-down assay
GST-tagged helicase proteins (200 nM) and TRIM proteins (200 nM) were incubated in PBS buffer containing 2 mM DTT at 4°C for 2 h, followed by the addition of Glutathione Magnetic Beads (Pierce). The samples were further incubated at 4°C for 2 h with gentle rotation, and the beads were washed with PBS containing 2 mM DTT three times. The bound proteins were eluted by adding the buffer containing 50 mM Tris-HCl pH 7.5, 300 mM NaCl, 2 mM DTT and 50 mM glutathione (Sigma-Aldrich). The eluted samples were boiled with SDS-loading buffer at 96°C for 3 min and analyzed by immunoblotting using anti-Strep-tag or anti-TRIM14.
Crystallization, X-ray data collection, processing and refinement
The purified human TRIM65PSpry (residues 312–504, C356L/C407S/C421S) protein was used for crystallization. Three solvent exposed cysteine residues (Cys356/Cys407/Cys421) were mutated to enhance crystallization. To further facilitate crystallization, the lysine residues of the protein were methylated, as previously described (Walter et al., 2006). In brief, the protein was incubated with the dimethylamine-borane (ABC) complex and formaldehyde in 50 mM HEPES pH 7.5, 150 mM NaCl at 4°C overnight. The methylated TRIM65PSpry protein was further purified by gel filtration chromatography on a Superdex 75 Increase column (GE Healthcare) and concentrated to 5 mg/ml using an Amicon Ultra-4 filter (10 kDa molecular-weight cutoff; Millipore). The methylated TRIM65PSpry was crystallized at 18°C by the hanging drop vapor diffusion method. Crystals were obtained by mixing 1 μL of protein solution (5 mg/ml TRIM65PSpry, 10 mM Tris-HCl pH 8.0, 150 mM NaCl and 3 mM 2-mercaptoethanol) and 1 μL reservoir solution (0.1 M Bicine, pH 9.0 and 20% PEG3,350). Crystals were cryoprotected in reservoir solution supplemented with 25% glycerol, and were flash-cooled in liquid nitrogen. The X-ray diffraction data was collected on 17-ID beamline at the NSLSII and processed using XDS (Kabsch, 2010). The structure was solved by molecular replacement with Phaser (McCoy et al., 2007). The search model was built by the Phyre 2 web server (Kelley et al., 2015), using the PSpry domain of TRIM25 (PDB ID 6FLN) as a template. The model was automatically built using Autobuild (Terwilliger et al., 2008), followed by manual model building using COOT (Emsley et al., 2010) and structural refinement using PHENIX (Liebschner et al., 2019). The structure was validated using MolProbity (Williams et al., 2018). Data collection and refinement statistics are summarized in Extended Data Table 1.
CryoEM sample preparation and data collection
TRIM65PSpry-MDA5ΔN (8.6 μM) was incubated with 1012 bp dsRNA (50 ng/μl) in 20 mM HEPES pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 2 mM DTT and 2 mM ADP:AlFx (ADP, AlCl3 and NaF in a molar ratio of 1:1:3) at 4°C for 2 h to form the protein-RNA filament. The filament sample was diluted 2-fold and 3.5 μL of the diluted sample was applied to a freshly glow-discharged C-flat 400 mesh copper grid (CF-1.2/1.3, Electron Microscopy Sciences) at 4°C at 100% humidity and plunged into liquid ethane after blotting for 3 s using a Vitrobot Mark IV (FEI). The grids were screened at Pacific Northwest Centre for Cryo-EM, OHSU using Talos Arctica microscope (FEI). The grids that showed a good sample distribution and ice thickness were subjected to data collection on a Titan Krios G3i microscope (Thermo Fisher 20 Scientific) operated at 300 kV and equipped with K3 summit direct electron detector (Gatan) at the Harvard Cryo-Electron Microscopy Center for Structural Biology (Harvard Medical School). A total of 12,248 micrographs were recorded from 2 independent sessions in counting mode at a magnification of 105,000x and magnified pixel size 0.85 Å using SerialEM software (Mastronarde, 2005). Each movie comprised of 50 frames at a dose rate of 35.604 e−/Å 2/s and an exposure time of 2.1 s resulting in a total dose of 74.8 e−/Å 2. The data was collected in a desired defocus range of −1.5 to −2.5 μm.
