Abstract
Background & objectives:
The rapid diagnosis of coronavirus disease 2019 (COVID-19) is a significant step towards the containment of the virus. The surge of COVID-19 cases in India and across the globe necessitates a rapid and sensitive molecular assay. Rapid point-of-care (PoC) assays (Truenat Beta CoV and Truenat SARS-CoV-2 assays) for the diagnosis of COVID-19 have been developed which are expected to shorten the turnaround time of reporting of results and also can be used for field investigations of COVID-19. The objectives of the study were to validate the performance of Truenat Beta CoV and Truenat SARS-CoV-2 PoC assays for the detection of SARS-CoV-2 infected cases with reference to analytical sensitivity, precision/inter-machine variation, clinical sensitivity and clinical specificity.
Methods:
The rapid PoC screening and confirmatory assays were prospectively validated at the State Level Virus Research and Diagnostic Laboratory at Bangalore Medical College and Research Institute, Bengaluru, under technical supervision by the Indian Council of Medical Research-National Institute of Virology (ICMR-NIV), Pune. Real-time reverse transcription-polymerase chain reaction (rRT-PCR) was considered as the reference standard against which the rapid assays were validated for all samples tested based on analytical sensitivity, precision/inter-machine variation, clinical sensitivity and clinical specificity.
Results:
Truenat Beta CoV and Truenat SARS-CoV-2 assays showed concordant results when compared with the reference standard rRT-PCR. These PoC assays exhibited 100 per cent sensitivity, specificity, positive predictive value and negative predictive value.
Interpretation & conclusions:
Truenat Beta CoV and Truenat SARS-CoV-2 assays showed concordance with the reference standard assay and may be recommended for screening and confirmation of SARS-CoV-2 in the field settings.
Keywords: COVID-19, point-of-care test, rRT-PCR, SARS-CoV-2, Truenat Beta CoV, Truenat SARS-CoV-2
Coronavirus disease 2019 (COVID-19) pandemic has resulted in 37,601,548 confirmed cases, with over 1,077,799 deaths globally, as of October 13, 20201. In the backdrop of a surge in cases of COVID-19 in India and 216 affected countries across the globe, the rapid diagnosis of cases is considered as a significant tool towards the containment of cases. Currently, real-time reverse transcription-polymerase chain reaction (rRT-PCR) has been globally accepted as the reference standard for the detection of SARS-CoV-2 which is being performed in COVID-19 testing centers identified by the Indian Council of Medical Research (ICMR) across the country using approved protocols and kits2,3. The ICMR has harnessed the potential of a network of Virus Research and Diagnostic Laboratories (VRDLs) present across the nation to address the issue of SARS-CoV-2 diagnosis4.
Timely diagnosis, effective treatment and future prevention are crucial to the management of COVID-19 cases. The current race to develop cost-effective point-of-care (PoC) test kits and efficient laboratory techniques for confirmation of SARS-CoV-2 infection has fueled a new frontier of diagnostic innovation. Benefits of PoC tests include case confirmation in field settings, guidance in early public health interventions like contact tracing, isolation of case and prophylaxis, apart from on-spot case confirmation of SARS-CoV-2, which is expected to improve case management. On these lines, rapid Truenat PoC assays for the diagnosis of COVID-19 have been developed by Molbio Diagnostics Private Limited, India, which are expected to shorten the turnaround time of reporting of results and also can be used for field investigations of COVID-19. Truenat platforms are light and portable indigenous chip-based rRT-PCR designed for rapid diagnosis of infectious diseases, including COVID-19. The processing of clinical specimens from RNA extraction to amplification can be achieved in <60 min. Availability of ready-made master mix prep and negating the need of clean biosafety cabinet, minimal training for field testing are additional benefits. Truelab workstation Real-Time micro PCR system is achieved through a combination of lightweight, portable, mains/battery operated Truelab Real-Time micro PCR analyzers, TruePrep AUTO universal cartridge-based SamplePrep device, room temperature stable Truenat microPCR chips and TruePrep AUTO SamplePrep tests.
