Abstract
In animal cells, nuclear envelope breakdown (NEBD) is required for proper chromosome segregation. Whereas mitotic kinases have been implicated in NEBD, how they coordinate their activity to trigger this event is unclear. Here, we show that both in human cells and Caenorhabditis elegans, the Polo-like kinase 1 (PLK-1) is recruited to the nuclear pore complexes, just prior to NEBD, through its Polo-box domain (PBD). We provide evidence that PLK-1 localization to the nuclear envelope (NE) is required for efficient NEBD. We identify the central channel nucleoporins NPP-1/Nup58, NPP-4/Nup54, and NPP-11/Nup62 as the critical factors anchoring PLK-1 to the NE in C. elegans. In particular, NPP-1, NPP-4, and NPP-11 primed at multiple Polo-docking sites by Cdk1 and PLK-1 itself physically interact with the PLK-1 PBD. We conclude that nucleoporins play an unanticipated regulatory role in NEBD, by recruiting PLK-1 to the NE thereby facilitating phosphorylation of critical downstream targets.
Keywords: Cell cycle, Development, Mitosis, Polo-like kinase
Introduction
Mitosis is a highly regulated process that ensures faithful segregation of the genetic material into daughter cells. In higher eukaryotes, rupture of the nuclear envelope (NE) during mitosis is essential for the capture of the chromosomes by microtubules and for proper chromosome segregation. Nuclear envelope breakdown (NEBD) is a step-wise process that involves Nuclear Pore Complex (NPC) disassembly, microtubule-dependent mechanical tearing of the NE and depolymerisation of the nuclear lamina (Ungricht and Kutay, 2017). All these steps are regulated by mitotic kinases, in particular the master mitotic kinase Cyclin-dependent kinase (Cdk1) and the Polo-like kinase 1 (Plk1).
NPCs consist of ~30 different nucleoporins that are present in multiple copies and are organized in subcomplexes (Hurt and Beck, 2015). Cdk1 plays a key role in NPC disassembly, notably by promoting, along with other mitotic kinases, the dispersal of the soluble Nup98 nucleoporin from the NPC, resulting in the loss of the NE permeability barrier (Mühlhäusser and Kutay, 2007; Laurell et al., 2011; Ungricht and Kutay, 2017). Several other nucleoporins are phosphorylated on Cdk1 consensus sites during mitosis but the functional significance of these phosphorylations is currently unknown (Blethrow et al., 2008). Cdk1 also phosphorylates Lamins and triggers the disassembly of the nuclear lamina (Ungricht and Kutay, 2017).
Likewise, Plk1 phosphorylates a number of NE-associated proteins, including subunits of the NPCs in human cells (Grosstessner-Hain et al., 2011; Santamaria et al., 2011; Kettenbach et al., 2011; Bibi et al., 2013) but the precise contribution of these phosphorylations for NPC disassembly is elusive. In addition, Plk1 phosphorylates p150Glued, a subunit of the dynein/dynactin complex that participates in microtubule-dependent tearing of the NE during mitosis (Li et al., 2010).
In C. elegans, PLK-1 (the orthologue of human Plk1) is also required for NEBD both in oocytes (Chase et al., 2000) and in early embryos (Rahman et al., 2015), indicating that the role of Plk1 in NEBD is evolutionarily conserved. After fertilization, the female and the male pronuclei, which are surrounded by a NE, meet at the posterior pole of the embryo. After rotation and centration of the nucleo-centrosomal complex, the NE breaks down in the vicinity of the centrosomes and between the juxtaposed pronuclei (Hachet et al., 2012; Rahman et al., 2015). This allows the capture of the chromosomes by the cytoplasmic microtubules and the merging of parental chromosomes (Figure S1A, a). The complete disassembly of the nuclear lamina occurs later at the metaphase to anaphase transition (Lee et al., 2000). PLK-1 inactivation, using either partial RNAi or a temperature-sensitive plk-1(or683ts) allele (O'Rourke et al., 2011), prevents NE disassembly and lamin depolymerisation in one-cell early C. elegans embryos such that parental chromosomes segregate without merging during mitosis, resulting in the formation of paired nuclei at the two-cell stage (Rahman et al., 2015) (Figure S1A, b). RNAi-mediated partial inactivation of several nucleoporins suppresses the paired nuclei phenotype of plk-1(or683ts) mutant embryos at semi-permissive temperature (Rahman et al., 2015). The underlying mechanism of this suppression is currently unclear but it is possible that partial inactivation of structural nucleoporins weakens the NE and facilitates NEBD in plk-1(or683ts) mutants at semi-permissive temperature.
Whereas Plk1 is clearly required for NEBD both in human cells and in C. elegans, how it contributes to this process is not understood. Plk1 localization is highly dynamic during mitosis: it associates with the centrosomes in prophase, and with the spindle poles and the kinetochores in prometaphase and metaphase. Plk1 is then recruited to the central spindle during anaphase and to the midbody during telophase (Schmucker and Sumara, 2014). This dynamic localization of Plk1 depends on its C-terminal Polo Box Domain (PBD) (Park et al., 2010). The PBD comprises two Polo boxes (PB1 and PB2) that bind to specific phosphorylated sequence motifs (PBD-docking sites) generated by other priming kinases, such as Cdk1 (non-self-priming), or are self-primed by Plk1 itself (Elia et al., 2003b; Elia et al., 2003a). This provides an efficient mechanism to selectively target Plk1 to the vicinity of its substrates in space and time (Neef et al., 2003; Kang et al., 2006; Neef et al., 2007; Burkard et al., 2009; Park et al., 2010).
Here we show that Plk1 is specifically recruited to the NE in prophase through its PBD, both in human cells and in C. elegans embryos. Using the early C. elegans embryo, we show that Cdk1 and PLK-1 phosphorylate central channel nucleoporins on multiple sites to prime their interaction with the PBD, and thereby to anchor PLK-1 to the NPC for the phosphorylation of critical downstream targets and efficient NEBD.
Results
Plk1 localizes to Nuclear Pore Complexes in prophase through its PBD in human cells
Plk1 has been implicated in NEBD but whether it needs to be recruited to the NE to perform this function is not clear. We used indirect immunofluorescence to analyze Plk1 localization during the cell cycle in HeLa cells. Consistent with previous studies (Schmucker and Sumara, 2014), we detected Plk1 at the mitotic centrosomes and kinetochores, at the spindle midzone and the midbody (not shown). In addition, we noticed that Plk1 accumulated as a rim around the nucleus in a fraction of cells with condensed DNA, suggesting that Plk1 is recruited to the nuclear envelope in prophase. Consistently, most of the late G2 and prophase HeLa cells (41 out of 47 cells, detected based on positive phospho-histone H3 staining) showed a clear Plk1 localization at the nuclear envelope (Figure 1A).
Figure 1: Plk1 localizes to the NPCs through its Polo-box domain in human prophase cells.
A- Wide-field images of HeLa cells stained with Plk1 (green) and Phospho-Histone H3 (purple) antibodies and counterstained with DAPI (blue).
B- Wide-field images of a representative prophase HeLa cell expressing GFP-Plk1-PBD (green), stained with mAb414 (red) and Phospho-Histone H3 (purple) antibodies and DAPI (blue). Insets are higher magnification of the boxed regions. *: to better visualize the discontinuous staining at the NE, the contrast within this inset was increased as compared to the entire cell below. Scale bar, 10 μm.
C- (a, b) Two distinct confocal sections through a representative prophase HeLa cells stained with Plk1 (green) and Nup62 (red) antibodies revealing a discontinuous labeling at the nuclear rim (a) and a punctate labeling of the NE surface (b). Scale bar, 5 μm. (c) Confocal section through a representative prophase HeLa cells stained with Plk1 (green) and Lamin A/C (red) antibodies. (d) Image cross-correlation statistical analysis of the distributions of Plk1 and Nup62 or Lamin A/C before (0°) and after rotation of the Nup62 or Lamin A/C images by 90°C (see methods).
In mammalian cells, the PBD has a crucial role in the proper subcellular localization of Plk1 (Park et al., 2010). We thus transiently transfected HeLa cells with a GFP-PBD construct to determine whether the Polo-box domain (PBD) was sufficient to localize to the NE in prophase. Similarly to the endogenous Plk1 kinase, the GFP-PBD accumulated at the nuclear envelope in prophase HeLa cells (as detected in 23 out of 24 phospho-histone H3 positive cells expressing the GFP-PBD transgene) (Figure 1B). Taken together, these results indicate that Plk1 is recruited to the NE in prophase in human cells and that the PBD is sufficient to support this localization. Higher resolution images acquired using confocal microscopy revealed that the NE localization of Plk1 had a punctate pattern (Figure 1C, a, b, c), reminiscent of a nuclear pore staining. Consistently, Plk1 partially colocalized with Nup62 (Figure 1C), a central channel nucleoporin but not with the Lamin A/C (Figure 1C, see Methods). Together, these observations indicate that Plk1 localizes to the NPCs during prophase in human cells.
PLK-1 is recruited to the nuclear envelope in prophase through its PBD in C. elegans oocytes and early embryos
To determine if PLK-1 also localizes to the NE in the early C. elegans embryo, we generated a superfolder (s)GFP Knock-in plk-1 allele using the CRISPR/Cas9 system (Dickinson et al., 2013) (Figure 2A, a, see Material and Methods). Western blot experiments using PLK-1 antibodies confirmed the expression of the PLK-1::sGFP fusion protein at the expected size, despite a reduction in its expression level (Figure 2A, b). Embryos expressing PLK-1::sGFP were fully viable (100%, n=1036) but in rare cases presented a paired nuclei phenotype (Figure S2B), possibly reflecting the reported sensitivity of NEBD to a reduction in PLK-1 levels (Rahman et al., 2015).
Figure 2: PLK-1 is recruited to the NE in prophase through its Polo-box domain in early C. elegans embryos.
A- (a) Schematic of PLK-1::sGFP fusion. PLK-1 N-terminal serine/threonine Kinase domain (KD, dark blue) and C-terminal Polo-box domain (PBD) containing two Polo Boxes (PB1 and PB2, orange) are represented. (b) Western blot analysis of embryonic extracts from Wild-type and PLK-1::sGFP expressing strain using PLK-1 (upper panel) and tubulin antibodies (lower panel, loading control). (c) Spinning disk confocal micrographs of early embryos expressing PLK-1::sGFP and mCherry::HIS-11 at the one-cell (P0) and two-cell stages (AB and P1). Insets are higher magnification of the boxed regions. Scale Bars, 10 and 5 μm.
B- Timing of PLK-1::sGFP recruitment to the NE (arrow) relative to NEBD defined as the time-point at which the nuclear envelope starts to deform (arrowhead). Scale bar, 5μm.
C- (a) Schematic of GFP::PBD fusion protein. (b) Western blot analysis of embryonic extracts from Wild-type and GFP::PBD expressing strains using PLK-1 (upper panel), GFP (middle) and tubulin (lower panel, loading control) antibodies. (c) Spinning disk confocal micrographs of early embryos expressing GFP::PBD at the one-cell and four-cell stages. Scale bars, 10 and 5 μm.
D- Model of the tridimensional structure of the C. elegans PLK-1 PBD. Residues H542 and K544 of the PB2 contacting the phosphopeptides and residue M547 that is mutated in the plk-1(or683ts) allele are highlighted in green and purple, respectively.
E- SPAT-1 interacts with PLK-1 PBD Wild-type but not with its H542A/K544M or M547K mutant forms in a yeast two-hybrid assay. Interactions were determined by assaying growth of yeast cells on selective medium (-L-W-H +3AT, 25 mM). ϕ: Empty Gal4 AD plasmid. Proper expression of the constructs was validated by Western blot (Figure S1D, b).
F- Western blot analysis of embryonic extracts from N2 animals (Wild-type) exposed to mock RNAi (ctrl), or plk-1(RNAi), and plk-1(or683ts) animals raised at 15°C or shifted 5 hours at 25°C using PLK-1 (upper panel) and tubulin (lower panel, loading control) antibodies. Quantification of the ratio of PLK-1 versus Tubulin signal intensity revealed that PLK-1 levels were reduced by 2.5 times in plk-1(or683ts) embryos shifted 5 hours at 25°C compared to the control.
G- Confocal images of fixed Wild-type and plk-1(or683ts) embryos shifted 5 hours at 25°C stained with PLK-1 antibodies (red) and counterstained with DAPI (blue). Insets are higher magnification of the boxed regions. Scale bar, 10 μm. The arrows point the maternal and paternal chromosomes segregating without merging during anaphase resulting in the formation of paired nuclei at the two-cell stage in plk-1(or683ts) embryos.
