Abstract
Background
Circular RNAs (circRNAs) are involved in various diseases and serve as biomarkers. The present study aimed to investigate unique expression profiles of circRNAs in colon tissues of Crohn's disease (CD) and search novel biomarkers for the diagnosis.
Methods
Differentially expressed (DE) circRNAs in biopsies from four CD patients, four ulcerative colitis (UC) patients, and four healthy controls (HC) were screened by microarray. Hsa_circ_0062142 and hsa_circ_0001666 were verified in another expanded validation cohort. Bioinformatics analysis was applied to predict the function of two DE circRNAs. Receiver operating characteristic (ROC) curves were constructed to evaluate the diagnostic value of CD.
Results
The top 10 upregulated circRNAs in CD compared with HC were hsa_circ_0000691, hsa_circ_0001666, hsa_circ_0004183, hsa_circ_0009024, hsa_circ RNA_405324, hsa_circ_0002003, hsa_circ_0085323, hsa_circ_0040994, hsa_circ_0062142, and hsa_circ_0048148; the top 10 downregulated circRNAs were hsa_circ_0049356, hsa_circ RNA_405443, hsa_circ RNA_403556, hsa_circ_0092328, hsa_circ_0003979, hsa_circ_0074491, hsa_circ_0023461, hsa_circ RNA_406237, hsa_circ_0034044, and hsa_circ RNA_400564 (fold change in descending order). The expression levels of hsa_circ_0001666 and hsa_circ_0062142 in CD were significantly higher than those in UC and HC (p < 0.01). ROC curves suggested the favorable diagnostic value of hsa_circ_0062142 and hsa_circ_0001666 (AUC = 0.803 and 0.858, respectively, p < 0.01). In silico analysis indicated that these circRNAs may be involved in the progress of CD.
Conclusion
Hsa_circ_0062142 and hsa_circ_0001666 may play critical roles in the pathogenesis and serve as potential biomarkers of CD.
Keywords: bioinformatics analysis, circular RNAs, Crohn's disease, microarray
We explored the comprehensive circRNA expression profiles in colon tissues of Crohn's disease (CD) compared to tissues of ulcerative colitis (UC) and healthy controls. We showed that two circRNAs (hsa_circ_0062142 and hsa_circ_0001666) were significantly upregulated in colon tissues of CD than those in UC and healthy controls, presenting AUC of 0.803 and 0.858, respectively. Bioinformatics analysis indicated that the two differentially expressed circRNAs might be involved in the progression of CD. Our findings suggested that circRNAs might play a crucial role in the pathogenesis of CD and might provide potential diagnostic biomarkers and therapeutic targets for CD.

1. INTRODUCTION
Crohn's disease (CD) and ulcerative colitis (UC) are two major subtypes of inflammatory bowel disease (IBD). CD is characterized by transmural inflammation that affects any segments of gastrointestinal tract. More than 50% of the patients with CD also present complications of stricturing and fistulas within 10 years, followed by significant morbidity and disability. The incidence and prevalence of CD have been increasing in Asia 1 , 2 ; however, the etiology of CD is not yet understood. It is widely accepted that multiple factors involving gene, environment, microbe, and the mucosal immune system interact in a complex mechanism. Although CD and UC possess different clinical, radiographical, endoscopic, genetic, histological and immunological characteristics, occasionally, clinical symptoms, and histological features are overlapping. Currently, common biomarkers in serum and feces, such as the anti‐Saccharomyces cerevisiae antibody (ASCA), antineutrophil cytoplasmic antibody (ANCA), and fecal calprotectin, have limited value in differentiating CD from UC. 3 Especially, when the inflammation is localized to the colon, differential diagnosis is challenging. It is estimated that about 10% of these colonic IBD patients are diagnosed with IBD unclassified (IBDU). 4 Thus, novel efficient diagnostic biomarkers for CD are required for accurate diagnosis, appreciate treatment, and gain new insights into the mechanisms.
Circular RNAs (circRNAs), a novel class of noncoding RNAs, are featured with a covalently closed loop that lacks either 5′–3′ polarity or poly‐adenylated tail. CircRNAs used to be regarded as the nonfunctional byproducts of mRNA splicing. However, with the development of RNA sequencing technology, circRNAs occur ubiquitously and show spatial and temporal specific expression. 5 , 6 , 7 Recent studies have shown that circRNAs regulate gene expression at the transcriptional or post‐transcriptional level by functioning as microRNA sponges, interacting with small nuclear RNA (snRNA) or RNA polymerase II in the nucleus, and binding to transcription factors. 8 , 9 Emerging evidence demonstrated that circRNAs were involved in various diseases including cancer, 10 , 11 diabetes, 12 Alzheimer's disease, 13 and atherosclerosis. 14 Moreover, circRNAs were reported to be used as diagnostic or prognostic biomarkers based on specific expression profiles and high biological stability. 15 , 16 , 17 , 18 , 19 , 20 Nevertheless, the expression profiles and potential roles of circRNAs in CD are limited.
In present study, we investigated the expression profiles of circRNAs in colon tissues of CD by microarray. Two differentially expressed circRNAs were verified by quantitative real‐time polymerase chain reaction (qRT‐PCR); also, their diagnostic value as potential biomarkers for CD was evaluated. Furthermore, the potential function of the two selected circRNAs in CD was predicted by bioinformatics analysis.
2. MATERIALS AND METHODS
2.1. Tissue samples
Colonic biopsies were obtained from CD patients, UC patients, and healthy controls (HC) undergoing colorectal cancer screening at Jinling Hospital and Drum Tower Hospital from February 2016 to March 2021. The diagnosis of CD or UC was confirmed according to clinical, endoscopic, and histological criteria. All patients were recently diagnosed without treatment. Demographic and clinical characteristics of the study cohort were shown in Table 1. There was no significant difference in age among three groups. In total, 94 pinch tissues from 34 CD patients, 24 UC patients, and 36 healthy controls were obtained via endoscopic pinch biopsies. Among them, 12 samples including four CD patients, four UC patients, and four healthy controls were conducted with microarray analysis. The remaining samples were used for validation with qRT‐PCR and for receiver operating characteristic (ROC) curve analysis. The pinch tissues were immediately submerged in RNAlater (Sigma) at 2–8°C overnight, then transferred to –80°C until use for microarray analysis and qRT‐PCR. The study protocol was approved by the Human Ethics Committees of Jinling Hospital and Drum Tower Hospital. The written informed consent was obtained from every participant prior to sample collection.