The present study was undertaken to validate the performance of Truenat Beta CoV (screening assay) and Truenat SARS-CoV-2 (confirmation assay) PoC diagnostic assays for the detection of SARS-CoV-2 with respect to analytical sensitivity, precision/inter-machine variation, clinical sensitivity and clinical specificity. The results were compared with reference standard rRT-PCR.
Material & Methods
The present study was undertaken during the months of April and May 2020 at the State Level VRDL, Bangalore Medical College and Research Institute (BMCRI), Bengaluru, India5,6. Known SARS-CoV-2 positive and negative nasopharyngeal (NP)/oropharyngeal (OP) archived samples in viral transport medium (VTM), blood, and NP/OP samples collected from suspected cases of SARS-CoV-2 were included in the study. Samples from other respiratory infections like bacterial pneumonia and tuberculosis were excluded. NP and OP swabs in VTM from patients with H1N1 and severe acute respiratory illness (SARI) and blood samples from SARS-CoV-2 positive and negative cases were used for validating the Truenat screening (Beta CoV) and confirmatory (SARS-CoV-2) assays. Institutional Ethics Committee approval (Vide No: BMCRI/PS/02/2020-21 dated 18.04.2020) was taken before undertaking the study. A total of 75 samples, including 30 confirmed SARS-CoV-2 positives, 45 confirmed SARS-CoV-2 negatives and six blood samples (3 each from SARS-CoV-2 positive and negative cases) were used.
Evaluation of analytical sensitivity of the test: Aliquot of one VTM sample with low Ct values for envelope protein (E) gene and RNA-dependent RNA polymerase (RdRp-2) was used for extraction by Trueprep Auto (as per manufacturer's protocol) and also by manual RNA extraction kit (QiAmp viral RNA extraction kit - mini prep, Qiagen, Germany). Ribonuclease P (RNase P), a human constitutive gene, was used as an internal control for all the assays. RNA extracted from the samples was diluted 10-fold from 10−1 (dilution 1) to 10−6 (dilution 6). Six dilutions (1:10) were made from both Trueprep elute and Qiagen elute. These were run in parallel in TaqMan rRT-PCR using ICMR-NIV protocol, considered as the reference standard and Truenat assays5,6 (Fig. 1).
Fig. 1.
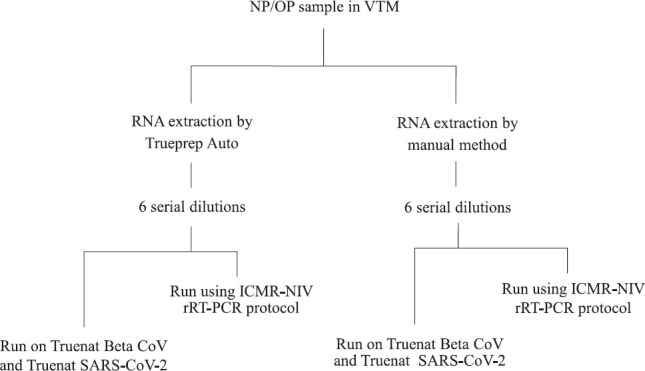
Flowchart showing evaluation of analytical sensitivity of the test. NP/OP nasopharyngeal/oropharyngeal swab; VTM, viral transport medium; rRT-PCR, real-time reverse transcription-polymerase chain reaction.
For E gene, strong positive: low Ct value: 20±1.5 (13 samples); medium positive: medium Ct value: 29±1.5 (10 Samples); weak positive: high Ct value: 32±1.5 (7 Samples) (used for validation of Truenat Beta CoV assay in this study).
For RdRp-2, strong positive: low Ct value: 27.5±2.5 (11 samples); medium positive: medium Ct value: 32.5±2.5 (14 samples); weak positive: high Ct value: 37.5±2.5 (5 samples). Both dilution series were run on Truenat SARS-CoV-2 chips as well as SARS-CoV-2 rRT-PCR systems in parallel. Log-linear curves were plotted to determine the linearity of assays.