RIPLETPSpry-RIG-IΔN (9.6 μM) was incubated with 112 bp dsRNA (48 ng/μl) at 37°C for 5 min in 20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, 2 mM DTT and 1 mM ATP to form the protein-RNA filament. The reaction was quenched with 5 mM ADP:AlFx and applied to the EM-grid without dilution and blotted for 2 s. The data for RIPLETPSpry-RIG-IΔN was collected at National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research using Titan Krios G3i microscope operated at 300 kV equipped with K3 summit direct electron detector (Gatan) in counting mode at 105,000x magnification and 0.855 Å pixel size. A total of 18,309 micrographs were recorded from 2 independent sessions with each movie comprising of 40 frames at a dose rate of 19.424 e−/ Å 2 per s and an exposure time of 2.58 s, resulting in a total dose of 50.0 e−/ Å 2. The data was collected in a desired defocus range of −0.7 to −1.4 μm.
CryoEM data processing and structure refinement
All image processing was performed in RELION3.08 or RELION3.1 (Zivanov et al., 2018). The dose-fractionated movies were motioncorrected using MotionCor2 (Zheng et al., 2017). The contrast transfer function (CTF) was estimated with CTFFIND 4.1 (Rohou and Grigorieff, 2015). Particles were picked using the Autopick function in RELION (Zivanov et al., 2018). For TRIM65PSpry-MDA5ΔN, helical segments were extracted in a box size equal to 300 pixel with an inter-box distance of 46 Å. These particles were subject to several rounds of 2D classifications and selection in RELION 3.1 to get rid of badly aligned particles. The selected class averages were used for 3D refinement with helical reconstruction using a featureless cylinder as the starting reference map. The resulting 3D map included three monomers of TRIM65PSpry-MDA5ΔN helically arranged on the central dsRNA duplex. The resulting 3D model and particle set were subjected to per-particle defocus refinement, beam-tilt refinement, Bayesian polishing (Zivanov et al., 2019) and 3D classification. The selected class containing 143,063 particles were subjected to another round of per-particle defocus refinement and Bayesian polishing. To deal with structural heterogeneity of TRIM65PSpry, no-align 3D classification was performed using a mask covering the central monomer of TRIM65PSpry-MDA5ΔN-dsRNA duplex. The best class contained 49,051 particles and the resulting 3D map was subjected to 3D refinement and postprocessing, yielding a map with a global resolution of 4.3 A, according to the Fourier shell correlation (FSC) = 0.143 criterion (Rosenthal and Henderson, 2003). For filamentous TRIM65PSpry-MDA5ΔN, no-align 3D classifications were performed for all three monomers of TRIM65PSpry-MDA5ΔN-dsRNA duplex. The resulting 15,306 particles yielded a map with a global resolution of 4.3 A after 3D refinement with helical reconstruction followed by imposing helical symmetry on the unfiltered half-maps.
For RIPLETPSpry-RIG-IΔN, helical segments were extracted in a box size equal to 296 pixel with an inter-box distance of 46 Å. After several rounds of 2D c lassifications, the particles were subjected to 3D refinement, per-particle defocus refinement, beam-tilt refinement, Bayesian polishing, and 3D classification. The selected classes containing 283,110 particles were further classified by no-align 3D classification using a mask covering the central monomer of RIPLETPSpry-RIG-IΔN-dsRNA duplex, yielding 114,824 particles. These particles were subjected to 3D refinement, per-particle defocus refinement, beam-tilt refinement, Bayesian polishing, and no-align 3D classification. The best class contained 74,079 particles and the resulting 3D model was subjected to 3D refinement and postprocessing, yielding a map with a global resolution of 3.9 A. For filamentous RIPLETPSpry-RIG-IΔN, no-align 3D classification was performed for all three monomers of RIPLETPSpry-RIG-IΔN-dsRNA duplex. The resulting 39,718 particles yielded a map with a global resolution of 4.2 A after 3D refinement with helical reconstruction followed by imposing helical symmetry on the unfiltered half-maps. The local resolution was estimated by RELION. The data processing scheme is summarized in Extended Data Figures 2 and 3. For TRIM65PSpry-MDA5ΔN-dsRNA duplex, the crystal structure of MDA5 complexed with 12 bp dsRNA (PDB ID: 4GL2) and TRIM65PSpry were used as starting atomic model. These models were docked as a rigid body into the postprocessed EM density map in UCSF Chimera (Pettersen et al., 2004), and built manually against the density map using COOT (Emsley et al., 2010). The model was refined using phenix.real_space_refine (Liebschner et al., 2019), with the restrains for side chain rotamer, Ramachandran, secondary structure and base pair/stacking. For the filamentous TRIM65PSpry-MDA5ΔN-dsRNA duplex, the non-crystallographic symmetry (NCS) restraints were applied between each monomer of TRIM65PSpry-MDA5ΔN in real space refinement. To generate the filamentous trimer model, 3 repeats of the 14 bp dsRNA were used since there was no information about the RNA sequence in the maps due to helical symmetry averaging.