Using spinning disk confocal microscopy, we recorded early embryos expressing PLK-1::sGFP and mCherry::HIS-11 as a DNA marker (Movie S1). In interphase of two-cell stage embryos, PLK-1::sGFP was exclusively cytoplasmic and enriched in the AB blastomere, as reported previously using PLK-1 antibodies (Budirahardja and Gonczy, 2008; Rivers et al., 2008). Concomitantly to chromosomes condensation, PLK-1::sGFP started to accumulate on chromosomes but could also be detected at the NE (Figure 2A, c). The PLK-1::sGFP signal at the NE became detectable 2.5-3 min prior to NEBD (Figures 2B and S1C, Movie S1). PLK-1::sGFP was similarly detected at the NE in oocytes with condensed chromosomes (Figure S1B, b). Using PLK-1 antibodies, we confirmed the localization of endogenous PLK-1 to the NE in wild-type oocytes and in early embryos (Figure S1B, a, d).
A strain expressing the PLK-1 PBD N-terminally fused to GFP has been generated previously (Nishi et al., 2008) (Figure 2C, a, b). Using this strain, we found that the GFP::PBD recapitulated the localization of the full-length PLK-1 kinase in oocytes and in early embryos (Figure 2C, c and S1B, c). We conclude that, as in human cells (Figure 1), PLK-1 is recruited to the NE in prophase through its PBD domain in C. elegans oocytes and embryos.
A mutation in the phosphopeptide-binding pocket of the PLK-1 PBD affects NEBD
A plk-1 temperature-sensitive (ts) allele has been identified by forward genetic approaches in C. elegans using a screen aimed at identifying novel ts allele of essential genes regulating cell division in early embryos (O'Rourke et al., 2011). At a semi-permissive temperature (23°C), plk-1(or683ts) embryos present a highly penetrant “paired nuclei” phenotype, which is a consequence of defects in nuclear envelope breakdown of the two pronuclei in the one-cell embryo (Audhya et al., 2007; Galy et al., 2008; Rahman et al., 2015). The plk-1(or683ts) allele harbours a mutation located in the second Polo-Box (PB2) that changes a methionine into a lysine residue (M547K) (Figure 2D). In a 3D model of the C. elegans PLK-1 PBD, this residue is located at the top of a β-sheet that contains the two conserved residues crucial for phosphopeptide binding (H538/K540 and H542/K544 in humans and C. elegans, respectively) (Elia et al., 2003b; Nishi et al., 2008) (Figure 2D and S1D). Consistent with this location, the M547K mutation disrupted the phosphorylation-dependent interaction of the PBD with SPAT-1, a well-defined PBD interactor (Figure 2E) (Noatynska et al., 2010; Tavernier et al., 2015). In embryos shifted to 25°C for 2-5 hours, this mutation reduced PLK-1 levels by 2.5-fold and abolished PLK-1 localization to centrosomes, kinetochores, and the anterior blastomere (Figure 2F, G). Collectively, these observations suggest that the phosphopeptide-binding ability of the PBD is important for the proper localization of PLK-1 in the early embryo and for its function in NEBD.
npp-1, npp-3, npp-4 and npp-11 positively regulate PLK-1 function in NEBD
We next sought to identify the nucleoporin(s) anchoring PLK-1 to the NPC (Figure 3A). To this end, we searched for genetic interactions between the plk-1(or683ts) allele and nucleoporins. Inactivation of a subset of nucleoporins suppresses the paired nuclei phenotype of the plk- 1(or683ts) embryos at restrictive temperature (Rahman et al., 2015) and Figure S2A). We reasoned that depletion of the nucleoporin(s) involved in PLK-1 recruitment to the NPC should instead enhance the paired nuclei phenotype of plk-1(or683ts) mutant embryos at permissive temperature (15°C) (Figure S2B upper panel). We thus specifically concentrated on the nucleoporins whose partial inactivation by RNAi did not suppress the paired nuclei phenotype of the plk-1(or683ts) embryos at 25°C (Figure S2A).
Figure 3: The NPP-1•NPP-4•NPP-11 complex recruits PLK-1 to the nuclear envelope for NEBD.
A- Schematic of human and C. elegans NPC. Relevant nucleoporins for this study are colored in NPP-1 (violet), NPP-4 (pink) and NPP-11 (blue). The other C. elegans nucleoporins studied in this Figure and their human counterparts are shaded in dark grey on the schematic of the NPC.
B- DIC micrographs and corresponding schematics of two-cell stage embryos treated with mock RNAi (ctrl) or plk-1(RNAi) and presenting 0 (green), 1 (orange) or 2-paired nuclei (red) (white arrows).
C- Percentage of plk-1(or683ts) embryos presenting 0 (green bars), 1 (orange bars) or 2 (red bars) paired nuclei at the two-cell stage upon exposure to mock RNAi (ctrl), npp-15, npp-13, npp-3, npp-1, npp-4 or npp-11 RNAi at 15°C. The number of embryos analyzed is indicated on the right and was generated by aggregation over more than 3 independent experiments.
D- 6xHis-NPP-11[C] (aa333-805), GST-NPP-4 and Strep-NPP-11 and were co-expressed in E. coli, and successively purified on Hi-TRAP columns and Glutathione beads. The fractions were analyzed by SDS-PAGE (T: Total extract, Ni2+: elution from Hi TRAP, GSH: elution from glutathione beads). The gel was either stained with Coomassie Brilliant Blue (upper panels) or immunoblotted with 6xHis, Strep or GST antibodies (lower panels). * marks an unspecific bands revealed by the GST antibody.
E- Spinning disk confocal micrographs of early 2-cell stage embryos expressing GFP::NPP-1 after exposure to mock RNAi (ctrl), or npp-1, npp-4, npp-11, npp-3 or npp-13 RNAi. Scale bar 2μm. The graph shows the quantification of GFP::NPP-1 signal intensity at the NE in embryos of the indicated genotype. The mean +/− standard error of the mean (sem) is presented. **** indicates statistically significant difference p<0.0001 with one-way ANOVA followed by Dunnett’s multiple comparisons test.
F- Spinning disk confocal micrographs of one-cell stage embryos expressing (a) PLK-1::sGFP; mCherry::HIS-11 or (b) GFP::NPP-19; mCherry::HIS-58 after exposure to mock RNAi (ctrl), npp-12/gp210, npp-5, npp-1, npp-4 and npp-11 RNAi. Scale bar, 5μm.
We inactivated npp-1, npp-3, npp-4, npp-11, npp-13 and npp-15 by RNAi in the plk-1(or683ts) strain at 15°C and scored the percentage of paired nuclei phenotype (Figure 3B, C). While less than 25% of plk-1(or683ts) embryos exposed to mock RNAi exhibited paired nuclei at the permissive temperature (15°C), more than 90% of plk-1(or683ts) embryos exhibited this phenotype upon partial RNAi of npp-1, npp-4 or npp-11 (Figure 3C). An intermediate phenotype was also observed upon depletion of npp-3 and npp-13 whereas inactivation of npp-15 had no significant effect (Figure 3C). Inactivation of npp-1, npp-4 or npp-11 similarly enhanced the mild paired nuclei phenotype of embryos with the plk-1::sgfp knock-in allele that express slightly reduced PLK-1 levels (Figure 2A, b and S2B lower panel). In contrast, inactivation of these nucleoporins in control N2 embryos did not lead to any paired nuclei (not shown).
RNAi of npp-1 or npp-11 had no effect on PLK-1 protein levels (Figure S2C), excluding that the synergy was due to reduced PLK-1 stability. We conclude that partial inactivation of npp-1, npp-4 and npp-11, and to a lesser extent npp-3 and npp-13, enhances the paired nuclei phenotype of embryos with reduced plk-1 function, indicating that these nucleoporins promote PLK-1 function in NEBD.
NPP-1, NPP-4 and NPP-11 are the C. elegans orthologues of human Nup54, Nup58 and Nup62 nucleoporins respectively (Figure 3A), as revealed by multiple protein alignments (Figure S3) (Schetter et al., 2006). These proteins contain unstructured barrier-forming Phenylalanine-Glycine (FG) repeats in their N-terminal regions that contribute to nucleo-cytoplasmic transport selectivity through the central NPC channel. In vertebrates, these three FG nucleoporins assemble via their C-terminal coiled regions into a trimeric complex. This hetero-trimer is anchored to the NPC scaffold via Nup93 (orthologous to C. elegans NPP-13) a constituent of the inner ring complex of the NPC that also includes Nup205 (homologous to C. elegans NPP-3) (Knockenhauer and Schwartz, 2016; Chug et al., 2015; Kosinski et al., 2016). Although yeast two-hybrid assays have revealed interactions between NPP-1, NPP-3, NPP-4, NPP-11 and NPP-13 (Schetter et al., 2006), their assembly had not been assessed in C. elegans.
Using double affinity purification, we found that NPP-1 (Strep), NPP-4 (GST) and amino acids (aa) 333-805 of NPP-11 (NPP-11[C]) (6xHis) co-purify in apparent stoichiometric amounts when co-expressed in E. coli (Figure 3D). Moreover, GFP::NPP-1 or NPP-11::GFP accumulation at the NE was severely reduced upon RNAi-mediated depletion of the other constituents of the NPP-1•NPP-4•NPP-11 complex (Figure 3E and S2D), suggesting that their anchorage within the NPC requires prior formation of the trimeric complex. Finally, the NE localization of GFP::NPP-1 or NPP-11::GFP was also altered in npp-3 and npp-13(RNAi) embryos (Figure 3E and S2D), which may explain why npp-3 and npp-13 inactivation enhanced the paired nuclei phenotype of plk-1(or683ts) at 15°C (Figure 3A, C).
Thus, C. elegans NPP-1, NPP-4 and NPP-11 form, as do their mammalian counterparts, a trimeric complex that is anchored to the central NPC channel by NPP-3 and NPP-13, which contributes to PLK-1 function in NEBD.
Inactivation of npp-1, npp-4 and npp-11 affects PLK-1::sGFP recruitment to the nuclear envelope
We next used spinning disk confocal microscopy to monitor the localization of PLK-1::sGFP following RNAi of npp-1, npp-4 or npp-11. In contrast to mock RNAi, PLK-1::sGFP failed to localize to the NE in npp-1, npp-4 and npp-11(RNAi) embryos whereas it was still recruited to the centrosomes and to the kinetochores (Figure 3F, a). Reduction of the PLK-1::sGFP signal at the NE was not merely a result of major alterations of the NE or NPC assembly because GFP::NPP-19/Nup53 was readily detected at the NE in npp-1, npp-4 and npp-11(RNAi) embryos (Figure 3F, b). PLK-1::sGFP localization at the NE specifically relies on the NPP-1•NPP-4•NPP-11 complex because inactivation of the structural nuclear pore component NPP-5/Nup107 did not prevent PLK-1::sGFP recruitment to the NE in prophase. Likewise, inactivation of npp-12/gp210, which leads to the formation of paired nuclei (Audhya et al., 2007; Galy et al., 2008), did not alter PLK-1::sGFP recruitment to the NE (Figure 3F, a). Taken together these results indicate that the central nuclear pore components NPP-1, NPP-4 and NPP-11 promote PLK-1 localization to the NE in prophase.
The role of the central channel nucleoporins in PLK-1 localization to the nuclear envelope is independent of their function in nuclear import
We next sought to identify the molecular mechanism by which the central channel nucleoporins complex localizes PLK-1::sGFP to the NE in prophase in C. elegans embryos. PLK-1 is exclusively cytoplasmic during interphase but rapidly enters the nucleus just prior to NEBD (Figure 2, Movie S1). The mechanism by which its timely import into the nucleus occurs is currently unknown but PLK-1 contains a predicted (PSORT, https://psort.hgc.jp/form2.html) and conserved nuclear localization sequence (NLS) (Figure 4A), previously shown to target human Plk1 to the nucleus in tissue culture cells (Taniguchi et al., 2002).
Figure 4: The importins IMA-2 and IMB-1 promote PLK-1::sGFP nuclear import but are dispensable for its targeting to the nuclear envelope.
A- Multiple protein alignments show the presence of a nuclear localization signal in the Kinase domain of C. elegans (C. e) PLK-1 and its homologues from H. s: Homo sapiens, R. n: Rattus norvegicus, M. m: Mus musculus, D. r: Danio rerio, D. m: Drosophila melanogaster. Sequences were aligned using Clustal W2 (Larkin et al., 2007) and visualized in Boxshade (Waterhouse et al., 2009). Potential NLS located in the Kinase domain are shaded in green.