TABLE 1.
Clinical characteristics of patients
| Microarray cohort | Validation cohort | |||||
|---|---|---|---|---|---|---|
| CD | UC | HC | CD | UC | HC | |
| n | 4 | 4 | 4 | 30 | 20 | 32 |
| Sex, n (M/F) | 2/2 | 3/1 | 2/2 | 17/13 | 9/11 | 21/11 |
| Age (years) | 39.25 ± 3.88 | 47.18 ± 3.81 | 46.00 ± 2.74 | 36.27 ± 12.55 | 42.20 ± 10.68 | 42.63 ± 7.01 |
Abbreviations: CD, Crohn´s disease; HC, healthy control; UC, ulcerative colitis.
2.2. RNA extraction and quality control
Total RNAs were isolated from pinch tissues using TRIzol reagent (Invitrogen Life Technologies) according to the standard protocol. The yield and purity were determined with a NanoDrop ND‐1000 (Agilent), and the integrity of RNAs was checked by 1% formaldehyde denaturing agarose gel electrophoresis.
2.3. Microarray hybridization
Microarray hybridization was performed using a Human 8 × 15 K circRNA Array based on the Arraystar's standard protocols (Arraystar Inc.). 20 Briefly, after removing linear RNAs by digested with RNAse R (Epicentre, Inc.), circRNAs in the sample were enriched. Next, the enriched circRNAs were amplified and transcribed into Cy3‐labeled cRNA with random primers and an Arraystar's Super RNA labeling system (Arraystar Super RNA Labeling Kit, Arraystar). The labeled‐cRNAs were then purified and fragmented. Subsequently, the Arraystar Human 8 × 15 K circRNA Array slides were hybridized with the labeled‐cRNAs and incubated for 17 h at 65°C in an Agilent Hybridization Oven (Agilent Technologies). Finally, the arrays were washed, fixed, and scanned with the Agilent Scanner G2505C (Agilent Technologies, Inc.). Scanned array images and data extraction were processed with Agilent Feature Extraction software (version 11.0.1.1). The R software limma package was used for quantile normalization of raw data and subsequent data analysis. The fold change of each circRNA between two groups was calculated. The circRNAs having fold changes ≥2 and Student's t test p‐values <0.05 were identified as differentially expressed (DE) circRNAs. The microarray work was performed by KangChen Bio‐tech (Shanghai, China).
2.4. Quantitative real‐time PCR
Complementary DNA was reversely transcribed from total RNAs with random primers and SuperScriptTM III Reverse Transcriptase (Invitrogen). qRT‐PCR was conducted using master mix (Arraystar) on ViiA 7 Real‐time PCR System (Applied Biosystems). The reaction conditions were as follows: 95°C for 10 min and 40 cycles of 95°C for 10 s, and 60°C for 60 s. All of the qRT‐PCR reactions were run in triplicate. β‐actin was used as the internal control. Divergent primers, rather than convergent primers, were designed to specifically amplify circRNAs. The sequences of the primers were listed in Table 2. The relative expression levels of circRNAs were normalized to β‐actin and calculated by using the method. The appearance of a single‐peak in the melt‐curve indicated that the amplification was specific.
TABLE 2.
The sequences of primers used in RT‐PCR for validation
| Target ID | Primers sequence | Product size(bp) |
|---|---|---|
| β‐actin | F:5′ GTGGCCGAGGACTTTGATTG3′ | 73 |
| R:5′ CCTGTAACAACGCATCTCATATT3′ | ||
| hsa_circ_0067185 | F:5′ CTCTCTCGGAATAAGACAGAGGG3′ | 78 |
| R:5′ AGCTCTTCATAGCGGCCACT3′ | ||
| hsa_circ_0001666 | F:5′ CTGCCTAGCTGTCAAGGAGTGG3′ | 102 |
| R:5′ TCCGGGAAAGGATCTGGAATG3′ | ||
| hsa_circ_0002003 | F:5′ GAAAGTTCTCTTCACCAAGGAG3′ | 99 |
| R:5′ AGTCTTTCTGCTAGTCCACCTC3′ | ||
| hsa_circ_0027774 | F:5′ GAAGTTATGGAGTCCTATGAAGTTG3′ | 66 |
| R:5′ GTCTGTTTGAACTTTTGCTTGAT3′ | ||
| hsa_circ_0005043 | F:5′ CCTTTGCCCAGGATGTTCG3′ | 75 |
| R:5′ CACAGATGCTGAACTCACAGGTG3′ | ||
| hsa_circ_0028912 | F:5′ TTCTGCGTTGGGAGTCTGGA3′ | 70 |
| R:5′ GGAATGTGGACTTCTGGGTCTG3′ | ||
| hsa_circ_0040994 | F:5′ CGTCACATCTGACCTCAAATGA3′ | 114 |
| R:5′ CAAGTGGAAGAACTGCTCGC3′ | ||
| hsa_circ_0004183 | F:5′ GTCCATTCCACGAGGTTCTC3′ | 112 |
| R:5′ CCTCTGACGCAGGGTTTC3′ | ||
| hsa_circ_0037274 | F:5′ AGCTGCCAGTTACTGAGTCGTG3′ | 67 |
| R:5′ GTCACCGATGAGCTGCTTGTT3′ | ||
| hsa_circ_0062142 | F:5′ TCGCCCGTAGTTTTGTTTCT3′ | 124 |
| R:5′ TTTCTTAATCTTGCTGCTGCAC3′ |
Abbreviations: F, Forward; R, Reverse.