In vitro transcribed (IVT) RNA dilutions: Serial dilutions of in vitro transcribed (IVT) RNA received from ICMR-NIV were done and compared on Truenat Beta CoV, Truenat SARS-CoV-2 and rRT-PCR. Ct values of these dilutions in assays performed by all the methods were noted7.
Evaluation of repeatability of the test (precision)/inter-machine variations: Repeatability of the PCR test is essential to ensure assay reproducibility and reliability. Three clinical elutes representing high, medium and low Ct values [Sample IDs: R1 (Ct: 30.24), R2 (Ct: 24.19), R3 (Ct: 20.97) for E gene detection by Truenat Beta CoV; R4 (Ct: 26.71), R5 (Ct: 20.75), R6 (Ct: 14.5) for RdRp-2 detection by Truenat SARS-CoV-2] were run on all four Truenat PCR devices under evaluation for precision/inter-machine variations.
Clinical sensitivity and specificity: Clinical sensitivity was tested by running confirmed positives samples of SARS-CoV-2 (n=30), representing high, medium, and low Ct values of RdRp-2 for testing and comparison with both the systems (Table I). To evaluate clinical specificity, known positive NP/OP samples of H1N1, SARI, blood samples from SARS-CoV-2 positive and negative cases, as well as confirmed COVID-19 negatives were used.
Table I.
Comparison of Ct values of 30 confirmed SARS-CoV-2 positive samples on Truenat and real-time PCR platforms
| Sample | Truenat Beta CoV (Ct) | Truenat SARS-CoV-2 (Ct) | rRT-PCR SARS-CoV-2 (Ct) | ||||
|---|---|---|---|---|---|---|---|
| RNase P | E gene | RNase P | RdRp-2 | RNase P | E gene | RdRp-2 | |
| 1 | 26.29 | 23.6 | 26.14 | 26.71 | 31.36 | 29.04 | 36.29 |
| 2 | 22.43 | 22 | 22.75 | 18 | 27.88 | 24.12 | 30 |
| 3 | 27 | 33.33 | 27.25 | 31.6 | 34.24 | 26 | 34 |
| 4 | 20.75 | 18.5 | 21.2 | 16.75 | 27.92 | 19.45 | 26.89 |
| 5 | 23.33 | 24 | 23.5 | 24 | 29.6 | 30.81 | 35 |
| 6 | 24.8 | 20.5 | 25.17 | 18.3 | 32.23 | 25.23 | 31.07 |
| 7 | 20.33 | 15 | 20.5 | 13 | 27.76 | 19.11 | 26.15 |
| 8 | 23.33 | 22 | 24.33 | 21 | 33.09 | 28.95 | 34.53 |
| 9 | 23.14 | 23.33 | 23.86 | 21.63 | 27.56 | 24.5 | 31 |
| 10 | 21.8 | 25.33 | 22.29 | 22.4 | 27.42 | 27.8 | 24.7 |
| 11 | 25 | 18.4 | 25.75 | 18.17 | 29.99 | 23.03 | 29.85 |
| 12 | 25 | 21.11 | 25.25 | 19.71 | 29.8 | 24.07 | 31.53 |
| 13 | 26.2 | 17.4 | 26.5 | 16.6 | 32.23 | 24.18 | 30 |
| 14 | 22.17 | 21.6 | 22.5 | 20.43 | 29.48 | 27.23 | 33 |
| 15 | 21.5 | 20.5 | 22.29 | 19 | 30.51 | 24.42 | 32.28 |
| 16 | 22 | 16 | 22.4 | 14.5 | 30.45 | 19.49 | 27.85 |
| 17 | 27.8 | 21 | 29.5 | 20.29 | 33.54 | 25.16 | 31.55 |
| 18 | 23 | 22.75 | 23 | 22 | 28.23 | 26.19 | 32.47 |
| 19 | 26 | 12.8 | 25 | 11.4 | 33.66 | 21.05 | 32.38 |
| 20 | 26.33 | 22.14 | 26.5 | 21.14 | 32.62 | 26.09 | 35.45 |