For RIPLETPSpry-RIG-IΔN-dsRNA duplex, the initial model of RIPLETPSpry was built by the Phyre 2 server (Kelley et al., 2015), using the PSpry domain of TRIM25 (PDB ID: 6FLN) as a template. The modeled RIPLETPSpry and the crystal structure of RIG-I complexed with 14 bp dsRNA (PDB ID: 5E3H) were docked into the postprocessed EM density map. The model building and structural refinement were performed as for TRIM65PSpry-MDA5ΔN-dsRNA duplex. The structure validation was performed using MolProbity (Williams et al., 2018) from the PHENIX package. The curve representing model versus full map was calculated, based on the final model and the full map. The statistics of the 3D reconstruction and model refinement are summarized in Extended Data Table 2. All molecular graphics figures were prepared with CueMol (http://www.cuemol.org) and UCSF Chimera (Pettersen et al., 2004).
Negative-stain electron microscopy
The samples of filamentous TRIM65Spry-MDA5ΔN and RIPLETPSpry-RIG-IΔN were prepared as described above. The samples were diluted 10-fold with 20 mM HEPES pH 7.5, 150 mM NaCl, 1.5 mM MgCl2 and 2 mM DTT, and was then immediately adsorbed to freshly glow-discharged carbon-coated grids (Ted Pella) and stained with 0.75% uranyl formate as described (Ohi et al., 2004). Images were collected using a JEM-1400 transmission electron microscope (JEOL) at 50,000x magnification.
Phylogenetic analysis
Sequence collection and phylogenetic analysis were performed iteratively, using phylogenies to select additional species to add to the analysis, and/or additional query sequences to diversify the range of blast searches. To identify sequences, we used a combination of HMMER (v.3.1b2; http://hmmer.org/) and BLASTP (Altschul et al., 1997) searches. BLASTP searches used diverse queries across the trees shown, against either local databases representing the downloaded proteomes of selected species, or against NCBI’s non-redundant (NR) database, using the ‘entrez_query’ option to focus output on species groups of interest. HMMER searches used the hmmsearch algorithm and PFAM’s Hidden Markov Models (HMMs) for the SPRY (PF00622) and PRY (PF13765) domains, as well as HMMs we generated using hmmbuild from more focused alignments of only the TRIM proteins of interest. The target databases for HMMER searches were downloaded proteomes of species of interest, obtained from NCBI, UCSC, Ensembl, FlyBase, SGD, and Wormbase.
For phylogenies, sequences were aligned using MAFFT (Katoh and Standley, 2013) with the ‘–leavegappyregion’ option. Alignments were manually trimmed to retain only well-aligned regions, poorly aligning sequences were removed, and the prottest (Darriba et al., 2011) algorithm was used to select an amino acid substitution model, which in both cases was the WAG model. Alignments were then used to generate a maximum likelihood phylogeny with the phyml (Guindon et al., 2010) algorithm, using empirical amino acid frequencies, and 100 bootstrap replicates to estimate the confidence of each node.
