Sir,
A novel SARS-CoV-2 causing the global coronavirus disease 2019 pandemic has affected most countries and territories around the world with a death toll of more than one million cases. As of November 19, 2020, a total of 56,674,523 cases have been reported for SARS-CoV-21. The entry mechanism of SARS-CoV-2 involves the binding of spike (S) protein to the angiotensin-converting enzyme 2 (ACE-2) receptor of host cell through the receptor-binding domain (RBD)2. The ectodomain of the S protein consists of S1 and S2 subunits. The S1 subunit contains the RBD, and is involved in recognition and binding, whereas the S2 subunit is associated with the fusion mechanism3.
The race for finding a potential antiviral agent and development of a protective vaccine against SARS-CoV-2 is still on. The RNA interference (RNAi)-based strategies can be a promising treatment option to combat SARS-CoV-24. RNAi is an evolutionary mechanism of gene regulation induced by small interfering RNA (siRNA) along with a specific endonuclease. Synthetic siRNAs are 19-23 nt long RNAs, containing a complementary sequence to a target region on the genome sequence that can block the protein translation by hybridizing to the target. Targeting the S gene for designing siRNAs would be effective since siRNA against this gene would inhibit its translation, reduce the protein availability for formation of functional infectious virions and, as a result, reduce the host cell infectivity. However, it is important that the design of the siRNAs should be restricted to the highly conserved regions in the S gene, to ensure that these will be effective against all strains of SARS-CoV-2.
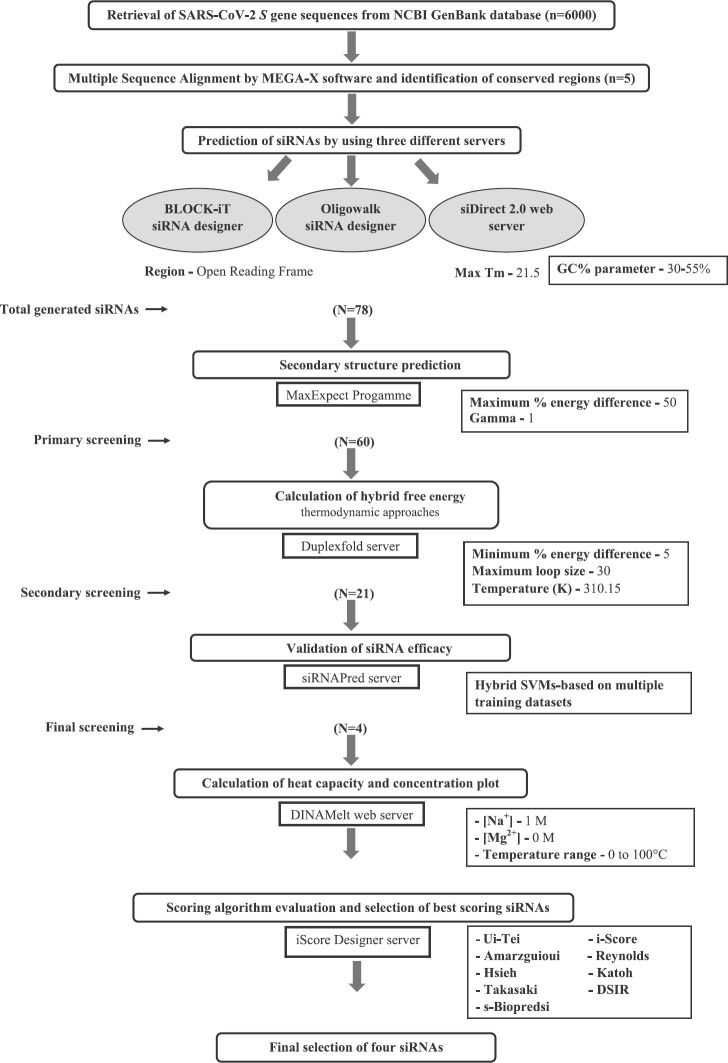
In this study conducted at ICMR-National Institute of Virology, Pune, India, a total of 6000 different S gene sequences of SARS-CoV-2 from different regions of the world were retrieved from the NCBI GenBank database as on May 25, 2020. The S gene of SARS-CoV-2 lies in the region from 21563 to 25384 nt. Using multiple sequence alignment, the conserved regions within the S gene sequences were identified using the MEGA-X software5. The conserved regions shorter than 30 nt were not incorporated for the study. The target sequences for siRNA binding were identified using the predictions from three different online siRNA designing servers: Block-iT RNAi designer6, OligoWalk siRNA designer server6 and siDirect 2.0 web server7. Block-iT RNAi designer (Thermo Fisher Scientific, USA) is an online siRNA design server requiring the user to mainly specify the minimum and maximum guanine-cytosine (GC) content. The server OligoWalk generates siRNAs through the calculation of the thermodynamic free energy of hybridization and the use of support-vector machines (SVMs)8. The server siDirect 2.0 generates efficient siRNAs by minimizing off-target effects through calculation of thermodynamic stabilities of the seed-target duplex which is formed between the nucleotides positioned at 2-8 from the 5' end of the siRNA guide strand and its target mRNA7. The selection of lower thermodynamic stability defined by the melting temperature (Tm) (benchmark Tm <21.5°C) is followed by the elimination of unrelated transcripts with nearly perfect match. A sequence with lower GC content is more preferred as a siRNA target because of its lesser probability to form secondary structures with strong bonds9. Hence, in all the three servers, the range for GC content of the target sequences was selected to be 30 to 55 per cent.
