Abstract
Objective
Circular RNA_0004913 (circ_0004913), circular RNA_0008160 (circ_0008160), and circular RNA_0000517 (circ_0000517) are shown to be dysregulated in HCC tissues and cell lines, and also show potential in regulating hepatocellular carcinoma (HCC) pathogenesis. This current study attempted to find possible associations of circ_0004913, circ_0008160, and circ_0000517 with clinical features and prognosis of HCC patients.
Methods
A hundred and fifty HCC patients who received hepatectomy were retrospectively reviewed, and their resected specimens including tumor tissues and paired adjacent tissues were obtained, in which the circ_0004913, circ_0008160, and circ_0000517 expressions were detected. Overall survival (OS) data were collected according to the clinical visit records.
Results
Circ_0004913 (p < 0.001) and circ_0008160 (p < 0.001) were downregulated, while circ_0000517 (p < 0.001) was upregulated in tumor tissue compared with paired adjacent tissue. Additionally, tumor circ_0004913 was negatively associated with largest tumor size (p = 0.009) and Barcelona clinic liver cancer (BCLC) stage (p = 0.020), while tumor circ_0008160 and circ_0000517 were not correlated with any clinicopathological features of HCC patients (all p > 0.05). Tumor circ_0004913 high expression was associated with prolonged OS in total HCC patients (p = 0.008) and in subgroup of patients with largest tumor size <5 cm (p = 0.008). Tumor circ_0008160 high expression was correlated with longer OS in patients with BCLC stage B (p = 0.026). Univariate Cox's analysis disclosed that tumor circ_0004913 high expression was correlated with longer OS; while after adjustment by multivariate Cox's analysis, it failed to predict OS independently.
Conclusion
Circ_0004913 was downregulated in tumor tissue and may serve as a biomarker for evaluating disease severity and prognosis in HCC patients.
Keywords: circular RNA 0004913, clinicopathological characteristics, hepatocellular carcinoma, overall survival, prognostic factor
Circular RNA_0004913 (circ_0004913), circular RNA_0008160 (circ_0008160), and circular RNA_0000517 (circ_0000517) are shown to be dysregulated in HCC tissues and cell lines, and also show potential in regulating hepatocellular carcinoma (HCC) pathogenesis. This current study attempted to find possible associations of circ_0004913, circ_0008160, and circ_0000517 with clinical features and prognosis of HCC patients. A hundred and fifty HCC patients who received hepatectomy were retrospectively reviewed, and their resected specimens including tumor tissues and paired adjacent tissues were obtained, in which the circ_0004913, circ_0008160, and circ_0000517 expressions were detected. Overall survival (OS) data were collected according to the clinical visit records. Circ_0004913 (p < 0.001) and circ_0008160 (p < 0.001) were downregulated, while circ_0000517 (p < 0.001) was upregulated in tumor tissue compared with paired adjacent tissue. Additionally, tumor circ_0004913 was negatively associated with largest tumor size (p = 0.009) and Barcelona clinic liver cancer (BCLC) stage (p = 0.020), while tumor circ_0008160 and circ_0000517 were not correlated with any clinicopathological features of HCC patients (all p > 0.05). Tumor circ_0004913 high expression was associated with prolonged OS in total HCC patients (p = 0.008) and in subgroup of patients with largest tumor size <5 cm (p = 0.008). Tumor circ_0008160 high expression was correlated with longer OS in patients with BCLC stage B (p = 0.026). Univariate Cox's analysis disclosed that tumor circ_0004913 high expression was correlated with longer OS; while after adjustment by multivariate Cox's analysis, it failed to predict OS independently. Circ_0004913 was downregulated in tumor tissue and may serve as a biomarker for evaluating disease severity and prognosis in HCC patients.

1. INTRODUCTION
Hepatocellular carcinoma (HCC) accounts for the majority of liver cancers, which is one of the most common and mortal malignancies in the world. 1 It is reported that the medical burden caused by HCC rises every year with an incidence exceeding 1 million per year as well as a very poor survival profile. 1 , 2 Only a few HCC patients could be treated by surgical resection, transplantation, or ablation due to a result of diagnosis delay in most patients, who are initially diagnosed as middle or advanced stage disease. 3 Many progresses have been achieved in the exploration of HCC molecular features, which has to some extent improves HCC patients' management. Nonetheless, translation of these molecular findings to the application of HCC precision medicine is still stalled at present, hence, seeking possible biomarkers which may facilitate diagnosis, surveillance or treatment efficacy becomes a growing concern in HCC studies.