2.5. Bioinformatics analysis
The circRNA/microRNA/mRNA network was conducted to predict the potential function of selected DE circRNAs with the Arraystar's homemade miRNA target prediction software based on TargetScan and miRanda. Targeted mRNAs of DE circRNAs identified by profiling data were subjected to gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using Gene Ontology (http://www.geneongoloty.org/) and KOBAS software (KEGG Orthology‐Based Annotation System). The bioinformatics analysis was carried out by KangChen Bio‐tech (Shanghai, China).
2.6. Statistical analysis
All statistical analyses were performed using SPSS 11.0 (SPSS Inc.). The expression level of each circRNA was represented as fold change using the method. Results were expressed as the mean ± standard deviations or medians (quartiles) when they fit. Differences were evaluated with one‐way analysis of variance (ANOVA) or Kruscal‐Wallis H test for multiple groups and Mann‐Whitney test for two groups, as appropriate. Receiver operating characteristic (ROC) curve was established to evaluate the diagnostic value.
3. RESULTS
3.1. General expression profiles of CircRNAs in CD
Pinch biopsies from four CD patients, four UC patients, and four healthy controls were used to explore the expression profiles of circRNAs by microarray. In total, 9200 out of 13,617 circRNAs were detected by the microarray platform. As illustrated in Figure 1, 182 circRNAs exhibited significantly differential expression between CD and HC groups (fold change ≥ 2.0, p < 0.05); among these, 51 were upregulated and 131 downregulated. Moreover, 152 differentially expressed circRNAs were identified between CD and UC: 72 were significantly upregulated and 80 were significantly downregulated. A total of 110 dysregulated circRNAs were determined between UC and HC: 39 upregulated and 71 downregulated. The differentially expressed circRNAs are shown in scatter plot, volcano plot, and heat map (Figure 1). The top 10 upregulated and downregulated circRNAs in CD patients compared with UC patients or HC are listed in Tables 3 and 4. A total of 18 circRNAs showed higher expression levels in CD than those in UC and HC and were further categorized into different types, including 14 exonic, 1 antisense, 1 intronic, and 2 sense overlapping circRNAs. In addition, 53 circRNAs showed lower expression levels in CD than those in UC and HC, comprising 40 exonic, 10 intronic, 2 sense overlapping, and 1 intergenic circRNAs.
FIGURE 1.
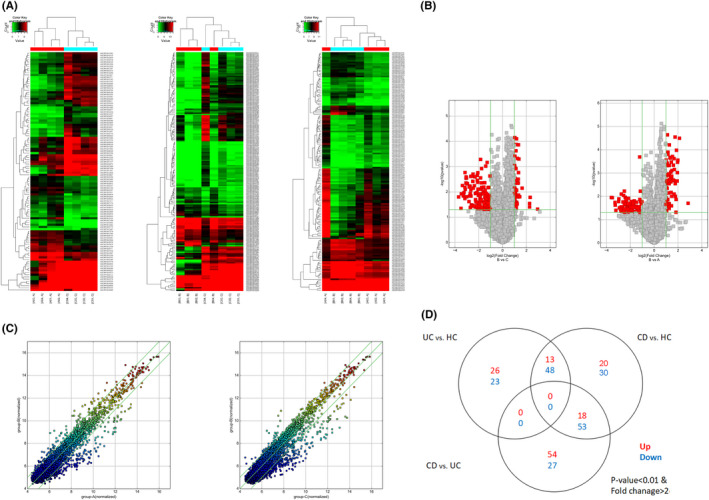
Overview of the microarray results. (A) Heat map of the circRNA microarray profiles. The expression of circRNAs was hierarchically clustered on the y‐axis, and tissue samples were hierarchically clustered on the x‐axis. Green indicated lower expression levels and red presented higher expression levels. (B) The volcano plots showing differentially expressed circRNAs. The green vertical line represented twofold changes, while the horizontal line marked a p value of 0.05. (C) Scatter plots of differentially expressed circRNAs. CircRNAs above and below the border green line were expressed more than twofold changes. (D) Venn diagram summarizing differentially expressed circRNAs shared by groups. Group A: UC, Group B: CD, Group C: healthy controls
TABLE 3.
Top 10 upregulated and downregulated circRNAs in CD patients compared with UC patients screened by microarray analysis
| circRNA (hsa_circRNA_) | Alias (hsa_circ_) | Fold change | p‐Value | FDR | Regulation | circRNA_type | chr | Best_transcript | GeneSymbol |
|---|---|---|---|---|---|---|---|---|---|
| 004183 | 0004183 | 7.21 | 0.020 | 0.2797 | Up | Exonic | chr10 | NM_018027 | FRMD4A |
| 405324 | 4.35 | 0.000 | 0.0264 | Up | Sense overlapping | chr15 | NM_020759 | STARD9 | |
| 000629 | 0000775 | 3.98 | 0.000 | 0.0612 | Up | Intronic | chr17 | ENST00000339151 | KIF18B |
| 051907 | 0051907 | 3.90 | 0.000 | 0.0654 | Up | Sense overlapping | chr19 | NM_001015 | RPS11 |
| 102207 | 0045881 | 3.86 | 0.001 | 0.0895 | Up | Exonic | chr17 | NM_001010982 | AFMID |
| 103107 | 0061251 | 3.82 | 0.002 | 0.1154 | Up | Exonic | chr21 | uc002yis.1 | TPTE |
| 101911 | 0040994 | 3.66 | 0.000 | 0.0456 | Up | Exonic | chr16 | NM_000135 | FANCA |
| 091419 | 0091419 | 3.59 | 0.013 | 0.2402 | Up | Exonic | chrX | ENST00000361575 | RPL39 |
| 406821 | 3.55 | 0.048 | 0.3969 | Up | Exonic | chr6 | NM_032131 | ARMC2 | |
| 406309 | 3.43 | 0.010 | 0.2102 | Up | Intronic | chr3 | ENST00000421999 | CMSS1 | |
| 102838 | 0056856 | 12.11 | 0.011 | 0.2175 | Down | Exonic | chr2 | NM_000888 | ITGB6 |
| 404595 | 11.47 | 0.011 | 0.2193 | Down | Intronic | chr1 | ENST00000295688 | CCT3 | |
| 066596 | 0066596 | 9.80 | 0.018 | 0.2692 | Down | Exonic | chr3 | NM_005233 | EPHA3 |
| 001350 | 0000253 | 9.68 | 0.024 | 0.2989 | Down | Intronic | chr10 | NR_047681 | BLNK |
| 400027 | 0092367 | 9.31 | 0.040 | 0.3741 | Down | Intronic | chr15 | uc001yxh.1 | SNURF‐SNRPN |
| 003997 | 0003997 | 7.92 | 0.017 | 0.2652 | Down | Exonic | chr11 | NM_024769 | CLMP |
| 400961 | 7.83 | 0.013 | 0.2358 | Down | Exonic | chr12 | NM_005653 | TFCP2 | |
| 001405 | 0001167 | 7.59 | 0.030 | 0.3342 | Down | Intronic | chr20 | ENST00000371941 | PREX1 |
| 000781 | 0000223 | 7.40 | 0.031 | 0.3377 | Down | Intronic | chr10 | ENST00000377524 | STAM |
| 404567 | 7.05 | 0.037 | 0.3614 | Down | Exonic | chr1 | NM_006608 | PHTF1 |
CircRNAs were selected by their Fold change (≥2) andp‐value (<0.05).