QUANTIFICATION AND STATISTICAL ANALYSIS
Average values and standard deviations were calculated using Microsoft excel. The values for n represent biological replicates for cellular experiments or individual samples for biochemical assays. For each figure, the number of replicates is indicated in the figure legends. Unless otherwise mentioned, cell culture assays were performed in 3–4 independent experiments. P values were calculated using the two-tailed unpaired Student’s t test. We consider values * p < 0.05, ** p < 0.01, ***p < 0.001 significant.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-MDA-5 antibody (D74E4) | Cell Signaling Technology | Cat# 5321; RRID: AB_10694490 |
| Rabbit anti-MDA5 antibody (AT113) | Enzo Life Sciences | Cat# ALX-210-935-C100; RRID: AB_2264475 |
| Rabbit anti-RIG-I antibody (AT111) | Enzo Life Sciences | Cat# ALX-210-932-C100; RRID: AB_2253427 |
| Rabbit anti-TRIM14 antibody (ab185349) | abcam | Cat# ab185349 |
| Mouse anti-TRIM25 antibody | BD Biosciences | Cat# 610570; RRID: AB_397919 |
| Mouse TRIM26 antibody | Santa Cruz | Cat# sc-393832 |
| Rabbit anti-TRIM41 antibody (ab111580) | abcam | Cat# ab111580; RRID: AB_10866520 |
| Mouse anti-Strep Tag antibody (GT661) | Invitrogen | Cat# MA5-17283; RRID: AB_2538749 |
| Anti-rabbit IgG-HRP | Cell Signaling Technology | Cat# 7074; RRID: AB_2099233 |
| Anti-mouse IgG-HRP | GE Healthcare | Cat# NA931V; RRID: AB_772210 |
| Anti-FLAG-HRP | Sigma-Aldrich | Cat# A8592 |
| Rabbit anti-HA tag | Cell Signaling Technology | Cat# 3724S |
| Rabbit anti-myc tag | Cell Signaling Technology | Cat# 2278S |
| Rabbit anti-TRIM65 | Invitrogen | Cat# PA5-54459 |
| Rabbit anti-STING | Cell Signaling Technology | Cat# 13647S |
| Rabbit anti-actin | Cell Signaling Technology | Cat# 8457L |
| Chemicals, peptides, and recombinant proteins | ||
| In-Fusion HD | Takara | Cat# 638909 |
| Polyethylenimine | Polyscience | Cat# 23966-1 |
| mammalian protease inhibitor cocktail | G-Biosciences | Cat# 786-433 |
| anti-FLAG M2 agarose beads | Sigma-Aldrich | Cat# A2220 |
| 3X FLAG peptide | Sigma-Aldrich | Cat# F4799 |
| Zeba desalting column, 40 kDa molecular-weight cutoff | Thermo Fisher Scientific | Cat# 87766 |
| HyClone SFX Insect cell culture media | GE Healthcare | Cat# SH30278.02 |
| Ni-NTA agarose | QIAGEN | Cat# 30250 |
| HRV 3C protease | Homemade | N/A |
| HiTrap Heparin | GE Healthcare | Cat# 17040601 |
| Superdex 200 Increase 10/300 | GE Healthcare | Cat# 28990944 |
| Strep-Tactin Superflow | IBA | Cat# 2-1206-025 |
| Ulp1 protease | Homemade | N/A |
| HiTrap Q | GE Healthcare | Cat# 17505301 |
| Resource Q | GE Healthcare | Cat# 17117701 |
| Resource S | GE Healthcare | Cat# 17117801 |
| Superdex 75 Increase 10/300 | GE Healthcare | Cat# 29148721 |
| Recombinant mouse E1 | Cadena et al., 2019 | N/A |
| Recombinant human Ubc13 | Cadena et al., 2019 | N/A |
| Recombinant human Uev1A | Cadena et al., 2019 | N/A |
| Recombinant human Ubiquitin (WT/K63R/K48R) | Cadena et al., 2019 | N/A |
| Cy5-labeled dsRNA | This paper | N/A |
| Calf intestinal alkaline phosphatase | New England BioLabs | Cat# M0290 |
| Bis-Tris native PAGE | Life Technologies | BN1003BOX |
| Glutathione Magnetic Beads | Pierce | 88821 |