Circular RNA (circRNA) is a promising group of non‐coding RNAs presenting with growing values in human diseases, although these values emerge in only recent years. 4 , 5 , 6 CircRNA carries many crucial functions in human body, such as sponging microRNAs (miRNAs), mediating mRNA activities, and participating in protein translation. 7 , 8 , 9 Encouragingly, circRNAs may also be critical regulators in multiple carcinomas as reported by increasing studies, which include HCC as well. 9 , 10 , 11 Previously, three circRNAs (circ_0004913, circ_0008160, and circ_0000517) are reported to be dysregulated in HCC tissues (GSE94508 and GSE97332), and these three circRNAs are also confirmed by qPCR to be dysregulated in both HCC tissues and HCC cells; more interestingly, two of them are revealed to be potential regulators of HCC pathogenesis as well. 12 , 13 , 14 Nevertheless, the possible clinical values of these three circRNAs remain to be elusive in HCC.
Hence, the current study attempted to explore possible associations of circ_0004913, circ_0008160, and circ_0000517 with clinical features and prognosis in HCC patients.
2. MATERIALS AND METHODS
2.1. Patients
After the study was approved by the Institutional Review Board, we retrospectively reviewed the medical documents of HCC patients who received hepatectomy from July 2015 to December 2017. Only the patients meeting the following criteria were analyzed in the current study: (a) had pathologically confirmed diagnosis of primary HCC, (b) underwent hepatectomy without previous treatment, (c) had refrigerated specimens of tumor and adjacent tissue, (d) clinicopathologic data and survival data were well‐documented, and (e) presented no history of other malignant diseases. Finally, 150 surgical HCC patients were eligible and included in the present study. Written informed consents were obtained from enrolled patients or their family members.
2.2. Data compilation
Data collection and compilation were conducted by reviewing the medical records in the included patients. The main clinicopathologic data covered basic demographics (age and gender), medical history (hepatitis B and liver cirrhosis), preoperative liver functional reserve (Child‐Pugh stage), tumor features (nodule number and largest tumor size), disease stage (Barcelona Clinic Liver Cancer (BCLC) stage), preoperative liver function indexes (alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), and total bilirubin (TBIL)), and preoperative tumor markers (carcino‐embryonic antigen (CEA), carbohydrate antigen (CA199), and alpha fetoprotein (AFP)). Moreover, overall survival (OS) data were collected based on the clinical visit records. In addition, the ethical approval was given by the Ethical Committee of our hospital. As for the follow‐up, the median follow‐up time was 52.0 (48.5–55.5) months, and the last follow‐up date was August 2020.
2.3. Specimen acquisition
After the surgical resection, the resected specimens from all patients were immediately frozen in liquid nitrogen and cryopreserved at −80°C in the specimen library. After the permission by Institutional Review Board was achieved, the frozen specimens including HCC tissue and normal adjacent tissue of each enrolled patient were obtained from the specimen library. The expressions of circ_0004913, circ_0008160, and circ_0000517 in the specimens were quantified via reverse transcription quantitative polymerase chain reaction (RT‐qPCR) assay.
2.4. RT‐qPCR assay
All procedures of RT‐qPCR were carried out based on the manufacturer's protocol, with the use of the following kits: RNeasy Protect Mini Kit (Qiagen) was applied for the isolation and purification of total RNA; RNase R Kit (Epicentre) was used for the linear RNA removal; iScript™ cDNA Synthesis Kit (with random primer) (Bio‐Rad) was applied for reversing transcription; QuantiNova SYBR Green PCR Kit (Qiagen) was applied for quantitative PCR analysis, during which, GAPDH functioned as the internal reference. The method was applied for calculation of relative expression of circRNAs. The circRNA and GAPDH primer sequences designed in a previous study were used in the current study. 15
2.5. Statistical analysis
All data were analyzed using SPSS 24.0 software (IBM). All diagrams were generated using GraphPad Prism 8.01 software (GraphPad Software Inc.). Qualitative data were described using number (proportion); quantitative data were characterized using mean value with standard deviation (SD), or median value and interquartile range (IQR). Difference of circRNA expressions between paired samples was evaluated by Wilcoxon matched‐pairs signed rank test. Receiver operating characteristic (ROC) curve was conducted to assess the value of circRNA expression for distinguishing tumor tissue from paired adjacent tissue. Correlation of circRNA expressions with clinical features was determined by chi‐square test. Correlation of circRNA expressions with OS was assessed by Log‐rank (Mantel‐Cox) test, which was elucidated with the use of Kaplan‐Meier curve. Prognostic value of variables was detected using univariable and multivariable Cox's proportional hazard regression model analyses, which was exhibited in the form of forest plot. Statistical significance was defined as p value below 0.05.
