Abstract
Background & objectives:
Since its first recognition in Wuhan, China, in December 2019, the SARS-CoV-2 has spread rapidly across the world. Though SARS-CoV-2 spreads mainly via the droplets of respiratory secretions, it was also detected in stool samples of patients, indicating active infection of the gastrointestinal tract. Presence of SARS-CoV-2 RNA in sewage samples was reported in February 2020, raising the possibility of using environmental water surveillance to monitor SARS-CoV-2 activity in infected areas. The aim of this study was to standardize the methodology for detection of SARS-CoV-2 from sewage and explore the feasibility of establishing supplementary surveillance for COVID-19.
Methods:
Sewage specimens were collected from six sites in Mumbai, India, using the grab sample method and processed using polyethylene glycol (PEG)-dextran phase separation method for virus concentration. Real-time reverse transcription-polymerase chain reaction (RT-PCR) assay was used to detect the presence of SARS-CoV-2 RNA.
Results:
A total of 20 sewage samples collected from six different wards in Mumbai city, before the spread of SARS-CoV-2 infections and during May 11-22, 2020, were processed using the phase separation method. The WHO two-phase PEG-dextran method was modified during standardization. SARS-CoV-2 was found to concentrate in the middle phase only. All samples collected before March 16, 2020 were SARS-CoV-2 negative. Viral RNA was detected in sewage samples collected during the ongoing COVID-19 pandemic in all the six wards.
Interpretation & conclusions:
PEG-dextran phase separation method was effectively used to concentrate SARS-CoV-2 from domestic waste waters to detection levels. It would be feasible to initiate sewage surveillance for SARS-CoV-2 to generate data about the viral transmission in various epidemiologic settings.
Keywords: COVID-19, RT-PCR, SARS-CoV-2, sewage, two-phase separation
The World Health Organization (WHO) declared COVID-19 a global pandemic on March 11, 20201. The virus spread quickly across countries due to its respiratory route of spread and also the presence of susceptible population globally2. The unique feature of SARS-CoV-2 infection is the prolonged shedding of virus in stools, with viral load being high in stool samples3. While the major focus remains on containment zones, it is critical to ensure adequate surveillance in apparently low-transmission zones to monitor the disease trends. Such surveillance includes serological assays to understand the seroprevalence of infection and in turn the disease transmission dynamics. However, because prolonged shedding of SARS-CoV-2 RNA has been reported in stool samples of both symptomatic and asymptomatic infected individuals3, sewage surveillance seems to be a useful tool for monitoring the trend of COVID-19 disease in high- as well as low-transmission zones. Sewage surveillance has been used successfully as a tool to monitor the shedding of SARS-CoV-2 in stool samples in The Netherlands4. Presence of live virus in stool and the subsequent implications on faeco-oral transmission are still debated. Here, we describe the methodology standardized at the ICMR-National Institute of Virology, Mumbai Unit, Mumbai, India, for sewage surveillance for COVID-19. It was further planned to explore the feasibility of establishing sewage surveillance for COVID-19.
Material & Methods
Sewage samples were collected from a total of six sites in Mumbai, India. One of the sites was a sewage pumping station and the remaining sites were large open drains. Samples collected before the COVID-19 pandemic were available from each site. Fresh sewage samples were collected between May 11-22, 2020. One litre grab sample of sewage was collected by lowering a bucket in the flowing sewage water. The sample was transferred to a glass bottle containing 100 ml chloroform and a stirrer bar. The bottle contents were mixed well at the site. Samples were collected between 1000 and 1130 h. The samples were transported to the testing laboratory on wet ice within two hours. Personal protection equipment including face shield were used for protection. Materials used for sample collection were autoclave sterilized in the laboratory.
Sample processing for viral concentration: The sewage samples with chloroform were mixed thoroughly using a magnetic stirrer for 30 min at 4oC. The samples were centrifuged at 3000 × g for 20 min at 4°C and supernatant was collected. To 500 ml clarified sewage sample, 39.5 ml of 22 per cent (w/w) dextran T40 (Sigma, USA), 287 ml of 29 per cent (w/w) PEG6000 (polyethylene glycol 6000) (Sigma, USA) and 35 ml of 5 M NaCl (Sigma, USA) were added sequentially with constant mixing. The mixture was stirred for one hour at 4°C before setting up phase separation. The sample was transferred to a one litre separating funnel and allowed to stand at 4°C for 16 h for partitioning of the virus into one of the phases5,6,7. After phase separation, the lower, middle/inter and upper phases were collected individually for SARS-CoV-2 detection by real-time reverse transcription-polymerase chain reaction (RT-PCR)8.