Abbreviations: CD, Crohn disease; Chr, chromosome; circRNA, circular RNA; FDR, false discovery rate; UC, ulcerative colitis.
TABLE 4.
Top 10 upregulated and downregulated circRNAs in CD patients compared with HC screened by microarray analysis
| circRNA (hsa_circRNA_) | Alias (hsa_circ_) | Fold change | p‐Value | FDR | Regulation | circRNA_type | chr | Best_transcript | GeneSymbol |
|---|---|---|---|---|---|---|---|---|---|
| 001729 | 0000691 | 7.70 | 0.048 | 0.3553 | Up | Antisense | chr16 | NM_014699 | ZNF646 |
| 104270 | 0001666 | 4.99 | 0.016 | 0.2351 | Up | Exonic | chr6 | NM_032448 | FAM120B |
| 004183 | 0004183 | 4.81 | 0.039 | 0.3332 | Up | Exonic | chr10 | NM_018027 | FRMD4A |
| 009024 | 0009024 | 4.80 | 0.029 | 0.2951 | Up | Exonic | chrY | NR_045128 | TXLNGY |
| 405324 | 3.12 | 0.007 | 0.1777 | Up | Sense overlapping | chr15 | NM_020759 | STARD9 | |
| 104616 | 0002003 | 2.72 | 0.015 | 0.2329 | Up | Exonic | chr8 | NM_001080394 | SPIDR |
| 085323 | 0085323 | 2.59 | 0.000 | 0.0774 | Up | Exonic | chr8 | NM_001568 | EIF3E |
| 101911 | 0040994 | 2.54 | 0.002 | 0.1219 | Up | Exonic | chr16 | NM_000135 | FANCA |
| 062142 | 0062142 | 2.53 | 0.013 | 0.2198 | Up | Exonic | chr22 | NR_001591 | TPTEP1 |
| 048148 | 0048148 | 2.52 | 0.000 | 0.0566 | Up | Exonic | chr19 | uc002lqu.3 | CNN2 |
| 102446 | 0049356 | 12.96 | 0.016 | 0.2337 | Down | Exonic | chr19 | NM_199141 | CARM1 |
| 405443 | 11.28 | 0.044 | 0.3448 | Down | Intronic | chr16 | ENST00000342673 | NDE1 | |
| 403556 | 11.05 | 0.012 | 0.2123 | Down | Exonic | chr6 | uc010jpp.1 | LINC00340 | |
| 400101 | 0092328 | 10.38 | 0.006 | 0.1664 | Down | Intronic | chr9 | ENST00000315731 | RPL7A |
| 102312 | 0003979 | 9.90 | 0.013 | 0.2159 | Down | Exonic | chr18 | NM_152352 | FAM210A |
| 074491 | 0074491 | 9.84 | 0.014 | 0.2262 | Down | Exonic | chr5 | NM_133263 | PPARGC1B |
| 023461 | 0023461 | 9.46 | 0.011 | 0.2067 | Down | Exonic | chr11 | NM_015242 | ARAP1 |
| 406237 | 8.98 | 0.018 | 0.2450 | Down | Exonic | chr3 | uc003cax.3 | OXNAD1 | |
| 101458 | 0034044 | 8.93 | 0.007 | 0.1742 | Down | Exonic | chr15 | uc001ytg.3 | HERC2P3 |
| 400564 | 8.89 | 0.002 | 0.1219 | Down | Exonic | chr10 | NM_001001330 | REEP3 |
CircRNAs were selected by their Fold change (≥2) and p‐value (<0.05).
Abbreviations: CD, Crohn disease; Chr, chromosome; circRNA, circular RNA; FDR, false discovery rate; UC, ulcerative colitis.
3.2. Validation of CircRNAs expression by qRT‐PCR
Firstly, we selected 10 DE circRNAs (hsa_circ_0067185, hsa_circ_0001666, hsa_circ_0027774, hsa_circ_0004183, hsa_circ_0028912, hsa_circ_0037274, hsa_circ_0062142, hsa_circ_0005043, hsa_circ_0040994, and hsa_circ_0002003) to confirm the microarray results using the same cohort in microarrays. The candidate circRNAs were shown to be upregulated in CD, exonic types, and non‐derivation from chromosome Y. The qRT‐PCR data were consistent with those from microarray. The expression levels of 6 circRNAs (hsa_circ_0067185, hsa_circ_0001666, hsa_circ_0027774, hsa_circ_0004183, hsa_circ_0028912, and hsa_circ_0062142) were significantly upregulated in CD than those in UC and HC. Among these, the level of hsa_circ_0001666 increased significantly and successively in HC, UC, and CD (Figure 2). In view of the significant difference, hsa_circ_0004183, hsa_circ_0001666, and hsa_circ_0062142 became the candidates for further study.