3. RESULTS
3.1. HCC patients' characteristic0073
Among the 150 surgical HCC patients in the present study, the mean age, numbers of male and female were 60.4 ± 8.7 years, 128 (85.3%) and 22 (14.7%), respectively (Table 1). Additionally, the numbers of patients with history of HB and liver cirrhosis were 121 (80.7%) and 111 (74.0%), respectively. There were 122 (81.3%) patients in Child‐Pugh stage A and 28 (18.7%) patients in Child‐Pugh stage B, respectively. The numbers of patients with unifocal tumor and those with multifocal tumor were 80 (53.3%) and 70 (46.7%), respectively. The numbers of patients who were in BCLC stage A and BCLC stage B were 60 (40.0%) and 90 (60.0%), respectively. Furthermore, ALT, AST, ALP, and TBIL levels were 27.0 (21.4–40.5) U/L, 37.8 (27.9–51.8) U/L, 99.8 (77.7–149.1) U/L, and 15.4 (10.8–27.4) μmol/L, respectively. The levels of tumor markers CEA, CA199, and AFP were 4.3 (3.0–6.9) ng/ml, 14.3 (6.1–26.6) U/ml, and 82.6 (7.3–978.7) ng/ml, respectively.
TABLE 1.
Characteristics of patients with surgical HCC
| Items | Patients (N = 150) |
|---|---|
| Age (years), mean ± SD | 60.4 ± 8.7 |
| Gender, No. (%) | |
| Male | 128 (85.3) |
| Female | 22 (14.7) |
| History of HB, No. (%) | 121 (80.7) |
| History of liver cirrhosis, No. (%) | 111 (74.0) |
| Child‐Pugh stage, No. (%) | |
| A | 122 (81.3) |
| B | 28 (18.7) |
| Tumor nodule number, No. (%) | |
| Unifocal | 80 (53.3) |
| Multifocal | 70 (46.7) |
| Largest tumor size, No. (%) | |
| <5 cm | 74 (49.3) |
| ≥5 cm | 76 (50.7) |
| BCLC stage, No. (%) | |
| A | 60 (40.0) |
| B | 90 (60.0) |
| Liver related indexes, median (IQR) | |
| ALT (U/L) | 27.0 (21.4–40.5) |
| AST (U/L) | 37.8 (27.9–51.8) |
| ALP (U/L) | 99.8 (77.7–149.1) |
| TBIL (μmol/L) | 15.4 (10.8–27.4) |
| Tumor Marker, median (IQR) | |
| CEA (ng/ml) | 4.3 (3.0–6.9) |
| CA199 (U/ml) | 14.3 (6.1–26.6) |
| AFP (ng/ml) | 82.6 (7.3–978.7) |
Abbreviation: AFP, alpha‐fetoprotein; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BCLC, Barcelona clinic liver cancer; CA199; carbohydrate antigen 199; CEA, carcinoembryonic antigen; HB, hepatitis B; HCC, hepatocellular carcinoma; TBIL, total bilirubin; IQR, interquartile range; SD, standard deviation.
3.2. Tissue circ_0004913, circ_0008160, and circ_0000517 expressions in HCC patients
The circ_0004913 (p < 0.001) (Figure 1A) and circ_0008160 (p < 0.001) (Figure 1B) expressions were reduced, however, the circ_0000517 (p < 0.001) (Figure 1C) expression was elevated in tumor tissue compared to paired adjacent tissue in surgical HCC patients. These findings were in line with previous data revealed by high‐throughput assays (GSE94508 and GSE97332). In addition, ROC curve showed that combination of circ_0004913, circ_0008160, and circ_0000517 expressions presented with good value for differentiating tumor tissue from paired adjacent tissue, and the AUC was 0.977 (95%CI: 0.965–0.991) (Figure 2).
FIGURE 1.
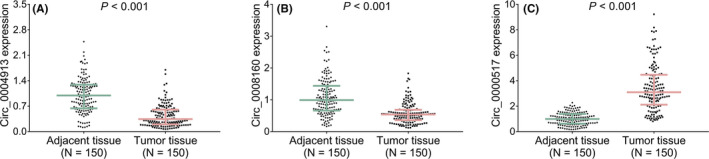
The three circRNA expressions in tumor tissue and paired adjacent tissue. The circ_0004913 (A), circ_0008160 (B), and circ_0000517 (C) expressions in tumor tissue and paired adjacent tissue of HCC patients. CircRNA, circular RNA; HCC, hepatocellular carcinoma
FIGURE 2.