The predicted sequences of the siRNAs were further screened for effectiveness by determining their secondary structure. The secondary structure of the selected siRNAs was generated using the MaxExpect10 programme in the RNA structure webserver (https://rna.urmc.rochester.edu/RNAstructureWeb/Servers/MaxExpect/MaxExpect html Maxexpect). The MaxExpect server generates a specified group of secondary structures from the given RNA sequence, each structure in the group containing base pairs which have the highest possible chance of being accurate. The gamma parameter value was kept as one to maintain a good balance in providing the weight on pairing and non-pairing bases during the secondary structure prediction, while the standard temperature of 37°C was selected10.
Next, the RNA-RNA interaction of the target site and siRNA was analyzed using the server DuplexFold11 which is based on folding the two given RNA sequences into their lowest hybrid free energy conformation. Default settings were considered for optimal specifications for the maximum permitted per cent energy difference, maximum number of output structures, window size, maximum loop size and temperature. Finally, an online siRNA validation server (siRNAPred)12 (http://crdd.osdd.net/raghava/sirnapred/algo.html) was used to validate the efficacy of the predicted siRNA. The siRNAPred incorporates hybrid SVM-based methods for predicting the actual efficacy of both 21-mer and 19-mer siRNAs with high accuracy.
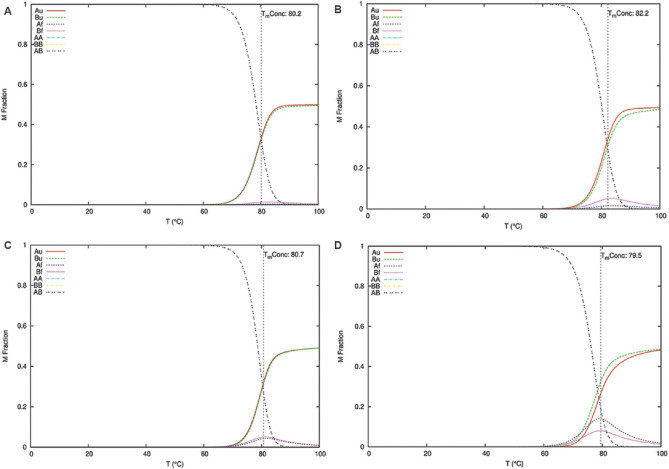
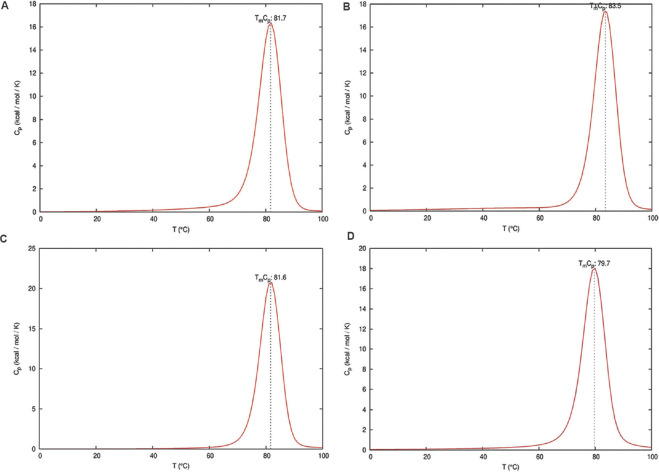
The ENDMEMO online server (http://www.endmemo.com/bio/gc.php) was used to determine the GC percentage of the predicted siRNAs. The determination of the entire equilibrium melting profile of the designed siRNAs is an important criterion for evaluation of its inhibition potency. The DINAMelt web server13 was used to determine the heat capacity plot and concentration plot for the designed siRNAs. The detailed heat capacity plot helps to determine the contribution of each species to the ensemble heat capacity (Cp). The concentration plot-Tm (Conc.) indicates the temperature at which the concentration of double-stranded siRNA molecules becomes one-half of its maximum value. The parameters such as temperature range and initial concentrations were kept at default value based on the standard described by Markham and Zuker13. The best antisense RNAs of the selected regions were evaluated by i-Score designer14,15 which evaluates nine different siRNA designing scores (Ui-Tei, Amarzguioui, Hsieh, Takasaki, s-Biopredsi, i-Score, Reynolds, Katoh and DSIR). Further monitoring of the target sequences of the final shortlisted siRNAs was done by analyzing the mutation information from the 2019nCoVR database16 (https://bigd.big.ac.cn/ncov/?lang=en).
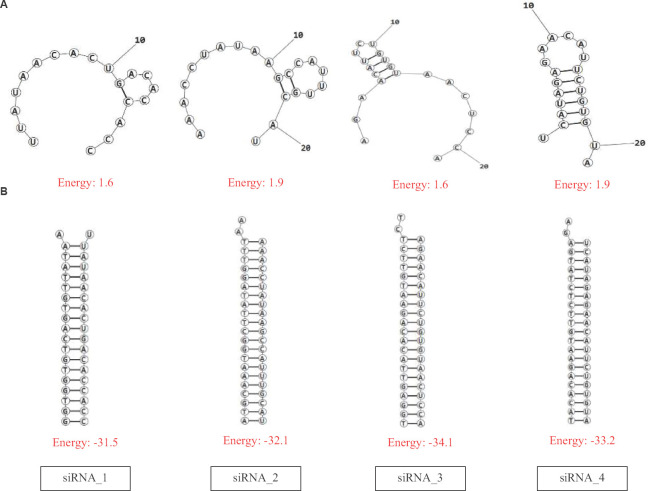