FIGURE 2.
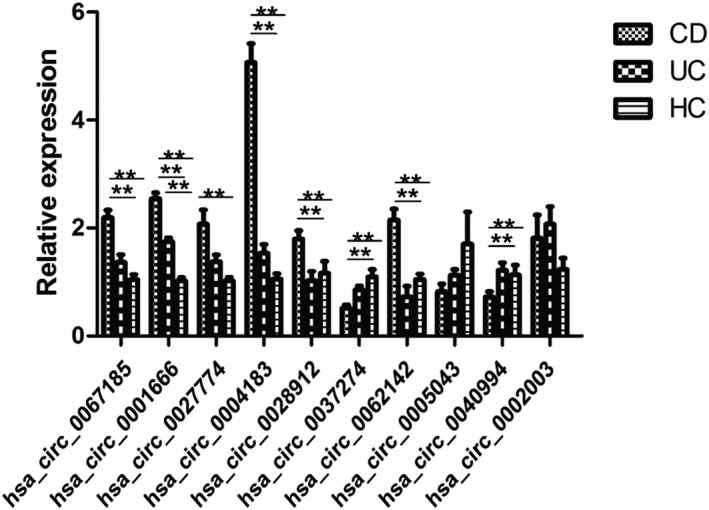
Validation of 10 differentially expressed circRNAs in microarray analysis samples by qRT‐PCR. The expression levels of hsa_circ_0067185, hsa_circ_0001666, hsa_circ_0027774, hsa_circ_0004183, hsa_circ_0028912, and hsa_circ_0062142 were upregulated significantly in CD than those in UC and HC, while hsa_circ_0037274 and hsa_circ_0040994 showed significant downregulation in CD than those in UC and HC. **: p < 0.01
Next, we validated whether the selected circRNAs show the differential expression in an independent cohort. The results showed significant differences in the expression levels of hsa_circ_0001666 and hsa_circ_0062142 among the three groups (χ2 = 30.758, p < 0.01 and χ2 = 20.749, p < 0.01, respectively). The expression levels (median, [P25, P75]) of hsa_circ_0001666 (2.437, [1.684,3.026]) and hsa_circ_0062142 (1.605, [1.213,3.348]) in the colon tissues of patients with CD increased significantly than those of UC (1.371, [0.743,2.031]; Z = −3.089, p < 0.01 and 0.856, [0.741,1.387]; Z = −3.505, p < 0.01, respectively) and HC (1.122, [0.670,1.550]; Z = −5.667, p < 0.01 and 0.957, [0.757,1.387]; Z = −4.169, p < 0.01, respectively). The levels of the two circRNAs did not differ in UC and HC (Z = −0.997, p = 0.319 and Z = −0.282, p = 0.778, respectively). However, the significant difference in the expression level of hsa_circ_0004183 was not observed among the groups (χ2 = 0.15, p = 0.928). The ROC curve analysis evaluated the diagnostic values of hsa_circ_0001666 and hsa_circ_0062142 for CD. The AUC, sensitivity, specificity, and Youden index of hsa_circ_0001666 and hsa_circ_0062142 were 0.858 (0.778–0.938), 0.833, 0.788, and 0.621, and 0.803 (0.701–0.905), 0.833, 0.673, and 0.506, respectively (Figure 3).
FIGURE 3.
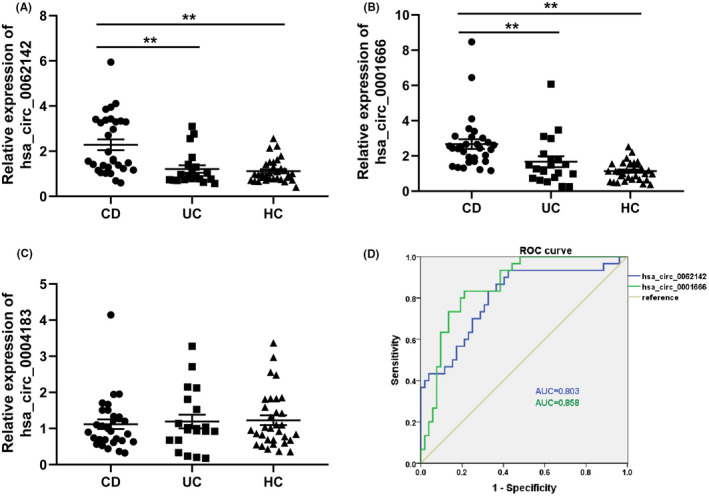
The expression levels of circular RNAs in CD, UC, and HC determined by qRT‐PCR. The expression levels of hsa_circ_0001666 and hsa_circ_0062142 in CD were significantly higher than those in UC and HC. (A) hsa_circ_0001666; (B) hsa_circ_0062142; (C) hsa_circ_0004183, **: p < 0.01; (D) ROC curves of hsa_circ_0001666 and hsa_circ_0062142
3.3. Bioinformatics analysis
Hsa_circ_0001666 and hsa_circ_0062142 were located on chromosome 6:170626457–170639638 and chromosome 22: 17117929–17128675, respectively. Gene ontology (GO) and KEGG pathway analysis were performed using target mRNAs of hsa_circ_0062142 and hsa_circ_0001666 (Figure 4). The top 10 significant GO terms of each subgroup (BP, CC, and MF) were displayed. The results revealed that several target genes were involved in the progression of CD. For example, epithelial cell differentiation and epidermis development showed significantly enriched GO terms in BP. KEGG analysis revealed that 23 pathways were associated with target genes of the two selected circRNAs. Among these, IL‐17 signaling pathway, Toll‐like receptor signaling pathway, TNF signaling pathway, and Th17 cell differentiation are implicated in the progression of CD.
FIGURE 4.