| glutathione | Sigma-Aldrich | G4251 |
| C-flat Holey Carbon 400 mesh copper grid (CF-1.2/1.3) | Electron Microscopy Sciences | CF413-50 |
| Carbon Support Film 400 mesh copper grids | Electron Microscopy Sciences | CF400H-CU |
| peptide (LPETGG) conjugated with fluorescein | Anaspec | Custom synthesized |
| Lipofectamine 2000 | Thermo Fisher | Cat# 11668027 |
| polyIC | Invivogen | Cat# tlrl-pic |
| TRIzol reagent | Thermo Fisher | Cat# 15596026 |
| Direct-zol RNA Miniprep Kits | Zymo Research | Cat# R2050 |
| High Capacity cDNA reverse transcription kit | Applied Biosystems | Cat# 4368814 |
| SYBR Green Master Mix | Applied Biosystems | Cat# 4309155 |
| DMEM, high glucose, pyruvate | Cellgro | Cat# 10-013-CV |
| Mammalian protease arrest | GBiosciences | Cat# 786-433 |
| Critical commercial assays | ||
| Bac-to-Bac baculovirus expression systems | Invitrogen | Cat# A11098 |
| Deposited data | ||
| TRIM65PSpry-MDA5-dsRNA Filament coordinates | This study | PDB: 7JL2 |
| TRIM65PSpry-MDA5-dsRNA Filament EM map | This study | EMDB: 22370 |
| TRIM65PSpry-MDA5-dsRNA Monomer coordinates | This study | PDB: 7JL0 |
| TRIM65PSpry-MDA5-dsRNA Monomer EM map | This study | EMDB: 22368 |
| RIPLETPSpry-RIG-I-dsRNA Filament coordinates | This study | PDB: 7JL3 |
| RIPLETPSpry-RIG-I-dsRNA Filament EM map | This study | EMDB: 22371 |
| RIPLETPSpry-RIG-I-dsRNA Monomer coordinates | This study | PDB: 7JL1 |
| RIPLETPSpry-RIG-I-dsRNA Monomer EM map | This study | EMDB: 22369 |
| TRIM65PSpry coordinates and map | This study | PDB: 7JL4 |
| Experimental models: cell lines | ||
| HEK293T (WT and TRIM65 KD) | This paper | N/A |
| HEK293T (Cas9-stable) | Laboratory of Dr. George Church | Chavez et al., 2015 |
| HEK293T (WT, RIG-I KO, and RIPLET KO) | Laboratory of Dr. Fajian Hou | Shi et al., 2017 |
| Experimental models: organisms/strains | ||
| E. coli Rosetta 2(DE3) | Novagen | Cat# 71397-3 |
| Oligonucleotides | ||
| TRIM65-specific guide RNA (Forward: TGCCGAGCGCCTCAAGCGCG) | This paper | N/A |
| TRIM65-specific guide RNA (Reverse: CGCGCTTGAGGCGCTCGGCA) | This paper | N/A |
| Human IFN-β qPCR primer (Forward: AAACTCA TGAGCAGTCTGCA) | This paper | N/A |
| Human IFN-β qPCR primer (Reverse: AGGAGA TCTTCAGTTTCGGAGG) | This paper | N/A |
| Human 18S qPCR primer (Forward: CGGCGACGACCCATTCGAAC) | This paper | N/A |
| Human 18S qPCR primer (Reverse: GAATCGAACCCTGATTCCCCGTC) | This paper | N/A |
| Recombinant DNA | ||
| pFastBac1 | Invitrogen | Cat# 10360014 |
| pFastBac1-LGP2 | This study | N/A |
| pFastBac1-TRIM65CC-PSpry | This study | N/A |
| pE-SUMO Kan vector | LifeSensors | Cat# 1001K |
| pE-SUMO-MDA5 | This study | N/A |
| pE-SUMO-MDA5ΔN | This study | N/A |
| pE-SUMO-RIG-IΔN | This study | N/A |
| pE-SUMO-TRIM65PSpry-MDA5ΔN | This study | N/A |
| pE-SUMO-MDA5ΔN-TRIM65PSpry | This study | N/A |
| pE-SUMO-RIPLETPSpry-RIG-IΔN | This study | N/A |
| pE-SUMO-TRIM14CC-PSpry | This study | N/A |
| pE-SUMO-N-His6-Strep-TRIM25 CC | This study | N/A |
| pE-SUMO-N-His6-Strep-TRIM25 PSpry | This study | N/A |
| pE-SUMO-N-His6-Strep-TRIM25 CC-PSpry | This study | N/A |
| pE-SUMO-N-His6-Strep-TRIM26 PSpry | This study | N/A |
| pE-SUMO-N-His6-Strep-TRIM26 CC-PSpry | This study | N/A |