Multiple sequence alignment of the S gene sequences (n=6000) identified five different conserved regions (nucleotide positions: 23,312-23,370; 23,474-23,535; 24,260-24,324; 24,575-24,619 and 24,242-24,347) which were considered for predicting the siRNA target regions. The three different siRNA prediction servers proposed a total of 78 different target sequences (Fig. 1). No common target regions were predicted by either of the servers. Further, the selection of more effective siRNAs was done based on the free energy of folding values obtained from the predicted secondary structures through MaxExpect. siRNAs with free energy of folding greater than zero indicate more efficient binding as it will be less prone to form a secondary structure17. A higher value of free energy of folding indicates lower folding probability and more efficient binding17. All the predicted siRNAs with a free energy of folding greater than 1.5 were selected (n=60). Further shortlisting of the siRNAs was done based on the free energy of binding of the antisense RNA towards the target sequence obtained from DuplexFold. Lower free energy of binding value signifies higher siRNA potency, as it will have efficient binding capability and better ability to inhibit the target sequence17. The siRNAs with lower free energy of binding were thus selected and considered for further validation of siRNA efficacy using siRNA validation server. The cut-off value of −30 was considered for further shortlisting of the siRNAs (n=21)17. Finally, based on the result of the validation server, siRNAPred at a cut-off value of 0.717, four different siRNA target regions were identified (Fig. 2) between 23339 and 24317 nt positions of the S gene sequences (Table). The siRNA_1, siRNA_2 and siRNA_4 have a validation score of more than 0.9, indicating higher inhibition efficacy12. Within the spike protein, the nucleotide region 225, 17-23, 185 nt is translated into the RBD, and hence, the target binding regions of the selected siRNAs are located outside the RBD. The proposed siRNA_2, siRNA_3 and siRNA_4 comprise 21-mer sequences, whereas the siRNA_1 is a 19 mer. All the four identified siRNAs of the target sequence were located in the S2 domain; three (siRNA_2, siRNA_3 and siRNA_4) in the region that is translated into the heptad repeat 1 (HR1), which plays a critical role in viral entry18. The siRNA_2, siRNA_3 and siRNA_4 were noted to be predicted from the siDirect 2.0 servers, while the siRNA_1 was predicted from Block-iT RNAi designer. The GC content of the siRNA molecule is an important parameter for its functionality. The predicted siRNAs in this study were found to possess GC content in the range of 33 to 42 per cent (Table). The Tm (Cp) and Tm (Conc.) values were calculated using the DINAMelt web server which defines the entire equilibrium melting profile of the siRNAs. All the four siRNAs had a Tm (Conc.) value between 79.5 and 82.2, whereas Tm (Cp) value ranged from 79.7 to 83.5 (Table). Values greater than 75 indicate higher effectiveness17,19. The graphical representation of the Tm values is shown in Figures. 