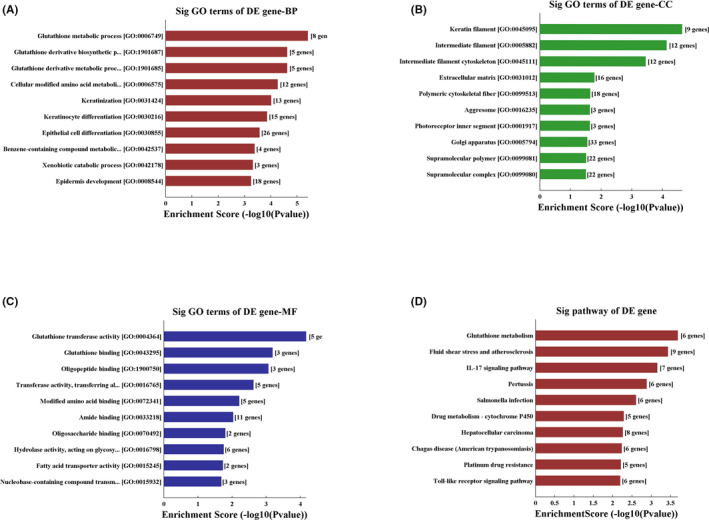
Gene ontology (GO) and KEGG pathway analysis for hsa_circ_0062142 and hsa_circ_0001666 target genes. (A) Enriched biological process in GO terms; (B) Enriched cellular component in GO terms; (C) Enriched molecular function in GO terms; (D) KEGG pathway analysis shows the top 10 enriched pathways related to hsa_circ_0062142 and hsa_circ_0001666
To predict the potential function of the CD‐associated circRNAs (hsa_circ_0062142 and hsa_circ_0001666), the circRNA/miRNA/mRNA network was conducted according to microRNA response elements (MREs) using Arraystar's homemade miRNA target prediction software based on TargetScan and miRanda. The likely potential miRNAs harbored by hsa_circ_0062142 include hsa‐miR‐940, hsa‐miR‐665, hsa‐miR‐20b‐3p, hsa‐miR‐574‐5p, hsa‐miR‐518a‐5p, hsa‐miR‐34b‐5p, hsa‐miR‐105‐5p, and hsa‐miR‐299‐3p. The putative target miRNAs for hsa_circ_0001666 consist of hsa‐miR‐30b‐5p, hsa‐miR‐30c‐5p, hsa‐miR‐518a‐5p, hsa‐miR‐326, hsa‐miR‐199b‐5p, hsa‐miR‐22‐3p, hsa‐miR‐125a‐5p, hsa‐miR‐125b‐5p, hsa‐miR‐661, hsa‐miR‐23b‐5p, hsa‐miR‐133b, hsa‐miR‐133a‐3p, and hsa‐miR‐330‐5p. The competing endogenous RNA (ceRNA) network exhibited complex interaction between selected circRNAs and mRNAs by competing for common microRNAs (Figure 5). Subsequently, we predicted 322 and 21 candidate ceRNAs of hsa_circ_0062142 and hsa_circ_0001666, respectively.
FIGURE 5.
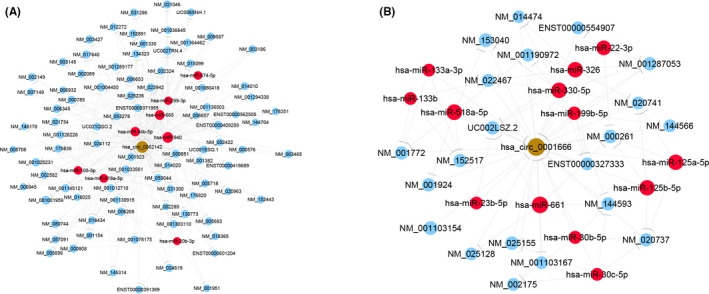
A snippet of detailed annotation for circRNA/miRNA/mRNAs interaction. (A) hsa_circ_0062142; (B) hsa_circ_0001666. Yellow represents circRNA, red represents miRNA, and blue represents mRNA
4. DISCUSSION
Circular RNAs have emerged as a hotspot and attracted increasing attention. The abundance and biological stability endow circRNAs with the advantage of ideal biomarkers. Accumulating evidence identified distinct expression patterns of circRNAs in many diseases. The differentially expressed circRNAs might be associated with the pathobiology, diagnosis, and treatment in diseases. For example, the downregulated expression of hsa_circ_0000140 was observed in gastric cancer. 21 Hsa_circ_0005075 22 and hsa_circ_0001649 23 are considered as novel biomarkers for hepatocellular carcinoma. Xia et al found that upregulated hsa_circ_0067934 might represent a potential biomarker for the diagnosis and prognosis of esophageal cancer. 16 Zhou et al discovered that downregulated expression of hsa_circ_0003906 in tissue might serve as a valuable biomarker for the diagnosis and treatment of colorectal cancer. 11 However, the correlation between circRNAs and CD is yet to be clarified.
Recently, Yu et al identified 218 differentially expressed circRNAs in the colon tissue between CD patients and HC using Arraystar Human CircularRNA Array (6 × 7 K; Arraystar, Inc.) consisting of 5396 probes, hsa_circRNA_102685 was upregulated in colon tissues in CD patients in the validation group comprising 10 patients and 10 healthy controls. 24 Ye et al reported that hsa_circRNA_103516 levels in peripheral blood mononuclear cells (PBMCs) increased in IBD and correlated positively with both disease activity and disease behavior, such as stricturing and penetrating. 25 Yin et al observed that 4 circRNAs (hsa_circRNA_092520, hsa_circRNA_102610, hsa_circRNA_004662, or hsa_circRNA_103124) in PBMCs were potential diagnostic biomarkers of CD. Additionally, hsa_circRNA_004662 might be served as a specific candidate for differentiating between CD and UC. 26
In the present study, UC patients were enrolled as patient controls and a Human 8 × 15 K circRNA Array consisting of 13,617 probes was selected to search the ideal biomarkers for CD. Next, we found that the expression levels of hsa_circ_0062142 and hsa_circ_0001666 were significantly upregulated in CD than in UC and HC, while the levels of both circRNAs did not differ in UC and HC. Therefore, we speculated that hsa_circ_0062142 and hsa_circ_0001666 are CD‐associated circRNAs. The AUC of hsa_circ_0001666 and hsa_circ_0062142 was 0.858 and 0.803, respectively, indicating their favorable diagnostic value. Thus, the current results suggested that hsa_circ_0062142 and hsa_circ_0001666 serve as novel biomarkers for CD.