B- Schematic of the nuclear import pathway: nuclear-targeted cargoes containing a basic nuclear localization sequence (NLS) (in green), comprised of a cluster of lysine and arginine residues, are recognized in the cytoplasm by a heterodimeric receptor consisting of an NLS recognition subunit, karyopherin/importin alpha (IMA-1/2/3 in C. elegans), and a pore targeting subunit, karyopherin/importin beta (IMB-1/2/3). In the nucleus, RAN GTP dissociates the cargo from the importins.
C- Spinning disk confocal micrographs of one-cell stage embryos expressing PLK-1::sGFP; mCherry::HIS-11 after RNAi treatments, either control (ctrl) or ima-1, ima-2, imb-1, imb-2, imb-3(RNAi). Scale Bar, 5μm.
As the primary function of the central channel nucleoporins is to regulate nucleocytoplasmic transport, they may aid localization of PLK-1::sGFP to the NE by regulating its nuclear import. To test this possibility, we analyzed embryos depleted of importins, which promote nuclear import of NLS-containing cargo. The importins alpha directly recognize NLS-containing cargos whereas importins beta mediate interaction with the NPC, particularly with FG nucleoporins.
The C. elegans genome encodes three importins alpha (IMA-1, IMA-2, IMA-3) and three importins beta (IMB-1, IMB-2 and IMB-3) (Figure 4B). We depleted 5 of these 6 importins and found that PLK-1::sGFP localization to kinetochores was abolished during prophase in ima-2(RNAi) and imb-1(RNAi) embryos (Figure 4C). However, PLK-1::sGFP was still localized to the nuclear envelope in these embryos (Figure 4C). These results demonstrate that the defect in nuclear envelope localization of PLK-1::sGFP in npp-1, npp-4 and npp-11(RNAi) embryos is not due to a defect in its nuclear import, suggesting that these central channel nucleoporins play a more direct role in PLK-1 localization to the NE.
PLK-1 interacts with NPP-1 and NPP-11 primed by Cdk1 phosphorylation
PLK-1 recruitment to the NE is concomitant with chromosome condensation and occurs exclusively in prophase, just prior to NEBD, when Cyclin B-Cdk1 activity increases. Furthermore, PLK-1 localizes to the NE through its PBD domain. These observations suggest that phosphorylation of NPP-1, NPP-4 or NPP-11 by Cyclin B-Cdk1 might prime their interaction with the PLK-1 PBD and thereby contribute to recruit PLK-1 to the NPCs. Based on the PBD-docking consensus motif (Figure 5A; (Elia et al., 2003b)), we identified four potential Cdk1-primed PBD binding sites in NPP-1 (T8, T49, T60, T105) and three in NPP-11 (T321, T393, T555, the last two being included in NPP-11[C]). To test if these sites mediate interaction with the PBD when phosphorylated, we performed in vitro phosphorylation using human Cyclin B-Cdk1 kinase followed by Far-Western ligand-binding assays with human GST-fused PBD (Wu et al., 2007). With a NPP-1•NPP-4•NPP-11[C] trimeric complex as substrate, Cyclin B-Cdk1 phosphorylated in vitro NPP-1, NPP-11[C] and NPP-4, albeit to a lower extent (Figure 5B, S4A). Both phosphorylated NPP-1 and NPP-11[C] in turn interacted with the human Plk1 (GST)-PBD in the far-Western assay (Figure 5C, S4B). Without pre-phosphorylation by Cyclin B-Cdk1, NPP-1 and NPP-11[C] showed no binding to the (GST)-PBD. Likewise, NPP-1 and NPP-11[C] phosphorylated by Cyclin B-Cdk1 showed no binding to the phosphate pincer (GST-PBD H538A/K540M) mutant (Figure 5C, S4B) (Elia et al., 2003b).
Figure 5: Cdk1 phosphorylates NPP-1 and NPP-11 at multiple Polo-docking sites to promote binding to the PLK-1 PBD.
A- The consensus sequence of the PBD-docking site and the sequence and the position of the putative PBD-docking sites primed by Cdk1 on NPP-1, NPP-4 and NPP-11 are presented schematically. ϕ: Hydrophobic residues. cc: coiled-coiled.
B- Radioactive in vitro kinase assay using human Cyclin B-Cdk1 kinase and the C. elegans NPP-1•NPP-4•NPP-11[C] (aa333-805) trimeric complex as substrate. Autoradiograph of the SDS-PAGE showing γ-[32P] incorporation into NPP-1, NPP-4 and NPP-11[C] (upper panel). Coomassie brillant blue (CBB) staining of the same SDS-PAGE (bottom panel). The full experiment is presented in Figure S4A.
C- In vitro kinase assay was performed with human Cyclin B-Cdk1 and the C. elegans NPP-1•NPP-4•NPP-11[C] trimeric complex as substrate. The samples were subjected to Phos-tag SDS-PAGE, followed by a Far-Western ligand-binding assay using GST-PBD Wild-type (upper panel) or the corresponding phosphate pincer (GST-PBD H538A/K540M) mutant (middle panel). Coomassie brillant blue (CBB) staining of the same SDS-PAGE (bottom panel). The full experiment is presented in Figure S4B.
D- E- 6xHis-NPP-1[N] (aal-194) or 6xHis-NPP-11[C1] (aa290–805) Wild-type or mutated in their PBD-docking sites were incubated with Cyclin B-Cdk1 kinase in the presence of γ- [32P]ATP. Autoradiograph of the SDS-PAGE showing γ-[32P] incorporation in 6xHis-NPP-1 and 6xHis-NPP-11 that is reduced in NPP-1 4A and NPP-11 3A (upper panels). Coomassie brillant blue (CBB) staining of the same SDS-PAGE (bottom panels).
F- G- In vitro kinase assay was performed with Cyclin B-Cdkl and the indicated proteins as substrates. The samples were subjected to Phos-tag SDS-PAGE, followed by a Far-Western ligand-binding assay using GST-PBD. Coomassie Brilliant Blue (CBB) staining or Western blot analysis using 6xHis antibodies showed protein loading of NPP-1[N] and NPP-11[C1] fragments respectively.
H- Representative spinning disk confocal micrographs of early embryos expressing PLK- 1::sGFP and mCherry::NPP-1 4A in (a) Wild-type or in (b) double npp-11(ok1599It96); npp- 1(syb207) mutant embryos. White arrows indicate the persistence of the nuclear envelope in double npp-11(ok1599It96); npp-1(syb207) mutant embryos. Note that a similar delay in NPP-1 4A removal was also observed with npp-11(ok1599It96) mutant alone (Figure S5c).
We next performed in vitro phosphorylation of fragments of NPP-1 and NPP-11 encompassing the predicted PBD-docking sites. Cyclin B-Cdk1 readily phosphorylated amino acids 1-194 of NPP-1 (NPP-1[N]) and aa 290-805 of NPP-11 (NPP-11[C1]) (Figure 5D, 5E). Phosphorylation of the same fragments harboring non-phosphorylatable alanines at the predicted PDB-binding sites was significantly reduced (Figure 5D, 5E). To test whether phosphorylation of these seven sites contributes to binding to the PBD, we combined the Far-Western ligand-binding assay with Phos-tag SDS-PAGE, to resolve the different phosphorylated forms of the NPP-1 and NPP-11 fragments. This analysis revealed a phosphorylation-dependent binding of GST-PBD to NPP-1[N] and NPP-11[C1] fragments. Notably binding was observed for several phosphorylated forms of NPP-1[N] and NPP-11[C1] indicating that the PBD is binding to multiple Cdk1-primed binding sites on NPP-1 and NPP-11 (Figure 5F, G). Consistent with this, in a NPP-11[C] fragment, mutating both T393 and T555 to alanine prevented binding to the PLK-1 PBD after phosphorylation by Cyclin B-Cdk1 whereas mutating only T555 did not prevent binding (Figure S4D).
To test if the 7 sites defined by sequence analysis accounted for PBD binding, we analyzed variants of NPP-1[N] (4A) or NPP-11[C1] (3A) in which all of the predicted PBD-docking sites were mutated. The introduced alanine mutations abolished PBD binding (Figure 5F, G) even though Cyclin B-Cdk1 still phosphorylated other sites on NPP-1[N] and induced a mobility shift on Phos-tag SDS-PAGE (Figure 5F). We confirmed the importance of most of these sites by analyzing a mutant NPP-1•NPP-11[C] dimer in which 6 out of 7 phosphorylation sites were altered to alanine (Figure S4C). Thus, seven Cdk1-primed sites on NPP-1 and NPP-11 mediate binding of the PBD of PLK-1 to this conserved nucleoporins complex in vitro.
Mutation of Cdk1-primed PLK-1 binding sites reveals a role for PLK-1 phosphorylation of NPP-1 and NPP-4 for self-priming and binding
The above data show that PLK-1 localizes to the NE prior to NEBD via its PBD, contributes functionally to NEBD in a PBD-dependent manner, and is recruited to the NE by NPP-1, NPP-4 and NPP-11 independently of its nuclear transport. In vitro biochemical analysis indicates that there are 7 Cdk1-primed binding sites for the PLK-1 PBD in the NPP-1•NPP-4 •NPP-11 complex. To investigate the contribution of these 7 sites to PLK-1 localization in vivo, we employed CRISPR/Cas9 to remove the region of NPP-1 containing its 4 Cdk1-primed sites (npp-1(syb207)) and combined an existing deletion mutant with a CRISPR/Cas9-engineered mutation to generate a version of NPP-11 lacking all 3 of its Cdk1-primed sites (npp-11(ok1599It96)) (Figure S5). We then crossed these two mutants to generate a double mutant lacking all 7 Cdk1-primed sites on NPP-1 and NPP-11 in vivo and introduced into this strain background PLK- 1::sfGFP. We also introduced into this double mutant background mCherry-fused NPP-1 with the 4 Cdk1-primed sites mutated to alanine (4A), to attribute potential defects to the Cdk1-primed sites, and not to the npp-1 deletion, and to monitor the dynamics of NPC disassembly. Surprisingly, we still observed robust PLK-1::sfGFP localization at the NE (Figure 5H). However, we noticed a consistent delay in removal of mCherry-fused NPP-1 4A (Figure 5H, b), a similar but milder delay was also observed with npp-11(ok1599It96) mutant alone (Figure S5, c). These observations suggest that the Cdk-primed PBD binding sites and/or the region deleted in the ok1599 allele of npp-11 contribute to the kinetics of NPC removal.
The fact that removing all 7 Cdk1-primed binding sites in NPP-1, NPP-4 and NPP-11 for the PLK-1 PBD did not prevent PLK-1 localization in vivo led us to consider a potential self-priming-based localization of PLK-1 to the NE, a mechanism previously shown to contribute to its targeting to various mitotic structures in human cells (Neef et al., 2003; Kang et al., 2006; Neef et al., 2007; Lénárt et al., 2007). To test this hypothesis, we used C. elegans PLK-1 kinase purified from Sf9 cells (Tavernier et al., 2015) and the trimeric NPP-1•NPP-4•NPP-11[C] complex as substrate for in vitro kinase and Far-western ligand-binding assays. Strikingly, PLK-1 readily phosphorylated NPP-4 (Figure 6A) and promoted its robust binding to the PBD in vitro whereas no binding was seen without phosphorylation by PLK-1 (Figure 6B). In addition, a minor phosphorylation, and subsequent binding of NPP-1 to the PBD was detected in this assay whereas NPP-11[C] showed no detectable binding to the PBD despite being phosphorylated by PLK-1 (Figure 6B, C).
Figure 6: Complementary mechanisms promote PLK-1 binding to phosphorylated nucleoporins of the central channel.
A- Radioactive in vitro kinase assay using PLK-1 and the NPP-1•NPP-4•NPP-11[C] trimeric complex as substrate. Autoradiograph of the SDS-PAGE showing γ-[32P] incorporation in NPP-1, NPP-4 and NPP-11[C] (upper panel). Coomassie brillant blue (CBB) staining of the same SDS-PAGE (bottom panel). Red asterisk marks autophosphorylated PLK-1. The full experiment is presented in Figure S4A.
B- In vitro kinase assay was performed with PLK-1 and the NPP-1•NPP-4•NPP-11[C] trimeric complex as substrate. The samples were separated by SDS-PAGE followed by a Far-Western ligand-binding assay using GST-PBD Wild-type (upper panel) or the corresponding phosphate pincer (GST-PBD H538A/K540M) mutant (middle panel). Coomassie Brilliant Blue (CBB) staining shows protein loading of NPP-1•NPP-4•NPP-11[C] trimeric complex. The full experiment is presented in Figure S4B.