RNA extraction and real-time RT-PCR: Viral RNA was extracted from the clarified sewage sample and the three phases using QIAamp Viral RNA Mini Kit (QIAGEN, Germany) according to the manufacturer's instructions. Presence of SARS-CoV-2 was inferred by detecting envelope (E) and RNA-dependent RNA polymerase (RdRp) genes amplification using LabGun COVID-19 real-time RT-PCR kit (LabGenomics, The Republic of Korea) as per the manufacturer's instructions. The kit manufacturer set a cut-off cycle threshold (Ct) for scoring a clinical sample positive for SARS-CoV-2 as Ct ≤40. The following reaction conditions were used: 50°C for 30 min, 45 cycles of 95°C for 15 min, 95°C for 15 sec and 60°C for one minute in a 7500 real-time PCR system (Applied Biosystems, USA). The RNA extracted from the clarified sewage sample and the lower, middle and upper phases was tested for SARS-CoV-2 RNA. Samples showing sigmoid curves of increasing fluorescence intensity in both the gene segments were considered positive for the virus.
Results
The six sewage collection sites were chosen for detecting SARS-CoV-2 because of the presence of large slum-like areas housing low socio-economic groups and substantial migrant populations. These same sites were being used for poliovirus environmental surveillance (ES). The Wadala pumping station (Ward F/N) was the only site where piped sewage was collected. Table I presents the date of the first COVID-19 case and the increasing intensity of the pandemic in each ward. The first COVID-19 case in Mumbai was reported on March 11, 2020. The selected wards experienced COVID-19 cases in the mid-March, and the case numbers increased from <20 in the early April to more than 1000 by the mid-May 2020. At least one sewage sample collected before the COVID-19 outbreak and two samples during the COVID-19 pandemic were used in the study.
Table I.
Confirmed COVID-19 cases in BMC wards where sewage samples were collected during May 11-22, 2020
| Sewage site | BMC ward | Population | COVID-19 cases as of date | ||
|---|---|---|---|---|---|
| Date of the first case in March 2020 | April 4 | May 22 | |||
| Wadala | F north | 551079 | 20 | 5 | 1534 |
| Dharavi | G north | 627744 | 30 | 4 | 1969 |
| Kurla | L | 928813 | 22 | 6 | 1450 |
| Shivaji Nagar | M east | 840461 | Not available | 17 | 1140 |
| Malad | P north | 983421 | 22 | 18 | 534 |
| Kanjur | S | 773365 | 15 | 11 | 1373 (May 26) |
As shown in (Fig. 1), PEG-dextran in water was separated in three phases, partitioning molecules as per their characteristics. If SARS-CoV-2 partitioned into lower or middle phase, it would get concentrated, enabling detection by PCR. The volume of middle phase varied from 2.2 to 4.5 ml (mean=2.7 ml) which was approximately 200-fold concentration if the virus partitioned into this phase and 7 to 10 ml (mean=8.3 ml) for the lower phase with a concentration factor of approximately 60-fold. In SARS-CoV-2 spiked sewage concentrates, no inhibitory effects of PEG/dextran T40 were observed (data not shown).
Fig. 1.
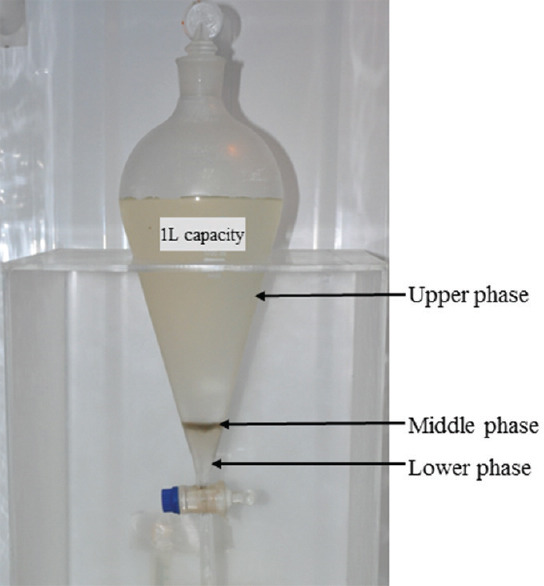
Separating funnel showing three phases of PEG6000-dextranT40-sewage sample mixture after 16 h at 4°C. The lower, middle and upper phases were collected individually for RNA extraction.