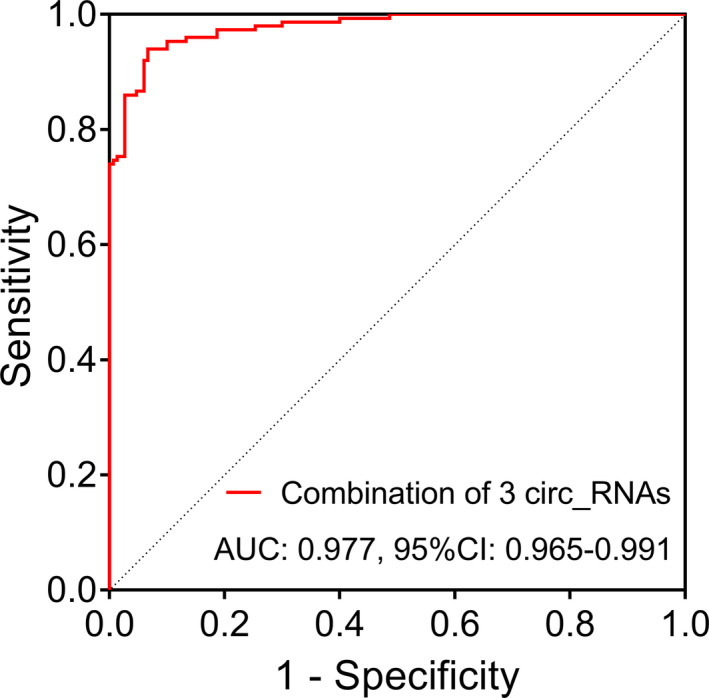
Value of the three circRNAs for differentiating tumor tissue from paired adjacent tissue. The ROC curve of combined circ_0004913, circ_0008160, and circ_0000517 expression value for differentiating tumor tissue from paired adjacent tissue. circRNA, circular RNA; CI, confidence interval; ROC, receiver operating characteristic
3.3. Correlations of circ_0004913, circ_0008160, and circ_0000517 with HCC patients' characteristics
Regarding the clinicopathological characteristics, circ_0004913 was negatively correlated with largest tumor size (p = 0.009) and BCLC stage (p = 0.020), however, its correlation with age (p = 0.102), gender (p = 0.166), history of HB (p = 0.535), history of cirrhosis (p = 0.094), Child‐Pugh stage (p = 0.675), or tumor nodule number (p = 0.326) did not exist in surgical HCC patients (Table 2). Moreover, circ_0008160 and circ_0000517 were not correlated with any clinicopathological characteristics (all p > 0.05). With regard to the hepatic function indexes and tumor markers, circ_0004913, circ_0008160, and circ_0000517 were not correlated with ALT, AST, ALP, TBIL, CEA, CA199, or AFP level (all p > 0.05) (Table 3).
TABLE 2.
Correlation of circ_0004913, circ_0008160, and circ_0000517 expressions with clinicopathological characteristics
| Items | Circ_0004913 expression | Circ_0008160 expression | Circ_0000517 expression | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | p value | Low | High | p value | Low | High | p value | |
| Age, No. (%) | |||||||||
| ≤60 years | 41 (54.7) | 31 (41.3) | 0.102 | 38 (50.7) | 34 (45.3) | 0.513 | 36 (48.0) | 36 (48.0) | 1.000 |
| >60 years | 34 (45.3) | 44 (58.7) | 37 (49.3) | 41 (54.7) | 39 (52.0) | 39 (52.0) | |||
| Gender, No. (%) | |||||||||
| Male | 61 (81.3) | 67 (89.3) | 0.166 | 63 (84.0) | 65 (86.7) | 0.644 | 61 (81.3) | 67 (89.3) | 0.166 |
| Female | 14 (18.7) | 8 (10.7) | 12 (16.0) | 10 (13.3) | 14 (18.7) | 8 (10.7) | |||
| History of HB, No. (%) | |||||||||
| No | 13 (17.3) | 16 (21.3) | 0.535 | 14 (18.7) | 15 (20.0) | 0.836 | 13 (17.3) | 16 (21.3) | 0.535 |
| Yes | 62 (82.7) | 59 (78.7) | 61 (81.3) | 60 (80.0) | 62 (82.7) | 59 (78.7) | |||
| History of liver cirrhosis, No. (%) | |||||||||
| No | 15 (20.0) | 24 (32.0) | 0.094 | 16 (21.3) | 23 (30.7) | 0.193 | 21 (28.0) | 18 (24.0) | 0.577 |
| Yes | 60 (80.0) | 51 (68.0) | 59 (78.7) | 52 (69.3) | 54 (72.0) | 57 (76.0) | |||
| Child‐Pugh stage, No. (%) | |||||||||
| A | 60 (80.0) | 62 (82.7) | 0.675 | 63 (84.0) | 59 (78.7) | 0.402 | 59 (78.7) | 63 (84.0) | 0.402 |
| B | 15 (20.0) | 13 (17.3) | 12 (16.0) | 16 (21.3) | 16 (21.3) | 12 (16.0) | |||
| Tumor nodule number, No. (%) | |||||||||
| Unifocal | 37 (49.3) | 43 (57.3) | 0.326 | 36 (48.0) | 44 (58.7) | 0.190 | 41 (54.7) | 39 (52.0) | 0.743 |
| Multifocal | 38 (50.7) | 32 (42.7) | 39 (52.0) | 31 (41.3) | 34 (45.3) | 36 (48.0) | |||
| Largest tumor size, No. (%) | |||||||||
| <5 cm | 29 (38.7) | 45 (60.0) | 0.009 | 35 (46.7) | 39 (52.0) | 0.514 | 42 (56.0) | 32 (42.7) | 0.102 |
| ≥5 cm | 46 (61.3) | 30 (40.0) | 40 (53.3) | 36 (48.0) | 33 (44.0) | 43 (57.3) | |||
| BCLC stage, No. (%) | |||||||||
| A | 23 (30.7) | 37 (49.3) | 0.020 | 27 (36.0) | 33 (44.0) | 0.317 | 34 (45.3) | 26 (34.7) | 0.182 |
| B | 52 (69.3) | 38 (50.7) | 48 (64.0) | 42 (56.0) | 41 (54.7) | 49 (65.3) | |||
Abbreviation: BCLC, barcelona clinic liver cancer; HB, hepatitis B.