| pE-SUMO-N-His6-Strep-TRIM41 CC | This study | N/A |
| pE-SUMO-N-His6-Strep-TRIM41 PSpry | This study | N/A |
| pE-SUMO-N-His6-Strep-TRIM41 CC-PSpry | This study | N/A |
| pGEX-6P-1 | GE healthcare | Cat# 28954648 |
| pGEX-N-His6-MDA5 helicase | This study | N/A |
| pGEX-N-His6-MDA5 α1 | This study | N/A |
| pGEX-N-His6-MDA5 Hel2i2 | This study | N/A |
| pGEX-N-His6-LGP2 Hel2i2 | This study | N/A |
| pGEX-N-His6-Dicer Hel2i2 | This study | N/A |
| pGEX-N-His6-TRIM65PSPry | This study | N/A |
| pGEX-N-His6-RIPLETPSPry | This study | N/A |
| pGEX-N-His6-TRIM14PSPry | This study | N/A |
| pGEX-N-His6-TRIM16PSPry | This study | N/A |
| pGEX-N-His6-TRIM47PSPry | This study | N/A |
| pGEX-N-His6-DDX41Hel12 | This study | N/A |
| pGEX-N-His6-DDX41Hel1 | This study | N/A |
| pGEX-N-His6-DDX41Hel2 | This study | N/A |
| pMal-c2x | NEB | N/A |
| pMal-N- His6-TRIM65PSpry | This study | N/A |
| pcDNA4-STINGV155M | Laboratory of Dr. Philip J. Kranzusch | N/A |
| Software and algorithms | ||
| XDS | Kabsch, 2010 | http://xds.mpimf-heidelberg.mpg.de |
| Phyre 2 | Kelley et al., 2015 | http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index |
| PHENIX | Liebschner et al., 2019 | https://www.phenix-online.org |
| COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| RELION (version 3.08/3.1) | Zivanov et al., 2018 | https://www3.mrc-lmb.cam.ac.uk/relion//index.php?title=Main_Page |
| MotionCor2 | Zheng et al., 2017 | https://emcore.ucsf.edu/ucsf-software |
| CTFFIND 4.1 | Rohou and Grigorieff, 2015 | https://grigoriefflab.umassmed.edu/ctffind4 |
| CueMol | N/A | http://www.cuemol.org/ja/ |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
Highlights.
TRIM65 selectively recognizes MDA5 filaments using PSpry bivalency
TRIM65 PSpry binds α1/α3 helices in the Hel2 domain of MDA5
RIPLET recognizes RIG-I using a similar epitope in the helicase domain
Distinct TRIM proteins recognize common epitopes in diverse helicases using bivalency
ACKNOWLEDGMENTS
We acknowledge Dr. Adam Frost (UCSF) for the guidance with cryo-EM analysis. K.K. was supported by an Uehara research fellowship and a Cancer Research Institute postdoctoral fellowship. Z.Z. was supported by the China Scholarship Council (2018032500003). J.M.Y. and H.S.M. are supported by grants from the Mathers Foundation and NIAID (HARC [HIV Accessory and Regulatory Complexes] Center, P50 AI082250, PI Nevan Krogan, subaward to H.S.M.) and an HHMI investigator award. S.H. acknowledges NIH grants R01AI154653, R01AI111784, and DP1AI152074). The cryo-EM grids were screened at PNCC at OHSU (U24GM129547), and cryo-EM data were collected at NCI-NCEF at the Frederick National Laboratory for Cancer Research (under contract HSSN261200800001E) and The Harvard Cryo-EM Center for Structural Biology at Harvard Medical School.
Footnotes
DECLARATION OF INTERESTS
H.S.M. is a member of the SAB for the Burroughs Wellcome Pathogenesis Award, Max Planck Institute for Evolutionary Biology, Institute for Molecular Biology-Academia Sinica, and Whitehead Institute. S.H. is a member of the SAB for IFM Therapeutics and Silicon Therapeutics.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.molcel.2020.11.047.