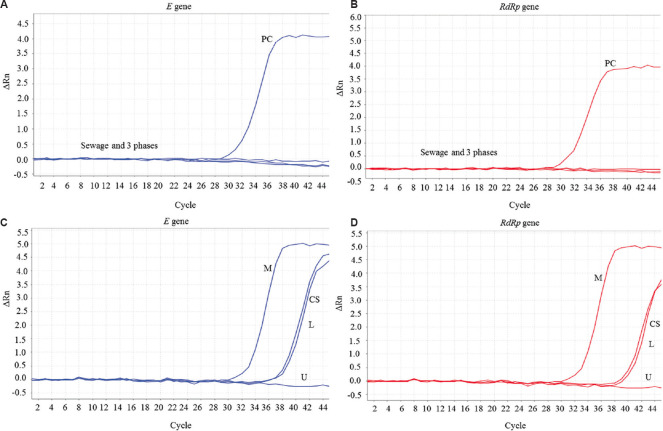
Real-time RT-PCR amplification curves of SARS-CoV-2 E and RdRp genes are presented in Figure 2. Figure 2A and B show completely flat curves for all the four RNA extracts (clarified sewage and that in lower, middle and upper phases) of a sewage sample collected in February 2020; figure 2C and D show the amplification curves of a sample collected in May 2020. RNA extracted from the middle phase gave the lowest Ct values of amplification for both the genes. The Ct values of middle phase RNA extracts were 3-9 points lower than the corresponding clarified sewage samples, confirming that SARS-CoV-2 partitioned only to the middle phase (Table II). Sewage samples with middle phase Ct values lower by 3 or more points were considered positive for SARS-CoV-2. Amplification curves (Ct >39) in the other phases could be due to a minor fraction of the viral particles or viral RNA that did not partition to the middle phase. Amplification curves with Ct >39 were observed in the RNA extracted directly from the sewage samples collected in May 2020 when the number of COVID-19 cases was increasing rapidly. Improved (lower) Ct values after concentration in the middle phase provided evidence of the presence of the virus.
Fig. 2.
Results of E and RdRp genes real-time RT-PCR of RNA extracted from clarified sewage and the three phases after phase separation. (A and B) SW-3713 (February 3, 2020); (C and D) SW-3737 (May 12, 2020). PC, positive control; CS, clarified sewage; L, lower phase; U, upper phase; M, middle phase. SARS-CoV-2 concentrated in to the middle phase. No amplification seen in SW-3713, the pre-COVID-19 sample.
Table II.
Results of real-time RT-PCR tests on RNA extracted from phase separation of sewage samples for SARS-CoV-2 detection
| Sample_ID | Site | SARS-CoV-2 real time RT-PCR result | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Ct values for gene E | Ct values for gene RdRp | ||||||||
| Clarified sewage | Lower phase | Middle phase | Upper phase | Clarified sewage | Lower phase | Middle phase | Upper phase | ||
| SW-3713 | Dharavi | UD | UD | UD | UD | UD | UD | UD | UD |
| SW-3731 | UD | UD | UD | UD | UD | UD | UD | UD | |
| SW-3737 | 38.64 | 39.4 | 33.02 | UD | 39.21 | 39.77 | 32.72 | UD | |
| SW-3741 | 39.4 | 37.72 | 33.15 | UD | 38.13 | 37.57 | 32.36 | UD | |
| SW-3714 | Wadala | UD | UD | UD | UD | UD | UD | UD | UD |
| SW-3732 | UD | UD | UD | UD | UD | UD | UD | UD | |
| SW-3738 | 38.74 | 37.64 | 33.58 | 40.86 | 38.59 | 37.5 | 33.25 | UD | |
| SW-3742 | 36.35 | 37.61 | 31.92 | 40.95 | 35.67 | 37.19 | 30.96 | UD | |
| SW-3730 | Shivaji Nagar | UD | UD | UD | UD | UD | UD | UD | UD |
| SW-3740 | 40.14 | 43.54 | 34.53 | UD | 40.18 | 41.38 | 38.76 | UD | |
| SW-3744 | 40.17 | UD | 36.27 | UD | 39.72 | 41.54 | 35.79 | UD | |
| SW-3734 | Kurla | UD | UD | UD | UD | UD | UD | UD | UD |
| SW-3739 | 40.6 | 41.07 | 41.94 | UD | 40.56 | 40.66 | 39.3 | UD | |
| SW-3743 | 40.52 | 41.52 | 35.37 | 41.94 | 38.28 | UD | 35.98 | UD | |
| SW-3729 | Kanjur | UD | UD | UD | UD | UD | UD | UD | UD |
| SW-3745 | 40.17 | 37.13 | 33.24 | UD | 39.32 | 38.1 | 33.5 | UD | |
| SW-3747 | 39.34 | 35.69 | 35.52 | UD | 35 | 34.1 | 32.85 | 37.42 | |
| SW-3733 | Malad | UD | UD | UD | UD | UD | UD | UD | UD |
| SW-3746 | 37.33 | 39.28 | 33.11 | 41.84 | 38.95 | 41.53 | 33.64 | 42.19 | |
| SW-3748 | 37.13 | 35.65 | 33.56 | 40.07 | 39.83 | 34.11 | 30.46 | 35.25 | |
UD, undetected, no amplification
The final results of the 20 samples collected from the six sites before and during the COVID-19 pandemic period are shown in (Table III). None of the sewage samples (from any site) collected before March 20, 2020 showed amplification of E and RdRp genes of SARS-CoV-2 RNA. In the absence of COVID-19 cases, the sewage concentrates (lower/middle phases) did not show any false-positive amplification in E and RdRp genes real-time RT-PCR. All sewage samples collected in May 2020 (except one sample from Kurla) were positive for SARS-CoV-2 (Ct values <36 for both E and RdRp genes for RNA extracted from the middle phase).