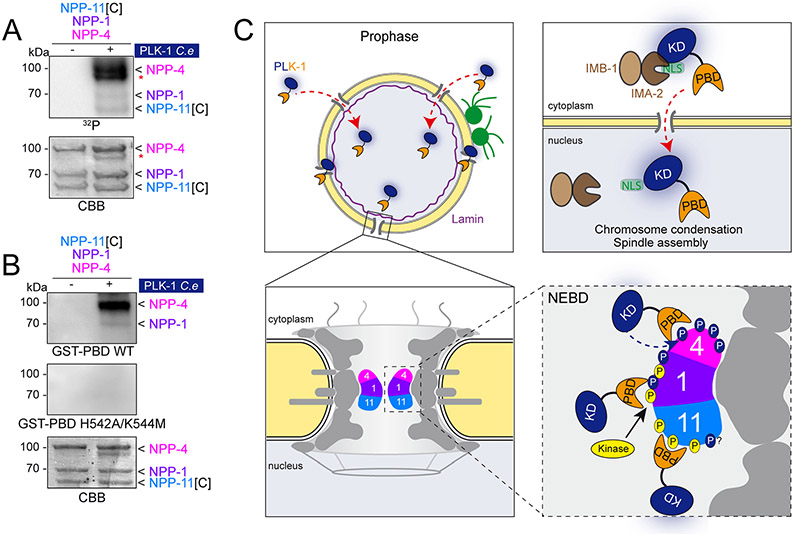
C- Schematic diagrams showing PLK-1 dynamics and targeting mechanisms during prophase (see discussion for details). PLK-1 is suddenly imported into the nucleus in prophase, via the importins IMA-2 and IMB-1 (brown) that most likely recognize the nuclear localization sequence located in the Kinase domain. PLK-1 accumulates in the NPC independently of nuclear import via its binding to phosphorylated motifs on the central channel nucleoporins generated by Cdk1 or PLK-1 itself.
Human Plk1 phosphorylates substrates on broad consensus motifs that do not account for all observed Plk1 substrates (Kettenbach et al., 2011; Santamaria et al., 2011). Previous work has shown that the minimal self-priming and binding site can be a phosphorylated serine or threonine residue preceded by a serine residue (S-Sp/Tp) with moderate similarity to the optimal PBD binding sites (e. g T602 in human PRC1: peptide GILNSTpN) (Neef et al., 2007). Based on this minimal (S-Sp/Tp) motif, there are more than twenty potential self-priming sites in each nucleoporin of the central channel. These sites are often repeated just before, or after, the FG repeats (Figure S6). Thus, we suggest that self and non-self-priming and binding contribute to PLK-1 recruitment to the NE in prophase (Figure 6C).
Discussion
Our results indicate that Plk1/PLK-1 is actively recruited to the nuclear pore complexes in prophase through its Polo-box domain to trigger NEBD, both in human cells and in C. elegans. Using the C. elegans embryo as model, we identified the nucleoporins NPP-1, NPP-4 and NPP-11, which form a trimeric complex localized in the central channel of the nuclear pore complexes, as the critical components anchoring PLK-1 to the NE in C. elegans embryos. Indeed, we show that Cdk1 and PLK-1 phosphorylates NPP-1, NPP-4 and NPP-11 on multiple PBD-docking sites in vitro to prime their interaction with the PLK-1 PBD. Consistently, we found that PLK-1::sGFP is no longer recruited to the NE under conditions that impair the NPC recruitment of NPP-1, NPP-4 or NPP-11. These conditions drastically enhanced the NEBD defects of plk-1(or683ts) embryos at permissive temperature, indicating that these nucleoporins positively regulate PLK-1 function in NEBD by recruiting this kinase to the NPCs for phosphorylation of critical downstream targets.
The importins IMA-2 and IMB-1 promote PLK-1 nuclear import in prophase but are dispensable for PLK-1 recruitment to the nuclear envelope
Using a unique C. elegans strain harboring plk-1 endogenously tagged with sGFP, we could monitor the dynamic of PLK-1::sGFP localization at mitotic entry, in space and time. In interphase, PLK-1::sGFP is exclusively cytoplasmic and is undetectable in the nucleus, even after inhibition of nuclear export (our unpublished data) indicating that PLK-1 is suddenly imported to the nucleus in prophase. PLK-1::sGFP then concomitantly accumulates on the condensing chromosomes and at the nuclear envelope 2.5-3 minutes before nuclear envelope breakdown (Figure 2 and Movie S1).
How is PLK-1 suddenly imported to the nucleus? Protein sequence analysis revealed the presence in the kinase domain of a conserved nuclear localization signal (Figure 4A), and we identified the importins alpha IMA-2 and beta IMB-1 as the critical factors promoting PLK-1::sGFP nuclear import in prophase (Figure 4B, C). Previous work established that ima-2 and imb-1(RNAi) embryos are unable to assemble a mitotic spindle (Askjaer et al., 2002) and we also noticed severe defects in chromosome condensation in these embryos (Figure 4C), consistent with the established role of PLK-1 in chromosome condensation and in kinetochore-MT attachments. Other proteins are likely imported to the nucleus by IMA-2 and IMB-1 to bring about chromosomes condensation and MT-kinetochore attachment. One candidate is HCP-4, which is cytoplasmic during interphase, but translocates to the nucleus during mitosis and localizes specifically to the centromere in a process requiring the centromeric histone HCP-3 (Oegema et al., 2001; Ferreira et al., 2017). Overall our results indicate that active nuclear import is essential for PLK-1 to access the chromosomes to regulate chromosome condensation and to promote mitotic spindle assembly. The sudden import of PLK-1 might result from a conformational change of the kinase upon its activation, thereby opening the structure and unmasking the NLS. Indeed, in the closed inactive conformation, residues of the NLS are engaged in interaction with residues of the PBD and are not accessible to the importins (Xu et al., 2013).
The mechanism of Plk1 recruitment to the nuclear envelope
Although active transport might also facilitate PLK-1 targeting to the NPC, its recruitment to the nuclear envelope is not altered in ima-2 and imb-1(RNAi) embryos indicating that PLK-1 can access the NPC without the need of an active transport.
We found that both in human cells and in C. elegans embryos, the PBD is sufficient to target Plk1/PLK-1 to the NPC and identified the central channel nucleoporins NPP-1, NPP-4 and NPP-11 as the key factors anchoring PLK-1 to the NPC. We identified seven sites on NPP-1 and NPP-11 primed by Cdk1 that enable binding to the PBD. Among these sites, large-scale phospho-proteomic analyses have shown that threonine 8 of NPP-1 and threonine 555 residues of NPP-11 are phosphorylated in worms (Zielinska et al., 2009; Gnad et al., 2011). The other sites are part of very large peptides generated by trypsin digestion that are not visible by tandem mass spectrometry.
Mutating the seven sites in NPP-1 and NPP-11 to non-phosphorylatable alanine abrogated binding to the PBD in vitro after phosphorylation by Cdk1 (Figure 5), indicating that these sites are the only sites primed by Cdk1 for binding to the PBD. However, deleting these seven sites in vivo was not sufficient to delocalize PLK-1::sGFP from the NE, indicating that complementary mechanism(s) contribute to PLK-1 localization at the NPC. It is very unlikely that depletion of npp-1, npp-4, npp-11 destabilize other nucleoporin(s) anchoring PLK-1 to the NE because these central channel nucleoporins are not core components of the nuclear pore complexes. We also unequivocally excluded the possibility that the defects in PLK-1 localization to the NE in npp-1, npp-4 and npp-11 embryos could stem from a defect in its nuclear import, as discussed above. Our results rather indicate that PLK-1 itself phosphorylates the central channel nucleoporins, particularly NPP-1 and NPP-4, but possibly also amino acids 1-332 of NPP-11 (NPP-11[N]), not tested in our assay, for self-priming and binding (Figure 6B). The main advantage of self-priming and binding is that it is a feed forward mechanism of amplification. Once a few molecules of PLK-1 are recruited to the NPC, they can generate additional PBD binding sites, thereby accelerating PLK-1 recruitment.
Likewise, the PBD is sufficient to target human Plk1 to the NPC in HeLa cells (Figure 1 and the accompanying paper by Linder et al.) indicating that the general mechanism of PLK-1/Plk1 recruitment to the NE is conserved between C. elegans embryos and human cells. However, the identity of the nucleoporins recruiting Plk1 to the NPC likely differs between C. elegans and human cells. The Polo-docking sites in NPP-1 and NPP-11 primed by Cdk1 are not conserved in their human orthologues, Nup58 and Nup62 respectively (Figure S3). Furthermore, the accompanying paper by Linder et al. indicates that Nup53 (NPP-19 in C. elegans) contributes to NPC localization of Plk1 in human cells. Inspired by this finding (U. Kutay personal communication) and consistent with a previous proteome-wide Y2H study (Boxem et al., 2008), we found that Plk1-PBD binds C. elegans NPP-19[N] (amino acids 1-301) upon in vitro phosphorylation by either Cyclin B-Cdk1, or PLK-1 (Figure S4E). While this indicates that PLK-1 interacts with phosphorylated NPP-19 in C. elegans, NPP-19 is likely a secondary PLK-1 binding site in C. elegans embryos. Indeed, although depleting the central channel nucleoporins is sufficient to delocalize PLK-1::sGFP from the NE, NPP-19 is still normally localized to the NE under these conditions (Figure 3F, b). Regulated binding of PLK-1 to phospho-NPP-19 might be required in a second step to promote NEBD in C. elegans embryos.
It is worth mentioning that the mechanism of NEBD is different between human cells and C. elegans. Whereas mitosis is open in human cells, it is semi-closed in C. elegans. Consequently the positioning of Plk1/PLK-1 in the vicinity of its substrates is expected to differ between the two species, although the general principle is identical. In both cases, Plk1 is specifically recruited to the NPC via the binding of the PBD to phosphorylated nucleoporins.
Substrates for PLK-1 in NEBD
What could be the role of PLK-1 at the NE? PLK-1 might trigger NPC disassembly by phosphorylating nucleoporins. Consistently, nuclear assembly in C. elegans relies on the specific docking of the PP1 phosphatase on the nucleoporin MEL-28 suggesting that PP1 might counteract PLK-1-dependent phosphorylation of nucleoporins (Hattersley et al., 2016). Furthermore, Plk1 has been shown to phosphorylate several nucleoporins in human cells (Grosstessner-Hain et al., 2011; Santamaria et al., 2011; Kettenbach et al., 2011; Bibi et al., 2013) and the accompanying manuscript by Linder et al. shows a critical role of Plk1 in NEBD through the phosphorylation of Nup53 and Nup98 in human cells. In C. elegans, PLK-1 might also regulate lamina disassembly, which occurs after Cyclin B-Cdk1 inactivation in C. elegans and therefore does not rely on Cyclin-Cdk1 complexes as in human cells (Lee et al., 2000). Consistently, the evolutionarily conserved Cdk sites present in all lamins are not present in the C. elegans LMN-1 (Machowska et al., 2015). It is likely that PLK-1 has multiple targets at the NE to orchestrate the different steps of NEBD. An important challenge for the future will be to identify these numerous targets.