TABLE 3.
Correlation of circ_0004913, circ_0008160, and circ_0000517 expressions with hepatic function index and tumor markers
| Items | Circ_0004913 expression | Circ_0008160 expression | Circ_0000517 expression | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | p value | Low | High | p value | Low | High | p value | |
| ALT, No. (%) | |||||||||
| <40 U/L | 57 (76.0) | 53 (70.7) | 0.460 | 53 (70.7) | 57 (76.0) | 0.460 | 52 (69.3) | 58 (77.3) | 0.268 |
| ≥40 U/L | 18 (24.0) | 22 (29.3) | 22 (29.3) | 18 (24.0) | 23 (30.7) | 17 (22.7) | |||
| AST, No. (%) | |||||||||
| <40 U/L | 39 (52.0) | 45 (60.0) | 0.324 | 41 (54.7) | 43 (57.3) | 0.742 | 46 (61.3) | 38 (50.7) | 0.188 |
| ≥40 U/L | 36 (48.0) | 30 (40.0) | 34 (45.3) | 32 (42.7) | 29 (38.7) | 37 (49.3) | |||
| ALP, No. (%) | |||||||||
| <128 U/L | 43 (57.3) | 52 (69.3) | 0.127 | 48 (64.0) | 47 (62.7) | 0.865 | 51 (68.0) | 44 (58.7) | 0.236 |
| ≥128 U/L | 32 (42.7) | 23 (30.7) | 27 (36.0) | 28 (37.3) | 24 (32.0) | 31 (41.3) | |||
| TBIL, No. (%) | |||||||||
| <19 μmol/L | 47 (62.7) | 44 (58.7) | 0.616 | 48 (64.0) | 43 (57.3) | 0.403 | 46 (61.3) | 45 (60.0) | 0.867 |
| ≥19 μmol/L | 28 (37.3) | 31 (41.3) | 27 (36.0) | 32 (42.7) | 29 (38.7) | 30 (40.0) | |||
| CEA, No. (%) | |||||||||
| <5 ng/ml | 36 (53.7) | 44 (65.7) | 0.159 | 37 (53.6) | 43 (66.2) | 0.139 | 44 (63.8) | 36 (55.4) | 0.323 |
| ≥5 ng/ml | 31 (46.3) | 23 (34.3) | 32 (46.4) | 22 (33.8) | 25 (36.2) | 29 (44.6) | |||
| CA199, No. (%) | |||||||||
| <27 U/ml | 53 (79.1) | 48 (71.6) | 0.316 | 49 (71.0) | 52 (80.0) | 0.228 | 51 (73.9) | 50 (76.9) | 0.686 |
| ≥27 U/ml | 14 (20.9) | 19 (28.4) | 20 (29.0) | 13 (20.0) | 18 (26.1) | 15 (23.1) | |||
| AFP, No. (%) | |||||||||
| <25 ng/ml | 24 (35.8) | 31 (46.3) | 0.219 | 24 (34.8) | 31 (47.7) | 0.129 | 26 (37.7) | 29 (44.6) | 0.415 |
| ≥25 ng/ml | 43 (64.2) | 36 (53.7) | 45 (65.2) | 34 (52.3) | 43 (62.3) | 36 (55.4) | |||
Abbreviation: AFP, alpha‐fetoprotein; ALP, alkaline phosphatase; ALT, Alanine aminotransferase; AST, aspartate aminotransferase; CA199, carbohydrate antigen 199; CEA, carcinoembryonic antigen; TBIL, total bilirubin.