Table III.
Results of SARS-CoV-2 detection in sewage samples collected from six wards in Mumbai before and during the COVID-19 pandemic
| Site | Pre-COVID-19 period | During COVID-19 period | ||||||
|---|---|---|---|---|---|---|---|---|
| Date of collection | Result | Date of collection in March 2020 | Result | Date of collection in May 2020 | Result | Date of collection in May 2020 | Result | |
| Dharavi | February 3, 2020 | Negative | 16 | Negative | 12 | Positive | 14 | Positive |
| Wadala | February 3, 2020 | Negative | 16 | Negative | 12 | Positive | 14 | Positive |
| Shivaji Nagar | X | X | 11 | Negative | 13 | Positive | 15 | Positive |
| Kurla | X | X | 18 | Negative | 13 | Positive | 15 | Positive |
| Kanjur | X | X | 11 | Negative | 16 | Positive | 22 | Positive |
| Malad | X | X | 18 | Negative | 16 | Positive | 22 | Positive |
X, samples not available for testing
Although it may be tempting to argue that the late amplification (Ct >39) could be SARS-CoV-2 RNA specific, but we suggest that a viral concentration step be included and improvement of Ct value by about 3 points be used for the interpretation of positive results until substantial additional data become available from several different SARS-CoV-2-infected and non-infected regions.
Discussion
A global emergency of pandemic was declared by the WHO on March 11, 2020 after the detection of SARS-CoV-2 in Wuhan, China, in December 20191. The virus was found to be transmitting via respiratory route. However, SARS-CoV-2 RNA was also found to be excreted in human faeces9. This suggested that the virus could be detected from sewage water. ES for poliovirus detection has played a critical role in the eradication of wild poliovirus globally. The WHO-Global Polio Laboratory Network (GPLN) has established supplemental surveillance for polioviruses by testing sewage samples in several countries10. In India, ES was first initiated in Mumbai city in 2001 by ICMR-National Institute of Virology, Mumbai Unit (formerly known as ICMR-Enterovirus Research Centre).
The GPLN-validated PEG-dextran two-phase separation method to concentrate poliovirus from sewage recommends collection of the entire lower and interphase sample and treat it with chloroform to inactivate microbial contamination to render it compatible for virus isolation in cell cultures7. We, therefore, decided to employ the same two-phase separation methodology to concentrate and detect SARS-CoV-2 in sewage samples.
Risk analysis showed that SARS-CoV-2, an enveloped virus, is inactivated by household bleach, detergents/soaps and lipid solvents11. Sewage samples were collected in chloroform-containing bottles to inactivate the virus to further mitigate the risk and to allow sample handling in BSL-2 (Biosafety level 2) laboratories. This became the first modification of the WHO two-phase protocol. In our initial experiments with spiked sewage samples, SARS-CoV-2 could not be effectively concentrated in the lower phase (lower and middle layers collected together) collected as per the WHO protocol7. The three layers of upper, middle and lower were collected separately and tested for SARS-CoV-2. The virus was found to be concentrated only in the middle phase. This was the second and major modification of the WHO two-phase separation method introduced during standardization.
PEG precipitation and PEG-dextran phase separation methods are widely used for concentrating viruses5,6,7. Pre-COVID-19 samples were collected for poliovirus ES in our laboratory. Sample collection was done during May 11-22, 2020 specifically for standardization of the method for detecting SARS-CoV-2 in sewage samples.
None of the pre-COVID-19 sewage samples showed amplification in any of the fractions (phase) tested, indicating that false amplification did not happen. All the samples collected in May 2020 were positive for SARS-CoV-2. Thus, the feasibility of using PEG-dextran phase separation method for ES of COVID-19 was established. The method may be further simplified by substituting gravity filtration for the centrifugation step and phase separation at ambient temperature. Other viral concentration methods may be compared to identify the most suitable one for long-term use.