It is known that circRNAs are able to regulate gene expression, especially by acting as miRNA sponge. CircRNAs are regarded as ceRNAs due to binding to miRNAs through MRE and relieving the inhibition of miRNA‐targeted mRNA expression. As a result, dysregulated circRNAs are speculated to play critical roles in the pathogenesis of diseases. Circular RNA sponge for miR‐7 (CiRS‐7) is a well‐known sponge for miR‐7 and is reported to be widely involved in cancers (such as hepatocellular carcinoma, 27 gastric cancer, 28 colorectal cancer, 29 and breast cancer 30 ) and Alzheimer's disease. 13 Based on the circRNA/microRNA/mRNA network and bioinformatics analysis of differentially expressed circRNAs, we observed that the predicted microRNAs of hsa_circ_0062142 and hsa_circ_0001666 are involved in several cellular processes, such as epithelial to mesenchymal transition (EMT), Th17 differentiation, and carcinogenesis.
Fibrosis is a common feature of CD. EMT might be a major contributor to the pathogenesis of fibrosis in CD, owing to activated fibroblasts being recruited in the inflamed intestinal tract. 31 Reportedly, hsa_circRNA_102610 is upregulated in PBMCs of CD patients and promotes intestinal epithelial cells proliferation and TGF‐β1‐induced EMT by sponging hsa‐miR‐130a‐3p. Thus, hsa_circRNA_102610 was inferred to participate in the mechanism of CD. 32 Among the predicted miRNAs of the two DE circRNAs, hsa‐miR‐199a‐5p, hsa‐miR‐34b‐5p, and hsa‐miR‐23b were previously reported to be associated with EMT. Giovannini et al. revealed that hsa‐miR‐199a‐5p and hsa‐miR‐199a‐3p downregulated Notch1 or E‐cadherin protein levels in hepatocellular carcinoma patients and influenced EMT. 33 Hsa‐miR‐34b‐5p was found to regulate the mRNAs of EMT‐transcription factors and play a role in the metastasis and progression of colorectal cancer. 34 Hsa‐miR‐23b was identified to suppress EMT and the metastasis of hepatocellular carcinoma. 35 CD patients suffer from dysregulated immune responses against the microorganisms of the intestinal flora. Th17 cells play a role in the pathogenesis of CD by exerting the dual roles in maintaining gut homeostasis and inducing inflammation lesions. 36 Moreover, the imbalance between Treg cells and Th17 cells is associated with the progression of CD. 37 In the potential target miRNAs, hsa‐miR‐30c and hsa‐miR‐20b are deemed as autoimmune‐deregulated miRNAs due to their positive and negative regulation of Th17 differentiation, respectively. 38 Hsa‐miR‐326 was identified to promote Th17 differentiation in multiple sclerosis. 39 Furthermore, as a chronic intestinal inflammation, CD possesses an increased risk for colorectal cancer. Hsa‐miR‐574‐5p, hsa‐miR‐133b, hsa‐miR‐133a‐3p, hsa‐miR‐30b‐5p, and hsa‐miR‐326 in the ceRNA network were proved to play suppressive roles in colorectal cancer by inhibiting cell proliferation, invasion, and migration. 40 , 41 , 42 , 43 , 44 , 45
Nevertheless, the present study has some limitations. First, the number of subjects is small, and the results need to be substantiated in a larger cohort. Second, the circRNA/miRNA/mRNA network was only predicted by bioinformatics analysis. Experimental research is required in the future.
In summary, the current study identified the comprehensive circRNA expression profiles in colon tissues of CD compared with tissues of UC and healthy controls. We showed that two circRNAs (hsa_circ_0062142 and hsa_circ_0001666) were significantly upregulated in colon tissues of CD than those in UC and healthy controls. Bioinformatics analysis indicated that hsa_circ_0062142 and hsa_circ_0001666 might be involved in the progression of CD. Together, these findings suggested that circRNAs play a crucial role in the pathogenesis of CD and might provide potential diagnostic biomarkers and therapeutic targets for CD.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Natural Science Foundation of China (No. 81470071), National Key Clinical Program of China (2014ZDZK003), and Jiangsu Provincial Special Program of Medical Science (No. BL2014072).
Hu and Zhu equal contributors.
DATA AVAILABILITY STATEMENT
The original data of this research are available from the corresponding author on request.