In summary, this work further illustrates the importance of nucleoporins in cell cycle control and their role in nuclear envelope breakdown, beyond their canonical function in nucleocytoplasmic transport.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by the lead contact author L. Pintard (lionel.pintard@ijm.fr)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains, HeLa Cells and bacterial strains used in this study are listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human Plk1 | Euromedex | Upstate Cat#05-844 Batch 27108 |
| Anti-mAb414 | Covance | Cat#MMS-120P-0500 Batch B203094 |
| Anti-Phospho-Histone H3 (Ser10) | Millipore | Upstate Cat#06-570 Batch 2517793 |
| Anti-Nup62 | (Cordes et al., 1991) | N/A |
| Anti-C. elegans PLK-1 | (Tavernier et al., 2015) | Monica Gotta |
| Anti-Tubulin – DM1A | Sigma | Cat#T9026 |
| Anti-Lamin A/C | (Chaudhary and Courvalin, 1993) | N/A |
| Anti-Gal4-AD | Clontech | Cat#630402 |
| Anti-Gal4-BD | Santa Cruz | Cat#sc-577 |
| Anti-6xHis | Novagen | Cat#70963 |
| Anti-GST | This Study | N/A |
| Anti-GFP | ThermoFisher | Cat#A-11122 |
| StrepTactin-HRP | IBA | Cat#2-1502-001 |
| Peroxidase Goat Anti-Mouse IgG (H+L) | Sigma | Cat#A9917 |
| Peroxidase Goat Anti-Rabbit IgG (H+L) | Sigma | Cat#A0545 |
| Goat Anti-Rabbit Alexa 568 | Invitrogen | Cat#A-11011 |
| Goat Anti-Mouse Alexa 488 | Invitrogen | Cat#A-11008 |
| Donkey Anti-Mouse Alexa 488 | Invitrogen | Cat #A-21202 Batch #55980A |
| Donkey Anti-Rabbit Cy3 | Jackson | IR #711-165-152 Batch #79935 |
| Donkey anti-Guinea pig Cy3 | Jackson | IR # 706-165-148 Batch #68252 |
| Donkey Anti-Mouse Cy5 | Jackson | IR #715-175-150 Batch #63788 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Coomassie R250 | Sigma | Cat#B014925G |
| Ponceau Red | Sigma | Cat#A1405 |
| VECTASHIELD Mounting Medium with DAPI | Eurobio | Cat#H-1200 |
| Poly-Lysine L-lysine hydrobromide | Sigma | Cat#P1524 |
| 3-Amino-1,2,4-Triazole (3AT) | Sigma | Cat#A8056 |
| IPTG | Euromedex | Cat#EU0008-B |
| Adenosine TriPhosphate (ATP) | Sigma | Cat#A2383 |
| γ-[32P] Adenosine TriPhosphate (ATP) 500 μCi 3000Ci/mmol | PerkinElmer | Cat#NEG002A500UC |
| Human Cyclin B-Cdk1 kinase | New England Biolabs | Cat#P6020L |
| Phos-Tag™ Acrylamide AAL-107 | WAKO-SOBIODA | Cat#W1W304-93521 |
| Imidazole | Sigma | Cat#I202 |
| Glutathione | Sigma | Cat#G4251 |
| Critical Commercial Assays | ||
| Hitrap Chelating HP 5x1mL | GE Healthcare | Cat#17-0408-01 |
| Glutathion Sepharose 4B Fast Flow | GE Healthcare | Cat#17-0756-01 |
| ECL reagent | Millipore | Cat#WBKLS0500 |
| ECL reagent SuperSignal West Pico PLUS | ThermoFisher Scientific | Cat#34579 |
| BP Clonase II Enzyme Mix (Gateway cloning) | Invitrogen | Cat#11789-020 |
| LR Clonase II Enzyme Mix (Gateway cloning) | Invitrogen | Cat#11791-020 |
| Pfu | Promega | Cat#M7741 |
| DpnI | Biolabs | Cat#R0176S |
| Lipofectamine 2000 | Invitrogen | Cat#11668-019 |
| Experimental Models: Cell Lines | ||
| HeLa Kyoto | Provided by J. Ellenberg | Wild type HeLa kyoto (HeLa-K) cell line was from Prof. Narumiya in Kyoto University (RRID: CVCL_1922), and the genome was sequenced previously (Landry et al., 2013) |
| Experimental Models: Organisms/Strains | ||
| C. elegans N2 Bristol | CGC | http://www.cgc.cbs.umn.edu/strain.php?id=10570 |
| plk-1(or683ts)III | (O'Rourke et al., 2011) | EU1441 |
| jjIs1092 [(pNUT1) npp-1::GFP + unc-119(+)]. ltIs37[(pAA64) Ppie-1::mCherry::his-58 + unc-119(+)]IV | (Golden et al., 2009) | OCF3 |
| unc-119(ed3)III bqIs07 [(Ppie-1::LAP::npp-19 unc-119(+)]. ltIs37 [(pAA64) Ppie-1::mCherry::his-58 + unc-119(+)]IV | (Ródenas et al., 2009) | BN14 |
| unc-119(ed3)III::ddls77[npp-11::TY1::EGFP::3xFLAG + unc-119(+)] | (Sarov et al., 2012) | TH239 |
| unc-119(ed3)III TeIs65 [(Ppie-1gfp::plk-1PBD unc-119(+)] | (Nishi et al., 2008) | TX773 |
| unc-119(ed3)III | CGC | HT1593 |
| [ttTi5605II; unc-119 (ed3)III; ox∣Ex1578] | CGC | EG6699 |
| plk-1(lt17) [plk-1::sgfp]loxp)III | This Study | OD2425 |
| Itls37 [(pAA64) pie-1p::mCherry::his-58 + unc-119(+)]IV qals3507 [pie-1::GFP::LEM-2 + unc-119(+)] | CGC | OD83 |
| npp-11(ok1599)I | CGC | RB1406 |
| npp-11(ok1599It96[npp-11 T555A])I | This Study | OD3650 |
| npp-1(syb207)IV | This Study (SunyBiotech) | WLP664 |
| npp-11(ok1599It96)I; plk-1(lt17) [plk-1::sgfp]loxp)III | This Study | WLP663 |
| npp-11(ok1599It96)I; plk-1(lt17) [plk-1::sgfp]loxp)III; npp-1(syb207)IV | This Study | WLP666 |
| leals32 [Pmex-5::mCherry::npp-1 tbb-2 3’UTR unc-119(+)]II | This Study | WLP634 |
| leals33 [Pmex-5::mCherry::npp-1 4A tbb-2 3’UTR unc-119(+)]II | This Study | WLP652 |
| leals32 [Pmex-5::mCherry::npp-1 tbb-2 3’UTR unc-119(+)]II; plk-1(lt17) [plk-1::sgfp]loxp)III | This Study | WLP639 |
| leals33 [Pmex-5::mCherry::npp-1 4A tbb-2 3’UTR unc-119(+)]II; plk-1(lt17) [plk-1::sgfp]loxp)III | This Study | WLP660 |
| npp-11(ok1599It96)I; leals33 [Pmex-5::mCherry::npp-1 4A tbb-2 3’UTR unc-119(+)]II; plk-1(lt17) [plk-1::sgfp]loxp)III; npp-1(syb207)IV | This Study | WLP672 |
| ijmSi31 [Pmex-5mCherry::his11:: tbb-2 3'UTR]II; unc-119(ed3)III | This Study | JDU128 |
| ijmSi31 [Pmex-5 mCherry::his11:: tbb-2 3'UTR]II; plk-1(lt17) [plk-1::sgfp]loxp)III | This Study | WLP552 |
| MATa, ura3-52, his3-200, lys2-801, ade2-101, trp 1-901, leu2-3, 112, gal4-542, gal80-538, LYS 2::GAL1UAS-GAL1TATA-HIS3, URA3::(GAL4 17-mers)3-Cyc1TATA-lacZ,cyhr2 | (Feilotter et al., 1994) | CG1945 |
| MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, met−, gal4D, gal80D, URA3::GAL1UAS-GAL1TATA-lacZ | (Harper et al., 1993) | Y187 |
| Recombinant DNA | ||
| pEGFP-Plk1 PBD H. s | Gift V. Mirouse | N/A |
| L4440 (RNAi Feeding vector) | (Kamath et al., 2001) | N/A |
| plk-1 cloned into L4440 | (Kamath et al., 2003) | Arhinger Library Clone III-4-E08 |
| npp-1 cloned into L4440 | (Galy et al., 2003) | N/A |
| npp-2 cloned into L4440 | (Galy et al., 2003) | N/A |
| npp-3 cloned into L4440 | (Galy et al., 2003) | N/A |
| npp-4 cloned into L4440 | (Galy et al., 2003) | N/A |
| npp-5 cloned into L4440 | (Galy et al., 2003) | N/A |
| npp-6 cloned into L4440 | (Galy et al., 2003) | N/A |
| npp-7 cloned into L4440 | (Galy et al., 2003) | N/A |
| npp-10 cloned into L4440 | (Galy et al., 2003) | N/A |
| npp-11 cloned into L4440 | (Galy et al., 2003) | N/A |
| npp-12/gp210 cloned into L4440 | (Galy et al., 2003) | N/A |
| npp-13 cloned into L4440 | (Galy et al., 2003) | N/A |
| npp-15 cloned into L4440 | (Galy et al., 2003) | N/A |
| ima-1 cloned into L4440 | (Kamath et al., 2003) | Arhinger library Clone V-7-L21 |
| ima-2 cloned into L4440 | (Kamath et al., 2003) | Arhinger library Clone I-3-C11 |
| imb-1 cDNA cloned into L4440 | This Study | pLP1956 |
| imb-2 gDNA cloned into L4440 | This Study | pLP1958 |
| imb-3 cDNA cloned into L4440 | This Study | pLP1959 |
| pDonR NPP-1 gDNA | This Study | pLP1813 |
| Pmex-5::mCherry::npp-1 tbb-2 3’UTR unc-119(+) | This Study | pLP1869 |
| Pmex-5::mCherry::npp-1 4A tbb-2 3’UTR unc-119(+) | This Study | pLP1886 |
| Peft-3 Mos1 transposase | (Frokjaer-Jensen et al., 2008) | pCFJ601 |
| Prab-3::mCherry | (Frokjaer-Jensen et al., 2008) | pGH8 |
| Pmyo-2::mCherry | (Frokjaer-Jensen et al., 2008) | pCFJ90 |
| Pmyo-3::mCherry | (Frokjaer-Jensen et al., 2008) | pCFJ104 |
| Peft-3::Cre | (Dickinson et al., 2013) | pDD104 Addgene Cat#47551 |
| Pmyo-2::mCherry::unc-54 | (Frokjaer-Jensen et al., 2008) | pCFJ90 Addgene Cat#19327 |
| Pmyo-3::mCherry::unc-54 | (Frokjaer-Jensen et al., 2008) | pCFJ104 Addgene Cat#19328 |
| Gal4 pDEST DB | (Noatynska et al., 2010) | pMG97 |
| Gal4 pDEST DB-PLK-1 PBD | (Noatynska et al., 2010) | pMG477 |
| Gal4 pDEST DB-PLK-1 PBD H542A, K544M | (Noatynska et al., 2010) | pMG538 |
| Gal4 pDEST DB-PLK-1 PBD M547K | This Study | pLP1605 |
| Gal4 pDEST AD | (Noatynska et al., 2010) | pMG164 |
| Gal4 pDEST AD-SPAT-1 | (Noatynska et al., 2010) | pMG460 |
| pGEX-4T (GST) | GE Healthcare | Cat#GE28-9545-49 |
| pFasTBAC Hta PLK-1 C. elegans | (Tavernier et al., 2015) | pLP871 |
| pGEX-6p1 GST-PLK-1 PBD H. s | Gift I. Sumara | N/A |
| pGEX-6p1 GST-PLK-1 PBD H538A/K540M H. s | Gift I. Sumara | N/A |
| pDonR201 | ThermoFisher | N/A |
| pDonR201 NPP-1 | This Study | pLP1727 |
| pDonR201 NPP-1 4A | This Study | pLP1833 |
| pDonR201 NPP-4 cDNA | This Study | pLP1811 |
| pDonR201 NPP-11 | This Study | pLP1601 |
| pDonR201 NPP-11 T555A | This Study | pLP1652 |
| pDonR201 NPP-11 T321A T393A T555A | This Study | pLP1653 |
| pDonR201 NPP-11[C] (aa333-805) | This Study | pLP1606 |
| pDonR201 NPP-11[C] (aa333-805) T555A | This Study | pLP1610 |
| pDonR201 NPP-11[C] (aa333-805) T393A T555A | This Study | pLP1655 |
| pDonR201 NPP-19[N] (aa1-301) | This Study | pLP1806 |
| pDEST15 | ThermoFisher | Cat#11802014 |
| pDEST15 GST-NPP-4 | This Study | pLP1821 |
| pDEST17 | ThermoFisher | Cat#11803012 |
| pDEST17 6xHis- NPP-19[N] (aa1-301) | This Study | pLP1861 |
| pET28b | Novagen | Cat#69864 |
| pET28b 6xHis-NPP-1[N] (aa1-194) | This Study | pLP1820 |
| pET28b 6xHis-NPP-1[N] (aa1-194) 4A | This Study | pLP1832 |
| pET28b 6xHis-NPP-11[C1] (aa290-805) | This Study | pLP1823 |
| pET28b 6xHis-NPP-11[C1] (aa290-805) 3A | This Study | pLP1824 |
| pET28b 6xHis-NPP-11[C] (aa333-805) | This Study | pLP1650 |
| pET28b 6xHis-NPP-11[C] (aa333-805) T555A | This Study | pLP1651 |
| pET28b 6xHis-NPP-11[C] (aa333-805) T393A, T555A | This Study | pLP1666 |