3 and 4. In the i-Score designer server, the selected siRNAs were scored using different algorithms and the score-based ranks according to i-Score, s-Biopredsi and DSIR indicate the probability of being the best siRNA20. All the four shortlisted siRNAs in this study showed rank one by all the above three scores. The 2019nCoVR database is maintained by the China National Center for Bioinformation and provides information regarding the sequence variability16. No such information of mutations of the target sequences was indicated by the server, indicating the suitability of the designed siRNAs.
Fig. 1.
Flow chart for prediction of potential small interfering RNAs (siRNAs) against SARS-CoV-2 spike gene. The successive siRNAs screened and shortlisted by different servers is denoted by the values of ‘N’ in the bracket. Different parameters selected for the various softwares are indicated in the rectangular boxes towards the right.
Fig. 2.
(A) Secondary structure prediction and free energy of folding of the predicted siRNAs. (B) Lowest free energy structure upon binding of the predicted siRNAs with the target sequences.
Table.
Details of the best predicted small interfering RNAs (siRNAs) against SARS-CoV-2
| Serial number | Target region | siRNA sequences | Free energy of folding | Free energy of binding | Validation score | GC % | Tm (concentration) | Tm (Cp) |
|---|---|---|---|---|---|---|---|---|
| siRNA_1 | 23339-23357 nt | Antisense - UUAUAACACUGACACCACC Sense - GGUGGUGUCAGUGUUAUAA |
1.6 | −31.5 | 0.917 | 42 | 80.2 | 81.7 |
| siRNA_2 | 24260-24282 nt | Antisense - AAACCUAUAAGCCAUUUGCAU Sense - GCAAAUGGCUUAUAGGUUUAA |
1.9 | −32.1 | 0.964 | 33 | 82.2 | 83.5 |
| siRNA_3 | 24289-24311 nt | Antisense - AGAACAUUCUGUGUAACUCCA Sense - GAGUUACACAGAAUGUUCUCU |
1.6 | −34.1 | 0.781 | 38 | 80.7 | 81.6 |
| siRNA_4 | 24295-24317 nt | Antisense - UCAUAGAGAACAUUCUGUGUA Sense - CACAGAAUGUUCUCUAUGAGA |
1.9 | −33.2 | 0.935 | 35 | 79.5 | 79.7 |
GC, guanine-cytosine; Tm, melting temperature
Fig. 3.
Equilibrium melting profile of the hybridized siRNAs in terms of concentration plot (Conc.). Concentration plots of siRNAs_1-4 are represented in (A-D), respectively. X-axis represents temperature in degree Celsius and Y-axis represents the mole fraction of each species of the siRNA strands. The red and green lines indicate the concentrations of the unfolded single strands (Au- A strand unfolded; Bu- B strand unfolded), and the blue and magenta lines show the folded single strands (Af- A strand folded; Bf- B strand folded). The yellow and cyan curves correspond to the two homodimers (AA & BB) and the black curve to the heterodimer (AB).
Fig. 4.
Equilibrium melting profile of the hybridized siRNAs in terms of heat capacity (Cp) plot. Heat capacity plots of siRNAs_1-4 are represented in (A-D), respectively. X-axis represents temperature in degree Celsius and Y-axis represents the heat capacity which is calculated by numerical differentiation of the ensemble free energy.