| 21 | 25.2 | 22.8 | 32 | 18.71 | 26.2 | 22.35 | 28.17 |
| 22 | 24.14 | 15.17 | 24.83 | 14.17 | 31.14 | 19.28 | 25.82 |
| 23 | 24.17 | 25 | 24.13 | 20.43 | 26.2 | 20.97 | 26.36 |
| 24 | 22.43 | 21.83 | 22 | 20.75 | 26.09 | 30.88 | 27.88 |
| 25 | 23.29 | 27.17 | 23.6 | 25.8 | 28.34 | 31.92 | 32.68 |
| 26 | 27.2 | 26.38 | 26.4 | 24.8 | 31.88 | 29.52 | 35.5 |
| 27 | 24.6 | 21.14 | 24.5 | 19.25 | 30.84 | 22.88 | 31.31 |
| 28 | 25 | 31.4 | 26 | 30.1 | 30.77 | 36.86 | 38.56 |
| 29 | 20.6 | 30 | 21 | 27.8 | 27.19 | 33.16 | 39.36 |
| 30 | 24.33 | 29.6 | 24 | 28 | 31.19 | 31.66 | 34.28 |
Cross-reactivity testing: Cross-reactivity was evaluated by testing 15 RNA samples from clinical specimens, previously tested for H1N1 (5 positive, 10 negative). To check cross-reactivity with SARI, 30 samples from confirmed SARI patients were used for the detection of the E gene and RdRp-2.
Evaluation of blood samples: RNA extracted from six blood samples (3 SARS-CoV-2 positive and 3 SARS-CoV-2 negative) using Trueprep Auto were run on Truenat SARS-CoV-2 assay. RNAs extracted by the manual method were processed using reference standard rRT-PCR protocol and the Ct values recorded were compared with that of Truenat assays.
Results
Reference standard assay rRT-PCR detected up to dilution 10−5 (D5) from the undiluted sample, with valid Ct values. Truenat Beta CoV/SARS-CoV-2 detected E gene and RdRp-2 targets up to dilution 106 (D6) with valid Ct value (Table II).
Table II.
Analytical sensitivity of Truenat point-of-care (PoC) assays compared to reference rRT-PCR method values
| Dilutions | Truenat Beta CoV | Truenat SARS-CoV-2 | SARS-CoV 2 rRT-PCR | ||||
|---|---|---|---|---|---|---|---|
| E gene | RNase P | RdRp-2 | RNase P | E gene | RdRp-2 | RNase P | |
| Neat | 15 | 20.33 | 13 | 20.5 | 19.29 | 24.49 | 26.45 |
| D1 | 16.33 | 23.75 | 16.17 | 24.43 | 22.05 | 26.87 | 29.63 |
| D2 | 20.6 | 28.11 | 18.5 | 27.86 | 25.78 | 30.28 | 33.01 |
| D3 | 24 | 31.6 | 22.25 | 30.75 | 29.22 | 33.76 | 35.55 (-ve) |
| D4 | 27.2 | 33.29 | 25 | 33.6 | 32.4 | 36.87 (-ve) | 38.6 (-ve) |
| D5 | 30.29 | ND | 28.8 | ND | 36.25 | 40.52 (-ve) | 39.8 (-ve) |
| D6 | 33.33 | ND | 32 | ND | 38.3 | ND | ND |
ND, not detected
Linearity of assay (Trueprep Auto extract): Using the dilution series from Trueprep Auto elutes run on Truenat Beta CoV and SARS-CoV-2, the log-linear curve was plotted to check the linearity of Ct values (Figs. 2 and 3). The slope of the line was −3.289 (R2 value: 0.9944 for Truenat Beta CoV; 0.9901 for Truenat SARS-CoV-2). The assay was observed to be linear over the range of dilutions tested, and PCR efficiency was found to be 99.57 per cent (Figs 2 and 3).