REFERENCES
- 1. Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142(1):46‐54.e42. [DOI] [PubMed] [Google Scholar]
- 2. Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population‐based studies. Lancet. 2017;390(10114):2769‐2778. [DOI] [PubMed] [Google Scholar]
- 3. Chen P, Zhou G, Lin J, et al. Serum biomarkers for inflammatory bowel disease. Front Med. 2020;7:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin J, Cao QI, Zhang J, et al. MicroRNA expression patterns in indeterminate inflammatory bowel disease. Mod Pathol. 2013;26(1):148‐154. [DOI] [PubMed] [Google Scholar]
- 5. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333‐338. [DOI] [PubMed] [Google Scholar]
- 6. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salzman J. Circular RNA expression: Its potential regulation and function. Trends Genet. 2016;32(5):309‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang C, Shan G. What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription. 2015;6(4):61‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA. 2017;8(2):e1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang M, He Y‐R, Liang L‐C, Huang Q, Zhu Z‐Q. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23(34):6330‐6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhuo F, Lin H, Chen Z, Huang Z, Hu J. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Onco Targets Ther. 2017;10:5187‐5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Liu J, Liu C, Naji A, Stoffers DA. MicroRNA‐7 regulates the mTOR pathway and proliferation in adult pancreatic beta‐cells. Diabetes. 2013;62(3):887‐895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet. 2013;4:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF‐associated non‐coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42‐51. [DOI] [PubMed] [Google Scholar]
- 16. Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6(1):35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zou M, Huang C, Li X, et al. Circular RNA expression profile and potential function of hsa_circRNA_101238 in human thoracic aortic dissection. Oncotarget. 2017;8(47):81825‐81837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang YU, Liang W, Zhang P, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Canc Res. 2017;36(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhuang Z‐G, Zhang J‐A, Luo H‐L, et al. The circular RNA of peripheral blood mononuclear cells: Hsa_circ_0005836 as a new diagnostic biomarker and therapeutic target of active pulmonary tuberculosis. Mol Immunol. 2017;90:264‐272. [DOI] [PubMed] [Google Scholar]
- 20. Ouyang Q, Wu J, Jiang Z, et al. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell Physiol Biochem. 2017;42(2):651‐659. [DOI] [PubMed] [Google Scholar]
- 21. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132‐136. [DOI] [PubMed] [Google Scholar]
- 22. Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine. 2016;95(22):e3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161‐169. [DOI] [PubMed] [Google Scholar]
- 24. Qiao YU, Cai C, Shen J, Zheng Q, Ran Z. Circular RNA expression alterations in colon tissues of Crohn's disease patients. Mol Med Rep. 2019;19:4500‐4506. [DOI] [PubMed] [Google Scholar]
- 25. Ye Y‐L, Yin J, Hu T, Zhang L‐P, Wu L‐Y, Pang Z. Increased circulating circular RNA_103516 is a novel biomarker for inflammatory bowel disease in adult patients. World J Gastroenterol. 2019;25(41):6273‐6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yin J, Hu T, Xu L, et al. Circular RNA expression profile in peripheral blood mononuclear cells from Crohn disease patients. Medicine. 2019;98(26):e16072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS‐7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143(1):17‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pan H, Li T, Jiang Y, et al. Overexpression of Circular RNA ciRS‐7 abrogates the tumor suppressive effect of miR‐7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119(1):440‐446. [DOI] [PubMed] [Google Scholar]
- 29. Weng W, Wei Q, Toden S, et al. Circular RNA ciRS‐7‐A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23(14):3918‐3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsiao Y‐C, Yeh M‐H, Chen Y‐J, Liu J‐F, Tang C‐H, Huang W‐C. Lapatinib increases motility of triple‐negative breast cancer cells by decreasing miRNA‐7 and inducing Raf‐1/MAPK‐dependent interleukin‐6. Oncotarget. 2015;6(35):37965‐37978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li C, Kuemmerle JF. Mechanisms that mediate the development of fibrosis in patients with Crohn's disease. Inflamm Bowel Dis. 2014;20(7):1250‐1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yin J, Ye YL, Hu T, et al. Hsa_circRNA_102610 upregulation in Crohn’s disease promotes transforming growth factor‐β1‐induced epithelial‐mesenchymal transition. World J Gastroenterol. 2020;26(22):3034‐3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giovannini C, Fornari F, Dallo R, et al. MiR‐199‐3p replacement affects E‐cadherin expression through Notch1 targeting in hepatocellular carcinoma. Acta Histochem. 2018;120(2):95‐102. [DOI] [PubMed] [Google Scholar]
- 34. Vu T, Datta PK. Regulation of EMT in colorectal cancer: A culprit in metastasis. Cancers. 2017;9(12):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cao J, Liu J, Long J, et al. microRNA‐23b suppresses epithelial‐mesenchymal transition (EMT) and metastasis in hepatocellular carcinoma via targeting Pyk2. Biomed Pharmacother. 2017;89:642‐650. [DOI] [PubMed] [Google Scholar]
- 36. Zhao J, Lu Q, Liu Y, et al. Th17 Cells in inflammatory bowel disease: Cytokines, plasticity, and therapies. J Immunol Res. 2021;2021:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xia S, Zhang D, Zheng S, et al. Association of Crohn's disease with Foxp3 gene polymorphisms and its colonic expression in Chinese patients. J Clin Lab Anal. 2019;33(4):e22835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Honardoost MA, Naghavian R, Ahmadinejad F, Hosseini A, Ghaedi K. Integrative computational mRNA–miRNA interaction analyses of the autoimmune‐deregulated miRNAs and well‐known Th17 differentiation regulators: An attempt to discover new potential miRNAs involved in Th17 differentiation. Gene. 2015;572(2):153‐162. [DOI] [PubMed] [Google Scholar]
- 39. Du C, Liu C, Kang J, et al. MicroRNA miR‐326 regulates TH‐17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10(12):1252‐1259. [DOI] [PubMed] [Google Scholar]
- 40. Cui Z, Tang J, Chen J, Wang Z. Hsa‐miR‐574‐5p negatively regulates MACC‐1 expression to suppress colorectal cancer liver metastasis. Cancer Cell Int. 2014;14(1):47‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li W, Chen A, Xiong L, et al. miR‐133a acts as a tumor suppressor in colorectal cancer by targeting eIF4A1. Tumor Biol. 2017;39(5):1‐9. [DOI] [PubMed] [Google Scholar]
- 42. Wu H, Wu R, Chen M, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs and construction of miR‐133b mediated ceRNA network in colorectal cancer. Oncotarget. 2017;8(13):21095‐21105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liao WT, Ye YP, Zhang NJ, et al. MicroRNA‐30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol. 2014;232(4):415‐427. [DOI] [PubMed] [Google Scholar]
- 44. Wu L, Hui H, Wang L‐J, Wang H, Liu Q‐F, Han S‐X. MicroRNA‐326 functions as a tumor suppressor in colorectal cancer by targeting the nin one binding protein. Oncol Rep. 2015;33(5):2309‐2318. [DOI] [PubMed] [Google Scholar]
- 45. Zhang G, Liu Z, Cui G, Wang X, Yang Z. MicroRNA‐486‐5p targeting PIM‐1 suppresses cell proliferation in breast cancer cells. Tumor Biol. 2014;35(11):11137‐11145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original data of this research are available from the corresponding author on request.