In a recent study by Chowdhury et al17, eight potential siRNAs were designed for SARS-CoV-2 using computational methods based on conserved sequences in the nucleocapsid phosphoprotein genes and the surface spike glycoprotein gene derived from a smaller sequence dataset of 139 strains. Among the siRNAs predicted in the S gene, two siRNA target regions were located within the RBD region, whereas one target region was located within the fusion peptide region. The other siRNA sequences predicted against the S gene were targeted towards the S2 domain in no specific functional region17. Another study by Chen et al21 also identified nine different siRNA target sequences for SARS-CoV-2 using a single reference sequence and computational approaches. The designed siRNAs were mainly located in Orf1ab, Orf1b, S gene, Orf3a, M gene and N gene. The target sequences for the S gene (n=1) were located within the S1 domain in no specific functional region. A study by Shi et al22 reported three different siRNAs against the earlier SARS-CoV-1 structural proteins (E, M and N) that reduced 80 per cent of the target gene expression. A total of 35 patent applications have been disclosed by Chemical Abstracts Service (CAS) for designed siRNAs against SARS-CoV-123. Most of the siRNAs were targeted towards the structural protein nucleotide sequences such as S, E, N and M genes23. CAS disclosed the siRNA target region information from patent application US20050004063, which was located within the nucleotide region 23165-23186 nt of the S gene, which was also translated into the RBD region23. The exact nucleotide information of the target regions is not available for the other four patent applications disclosed by CAS, which also target the S gene. However, as there is a considerable difference between the genomes24 and specifically the S gene sequences of SARS-CoV-1 and SARS-CoV-2, it is less likely that siRNAs designed for SARS-CoV-1 would be effective for SARS-CoV-223.
The four siRNAs designed in our study are based on the conserved regions identified from 6000 different SARS-CoV-2 sequences and considering the prediction from multiple prediction servers. This enabled a larger initial dataset for screening of potential siRNAs to increase the probability of designing highly functional siRNAs against the SARS-CoV-2. Even though the regions targeted by these siRNAs were noted to be devoid of mutations based on the large initial global dataset and the 2019nCoVR database, continuous monitoring of the variability in the target regions is mandated. Three of the siRNAs predicted in this study were targeted towards the HR1 nucleotide region, while the other target sequence was not found to be located in a specific functional region. Heptad repeats are common in both SARS-CoV-1 and SARS-CoV-2, though the nucleotide sequence and the translation pattern are different in both the viruses. The target siRNA sequences are unique in SARS-CoV-2, which indicates the novelty from previously designed siRNAs for SARS-CoV-122,23. The RNAi technology has the potential to combat viral pathogens as these are highly specific towards the target sequence and are also flexible for targeting multiple strains of the virus25. In SARS-CoV-2-mediated infection, the ciliated cells of lungs are the primary site for viral entry26. Several potential therapies against the SARS-CoV-2 are currently under experimental and developmental stages. The predicted siRNAs, having met the criteria of standard siRNA molecules, may therefore, be attempted as an alternative therapeutic/antiviral approach against SARS-CoV-2.
siRNA-based strategy for use against SARS-CoV-2 has to overcome many challenges such as high susceptibility to degradation, off-target gene silencing and activation of immune response. Further, the proper delivery of a targeted molecule into the host cell can provide better results towards reduction of viral copy number through the mechanism of gene silencing. A recent study27 presented in vivo data in a mice model, in which aerosolized delivery of siRNA via pressurized syringe to the selected organ was found to be effective for respiratory infections. Hence, the siRNAs predicted in this study would further need to be validated by in vitro studies, and later in vivo approaches can also be considered.
In conclusion, the present study predicted four potential siRNAs based on the evaluation of predictions from three different siRNA prediction servers and additional validation from other in silico tools to ensure that the predicted siRNAs would have the ability to interact efficiently with the target sequence with minimal non-specific binding. The predicted siRNAs may be useful in developing RNAi-based therapeutics against SARS-CoV-2 if found effective by in vitro and in vivo studies.
Acknowledgment:
Authors acknowledge Shri Santosh Jadhav for inputs to the gene sequence data set.
Footnotes
Conflicts of Interest: None.