3.4. Correlations of circ_0004913, circ_0008160, and circ_0000517 with HCC patients’ prognosis
The OS was more favorable in surgical HCC patients with circ_0004913 high expression compared to patients with circ_0004913 low expression (p = 0.008) (Figure 3A). However, the OS was similar between patients with circ_0008160 high expression and those with circ_0008160 low expression (p = 0.262) (Figure 3B), and between patients with circ_0000517 high expression and those with circ_0000517 low expression (p = 0.125) (Figure 3C). Furthermore, subgroup analyses revealed that, in patients with largest tumor size <5 cm, circ_0004913 low expression (p = 0.008) (Figure 4A) but not circ_0008160 expression (p = 0.052) (Figure 4B) or circ_0000517 expression (p = 0.648) (Figure 4C) was associated with better OS. In addition, in patients with largest tumor size ≥5 cm, circ_0004913 (p = 0.851) (Figure 4D), circ_0008160 (p = 0.601) (Figure 4E), or circ_0000517 expression (p = 0.148) (Figure 4F) was not correlated with OS. In BCLC stage A patients, circ_00004913 (p = 0.089) (Figure 4G), circ_0008160 (p = 0.266) (Figure 4H), or circ_0000517 (p = 0.511) (Figure 4I) expression was not correlated with OS, either. As for the BCLC stage B patients, the OS was more prolonged in patients with circ_0008160 low expression compared to those with circ_0008160 high expression (p = 0.026) (Figure 4K), while did not differ between patients with circ_0004913 high and those with circ_0004913 low expression (p = 0.085) (Figure 4J), nor between patients with circ_0000517 high expression and those with circ_0000517 low expression (p = 0.223) (Figure 4L).
FIGURE 3.
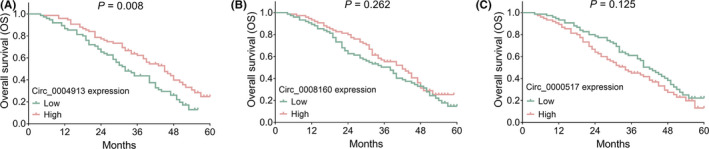
Associations of the three circRNAs with OS. The associations of circ_0004913 (A), circ_0008160 (B), and circ_0000517 (C) with OS in HCC patients. CircRNA, circular RNA; HCC, hepatocellular carcinoma; OS, overall survival
FIGURE 4.
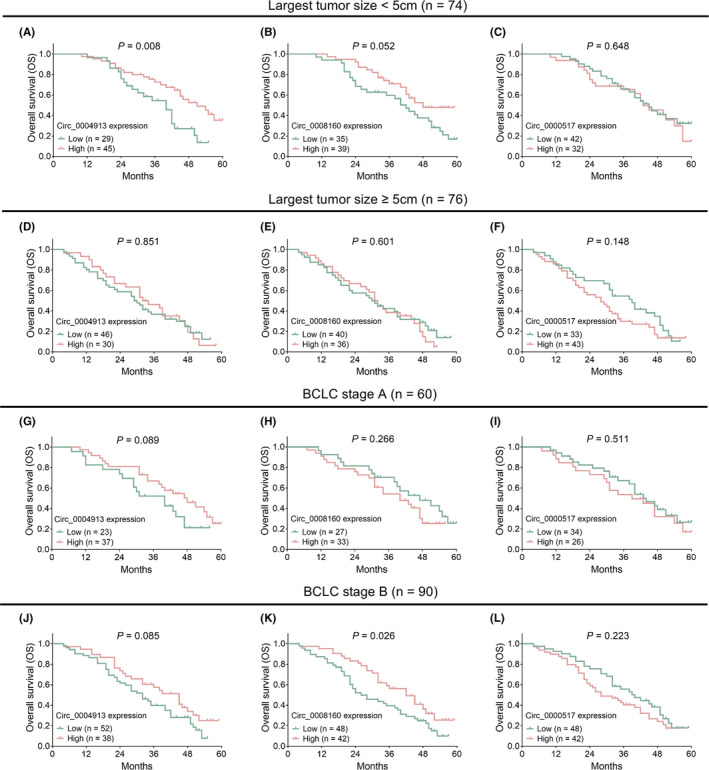
Subgroup analyses of the correlations of the three circRNAs with OS. The associations of circ_0004913, circ_0008160, and circ_0000517 with OS in HCC patients with largest tumor size <5 cm (A–C), in HCC patients with largest tumor size ≥5 cm (D–F), in BCLC stage A HCC patients (G–I) and in BCLC stage B HCC patients (J–L). BCLC, Barcelona Clinic Liver Cancer; CircRNA, circular RNA; HCC, hepatocellular carcinoma; OS, overall survival
3.5. Prognostic factor analyses
According to the univariate Cox's regression analysis, circ_0004913 high expression could predict increased OS in surgical HCC patients (p = 0.009) (Figure 5). In addition, Child‐Pugh stage (B vs. A) (p = 0.001), largest tumor size ≥5 cm (p = 0.001), and AST level ≥40 U/L (p = 0.014) could predict worse OS. As for the results from multivariate Cox's regression analysis, it was observed that circ_0004913 high expression (p = 0.098) was not independently associated with longer OS, while Child‐Pugh stage (B vs. A) (p < 0.001) and largest tumor size ≥5 cm (p = 0.002) were independently correlated with shorter OS.