Angiotensin-converting enzyme 2 (ACE2) was identified as the cellular receptor of SARS-CoV-212,13. ACE2 is expressed on the cells of several different organs including lungs, kidneys and intestine. About 107 SARS-CoV-2 RNA copies/g of faeces may be excreted by symptomatic patients during the first week of the disease9,14. SARS-CoV-2 in sewage was first detected in The Netherlands within weeks after the reports of COVID-19 cases4. Viral concentration was achieved by ultra-filtration technique. Sewage samples were found to be negative for SARS-CoV-2 in the area where the prevalence of COVID-19 was 2.9 cases/100,000 population, with partial positivity at 3.5 cases/100,000 and high positivity when the prevalence reached 12.9 cases/100,000 population4.
SARS-CoV-2 detection in waste waters could be used to understand the epidemiology of COVID-19. For example, (i) decreasing concentration or absence of virus at previously SARS-CoV-2-positive sewage sampling sites may indicate successful implementation of COVID-19 control strategies, and (ii) it may provide evidence of the presence or absence of SARS-CoV-2-infected populations and confirmation of COVID-19-free zones.
To conclude, SARS-CoV-2 was detected in sewage samples collected from six different sites in Mumbai during the ongoing pandemic of COVID-19. PEG6000-dextran T40 phase separation method was adopted to concentrate SARS-CoV-2 from sewage samples. The virus was found to partition and concentrate specifically at the middle phase. RNA extracted from the middle phase was suitable for SARS-CoV-2 detection by real-time RT-PCR.
Acknowledgment:
Authors acknowledge Prof. (Dr) Balram Bhargava, Secretary, Department of Health Research, Ministry of Health and Family Welfare Government of India, and Director-General, Indian Council of Medical Research (ICMR), New Delhi; Dr Raman Ganagakhedkar, National Chair, ICMR, New Delhi, and Prof (Dr) Priya Abraham, Director ICMR-National Institute of Virology, Pune, for encouragement, guidance and support. Authors also thank Dr Shailesh D. Pawar, Officer-in-Charge, ICMR-National Institute of Virology, Mumbai Unit, for his kind support.
Footnotes
Financial support & sponsorship: This study was financially supported by ICMR intramural grant.
Conflicts of Interest: None.
References
- 1.Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–60. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng J. SARS-CoV-2: An emerging coronavirus that causes a global threat. Int J Biol Sci. 2020;16:1678–85. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amirian ES. Potential fecal transmission of SARS-CoV-2: Current evidence and implications for public health. Int J Infect Dis. 2020;95:363–70. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-CoV-2 in Sewage. medRxiv. 2020 doi: 10.1021/acs.estlett.0c00357. doi: 10.1101/2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- 5.Deshpande JM, Shetty SJ, Siddiqui ZA. Environmental surveillance system to track wild poliovirus transmission. Appl Environ Microbiol. 2003;69:2919–27. doi: 10.1128/AEM.69.5.2919-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hovi T, Shulman LM, van der Avoort H, Deshpande J, Roivainen M, De Gourville EM, et al. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol Infect. 2012;140:1–3. doi: 10.1017/S095026881000316X. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization. Guidelines for environmental surveillance of poliovirus circulation. WHO/V&B/03.03. [accessed on March 1, 2020]. Available from: http://polioeradication.org/wp-content/uploads/2016/07/WHO_V-B_03.03_eng.pdf .
- 8.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–69. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang N, Gong Y, Meng F, Shi Y, Wang J, Mao P, et al. Comparative study on virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. Sci China Life Sci. 2020:1–3. doi: 10.1007/s11427-020-1783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asghar H, Diop OM, Weldegebriel G, Malik F, Shetty S, El Bassioni L, et al. Environmental surveillance for polioviruses in the Global Polio Eradication Initiative. J Infect Dis. 2014;210(Suppl 1):S294–303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization. Annex G, use of disinfectants: Alcohol and bleach. Infection prevention and control of epidemic- and pandemic-prone acute respiratory infections in health care. Geneva: WHO; 2014. [PubMed] [Google Scholar]
- 12.Zhou P, Yang P, Wang XG, Hu B, Zheng L, Zheng W, et al. A pneumonia outbreak with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woelfel R, Corman M, Guggemos W, Seilmaier M, Zange S, Mueller MA, et al. Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. medRxiv. 2020 doi: 10.1101/2020.03.05.20030502. [Google Scholar]