Fig. 2.
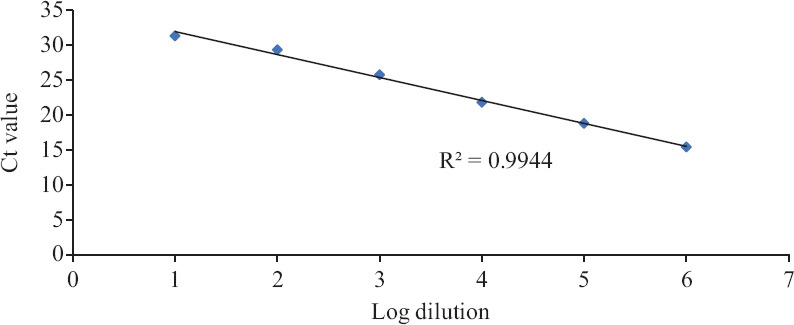
Linearity and PCR efficiency on Truenat Beta CoV. Y-axis indicates Ct values, and X-axis is arbitrary log numbers indicating dilutions.
Fig. 3.
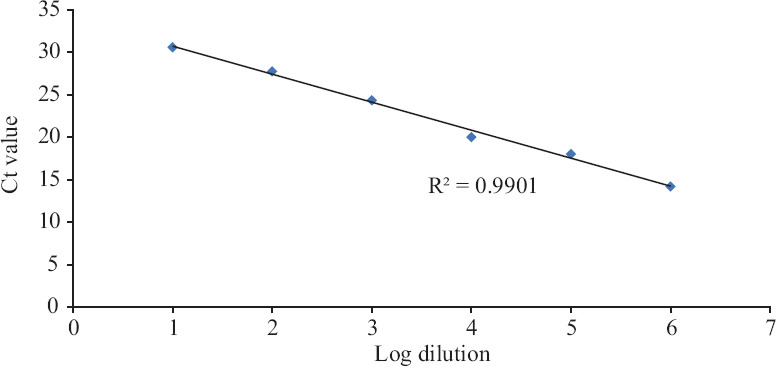
Linearity and PCR efficiency on Truenat SARS-CoV-2. Y-axis indicates Ct values, and X-axis is arbitrary log numbers indicating dilutions.
Serially diluted IVT RNA assayed for E gene, and RdRp-2 targets showed better sensitivity with Truenat Beta CoV and Truenat SARS-CoV-2 assays in comparison to rRT-PCR. Valid Ct values were detected by Truenat assays in log IVT dilutions higher than that of rRT-PCR, and these assays had a limit of detection (LOD) of 102 copies/μl (D6) for these targets, indicating higher sensitivity as compared to rRT-PCR assay, which had a LOD of 103 (1000) copies/μl (Table III).
Table III.
Analytical sensitivity of Truenat PoC assays using in vitro transcribed (IVT) RNA
| Dilutions (copies) | Truenat SARS-CoV-2 (Ct values) RdRp-2 | rRT-PCR (Ct values) RdRp-2 |
|---|---|---|
| 10−5 (107) | 17.0 | 25.65 |
| 10−6 (106) | 19.67 | 30.63 |
| 10−7 (105) | 23.5 | 34.08 |
| 10−8 (104) | 26.5 | 39.72 |
| 10−9 (103) | 29.83 | ND |
| 10−10 (102) | 32.18 | ND |
ND, Not Detected
Precision/inter-machine variations: The results of Truenat assays were found to be reproducible with a coefficient of variation in Ct values significantly <10 per cent, across samples. All positive samples were detected by both screening (Truenat Beta CoV, E gene) and confirmatory (Truenat SARS-CoV-2, RdRp-2) assays. These results were 100 per cent concordant with rRT-PCR results as represented in Table IV.
Table IV.