FIGURE 5.
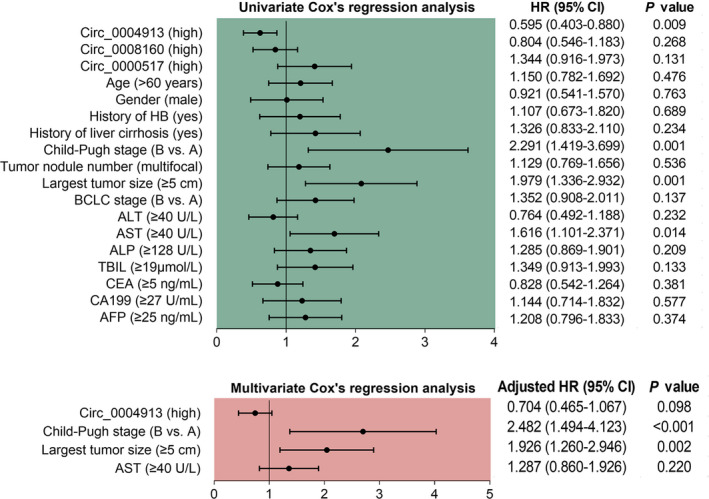
Univariate and multivariate Cox's regression analyses for OS predictive factors in HCC patients. AFP, alpha‐fetoprotein; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BCLC, Barcelona clinic liver cancer; CA199, carbohydrate antigen 199; CEA, carcinoembryonic antigen; HB, hepatitis B; HR, hazard ratio; OS, overall survival; TBIL, total bilirubin
4. DISCUSSION
As is the case with many other carcinomas, there is now extensive interest in the possible mechanistic and clinical value of circRNAs in HCC. For instance, a study elucidates that circRNA methionine adenosyltransferase 2A (MAT2B) increases glycolysis activities and promotes malignant behaviors of HCC under hypoxia condition in vivo and in vitro. 16 Another study exhibits that circ_0001955 represses cell proliferation in vitro and inhibits tumor growth in vivo via sponging miR‐516a‐5p and the subsequent release of TNF receptor associated factor 6 (TRAF6) and mitogen‐activated protein kinase 11 (MAPK11) in HCC. 17 In addition, a study discloses that circRNA MET enhances epithelial to mesenchymal transition (EMT) and immunosuppressive tumor microenvironment in vivo and in vitro through mediating the miR‐30‐5p/snail/DPP4 axis. 18 These studies reveal that circRNAs serve as mediators of HCC pathogenesis, and there are also several studies uncovering circRNAs as potential biomarkers for HCC management. For example, circ_0091579 was identified as a differentially expressed circRNA in HCC by microarray, and further analysis illuminates that it displays the same expression trend as that in microarray (upregulated), furthermore, it reveals with considerable specificity and sensitivity in predicting HCC risk, meanwhile is associated with dismal OS. 19 In terms of the three circRNAs assessed in our study, there are only a few studies that could be referred to, and circ_0008169 has not been reported yet. Previously, a study reports that circ_0004913 represses HCC cell growth, invasion, glycolysis, EMT, and inhibits tumor growth of HCC animal model via regulating hepcidin (HAMP) through sponging miR‐184. 12 Besides, circ_0000517 knockdown decreases HCC cell viability, colony formation, cell migration, cell invasion, and glycolysis by mediating the miR‐326/ insulin‐like growth factor 1 receptor (IGF1R) axis. 13 Although existing evidence is quite insufficient, these experimental studies of circ_0004913 and circ_0000517 may explain their potential clinical values found in our study to some extent.