SUPPORTING CITATIONS
The following references appear in the supplemental information: Kumar et al. (2017); Metzger et al. (2014); Zou et al. (2009)
REFERENCES
- Aguado LC, Schmid S, May J, Sabin LR, Panis M, Blanco-Melo D, Shim JV, Sachs D, Cherry S, Simon AE, et al. (2017). RNase III nucleases from diverse kingdoms serve as antiviral effectors. Nature 547, 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, Zhang CZ, and Hur S (2018). Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation. Cell 172, 797–810.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antos JM, Chew GL, Guimaraes CP, Yoder NC, Grotenbreg GM, Popp MW, and Ploegh HL (2009). Site-specific N- and C-terminal labeling of a single polypeptide using sortases of different specificity. J. Am. Chem. Soc. 131, 10800–10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, and Horvath CM (2015). LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling. Cytokine 74, 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena C, Ahmad S, Xavier A, Willemsen J, Park S, Park JW, Oh SW, Fujita T, Hou F, Binder M, et al. (2019). Ubiquitin-Dependent and -Independent Roles of E3 Ligase RIPLET in Innate Immunity. Cell 177, 1187–1200.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. (2015). Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz AA, Babon JJ, Norton RS, Nicola NA, and Nicholson SE (2013). Structure and function of the SPRY/B30.2 domain proteins involved in innate immunity. Protein Sci. 22, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, and Posada D (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito D, Koliopoulos MG, and Rittinger K (2017). Structural determinants of TRIM protein function. Biochem. Soc. Trans. 45, 183–191. [DOI] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, and Jung JU (2007). TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, and Gascuel O (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Hayman TJ, Hsu AC, Kolesnik TB, Dagley LF, Willemsen J, Tate MD, Baker PJ, Kershaw NJ, Kedzierski L, Webb AI, et al. (2019). RIPLET, and not TRIM25, is required for endogenous RIG-I-dependent antiviral responses. Immunol. Cell Biol. 97, 840–852. [DOI] [PubMed] [Google Scholar]
- Hur S (2019). Double-Stranded RNA Sensors and Modulators in Innate Immunity. Annu. Rev. Immunol. 37, 349–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LC, Keeble AH, Khan Z, Rhodes DA, and Trowsdale J (2007). Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl. Acad. Sci. USA 104, 6200–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang G-Q, Gale M Jr., Patel SS, and Marcotrigiano J (2011). Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, and Chen ZJ (2012). Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36, 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Zhu Y, Liu ZJ, and Ouyang S (2017). The emerging roles of the DDX41 protein in immunity and diseases. Protein Cell 8, 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamanova J, Sun H, Lara-Tejero M, and Galán JE (2016). The Salmonella Effector Protein SopA Modulates Innate Immune Responses by Targeting TRIM E3 Ligase Family Members. PLoS Pathog. 12, e1005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, and Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJ (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, and Siomi MC (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139. [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, and Cusack S (2011). Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147, 423–435. [DOI] [PubMed] [Google Scholar]
- Krause CD, and Pestka S (2005). Evolution of the Class 2 cytokines and receptors, and discovery of new friends and relatives. Pharmacol. Ther. 106, 299–346. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, and Hedges SB (2017). TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 34, 1812–1819. [DOI] [PubMed] [Google Scholar]
- Lang X, Tang T, Jin T, Ding C, Zhou R, and Jiang W (2017). TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J. Exp. Med. 214, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, and Pyle AM (2011). Structural insights into RNA recognition by RIG-I. Cell 147, 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Kohlway A, and Pyle AM (2013). Duplex RNA activated ATPases (DRAs): platforms for RNA sensing, signaling and processing. RNA Biol. 10, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, van der Veen AG, Poirier EZ, and Reis e Sousa C (2019). Slicing and dicing viruses: antiviral RNA interference in mammals. EMBO J. 38, e100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Pruneda JN, Klevit RE, and Weissman AM (2014). RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, Conzelmann KK, and Hopfner KP (2013). Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science 339, 690–693. [DOI] [PubMed] [Google Scholar]
- Mu X, Greenwald E, Ahmad S, and Hur S (2018). An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 46, 5239–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, and Walz T (2004). Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol. Proced. Online 6, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura H, Oikawa D, Nakane T, Kato M, Ishii R, Ishitani R, Tokunaga F, and Nureki O (2016). Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide. Sci. Rep. 6, 34756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H., Miyashita M., Matsumoto M., and Seya T. (2013). A distinct role of Riplet-mediated K63-Linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 9, e1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, and Morse HC 3rd (2008). TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, et al. (2012). The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 13, 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, and Hur S (2011). Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc. Natl. Acad. Sci. USA 108, 21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Yao H, Walz T, and Hur S (2013). RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol. Cell 51, 573–583. [DOI] [PubMed] [Google Scholar]