Precision of Truenat SARS-CoV-2 assay
| Equipment ID | Truenat SARS CoV-2 RdRp-2 (Ct) | ||
|---|---|---|---|
| ID:383 | ID:1263 | ID:885 | |
| TLDU0401 | 26.71 | 20.75 | 14.67 |
| TLDU1308 | 26.14 | 21 | 14.5 |
| TLDU1306 | 26.2 | 21.17 | 15 |
| TLQU0001 | 26.14 | 20.75 | 14.5 |
| Mean | 26.30 | 20.92 | 14.67 |
| Standard deviation | 0.28 | 0.21 | 0.24 |
| Per cent CV | 1.1 | 1.0 | 1.6 |
TLDU, Truelab™ Duo Real Time Quantitative micro PCR Analyzer; TLQU,Truelab™ Quattro Real Time Quantitative micro PCR Analyzer; ID, sample identification number; CV, coefficient of variation
Specificity, cross-reactivity and diagnostic performance: The PoC assays did not show cross-reactivity with known positive and negative samples of SARS-CoV-2 (n=75) used for evaluation and exhibited 100 per cent concordance with rRT-PCR results (Table V). Internal control RNase P was amplified in all samples with no amplification of E gene and RdRp-2 gene targets. For performance evaluation of Truenat Beta CoV and SARS-CoV-2 assays, a total of 75 samples (30 confirmed SARS-CoV-2 positives and 45 confirmed SARS-CoV-2 negatives) were tested on the four Truenat platforms under evaluation. A comparison of results from rRT-PCR and Truenat SARS-CoV-2 was done using a 2×2 matrix. Clinical sensitivity, clinical specificity and overall concordance were determined to be 100 per cent (Table V).
Table V.
Calculation of diagnostic performance
| Truenat Beta CoV and Truenat SARS-CoV-2 (n=75) | SARS-CoV-2 rRT-PCR (n=75) | ||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 30 (TP) | 0 (FP) | 30 |
| Negative | 0 (FN) | 45 (TN) | 45 |
| Total | 30 | 45 | 75 |
TP, true positive; TN, true negative; FP, false positive; FN, false negative, PPV, positive predictive value; NPV, negative predictive value. Sensitivity=TP/TP+FN; Specificity=TN/ TN+FP; PPV=TP/TP+FP; NPV=TN/TN+FN; Sensitivity: 100 per cent; Specificity: 100 per cent; PPV: 100 per cent; NPV: 100 per cent; Clinical sensitivity: 100 per cent; Clinical specificity: 100 per cent; Overall concordance: 100 per cent
Discussion
Conventionally, the preferred targets of coronavirus RT-PCR assays included the conserved and/or abundantly expressed genes such as the structural S and N genes and the nonstructural RdRp and replicase open reading frame (ORF) 1a/b genes. The reference standard rRT-PCR for SARS-CoV-2 employs E gene for screening and RdRp, as well as ORF for confirmation of cases. Molecular assays on PoC platforms have the added advantage of higher applicability in field settings for rapid screening and confirmation of SARS-CoV-2 cases without compromising the diagnostic parameters8,9,10,11,12,13,14. Truenat Beta CoV and Truenat SARS-CoV-2 are indigenous chip-based rRT-PCR assays for semi-quantification of SARS-CoV-2. E gene is employed for screening by Truenat Beta CoV assay and RdRp-2 for confirmation by Truenat SARS-CoV-2 assay.
The present study was done to evaluate the performance of Truenat PoC assays for screening and confirmation of SARS-CoV-2. The results indicated that the Truenat Beta CoV and SARS-CoV-2 assays were highly sensitive and specific for the detection of SARS-CoV-2 RNA. The Truenat PoC assays were also evaluated for performance based on the parameters of analytical sensitivity, precision/inter-machine-variations, clinical sensitivity and specificity. Cross-reactivity with other respiratory viruses like H1N1 was also evaluated.