In the present study, due to that three circRNAs, circ_0004913, circ_0008160, and circ_0000517 were discovered as dysregulated circRNAs in HCC tissues and HCC cells according to two datasets GSE94508 and GSE97332, in order to further evaluate their clinical values in HCC patients, the present study was conducted. 15 Subsequently, we discovered that (1) circ_0004913 and circ_0008160 were decreased while circ_0000517 was increased in tumor tissue compared with paired adjacent tissue in HCC patients. (2) circ_0004913 was correlated with largest tumor size, BCLC stage and worse OS. (3) Subgroup analyses disclosed that circ_0004913 correlated with decreased OS in patients with largest tumor size <5 cm, and circ_0008169 correlated with shorter OS in patients in BCLC B stage. In addition, univariate Cox's regression analysis revealed that circ_0004913 high expression could predict worse OS in HCC patients. Based on the several mechanistic studies in HCC, we presumed that the possible explanations to our findings may include the following speculations. In regard to circ_0004913, it was probable that circ_0004913 could repress the progression of HCC tumor via modulating multiple related factors as reported previously, resulting in its downregulation in HCC tissues and its negative correlation with clinical features of HCC patients; then the delay in progression caused a more prolonged survival time, hence its high expression could predict favorable OS in patients. 12 In terms of circ_0000517, it is able to promote the malignant behaviors in HCC via mediating factors such as the miR‐326/IGF1R axis, which then contributed to its upregulation in HCC tissues. 13 , 14 As for circ_0008160, we hypothesized that it might act as an inhibitor of tumor progression of HCC, nonetheless, no experimental findings could be referred to currently.
In our current study, there were several limitations that should be discussed. First, only patients who had received resection before were analyzed in this study, therefore the clinical values of these three circRNAs in unresectable HCC patients were not assessed. Second, the sample size of 150 HCC patients was relatively small. Third, peripheral expressions of these three circRNAs in HCC patients were not evaluated, and the tissue samples might be relatively hard to access for clinical application, especially for non‐surgical patients. However, in the clinical setting, the circRNA expressions are more easily to detect compared to their expressions in the peripheral blood, due to that circRNA expression is more abundant in the tissue.
In summary, circ_0004913 is downregulated in tumor tissue and may serve as a biomarker for the evaluation of disease severity and prognosis in HCC patients.
CONFLICT OF INTEREST
No potential conflict of interest was reported by the authors.
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in this article.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Massarweh NN, El‐Serag HB. Epidemiology of Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. Cancer Control. 2017;24(3):1073274817729245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ayuso C, Rimola J, Vilana R, et al. Diagnosis and staging of hepatocellular carcinoma (HCC): current guidelines. Eur J Radiol. 2018;101:72‐81. [DOI] [PubMed] [Google Scholar]
- 4. Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10(21):5015‐5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Altesha MA, Ni T, Khan A, et al. Circular RNA in cardiovascular disease. J Cell Physiol. 2019;234(5):5588‐5600. [DOI] [PubMed] [Google Scholar]
- 6. Akhter R. Circular RNA and Alzheimer's Disease. Adv Exp Med Biol. 2018;1087:239‐243. [DOI] [PubMed] [Google Scholar]
- 7. Liang HF, Zhang XZ, Liu BG, et al. Circular RNA circ‐ABCB10 promotes breast cancer proliferation and progression through sponging miR‐1271. Am J Cancer Res. 2017;7(7):1566‐1576. [PMC free article] [PubMed] [Google Scholar]
- 8. Wang S, Zhang Y, Cai Q, et al. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol Cancer. 2019;18(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Legnini I, Di Timoteo G, Rossi F, et al. Circ‐ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22‐37.e29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chase‐Lubitz JF, Alexander JM. The new Rhode Island AIDS statute. R I Med J. 1988;71(11):437‐443. [PubMed] [Google Scholar]
- 12. Wu M, Sun T, Xing L. Circ_0004913 inhibits cell growth, metastasis, and glycolysis by absorbing miR‐184 to regulate HAMP in hepatocellular carcinoma. Cancer Biother Radiopharm. 2020. 10.1089/cbr.2020.3779 [DOI] [PubMed] [Google Scholar]
- 13. He S, Yang J, Jiang S, et al. Circular RNA circ_0000517 regulates hepatocellular carcinoma development via miR‐326/IGF1R axis. Cancer Cell Int. 2020;20:404. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Zang H, Li Y, Zhang X, et al. Circ_0000517 Contributes to hepatocellular carcinoma progression by upregulating TXNDC5 via sponging miR‐1296‐5p. Cancer Manag Res. 2020;12:3457‐3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang F, Xu X, Zhang N, et al. Identification and integrated analysis of hepatocellular carcinoma‐related circular RNA signature. Ann Transl Med. 2020;8(6):294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Q, Pan X, Zhu D, et al. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR‐338‐3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298‐1316. [DOI] [PubMed] [Google Scholar]
- 17. Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR‐516a‐5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10(12):945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang XY, Zhang PF, Wei CY, et al. Circular RNA circMET drives immunosuppression and anti‐PD1 therapy resistance in hepatocellular carcinoma via the miR‐30‐5p/snail/DPP4 axis. Mol Cancer. 2020;19(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang C, Zhang C, Lin J, et al. Circular RNA Hsa_Circ_0091579 serves as a diagnostic and prognostic marker for hepatocellular carcinoma. Cell Physiol Biochem. 2018;51(1):290‐300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that supports the findings of this study are available in this article.