| pRSFDuet-1 | Novagen | Cat#71341 |
| pRSFDuet-1 6xHis-NPP-11[C] (aa333-805) | This Study | pLP1692 |
| pRSFDuet-1 Strep-NPP-1, 6xHis-NPP-11[C] (aa333-805) | This Study | pLP1734 |
| pRSFDuet-1 Strep-NPP-1 4A, 6xHis-NPP-11[C] (aa333-805) 2A | This Study | pLP1843 |
| Sequence-Based Reagents | ||
| Alt-R® CRISPR-Cas9 tracrRNA, 100nmol | IDT | Cat#1072534 |
| Primers for cloning and site-directed mutagenesis (see oligonucleotide sequences table) | This Study | N/A |
| Software and Algorithms | ||
| Clustal W2 | (Larkin et al., 2007) | N/A |
| Jalview | (Waterhouse et al., 2009) | N/A |
| USCF Chimera version | (Pettersen et al., 2004) | N/A |
| Adobe Illustrator CS6 | Adobe | N/A |
| Adobe Photoshop CS4 | Adobe | N/A |
| Image J | (Schneider et al., 2012) | N/A |
| ZEN | Zeiss | N/A |
| PRISM | Graphpad | N/A |
| Metamorph | N/A | |
| Other | ||
| DM6000B | Leica | N/A |
| Typhoon | GE Healthcare | N/A |
| Spinning disk confocal Microscope | Nikon | N/A |
| DIC Videomicroscope | Zeiss | N/A |
| LSM710 and LSM780 Confocal Microscopes | Zeiss | N/A |
| Oligonucleotide sequences | ||
| GTTCGGAGCACCAACATCTGCTCCTTCGTCCGCTCCAGCCACTCTTGGACTTGGCCTAGGATTGACGTCAGCACCATTAGCTAAGGGAACTGGGTACATTTTGGTTTACTTTTTAAAAATATAAAAAAAACTCTTAATT | ocrDC03 | Repair template ssODN for T555A CRISPCR/Cas9mutation |
| CACTTGAACTTCAATACGGCAAGATGAGAATGACTGGAAACCGTACCGCATGCGGTGCCTATGGTAGCGGAGCTTCACATGGCTTCAGACCAACAGCCTAT | OLP1488 | dpy-10 ssODN |
| GGGGACAAGTTTGTACAAAAAAGCAGGCTTCAGGAGCTCCAAATCCAAAATG | OLP1855 | imb-1 cDNA for L4440 |
| GGGGACCACTTTGTACAAGAAAGCTGGGTCTGAATTGAGGCAAGCTTCTCT | OLP1856 | imb-1 cDNA for L4440 |
| GGGCGGCCGCATTCGTGCGACAGTAGGTATC | OLP1872 | imb-2 gDNA for L4440 |
| GGGGTCGACATTCTAAATTCTCACATTGGCG | OLP1873 | imb-2 gDNA for L4440 |
| GGGCGGCCGCTATCCCAATCATCGCCGAAAG | OLP1874 | imb-3 cDNA for L4440 |
| GGGGTCGACTGTTGAAGATATTGAAGTCTGG | OLP1875 | imb-3 cDNA for L4440 |
| AGGATCATGTCAAGAAGATGATGTGCCC | OLP1201 | Mutagenesis of the PBD M547K |
| GGGCACATCATCTTCTTGACATGATCCT | OLP1202 | Mutagenesis of the PBD M547K |
| GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGTTTGGAGGATCTGCACC | OLP1275 | NPP-1 into pDONR201 |
| GGGGACCACTTTGTACAAGAAAGCTGGGTCTTATCCTTCTAGAGCCCTTCTACAAGC | OLP1277 | NPP-1 into pDONR201 |
| GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGTCGCTTTTCGGTACAAGTACCACGGCT | OLP1544 | NPP-4 cDNA in pDONR201 |
| GGGGACCACTTTGTACAAGAAAGCTGGGTCTTATTTCTTTGAACTGAACAAAAGACCACTGG | OLP1545 | NPP-4 cDNA in pDONR201 |
| GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGTTTGGAGGATCTGCACCAAAACCGTCG | OLP1179 | NPP-11 in pDONR201 |
| GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAGGCCTTCGACGAGTTGTTCG | OLP1180 | NPP-11 in pDONR201 |
| CGGGTCATCTGCTCCCCAAAAACAGGC | OLP1300 | Mutagenesis of NPP-1 T8A |
| GCCTGTTTTTGGGGAGCAGATGACCCG | OLP1301 | Mutagenesis of NPP-1 T8A |
| GCTCAGGCTTCATCAGCGCCTTCGCTTTTTGGAACAACAAATACCTCTGCTCCTTCTGGAGGATTGTTTGG | OLP1721 | Mutagenesis of NPP-1 T49A and T60A |
| CCAAACAATCCTCCAGAAGGAGCAGAGGTATTTGTTGT TCCAAAAAGCGAAGGCGCTGATGAAGCCTGAGC | OLP1722 | Mutagenesis of NPP-1 T49A and T60A |
| GCTCAATTTTCAGGATCGGCCCCATTCGGAGCAGCGTCG | OLP1723 | Mutagenesis of NPP-1 T105A |
| CGACGCTGCTCCGAATGGGGCCGATCCTGAAAATTGAGC | OLP1724 | Mutagenesis of NPP-1 T105A |
| GGGCTCACTTCAGCCCCATTGGCTAAGGG | OLP1207 | NPP-11 T555A in pDONR201 |
| CCCTTAGCCAATGGGGCTGAAGTGAGCCC | OLP1208 | NPP-11 T555A in pDONR201 |
| GCTTCAGCATCCGCTCCATCTGTTGGATTGTTCGG | OLP1255 | NPP-11 T321A in pDONR201 |
| CCGAACAATCCAACAGATGGAGCGGATGCTGAAGC | OLP1256 | NPP-11 T321A in pDONR201 |
| GGTCTCTTCGGATCGTCAGCTCCTGCTAAGACCC | OLP1257 | NPP-11 T393A in pDONR201 |
| GGGTCTTAGCAGGAGCTGACGATCCGAAGAGACC | OLP1258 | NPP-11 T393A in pDONR201 |
| GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGACAACCGCTGCGGCAGCCCC | OLP1205 | NPP-11[C] (aa333-805) in pDONR201 |
| GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAGGCC TTCGACGAGTTGTTCG | OLP1206 | NPP-11[C] (aa333-805) in pDONR201 |
| GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGTTCTCGCATCTTAACC | OLP1290 | NPP-19[N] (aa1-301) in pDONR201 |
| GGGGACCACTTTGTACAAGAAAGCTGGGTCTTATGAG TTATACATGGAAGCAGATCGGT | OLP1528 | NPP-19[N] (aa1-301) in pDONR201 |
| CTAGCTAGCATGTCACTTTTCGGGTCATCTACTCCCCA | OLP1524 | NPP-1[N] (aa1-194)in pET28b |
| CAGCGGCCGCTTAGGCAGCAACTTGATTCAACTTAGCA | OLP1549 | NPP-1[N] (aa1-194)in pET28b |
| GCTGCGCTAGCGAATTCGCTCCAGCCACTGGTGGTCT T | OLP1554 | NPP-11[C1] (aa290-805) WT and 3A in pET28b |
| CGAAGCTTCTAGGCCTTCGACGAG | OLP1251 | NPP-11[C1] (aa290-805) WT and 3A in pET28b |
| CTAGCTAGCACAACCGCTGCGGC | OLP1250 | NPP-11[C] (aa333-805) WT, T555A and T393A T555A mutants in pET28b |
| CGAAGCTTCTAGGCCTTCGACGAG | OLP1251 | NPP-11[C] (aa333-805) WT, T555A and T393A T555A mutants in pET28b |
| CGGAGATCTCATGGCTAGCTGGAGCCACCCGCAGTTCGAAAAAGGATCTGGTGGTGGTGGTGGTGAAAACCTGTACTTCCAGGGAATGTCACTTTTCGGGTCATCTACTCC | OLP1360 | Strep-NPP-1 with TEV cleavage site in pRSFDuet-1 6xHis-NPP11[C] (aa333-805) |
| CGCGGTACCTTATCCTTCTAGAGCCCTTCTACAAGC | OLP1361 | Strep-NPP-1 with TEV cleavage site in pRSFDuet-1 6xHis-NPP11[C] (aa333-805) |
| ATCGTGGTGTCACGCTCGTCGTTTGGTATGG | OLP870 | Gibson cloning of npp-1 gDNA into MosSCI vector |
| ATACCAAACGACGAGCGTGACACCACGATGC | OLP871 | Gibson cloning of npp-1 gDNA into MosSCI vector |
| AGGAGGAGGAGGTGGAATGTCACTTTTCGGGTCATCTACTCC | OLP1614 | Gibson cloning of npp-1 gDNA into MosSCI vector |
| ACCCGAAAAGTGACATTCCACCTCCTCCTCCTCCCTTATAC | OLP1615 | Gibson cloning of npp-1 gDNA into MosSCI vector |
| GGGCTCTAGAAGGATAAATGCAAGATCCTTTCAAGCATTCCC | OLP1616 | Gibson cloning of npp-1 gDNA into MosSCI vector |
| GAAAGGATCTTGCATTTATCCTTCTAGAGCCCTTCTACAAGC | OLP1617 | Gibson cloning of npp-1 gDNA into MosSCI vector |
METHOD DETAILS
Human Cell cultures and transfection
HeLa cells were grown at 37°C in DMEM (Life Technologies) supplemented with 10% fetal calf serum, 1% L-glutamine, 100μg/ml streptomycin, and 100 U/ml penicillin. Transfection was performed using Lipofectamine 2000 (Invitrogen).
Nematode strains and RNAi
C. elegans strains were cultured and maintained using standard procedures (Brenner, 1974). RNAi was performed by the feeding method using HT115 bacteria essentially as described (Kamath et al., 2001), except that 2 mM of IPTG was added to the NGM plates and in the bacterial culture just prior seeding the bacteria. As control, animals were exposed to HT115 bacteria harboring the empty feeding vector L4440 (mock RNAi). RNAi clones targeting nucleoporins were obtained from V. Galy (Galy et al., 2003). The other RNAi clones were obtained from the Arhinger library (Open Source BioScience) or were constructed.
Feeding RNAi in N2, GFP::NPP-1, NPP-11::GFP and in the PLK-1::sGFP animals was performed by feeding L4 animals 24-48 h at 20°C. In Figure 4C, PLK-1::sGFP L4 or young adults animals were fed at 20°C with RNAi bacteria as follows: mock RNAi (ctrl) 12-48 h, ima-1(RNAi) 20-30 h, ima-2(RNAi) 15-22 h, imb-1(RNAi) 16 h, imb-2(RNAi) 20-30 h, and imb-3(RNAi) 17-48 h. In Figure S2A, plk-1(or683ts) L4 animals were fed with RNAi bacteria for various amount of time at 15°C and then shifted 2-5 hours to 25°C before analysis. The feeding at 15°C was performed as follows: mock RNAi (ctrl) 24-48 h, lmn-1(RNAi) 27-53 h, npp-4(RNAi) and npp-11(RNAi) 24-53 h, npp-5(RNAi) 17-50 h, npp-7(RNAi) 15-17 h, npp-10(RNAi) 25-26 h, npp-6(RNAi) 24-40 h, npp-2(RNAi), npp-15(RNAi) and npp-13(RNAi) 24-50 h, npp-1(RNAi) and npp-3(RNAi) 15-53 h. In Figure S2B, RNAi was performed by feeding plk-1(or683ts) L4 animals at 15°C as follows: mock RNAi (ctrl) 45-50 h, npp-1(RNAi) and npp-11(RNAi) 40-46 h, npp-3(RNAi), npp-4(RNAi) and npp-15(RNAi) 52-54 h.
Genome editing
A transposon-based strategy (Frokjaer-Jensen et al., 2008) was used to insert mCherry::npp-1 Wild-type and non-phosphorylatable 4A transgenes in single copy at defined genomic location on Chr II. The correct insertion and expression of the transgenes was confirmed by PCR, fluorescence and Western blot analysis.
The strain npp-11(ok1599)I was generated in Robert Barstead’s laboratory as part of the C. elegans Gene Knockout Project. We pinpointed the deletion from 1000 to 1753 base pair of the npp-11 gene (Figure S5A). This deletion removes 251 amino acids including two Polo-docking sites primed by Cdk1 (T321 and T393 residues). We mutated the third Polo-docking site to non-phosphorylatable alanine (T555A) using CRISPR/Cas9 as described (Paix et al., 2015) (Figure S5A). The mix of injection was composed of the purified Cas9 protein, the tracrRNA Cat#1072534 from IDT, the repair template ssODN for npp-11 (Key resource table), the repair template ssODN for dpy-10 (Key resource table), the dpy-10 crRNA (GCTACCATAGGCACCACGAG) and the targeted gene crRNA (AGGGCTCACTTCAACACCAT). After 15min at 37°C, the mix was injected into the gonads of young adult N2 animals, and each injected hermaphrodite was separated into an individual plates at 20°C. 5 days later, 40 rollers from the plates that had the most roller worms, were again separated into individual plates. After 24 hours, the worms were lysed and genotyped by PCR and enzymatic digestion with AatII (AatII TTGACGTCA site was added in the repair template without changing the translation into amino acids). The generated strain was verified by sequencing.