- Peisley A, Wu B, Xu H, Chen ZJ, and Hur S (2014). Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfetto L, Gherardini PF, Davey NE, Diella F, Helmer-Citterich M, and Cesareni G (2013). Exploring the diversity of SPRY/B30.2-mediated interactions. Trends Biochem. Sci. 38, 38–46. [DOI] [PubMed] [Google Scholar]
- Peters D, Radine C, Reese A, Budach W, Sohn D, and Jänicke RU (2017). The DEAD-box RNA helicase DDX41 is a novel repressor of p21WAF1/CIP1 mRNA translation. J. Biol. Chem. 292, 8331–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Polprasert C, Schulze I, Sekeres MA, Makishima H, Przychodzen B, Hosono N, Singh J, Padgett RA, Gu X, Phillips JG, et al. (2015). Inherited and Somatic Defects in DDX41 in Myeloid Neoplasms. Cancer Cell 27, 658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quynh NT, Hikima J, Kim YR, Fagutao FF, Kim MS, Aoki T, and Jung TS (2015). The cytosolic sensor, DDX41, activates antiviral and inflammatory immunity in response to stimulation with double-stranded DNA adherent cells of the olive flounder, Paralichthys olivaceus. Fish Shellfish Immunol. 44, 576–583. [DOI] [PubMed] [Google Scholar]
- Rice GI, Del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW, et al. (2014). Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 46, 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, and Henderson R (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Desalle R, and Fisher PB (2008). Evolution of MDA-5/RIG-I-dependent innate immunity: independent evolution by domain grafting. Proc. Natl. Acad. Sci. USA 105, 17040–17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, and Takeuchi O (2010). LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. USA 107, 1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yuan B, Zhu W, Zhang R, Li L, Hao X, Chen S, and Hou F (2017). Ube2D3 and Ube2N are essential for RIG-I-mediated MAVS aggregation in antiviral innate immunity. Nat. Commun. 8, 15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, and Wigley DB (2007). Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50. [DOI] [PubMed] [Google Scholar]
- Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, McMillan A, Diamond MS, Clark EA, Bevan MJ, and Gale M Jr. (2012). The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity 37, 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P, He L, Cui J, Qian C, Cao X, Lin M, Zhu Q, Li Y, Xing C, Yu X, et al. (2017). Assembly of the WHIP-TRIM14-PPP6C Mitochondrial Complex Promotes RIG-I-Mediated Antiviral Signaling. Mol. Cell 68, 293–307.e5. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, and Adams PD (2008). Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa E, Lethier M, Malet H, Brunel J, Gerlier D, and Cusack S (2016). Structural Analysis of dsRNA Binding to Anti-viral Pattern Recognition Receptors LGP2 and MDA5. Mol. Cell 62, 586–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, and Barber GN (2007). Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178, 6444–6455. [DOI] [PubMed] [Google Scholar]
- Versteeg GA, Benke S, García-Sastre A, and Rajsbaum R (2014). InTRIMsic immunity: Positive and negative regulation of immune signaling by tripartite motif proteins. Cytokine Growth Factor Rev. 25, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter TS, Meier C, Assenberg R, Au KF, Ren J, Verma A, Nettleship JE, Owens RJ, Stuart DI, and Grimes JM (2006). Lysine methylation as a routine rescue strategy for protein crystallization. Structure 14, 1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JS, Suh HY, Park SY, and Oh BH (2006). Structural basis for protein recognition by B30.2/SPRY domains. Mol. Cell 24, 967–976. [DOI] [PubMed] [Google Scholar]
- Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, and Hur S (2013). Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152, 276–289. [DOI] [PubMed] [Google Scholar]
- Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, and Hur S (2014). Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol. Cell 55, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, and Fujita T (2010). Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 20, 4–22. [DOI] [PubMed] [Google Scholar]
- Yu Q, Qu K, and Modis Y (2018). Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis. Mol. Cell 72, 999–1012.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, and Chen ZJ (2010). Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141, 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, and Liu YJ (2011). The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, and Gao C (2019). E3 ubiquitin ligases, the powerful modulator of innate antiviral immunity. Cell. Immunol. 340, 103915. [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Jia X, Xue Q, Dou Z, Ma Y, Zhao Z, Jiang Z, He B, Jin Q, and Wang J (2014). TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I-like receptor-mediated innate immune response. Proc. Natl. Acad. Sci. USA 111, E245–E254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, and Scheres SHW (2019). A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Chang M, Nie P, and Secombes CJ (2009). Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol. Biol. 9, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps included in this study have been deposited in the Electron Microscopy Data Bank with accession codes EMDB: EMD-22368, EMD-22369, EMD-22370 and EMD-22371. The atomic coordinates have been deposited in the Protein Data Bank with accession codes PDB: 7JL0, 7JL1, 7JL2, 7JL3, and 7JL4.