The results of precision and inter-machine variation for Truenat screening and confirmatory assays were reproducible with per cent co-efficient of variation in Ct values being <10 per cent. The PoC tests under evaluation showed concordant results with a reference standard assay for cross-reactivity, and when elutes from blood samples of SARS-CoV-2 positive and negative cases were run in duplicates for both targets. SARS-CoV-2 positive samples categorized into three categories of the low, medium and high Ct values were run in parallel on the Truenat platform and reference standard assay. The results were found to be 100 per cent concordant and LOD was 100 copies. Diagnostic performance of the PoC tests in terms of sensitivity, specificity, positive and negative predictive values were determined to be 100 per cent. These PoC assays are expected to be of value in the field settings as these require minimal laboratory setup, workforce, and skill to perform the same. In turn, this would reduce the burden on testing sites performing RT-PCR and shorten the turnaround time of reporting of results. The extraction of RNA using Trueprep takes 20 min and each of the assays requires 45 minutes. This is quicker as compared to rRT-PCR, which takes around 4-6 h for the entire process. Owing to the urgent requirement of PoC assays validation for use in field settings, the present evaluation was done with a limited sample size, which was a limitation of the present study. Studies with a larger sample size performed in field settings are required to further validate the tests. However, the present study can be considered as a preliminary finding which needs to be tested independently in the field for further validation of results with large samples using the method of blinding to further strengthen the hypothesis.
Truenat Beta CoV and Truenat SARS-CoV-2 PoC assays, targeting E and RdRp-2 genes may be recommended for screening and confirmation, respectively, of suspected cases of COVID-19. These assays would be of value in rapid confirmation of COVID-19 cases in field settings.
Acknowledgment:
Authors acknowledge Dr Varsha A. Potdar, Influenza Group, ICMR-NIV, Pune, for providing IVT RNA for use and technical assistance provided by Ms. Asha Munegowda, Research Assistant, State Level Virus Research and Diagnostic Laboratory (VRDL), Bangalore Medical College and Research Institute (BMCRI), Bengaluru.
Footnotes
Financial support & sponsorship: Authors acknowledge the funding received from the Department of Health Research (DHR) and ICMR for the establishment of State Level VRDL at BMCRI, where the present study was undertaken.
Conflicts of Interest: None.
References
- 1.World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. [accessed on October 13, 2020]. Available from: https://covid19.who.int/
- 2.Chan JF, Yip CC, To KK, Tang TH, Wong SC, Leung KH, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58:e00310–20. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broedersa S, Huberb I, Grohmannc L, Berbend G, Tavernierse I, Mazzaraf M, et al. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci Technol. 2014;37:115–26. [Google Scholar]
- 4.Gupta N, Potdar V, Praharaj I, Giri S, Sapkal G, Yadav P, et al. Laboratory preparedness for SARS-CoV-2 testing in India: Harnessing a network of virus research; diagnostic laboratories. Indian J Med Res. 2020;151:216–25. doi: 10.4103/ijmr.IJMR_594_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ICMR-National Institute of Virology. Standard Operating Procedure for Detection of 2019 novel coronavirus (2019-nCoV) in suspected human cases by rRT-PCR: First Line Screening assay. Pune: ICMR-National Institute of Virology; 2020. pp. 1–7. [Google Scholar]
- 6.ICMR-National Institute of Virology. Standard operating procedure for detection of 2019 novel coronavirus (2019-nCoV) in suspected human cases by rRT-PCR: Confirmation Assay. Pune: ICMR-National Institute of Virology; 2020. pp. 1–7. [Google Scholar]
- 7.Choudhary ML, Vipat V, Jadhav S, Basu A, Cherian S, Abraham P, et al. Development of in vitro transcribed RNA as positive control for laboratory diagnosis of SARS-CoV-2 in India. Indian J Med Res. 2020;151:251–4. doi: 10.4103/ijmr.IJMR_671_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet. 2020;395:514–23. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–36. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–74. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Liu W, Zhang Q, Xu K, Ye G, Wu W, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9:313–9. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66:549–55. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. Laboratory testing for coronavirus disease (COVID-19) in suspected human cases: Interim guidance, 19 March 2020. [accessed on June 1, 2020]. Available from: https://apps.who.int/iris/handle/10665/331501 .