References
- 1.Coronavirus. [accessed on November 19, 2020]. Available from: www.worldometers.info/coronavirus/
- 2.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:11727–34. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARSCoV-2 spike glycoprotein. Cell. 2020;181:281–92.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uludağ H, Parent K, Aliabadi HM, Haddadi A. Prospects for RNAi therapy of COVID-19. Front Bioeng Biotechnol. 2020;8:916. doi: 10.3389/fbioe.2020.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villegas-Rosales PM, Méndez-Tenorio A, Ortega-Soto E, Barrón BL. Bioinformatics prediction of siRNAs as potential antiviral agents against dengue viruses. Bioinformation. 2012;8:519–22. doi: 10.6026/97320630008519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naito Y, Yoshimura J, Morishita S, Ui-Tei K. siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics. 2009;10:1–8. doi: 10.1186/1471-2105-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu ZJ, Mathews DH. OligoWalk: An online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 2008;36:W104–8. doi: 10.1093/nar/gkn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Chang Y, Zhang C, Wei Q, Chen J, Chen H, et al. Influence of mRNA features on siRNA interference efficacy. J Bioinformatics Comput Biol. 2013;11:1341004. doi: 10.1142/S0219720013410047. [DOI] [PubMed] [Google Scholar]
- 10.Bellaousov S, Reuter JS, Seetin MG, Mathews DH. RNA structure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013;41:W471–4. doi: 10.1093/nar/gkt290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reuter JS, Mathews DH. RNA structure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar M, Lata S, Raghava GPS. Proceedings of the OSCADD-2009: International Conference on Open Source for Computer Aided Drug Discovery; 2009 Mar 22-26. Chandigarh: IMTECH; 2009. siRNApred: SVM based method for predicting efficacy value of siRNA. [Google Scholar]
- 13.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–81. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ichihara M, Murakumo Y, Masuda A, Matsuura T, Asai N, Jijiwa M, et al. Thermodynamic instability of siRNA duplex is a prerequisite for dependable prediction of siRNA activities. Nucleic Acids Res. 2007;35:e123. doi: 10.1093/nar/gkm699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Hefnawi M, Hassan N, Kamar M, Siam R, Remoli AL, El-Azab I, et al. The design of optimal therapeutic small interfering RNA molecules targeting diverse strains of influenza A virus. Bioinformatics. 2011;27:3364–70. doi: 10.1093/bioinformatics/btr555. [DOI] [PubMed] [Google Scholar]
- 16.Zhao WM, Song SH, Chen ML, Zou D, Ma LN, Ma YK, et al. The 2019 novel coronavirus resource. Yi Chuan. 2020;42:212–21. doi: 10.16288/j.yczz.20-030. [DOI] [PubMed] [Google Scholar]
- 17.Chowdhury UF, Sharif Shohan MU, Hoque KI, Beg MA, Moni MA, Sharif Siam MK. A computational approach to design potential siRNA molecules as a prospective tool for silencing nucleocapsid phosphoprotein and surface glycoprotein gene of SARS-CoV-2. bioRxiv. 2020 doi: 10.1016/j.ygeno.2020.12.021. doi: 10.1101/2020.04.10.036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17:765–7. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nur SM, Hasan MA, Amin MA, Hossain M, Sharmin T. Design of potential RNAi (miRNA and siRNA) molecules for middle east respiratory syndrome coronavirus (MERS-CoV) gene silencing by computational method. Interdiscip Sci. 2015;7:257–65. doi: 10.1007/s12539-015-0266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–30. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 21.Chen W, Feng P, Liu K, Wu M, Lin H. Computational identification of small interfering RNA targets in SARS-CoV-2. Virol Sin. 2020;35:359–61. doi: 10.1007/s12250-020-00221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y, Yang DH, Xiong J, Jia J, Huang B, Jin YX. Inhibition of genes expression of SARS coronavirus by synthetic small interfering RNAs. Cell Res. 2005;15:193–200. doi: 10.1038/sj.cr.7290286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020;6:315–31. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–36. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qureshi A, Tantray VG, Kirmani AR, Ahangar AG. A review on current status of antiviral siRNA. Rev Med Virol. 2018;28:e1976. doi: 10.1002/rmv.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh S, Firdous SM, Nath A. siRNA could be a potential therapy for COVID-19. EXCLI J. 2020;19:528–31. doi: 10.17179/excli2020-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodgson J. The pandemic pipeline. Nat Biotech. 2020;38:523–32. doi: 10.1038/d41587-020-00005-z. [DOI] [PubMed] [Google Scholar]