The npp-1(syb207)IV allele was generated by SunyBiotech. It contains an in-frame deletion in the npp-1 gene from base pairs 18 to 1622, encompassing the four Polo-docking sites primed by Cdk1 (Figure S5A). The strain was verified by DNA sequencing.
Endogenous superfolder (s)GFP tagging of the plk-1 locus was performed using CRISPR/Cas9, as described (Dickinson et al., 2013). The repair template for plk-1::sgfp included the 3’ end (1.6 kb) of plk-1, fused in-frame to sgfp (Pédelacq et al., 2006) with a short linker (GGRAGSG) in between. The sgfp was followed by the plk-1 3′ UTR (480 bp), an unc-119(+) selectable marker flanked by loxp sequence on either side and 1.5kb of downstream genomic sequence. To modify the plk-1 locus using Cas9-triggered homologous recombination, the repair template (50ng/μL) was co-injected with a plasmid (40ng/μL) modified from pDD162 containing Cas9 and the guide RNA sequence (5’- ctgctcgttcagcttcggc −3’) and three plasmids encoding fluorescent markers (Key resource table) for negative selection into the strain HT1593. Recombinant strains were identified by selecting moving worms without fluorescent markers, and sGFP integration was confirmed by PCR and sequencing. Later, the Cb-unc-119 selection marker was deleted using Cre recombinase-mediated excision. An injection mix containing pDD104 (Peft-3::Cre, 50 ng/μL) and co-injection markers was injected into the gonads of unc-119(+) young adult plk-1::sgfp animals. Cre-excised animals were identified by selecting for the Unc phenotype and were confirmed by PCR. The unc-119 mutation was subsequently removed by mating with N2 males. The final strain was crossed with mCherry::HIS-11 males to allow the visualization of the DNA.
Molecular Biology
Details of plasmids constructions are available upon request. The plasmids and oligonucleotides used in this study are listed in the Key resource table. Gateway cloning was performed according to the manufacturer’s instructions (Invitrogen). All the constructs were verified by DNA sequencing (GATC-Biotech).
Yeast two-hybrid analysis
Yeast two-hybrid analyses were performed using a GAL-4-based system (Gateway) in diploid cells obtained by mating Y187 MAT alpha transformed with the bait PLK-1 PBD WT or mutated versions (C-terminal 309 amino acids of PLK-1) (Noatynska et al., 2010) and CG-1945 MATa strain transformed with the prey SPAT-1. Diploid cells hosting the prey-bait positive interaction were selected on selective medium lacking leucine, tryptophan and histidine (-L-W-H) and containing 3-amino-1, 2, 4-triazole drug (3AT, 25 mM).
To verify the expression of the yeast two-hybrid constructs by Western blot, 1.5 OD of exponentially growing yeast cells were collected by centrifugation for 2 min at 1 500 g and washed once in water, resuspended in 200 μl of 0.1 M NaOH and incubated 10 min at room temperature. After centrifugation 2 min at 16 100 g, the pellet was resuspended in 50 μl Laemmli 3X and analyzed by SDS-PAGE.
Biochemical assays
Western blot analysis was performed using standard procedures.
Protein Purification:
The expression of recombinant proteins was induced by the addition of 0.5 mM isopropyl β-D-thiogalactopyranoside to 1-liter cultures of exponentially growing E. coli BL21 (OD=0.5-0.8) before incubation 3 h at 25°C. After pelleting, the bacteria were resuspended in 0.5 M NaCl, 5% glycerol, 50 mM Tris/HCl pH 8, before lysis by sonication. The soluble portion of the lysate was loaded on a 1-ml Hi-Trap HP Column (GE Healthcare). The column was washed with ten volumes of lysis buffer, and bound proteins were eluted in lysis buffer containing between 70 and 600 mM imidazole. Imidazole was subsequently removed by dialysis against the lysis buffer. Proteins were aliquoted and flash-frozen with liquid nitrogen and stored at −80°C.
Purification of the human GST-PLK-1 PBD WT or phosphate pincer (GST-PBD H538A/K540M) mutant fusion proteins on glutathione beads was performed according to the manufacturer’s instructions (GE Healthcare). Bacterial cultures were induced 3 h at 25°C with 1mM IPTG and then lysed by sonication as described in Lysis Buffer (10 mM Tris pH 8, 150 mM NaCl, 1 mM EDTA, 5 mM DTT, 0.05 % NP40, 1 mM PMSF, 1X protease inhibitors (cOmplete cocktail from Roche). Proteins were eluted in Elution Buffer (100 mM Tris pH 8, 120 mM NaCl, 0.01 % triton X100, 1 mM DTT, 20 mM Glutathione pH 8), aliquoted, flash-frozen in liquid nitrogen and stored at −80°C.
In vitro kinase assays were performed as described (Tavernier et al., 2015). Briefly, equal amount of Wild-type and mutant proteins were phosphorylated by 1 μl (corresponding to 20 units) of Cyclin B-Cdk1 (P6020L from NEB) in 30 μl total volume of the kinase buffer (1X protease inhibitors (cOmplete cocktail from Roche), 1X phosphatase inhibitors (PhosSTOP cocktail from Roche), 2 mM ATP, 50 mM HEPES pH 7.6, 10 mM MgCl2) for 40 min at 30°C under 300 rpm agitation. For the radioactive in vitro kinase assay, we used a mix of 1.7 mM of cold ATP and 5 μCi of γ-[32P]ATP (PerkinElmer). The kinase reaction was stopped by the addition of 15 μl of Laemmli 3X, the samples were boiled and loaded on a SDS-PAGE. The gel was dried and exposed for autoradiography (Typhoon, GE Healthcare). For the Far-Western ligand-binding assay of the NPP-1•NPP-4•NPP-11[C] trimer, samples were loaded on a 12 % SDS-PAGE.
Phos-tag gels and Far-Western ligand binding assay
Far-Western ligand binding assays were performed essentially as described (Wu et al., 2007) on Phos-tag™ 100 μM SDS-PAGE 6% for NPP-11[C] (aa333-805) and 12% for NPP-1[N] (aa1-194). After washing 3 times the Phos-tag gel with 10 mM EDTA in transfer buffer (48 mM Tris-base, 39 mM Glycine, 20% ethanol, 0.03% SDS), each gel was transferred to a PVDF membrane 0.45 μm during 1h30 at 90V. Only for NPP-1[N], 0.2μm PVDF was used with fast transfer (iBlot 2, 5 min at 23V).
The membranes were then incubated overnight at 4°C in blocking solution (4% milk in TBS-Tween 0.1%). The next day, each membrane was incubated with 2 μg of GST-PBD WT or H538A/K540M of Plk1 during 5-6 h at 4°C, then washed extensively with the blocking solution. The human Plk1 antibody (1/1000) was then incubated on the membrane overnight at 4°C in a typical Western blot experiment to reveal the GST-PBD.
Immunofluorescence and Microscopy
Fixation and indirect immunofluorescence of C. elegans oocytes and embryos were performed essentially as described (Gönczy et al., 1999) on subbing solution-coated slides. After Freeze-crack and fixation with cold dehydrated methanol, slides were washed 3×5 min, blocked for 1 h in PBS 3% BSA and incubated overnight at 4°C with primary antibodies diluted in PBS 3% BSA. Working dilutions for the primary antibodies were 1/1000 for rabbit PLK-1 antibodies (M. Gotta). Slides were later incubated for 30 min at room temperature with secondary antibodies (anti-Rabbit) coupled to the Alexa 568 fluorophore. Next, embryos were mounted in Vectashield Mounting Medium with DAPI (Vector). Fixed embryos were imaged using a LSM 710 confocal microscope (Zeiss) with 63× N/A 1.4 objectives. Captured images were processed using ImageJ and Adobe Photoshop.
For the analysis of the paired nuclei phenotype in live specimens by Differential interference contrast (DIC) microscopy, embryos were obtained by cutting open gravid hermaphrodites using two 21-gauge needles. Embryos were handled individually and mounted on a coverslip in 3 μl of M9 buffer. The coverslip was placed on a 3% agarose pad. (DIC) images were acquired by an Axiocam Hamamatsu ICcI camera (Hamamatsu Photonics, Bridgewater, NJ) mounted on a Zeiss AxioImager A1 microscope equipped with a Plan Neofluar 100×/1.3 NA objective (Carl Zeiss AG, Jena, Germany), and the acquisition system was controlled by Axiovision software (Carl Zeiss AG, Jena, Germany). Images were acquired at 5-sec intervals.
Fluorescent live cell imaging was performed at 20°C using a spinning disc confocal head (CSU-X1; Yokogawa Corporation of America) mounted on a Ti-E inverted microscope (Nikon) equipped with 491nm and 561nm lasers (Roper Scientific) and a charge-coupled device camera (Coolsnap HQ2; Photometrics). Acquisition parameters were controlled by MetaMorph software (Molecular Devices). In all cases a 60×, 1.4 NA PlanApochromat lens with 2 × 2 binning was used, and six z-sections were collected at 0.5-μm intervals every 10 s.
Plk1 localization at the NE in human cells was best detected upon pre-extraction of cells followed by a fixation-extraction procedure as previously described (Morchoisne-Bolhy et al., 2015). Wide-field fluorescence images were acquired using a DM6000B Leica microscope with a 100x, NA 1.4 (HCX Plan-Apo) oil immersion objective and a charge-coupled device camera (CoolSNAP HQ; Photometrics). Alternatively, human cell nuclei were imaged using a LSM 780 confocal microscope (Zeiss) with 63× N/A 1.4 objectives. Confocal images correspond to a single focal plane spanning 0.35 μm. Captured images were processed using ImageJ and Adobe Photoshop.
QUANTIFICATION AND STATISTICAL ANALYSIS
For the cross-correlation analysis of the distributions of Plk1 and Nup62 or Lamin A/C the Pearson’s Correlation Coefficient (r) was measured on confocal images of the NE surface from prophase HeLa cells stained with Plk1 and Nup62 or Lamin A/C antibodies. The coefficients were measured before (0°) and after rotation of the Nup62 or Lamin A/C cells images by 90°C in seven or twelve cells respectively. Two 75 pixels-wide squares per cell in a region without kinetochores were selected for Pearson coefficient measurement.
For the quantification of GFP::NPP-1 or NPP-11::GFP signal at the nuclear envelope, the average signal intensity at the NE was measured using the Image J software in control and RNAi conditions after background subtraction. To allow for direct comparison between control and RNAi conditions, the average signal intensity at the NE in control embryos was arbitrarily defined as 1. One-way ANOVA followed by Dunnett’s multiple comparisons test was performed using GraphPad Prism version 6.00 for Mac OS X, GraphPad Software, La Jolla California USA, www.graphpad.com.
Supplementary Material
Highlights:
PLK-1 localizes to the NPC through its PBD in human cells and in C. elegans embryos
Central channel Nucleoporins positively regulate PLK-1 function during C. elegans NEBD
The NPP-1•NPP-4•NPP-11 complex of the central channel recruits PLK-1 to the NPC
PLK-1 binds directly to NPP-1, NPP-4 and NPP-11 in a phosphorylation-dependent manner
Acknowledgements
We thank V. Cordes, J.C. Courvalin, R. Lin, I. Sumara, M. Gotta, V. Galy, V. Mirouse and P. Askjaer for strains and reagents, A. Santamaria for her help with Far-Western ligand binding assay. We thank U. Kutay for sharing results prior to publication. We thank C. Jackson, B. Ossareh-Nazari and Y. Thomas for critical reading of the manuscript. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). D.K.C. and A.D. are supported by an NIH grant (GM074215). LM is supported by a PhD fellowship from the Ministry of Research. Work in the laboratories of LP and VD is supported by the French National Research Agency under grant no. ANR-2012-BSV2-0001-01 and ANR-2012-BSV2-0008-01 and by the "Fondation pour la Recherche Médicale” (Foundation for Medical Research) “Equipes FRM DEQ20140329538 and DEQ20150734355". LP and VD teams are supported by the Labex “Who am I?” Laboratory of Excellence No. ANR-11-LABX-0071, the French Government through its Investments for the Future program operated by the French National Research Agency (ANR) under Grant no. ANR-11-IDEX-0005-01. We acknowledge the ImagoSeine core facility of the Institut Jacques Monod, member of IBiSA and the France-BioImaging (ANR-10-INBS-04) infrastructure.
Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Footnotes
DATA AND SOFTWARE AVAILABILITY
N/A
References
- Askjaer P, Galy V, Hannak E, and Mattaj IW (2002). Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol Biol Cell 13, 4355–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Desai A, and Oegema K (2007). A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol 178, 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibi N, Parveen Z, and Rashid S (2013). Identification of potential Plk1 targets in a cell-cycle specific proteome through structural dynamics of kinase and Polo box-mediated interactions. PLoS One 8, e70843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blethrow JD, Glavy JS, Morgan DO, and Shokat KM (2008). Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci U S A 105, 1442–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul JD, van de Peut D, Devos M, Simonis N, Yildirim MA, Cokol M, Kao HL, de Smet AS, Wang H, Schlaitz AL, Hao T, Milstein S, Fan C, Tipsword M, Drew K, Galli M, Rhrissorrakrai K, Drechsel D, Koller D, Roth FP, Iakoucheva LM, Dunker AK, Bonneau R, Gunsalus KC, Hill DE, Piano F, Tavernier J, van den Heuvel S, Hyman AA, and Vidal M (2008). A protein domain-based interactome network for C. elegans early embryogenesis. Cell 134, 534–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budirahardja Y, and Gönczy P (2008). PLK-1 asymmetry contributes to asynchronous cell division of C. elegans embryos. Development 135, 1303–1313. [DOI] [PubMed] [Google Scholar]
- Burkard ME, Maciejowski J, Rodriguez-Bravo V, Repka M, Lowery DM, Clauser KR, Zhang C, Shokat KM, Carr SA, Yaffe MB, and Jallepalli PV (2009). Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol 7, e1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase D, Serafinas C, Ashcroft N, Kosinski M, Longo D, Ferris DK, and Golden A (2000). The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis 26, 26–41. [DOI] [PubMed] [Google Scholar]
- Chug H, Trakhanov S, Hülsmann BB, Pleiner T, and Görlich D (2015). Crystal structure of the metazoan Nup62•Nup58•Nup54 nucleoporin complex. Science 350, 106–110. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, and Goldstein B (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods 10, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia AE, Cantley LC, and Yaffe MB (2003a). Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 299, 1228–1231. [DOI] [PubMed] [Google Scholar]
- Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, and Yaffe MB (2003b). The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115, 83–95. [DOI] [PubMed] [Google Scholar]
- Ferreira J, Stear JH, and Saumweber H (2017). Nucleoporins NPP-10, NPP-13 and NPP-20 are required for HCP-4 nuclear import to establish correct centromere assembly. J Cell Sci 130, 963–974. [DOI] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, and Jorgensen EM (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40, 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Antonin W, Jaedicke A, Sachse M, Santarella R, Haselmann U, and Mattaj I (2008). A role for gp210 in mitotic nuclear-envelope breakdown. J Cell Sci 121, 317–328. [DOI] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, and Askjaer P (2003). Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell 14, 5104–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Gunawardena J, and Mann M (2011). PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res 39, D253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P, Schnabel H, Kaletta T, Amores AD, Hyman T, and Schnabel R (1999). Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J Cell Biol 144, 927–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosstessner-Hain K, Hegemann B, Novatchkova M, Rameseder J, Joughin BA, Hudecz O, Roitinger E, Pichler P, Kraut N, Yaffe MB, Peters JM, and Mechtler K (2011). Quantitative phospho-proteomics to investigate the polo-like kinase 1-dependent phospho-proteome. Mol Cell Proteomics 10, M111.008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet V, Busso C, Toya M, Sugimoto A, Askjaer P, and Gonczy P (2012). The nucleoporin Nup205/NPP-3 is lost near centrosomes at mitotic onset and can modulate the timing of this process in Caenorhabditis elegans embryos. Mol Biol Cell 23, 3111–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley N, Cheerambathur D, Moyle M, Stefanutti M, Richardson A, Lee KY, Dumont J, Oegema K, and Desai A (2016). A Nucleoporin Docks Protein Phosphatase 1 to Direct Meiotic Chromosome Segregation and Nuclear Assembly. Dev Cell 38, 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, and Beck M (2015). Towards understanding nuclear pore complex architecture and dynamics in the age of integrative structural analysis. Curr Opin Cell Biol 34, 31–38. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, and Ahringer J (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2, RESEARCH0002, doi: 10.1186/gb-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YH, Park JE, Yu LR, Soung NK, Yun SM, Bang JK, Seong YS, Yu H, Garfield S, Veenstra TD, and Lee KS (2006). Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol Cell 24, 409–422. [DOI] [PubMed] [Google Scholar]
- Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, and Gerber SA (2011). Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal 4, rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockenhauer KE, and Schwartz TU (2016). The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell 164, 1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, Hurt E, and Beck M (2016). Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352, 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, and Higgins DG (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, and Kutay U (2011). Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 144, 539–550. [DOI] [PubMed] [Google Scholar]
- Lee KK, Gruenbaum Y, Spann P, Liu J, and Wilson KL (2000). C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell 11, 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lénárt P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, and Peters JM (2007). The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol 17, 304–315. [DOI] [PubMed] [Google Scholar]
- Li H, Liu XS, Yang X, Song B, Wang Y, and Liu X (2010). Polo-like kinase 1 phosphorylation of p150Glued facilitates nuclear envelope breakdown during prophase. Proc Natl Acad Sci U S A 107, 14633–14638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machowska M, Piekarowicz K, and Rzepecki R (2015). Regulation of lamin properties and functions: does phosphorylation do it all. Open Biol 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morchoisne-Bolhy S, Geoffroy MC, Bouhlel IB, Alves A, Audugé N, Baudin X, Van Bortle K, Powers MA, and Doye V (2015). Intranuclear dynamics of the Nup107-160 complex. Mol Biol Cell 26, 2343–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhäusser P, and Kutay U (2007). An in vitro nuclear disassembly system reveals a role for the RanGTPase system and microtubule-dependent steps in nuclear envelope breakdown. J Cell Biol 178, 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, and Barr FA (2007). Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol 9, 436–444. [DOI] [PubMed] [Google Scholar]
- Neef R, Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU, and Barr FA (2003). Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol 162, 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi Y, Rogers E, Robertson SM, and Lin R (2008). Polo kinases regulate C. elegans embryonic polarity via binding to DYRK2-primed MEX-5 and MEX-6. Development 135, 687–697. [DOI] [PubMed] [Google Scholar]
- Noatynska A, Panbianco C, and Gotta M (2010). SPAT-1/Bora acts with Polo-like kinase 1 to regulate PAR polarity and cell cycle progression. Development 137, 3315–3325. [DOI] [PubMed] [Google Scholar]
- O’Rourke SM, Carter C, Carter L, Christensen SN, Jones MP, Nash B, Price MH, Turnbull DW, Garner AR, Hamill DR, Osterberg VR, Lyczak R, Madison EE, Nguyen MH, Sandberg NA, Sedghi N, Willis JH, Yochem J, Johnson EA, and Bowerman B (2011). A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS One 6, e16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Desai A, Rybina S, Kirkham M, and Hyman AA (2001). Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol 153, 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, and Seydoux G (2015). High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics 201, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Soung NK, Johmura Y, Kang YH, Liao C, Lee KH, Park CH, Nicklaus MC, and Lee KS (2010). Polo-box domain: a versatile mediator of polo-like kinase function. Cell Mol Life Sci 67, 1957–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, and Waldo GS (2006). Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24, 79–88. [DOI] [PubMed] [Google Scholar]
- Rahman MM, Munzig M, Kaneshiro K, Lee B, Strome S, Müller-Reichert T, and Cohen-Fix O (2015). C. elegans polo-like kinase PLK-1 is required for merging parental genomes into a single nucleus. Mol Biol Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers DM, Moreno S, Abraham M, and Ahringer J (2008). PAR proteins direct asymmetry of the cell cycle regulators Polo-like kinase and Cdc25. J Cell Biol 180, 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, Schmidt A, Sillje HH, Korner R, and Nigg EA (2011). The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics 10, M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetter A, Askjaer P, Piano F, Mattaj I, and Kemphues K (2006). Nucleoporins NPP-1, NPP-3, NPP-4, NPP-11 and NPP-13 are required for proper spindle orientation in C. elegans. Dev Biol 289, 360–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker S, and Sumara I (2014). Molecular dynamics of PLK1 during mitosis. Mol Cell Oncol 1, e954507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi E, Toyoshima-Morimoto F, and Nishida E (2002). Nuclear translocation of plk1 mediated by its bipartite nuclear localization signal. J Biol Chem 277, 48884–48888. [DOI] [PubMed] [Google Scholar]
- Tavernier N, Noatynska A, Panbianco C, Martino L, Van Hove L, Schwager F, Léger T, Gotta M, and Pintard L (2015). Cdk1 phosphorylates SPAT-1/Bora to trigger PLK-1 activation and drive mitotic entry in C. elegans embryos. J Cell Biol 208, 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R, and Kutay U (2017). Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol 18, 229–245. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, and Barton GJ (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li Q, and Chen XZ (2007). Detecting protein-protein interactions by Far western blotting. Nat Protoc 2, 3278–3284. [DOI] [PubMed] [Google Scholar]
- Xu J, Shen C, Wang T, and Quan J (2013). Structural basis for the inhibition of Polo-like kinase 1. Nat Struct Mol Biol 20, 1047–1053. [DOI] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Jedrusik-Bode M, Wisniewski JR, and Mann M (2009). Caenorhabditis elegans has a phosphoproteome atypical for metazoans that is enriched in developmental and sex determination proteins. J Proteome Res 8, 4039–4049. [DOI] [PubMed] [Google Scholar]
References
- Chaudhary N, and Courvalin JC (1993). Stepwise reassembly of the nuclear envelope at the end of mitosis. J Cell Biol 122, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes V, Waizenegger I, and Krohne G (1991). Nuclear pore complex glycoprotein p62 of Xenopus laevis and mouse: cDNA cloning and identification of its glycosylated region. Eur J Cell Biol 55, 31–47. [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, and Goldstein B (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods 10, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilotter HE, Hannon GJ, Ruddell CJ, and Beach D (1994). Construction of an improved host strain for two hybrid screening. Nucleic Acids Res 22, 1502–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, and Jorgensen EM (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40, 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, and Askjaer P (2003). Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell 14, 5104–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A, Liu J, and Cohen-Fix O (2009). Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Sci 122, 1970–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, and Elledge SJ (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, and Ahringer J (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2, RESEARCH0002, doi: 10.1186/gb-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, and Ahringer J (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, and Higgins DG (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Nishi Y, Rogers E, Robertson SM, and Lin R (2008). Polo kinases regulate C. elegans embryonic polarity via binding to DYRK2-primed MEX-5 and MEX-6. Development 135, 687–697. [DOI] [PubMed] [Google Scholar]
- Noatynska A, Panbianco C, and Gotta M (2010). SPAT-1/Bora acts with Polo-like kinase 1 to regulate PAR polarity and cell cycle progression. Development 137, 3315–3325. [DOI] [PubMed] [Google Scholar]
- O’Rourke SM, Carter C, Carter L, Christensen SN, Jones MP, Nash B, Price MH, Turnbull DW, Garner AR, Hamill DR, Osterberg VR, Lyczak R, Madison EE, Nguyen MH, Sandberg NA, Sedghi N, Willis JH, Yochem J, Johnson EA, and Bowerman B (2011). A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS One 6, e16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Ródenas E, Klerkx EP, Ayuso C, Audhya A, and Askjaer P (2009). Early embryonic requirement for nucleoporin Nup35/NPP-19 in nuclear assembly. Dev Biol 327, 399–409. [DOI] [PubMed] [Google Scholar]
- Sarov M, Murray JI, Schanze K, Pozniakovski A, Niu W, Angermann K, Hasse S, Rupprecht M, Vinis E, Tinney M, Preston E, Zinke A, Enst S, Teichgraber T, Janette J, Reis K, Janosch S, Schloissnig S, Ejsmont RK, Slightam C, Xu X, Kim SK, Reinke V, Stewart AF, Snyder M, Waterston RH, and Hyman AA (2012). A Genome-Scale Resource for In Vivo Tag-Based Protein Function Exploration in C. elegans. Cell 150, 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernier N, Noatynska A, Panbianco C, Martino L, Van Hove L, Schwager F, Léger T, Gotta M, and Pintard L (2015). Cdk1 phosphorylates SPAT-1/Bora to trigger PLK-1 activation and drive mitotic entry in C. elegans embryos. J Cell Biol 208, 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, and Barton GJ (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.